Abstract
Parkinson’s disease (PD) is a common progressive neurodegenerative disorder characterized by loss of striatal-projecting dopaminergic neurons of the ventral forebrain, resulting in motor and cognitive deficits. Despite extensive efforts in understanding PD pathogenesis, no disease-modifying drugs exist. Recent advances in cell reprogramming technologies have facilitated the generation of patient-derived models for sporadic or familial PD and the identification of early, potentially triggering, pathological phenotypes while they provide amenable systems for drug discovery. Emerging developments highlight the enhanced potential of using more sophisticated cellular systems, including neuronal and glial co-cultures as well as three-dimensional systems that better simulate the human pathophysiology. In combination with high-throughput high-content screening technologies, these approaches open new perspectives for the identification of disease-modifying compounds. In this review, we discuss current advances and the challenges ahead in the use of patient-derived induced pluripotent stem cells for drug discovery in PD. We address new concepts implicating non-neuronal cells in disease pathogenesis and highlight the necessity for functional assays, such as calcium imaging and multi-electrode array recordings, to predict drug efficacy. Finally, we argue that artificial intelligence technologies will be pivotal for analysis of the large and complex data sets obtained, becoming game-changers in the process of drug discovery.
1. Introduction
Parkinson’s disease (PD) is the second most common neurodegenerative disorder, with an increasing incidence in aged people, while an estimated 4% are diagnosed before the age of 50. Due to the high prevalence of PD and the increase in the proportion of aging population resulting from extended life expectancy, the disease has become a rapidly growing area of concern. PD is characterized by progressive loss of striatal-projecting midbrain dopaminergic neurons of the substantia nigra pars compacta [1], leading to motor symptoms, such as bradykinesia, rigidity and resting tremor, with a wide range of severity [2]. Although considered a classic movement disorder, PD is associated with a broad spectrum of non-motor symptoms, including psychiatric and cognitive manifestations such as depression, dementia and hallucinosis, as well as sleep and sensory disturbances. These persistent symptoms may appear long before motor dysfunction becomes apparent while they remain unaffected by currently available therapeutics, severely impairing patients’ quality of life [3].
Most PD cases are sporadic with unknown etiology; however, an approximate 10% represent familial cases (reviewed in [4]). Among the genes linked to heritable monogenic PD, mutations in the α-synuclein gene SNCA (PARK1/4) [5] and the leucine-rich repeat kinase 2 gene LRRK2 (PARK8) [6] are accountable for autosomal-dominant PD forms, while mutations in PINK1 (PTEN-induced putative kinase 1; PARK6) [7], Parkin (PARK2) [8], protein deglycase DJ-1 (PARK7) [9], ATP13A2 (PARK9) [10] and β-glucocerebrosidase (GBA) [11] are responsible for an autosomal recessive mode of inheritance.
Currently, only symptomatic or palliative treatments are available with none capable to prevent or slow down PD progression and revert neurological disabilities. Regardless of the divergent pathologies and clinical manifestations, dopamine-replacement drugs such as Levodopa (L-dopa), which was identified 53 years ago, are used as a first-line treatment to address the cardinal motor issues arising from nigrostriatal degeneration [12]. For most patients though, oral dopaminergic medication falls short after a few years and advanced therapies such as deep brain stimulation, continuous intestinal gel levodopa/carbidopa, and subcutaneous apomorphine administration, are considered as alternative options [13]. However, such treatments are administered only in a small subset of patients as they have limited benefit while they are associated with serious side effects, including deterioration of non-motor symptoms [14]. Therefore, although the field of therapies for PD continues to expand, there is still no therapeutic strategy capable to change or reverse the disease course.
Most present-day efforts in identifying novel PD drugs target the aggregation of misfolded α-synuclein (αSyn) as the major pathogenic factor that causes cellular toxicity (reviewed in [15]). αSyn is a presynaptic neuronal protein linked genetically and neuropathologically to PD [16,17,18]. It accumulates abnormally in the PD brain and is the main component of intracellular neuronal inclusions, termed Lewy bodies or Lewy neurites, which represent the histopathological hallmark of PD. The pioneering discovery that αSyn is the major gene associated with both sporadic and genetic PD [16,19,20] spurred intensive research over the past two decades for understanding the mechanisms of αSyn misfolding, oligomerization and aggregation. These studies resulted in deciphering the molecular pathways involved (reviewed in [21]) and developing drugs aiming to halt or even clear αSyn aggregation. A number of promising compounds have been developed targeting pathological αSyn in animal models of PD (reviewed in [22]), but have largely failed when tested in early phase clinical trials, causing disappointment. Several limitations lie behind these unsuccessful endeavors. Notably, despite the flood of studies that have revealed numerous disease-associated processes, fundamental knowledge gaps still exist in PD pathophysiology. Our understanding of the disease covers processes such as mitochondrial impairment resulting in energy failure and increased oxidative stress; lysosomal deficiency and misfolded protein accumulation in large inclusions impeding axonal transport; distorted synaptic vesicle trafficking affecting neurotransmission and synaptic connections. Yet the spatiotemporal sequence of events leading to PD and, most importantly, the initial disease-triggering factors still remain elusive [2,23]. Strikingly, a recent study in which cutting-edge imaging technologies were applied indicated that αSyn immunopositive Lewy bodies and Lewy neurites consist mainly of lipid vesicle clusters instead of the long-assumed proteinacious core [24], pointing to a paradigm shift with important implications for understanding and treating neurodegeneration in PD.
Another drawback in the course of PD drug development is that assessment of novel therapeutics has been performed in animal models that fail by large to recapitulate the human condition. Last, but not least, the traditionally held conviction that the basal ganglia is the main brain region implicated in PD, has been recently challenged. It is now acknowledged that PD is a system-level disorder that affects overlapping neural circuits comprising the entire basal ganglia-cortico-cerebellar axis, which works in concert to mediate motor and cognitive functions [25]. Embracing such a system-level perspective together with using human cell-based disease models for understanding and treating PD has higher potential for the development of innovative treatments.
In this review, we discuss how revolutionary human cell-derived systems based on induced pluripotent stem cell (hiPSC) technology offer an unprecedented opportunity to study PD in models that exhibit clinically relevant disease phenotypes. Such systems provide powerful platforms for understanding pathology and, in combination with recent cutting-edge drug discovery technologies, open up new perspectives for identification of disease-modifying compounds. Yet, there are still limitations and hurdles to face ahead before these young technologies come to fruition and fulfill their promise, particularly in reducing the number of drug failures in clinical trials.
2. Identification of Disease-Relevant Phenotypes in hiPSC-Derived Models of PD: A Glimpse into Human Pathology
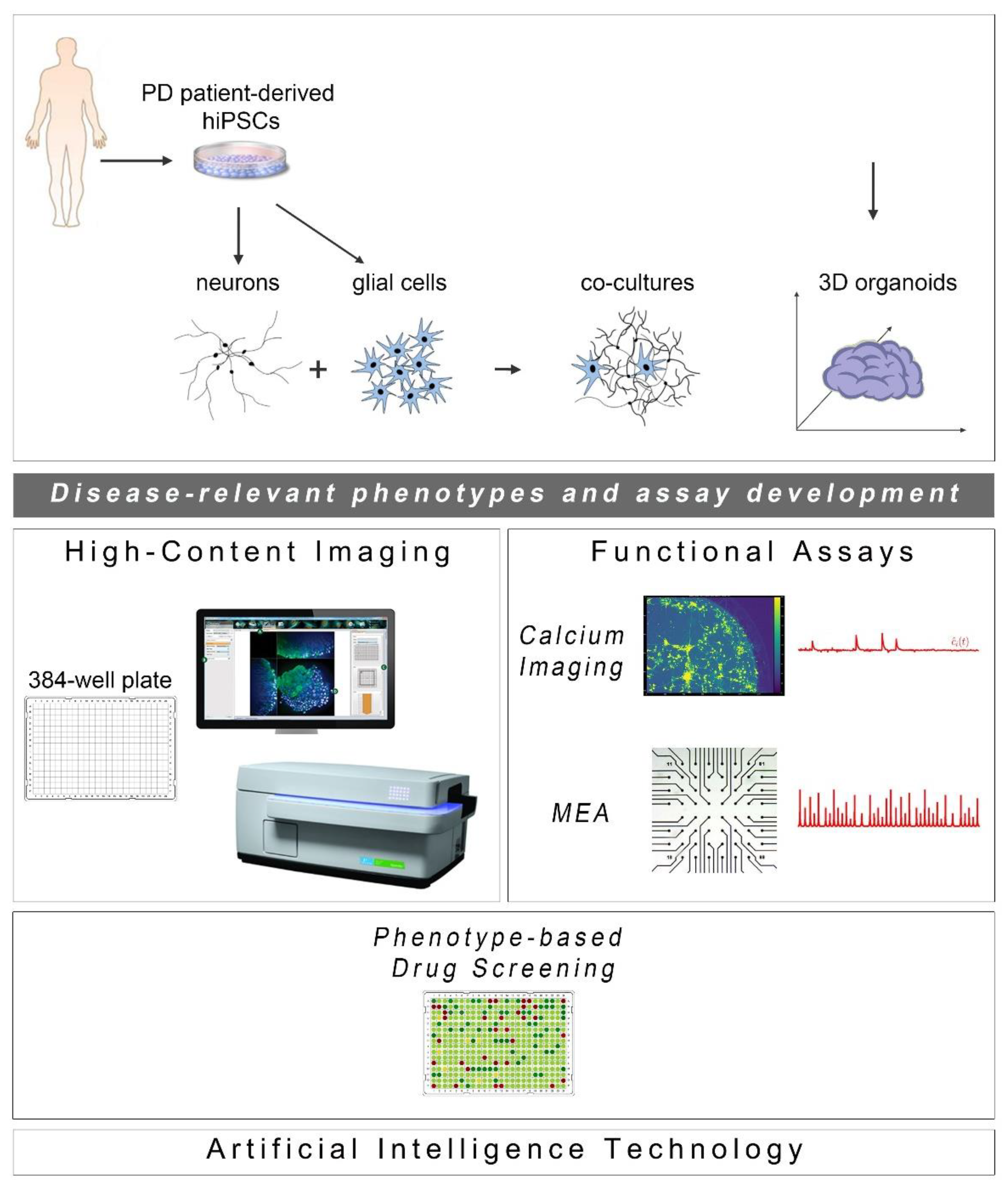
Since the initial discovery of human cell reprogramming technology in 2007 [26], a new exciting era for the field of drug discovery has been ushered. hiPSCs have been widely used to generate “disease-in-a-dish” models for numerous neurodegenerative diseases from which selected cellular systems have been applied in drug evaluation for efficacy and toxicity assessment (Figure 1). Compared to other traditional cellular systems and existing animal models, hiPSC-based platforms offer multiple advantages: human origin in a personalized manner, easy accessibility and expandability, capacity to give rise to many different cell types and avoidance of ethical concerns associated with the use of human embryonic stem cells. Moreover, scalability and manipulation of hiPSC lines for obtaining target cells in large quantities and desirable purity is also possible at GMP grade. In the PD field, intensive efforts have been made to generate high-purity human ventral midbrain dopaminergic neurons for in vitro maturation, but mainly for intracerebral transplantation that has long been an attractive prospect for PD treatment (reviewed in [27,28]). To date, a number of sporadic and familial PD hiPSC-based models have been created displaying a variety of disease-relevant characteristics that could be exploited in either target-based or unbiased phenotypic screens. The percentage of dopamine neurons in these models varies as validated by the expression of markers associated with neuronal type and subtype specificity. However, this may not be a problem, but rather an advantage, in view of the realization that PD is more likely a system-level disorder rather than a dysfunction of the nigrostriatal dopaminergic system.
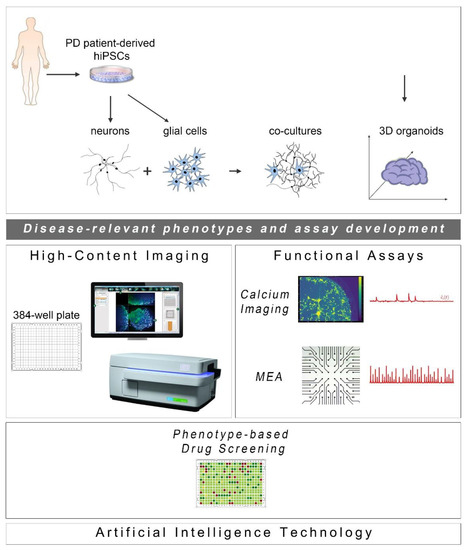
Figure 1.
Cell reprogramming technologies allowed the generation of Parkinson’s disease (PD) patient-derived hiPSCs that are further differentiated into neuronal and glial cell populations (astrocytes (shown), but also oligodendrocytes or microglia (not shown)) studied separately or in co-culture with neurons. hiPSC-derived brain organoids are being created to better simulate the human disease. The PD-relevant phenotypes identified in these cellular systems form the foundations for the development of drug discovery platforms encompassing high-content imaging/chemical library screening as well as functional assays, such as calcium imaging and high-resolution multi-electrode array (MEA) recordings (phenotypic drug screening). The application of artificial intelligence technologies will be critical for analysis of the resulting large and complex data sets.
The first PD-patient specific iPSC lines were generated in 2008 by Park et al. [29] followed by a second group [30]. Both teams provided proof-of-principle that it is feasible to produce hiPSC-derived dopaminergic neurons from patients with sporadic PD, but did not proceed further to identify disease-associated phenotypes. Since then, the majority of hiPSC-based PD models have been developed mostly from patients carrying various genetic mutations (reviewed in [31]). Most studied lines carry the G2019S mutation in the LRRK2 gene, one of the most prominent monogenetic risk factors for PD linked to both familial and sporadic forms of the disease (reviewed in [32]). hiPSC-derived cultures from patients bearing point mutations in the SNCA gene (A53T, A30P and E46K) or SNCA multiplications have also been extensively studied, while hiPSC-derived neurons carrying Parkin (PARK2), PINK1 or GBA mutations have been analyzed to a lesser extent (reviewed in [33]). Overall morphological, biochemical and functional analyses have revealed a battery of disease-associated phenotypes (Table 1), initially after induction of cellular stress and later—upon careful observations—at basal conditions. Some of these phenotypes were previously described in post-mortem PD brains or in relevant animal models, while a number were identified for the first time in hiPSC-based models.
2.1. α-Synuclein Accumulation
As discussed, αSyn is the major protein associated with both sporadic and genetic PD [19,20]. It is a small neuronal protein widely distributed in the brain with preferential localization at pre-synaptic terminals and direct association with synaptic vesicles, suggesting possible roles in the regulation of neurotransmitter release, synaptic function and plasticity [21]. Even though the physiological function of αSyn is not well understood [34], its involvement in neurodegeneration is well established [35]. αSyn accumulation and/or aggregation has been shown in hiPSC-derived neurons generated from PD patients carrying disease-associated mutations. In particular, increased αSyn protein levels were detected in hiPSC-derived neurons carrying the A53T (G209A) point mutation [5,36,37,38,39] or SNCA duplication [40] and triplication [39,41,42,43,44,45,46]. Accumulation of αSyn has also been described in several other hiPSC models of familial PD, including LRRK2, Parkin, PINK1 and hiPSC-derived neurons from GBA patients, as well as, in hiPSC models of sporadic PD (reviewed in [33]). Detection of the pathological species relies largely on the presence of phosphorylated αSyn, particularly at serine 129, but also on the presence of specific oligomeric and fibrillar forms as aggregation proceeds. The presence of cellular protein aggregates has also been observed using fluorescence-based assays. In particular, our team [38] and Ryan et al. [36] have shown discrete αSyn accumulations co-localizing with Tau and ubiquitin in both the soma and axons of hiPSC-derived neurons from patients bearing the A53T mutation.
2.2. Mitochondrial Defects
Mitochondrial impairment is thought to be a key factor in PD pathology, with PD-associated mutations in Parkin, PINK1, DJ-1, GBA, LRRK2 and SNCA being linked to distortions in mitochondrial function [47]. Studies in hiPSC-derived neurons from PD patients have revealed mitochondrial defects, including morphological and functional alterations (reviewed in [33]). In particular, fragmented mitochondria or mitochondria with abnormal morphology have been observed in hiPSC-derived neurons carrying LRRK2-G2019S [48], Parkin [49,50,51,52,53], PINK1 [51,54] or GBA mutations [55], but also SNCA-G209A or triplication [39]. Others have reported decreased mitochondrial content in patient neurons [56,57,58]. Altered mitochondrial functionality has also been demonstrated [52,55,57], with decreased ATP production [43,48], reduced membrane potential [59] or dysfunctional mobility [60], affecting multiple cellular processes and altering the redox status of the neuron.
2.3. Oxidative Stress
As mitochondrial respiration is the major source of reactive oxygen species (ROS) in the cell, mitochondrial defects should result in an increase in oxidative stress within patient neurons. A number of studies have demonstrated changes in mitochondrial and oxidative stress-related proteins observed in the human parkinsonian brain (reviewed in [61]). Similarly, oxidative stress phenotypes, such as increased ROS and carbonylated proteins or upregulation of proteins involved in dopamine oxidation, have been observed in several hiPSC-based models carrying LRRK2 [62,63], Parkin [49,50,64] and PINK1 [60,65] mutations as well as in hiPSC-derived neurons with SNCA-triplication [41,43,46].
2.4. ER Stress and Autophagy-Related Phenotypes
Consistent with protein aggregation and cellular stress, studies in hiPSC models reveal that endoplasmic reticulum (ER) dysregulation and increased ER stress may be involved in PD pathogenesis promoting neuronal cell death [37,45,66]. Additionally, altered function of other cellular organelles, such as lysosomes, together with autophagy impairment, has been observed in LRRK2 [48,63,67], SNCA [44] and GBA [68,69] mutant hiPSC-derived neurons, as well as in a hiPSC-based model of sporadic PD [67], resulting in defective clearance of aggregated proteins. Finally, several studies have shown that genetically diverse PD patient-derived neurons exhibit increased susceptibility to various forms of cellular stress, including proteotoxic [38,62,70] or nitrosative stress [36,37], linking specific cellular pathways to disease pathology that have not been previously described in other systems.
2.5. Compromised Neuritic Growth, Axonal Degeneration and Decreased Synaptic Connectivity
Interestingly, studies on LRRK2-G2019S [67,71,72,73,74], on Parkin-mutated [75] and on SNCA-mutated [38,44] hiPSC-derived neurons have revealed previously unrecognized morphological changes, including compromised neuritic outgrowth, reduced neurite complexity and axonal degeneration, resulting in impaired synaptic connectivity and network function. In particular, LRRK2-G2019S mutant neurons displayed shorter and less complex processes, reminiscent of immature neurons. Over time, these cells exhibited clear signs of degeneration, including very short or absent neurites, vacuolated soma, fragmented nucleus and positive staining for cleaved caspase-3. In the case of SNCA mutations, αSyn overexpression due to triplication of the gene led to poor formation of the neuronal network that correlated with significantly lower generation of action potentials in response to current injections [44]. In a later study performed by our team, A53T-αSyn neurons had a profound downregulation of mRNAs associated with synaptic formation, maintenance and function. The neuronal network formed was also less complex with neurite number, length and morphology being compromised when compared to control cells [38]. It will be interesting to investigate further whether the impaired synaptic connectivity and aberrant electrophysiological recordings noted in PD neurons represent early events in disease pathogenesis and how they are linked to distorted mitochondrial and/or axonal transport and ER stress. In any case it is an important finding that should be taken into account when developing new therapeutic or disease-intervening strategies.

Table 1.
Major phenotypes observed in PD hiPSC-derived neurons. This table summarizes key pathogenic phenotypes associated with PD mutant neurons as described in the cited publications.
Table 1.
Major phenotypes observed in PD hiPSC-derived neurons. This table summarizes key pathogenic phenotypes associated with PD mutant neurons as described in the cited publications.
| Major PD-Relevant Phenotypes | Patient-Derived iPSC-Based Models in PD | |||||
|---|---|---|---|---|---|---|
| Gene Mutations | ||||||
| SNCA | LRKK2 | PARKIN | PINK1 | GBA | OPA1 | |
| αSyn accumulation and/or aggregation; increased phosphorylated αSyn (Ser 129); presence of oligomeric and fibrillar αSyn forms | G209A [5,36,37,38,39] Duplication [40] Triplication [39,41,42,43,44,45,46] | G2019S [62,67,71,72,73] | Ex2–4 del and Ex6–7 del [49] Ex3del, R42P, Ex3–4del, 1-BP del 255A, R275W, R42P [56] V324A [51] c.255delA [72] | Q456X [51] | L444P [68] N370S [69] | |
| Mitochondrial defects: fragmented mitochondria or mitochondria with abnormal morphology; decreased mitochondrial content; decreased ATP production; reduced membrane potential; dysfunctional mitochondrial mobility | G209A Triplication [39] | G2019S [48,57] | Ex2–4 del and Ex6–7 del [49] Ex3, 5, and 6 del [50] V324A [51] c.1072delT, p.A324fsX110 [52] Ex2–4 del and Ex6–7 del [53] | Q456X [51] G309D; Ex7/del [54] | N370S, L444P, and RecNcil [55] | p.G488R and p.A495V [58] |
| Oxidative stress: increased ROS and carbonylated proteins; upregulation of proteins involved in dopamine oxidation | Triplication [41,43,46] | G2019S [62] I2020T [63] | Ex2–4 del & Ex6–7 del [49] Ex3, 5, and 6 del [50] Ex3 del/Ex5 del and Ex3 del/Ex 3 del [64] | Q456X [60,65] | ||
| ER dysregulation; increased ER stress; autophagy impairment | G209A [37,66] Triplication [44,45] | G2019S [48,67] I2020T [63] | RecNcil, L444P and N370S [68] N370S [69] | |||
| Compromised neurite growth & complexity; neurite swellings; axonal degeneration; decreased synaptic connectivity; impaired axonal transport | G209A [38] Triplication [44] | G2019S [67,71,72,73] | Ex3 del/Ex5 del and Ex3 del/Ex 3 del [75] | |||
3. Rescue of Disease-Related Phenotypes in hiPSC-Derived Models of PD: Setting the Foundations towards Drug Discovery
Contrary to the initial skepticism regarding the potential of hiPSC-based systems to recapitulate age-related diseases such as PD, the identification of a wealth of cellular and molecular phenotypes, either previously known or novel, has prompted the use of such systems for early drug discovery. A number of candidate drugs have been tested for their ability to restore disease-related phenotypes in hiPSC-derived models of PD with promising results (Table 2). These studies have laid the groundwork for future application of hiPSC models in larger drug screening campaigns, assisting in the development of robust assays.

Table 2.
Summary of hiPSC-based models of PD that have been used for drug testing. This table briefly describes the rescue of disease-related phenotypes in PD hiPSC-derived neurons by selected compounds.
Analyses of hiPSC-derived neurons from patients with different PD-linked mutations have shown that αSyn accumulation presents a common theme regardless of the gene mutated, suggesting that it could serve as a read-out when testing novel therapeutics. Accordingly, in hiPSC-derived dopaminergic neurons from sporadic PD patients, as well as from patients carrying mutations in GBA, LRRK2, DJ-1, Parkin and SNCA (A53T, triplication) with decreased lysosomal β-glucocerebrosidase (GCase) activity, treatment with small-molecule enhancers of GCase has resulted in reduced αSyn levels and associated toxicity. These observations suggest that non-inhibitory small-molecule chaperones of GCase may prove promising for treatment of PD and related synucleinopathies [76,77,78]. Additionally, recent data in GBA-linked PD patient-derived dopaminergic neurons have implicated increased acid ceramidase activity in the context of decreased GCase. This led to intracellular accumulation of αSyn, the levels of which were reduced upon inhibition of acid ceramidase by carmofur [79]. In the same lines, Burbulla et al. have applied treatment with mitochondrial antioxidants to smooth out a pathological cascade instigated by mitochondrial oxidant stress causing lysosomal dysfunction and αSyn accumulation in dopaminergic neurons derived from patients with sporadic and familial PD [80].
Alterations in neuronal morphology, neurite outgrowth and complexity as well as axonal degeneration have been used as parameters to evaluate the potential efficacy of compounds in rescuing pathological phenotypes. One study has demonstrated that LRRK2-G2019S hiPSC-derived neurons display neurite shortening and fewer neurites compared to wild-type controls [71], a PD-associated phenotype previously linked to ERK signaling [81]. Interestingly, treatment with an inhibitor of ERK phosphorylation or a selective inhibitor of LRRK2 kinase activity increased neurite growth and rescued mutant cultures from degeneration [71]. Similarly, Korecka et al. demonstrated that exposure to a LRRK2 kinase inhibitor or application of a LRRK2-specific antisense oligonucleotide (ASO) rescued neurite collapse, also in a model of LRRK2-G2019S hiPSC-derived neurons [74].
In the patient-derived SNCA-G209A (A53T) hiPSC-based model that we developed, PD neurons exhibited distinct morphological features characterized by extensive neuritic pathology and degeneration [38]. By immunostaining for βIII-tubulin, we observed contorted or fragmented axons with swollen varicosities and spheroid inclusions containing Tau and αSyn. Under morphological examination using a lentiviral vector for expression of the red fluorescent protein DsRed under the control of human synapsin 1 promoter (LV. SYN1.DsRed) to facilitate imaging of single neurons, we also observed a significant reduction in both total neurite length and the number of neurites extending from the soma. Three de novo in silico-designed compounds [82,83] that all interact with and reduce αSyn toxicity by interfering with αSyn oligomer formation could restore neurite length and rescue axonal pathology [38]. Interestingly, a small variation of one of these small molecules, NPT200-11, that was developed by the company Neuropore Therapies in collaboration with UCB Biopharma, is the only αSyn-inhibiting compound that has reached clinical trials successfully completing phase I (https://clinicaltrials.gov/ct2/results?cond=&term=NPT200-11&cntry=&state=&city=&dist=, Study Completion Date: February 2016).
Several studies with hiPSC-derived neurons have revealed dysregulation in the expression of genes involved in numerous cellular processes, rendering cells vulnerable to stressors that activate or modulate these pathways. Oxidative stress, mitochondrial impairment and proteasome inhibition are key factors that cause increased susceptibility and cell death of patient-derived neurons (reviewed in [33,84]). The first study recapitulating PD-associated phenotypes has revealed that dopaminergic neurons derived from LRRK2-G2019S hiPSCs displayed increased expression of key oxidative stress-response genes. Moreover, these cells were highly sensitive to cell death caused by exposure to hydrogen peroxide, the proteosomal inhibitor MG-132 or the neurotoxin 6-hydroxydopamine (6-OHDA) [62]. In a similar manner, Reinhardt et al. have confirmed that LRRK2-G2019S hiPSC-derived dopaminergic neurons are susceptible to oxidative stress induced by the mitochondrial complex 1 inhibitor rotenone or the neurotoxin 6-OHDA resulting in increased apoptosis, preferentially of dopaminergic neurons [71]. The incurred cytotoxicity was rescued in the presence of the small molecule inhibitor of LRRK2 kinase, LRRK2-IN1, which increased the survival of dopaminergic neurons.
An association between PD and exposure to mitochondrial toxins, including rotenone, has also been reported in SNCA-G209A (A53T) mutant neurons [36]. Microarray analysis of SNCA-G209A hiPSC-derived dopaminergic neurons has highlighted a pathway where toxin-induced nitrosative/oxidative stress results in S-nitrosylation of the transcription factor MEF2C. High-throughput screening of a chemical library for small molecules capable of targeting the MEF2C-PGC1α pathway pinpointed isoxazole as a potential therapeutic, protecting SNCA-G209A neurons from apoptosis induced by mitochondrial toxins [36].
When exposing the A53T-αSyn hiPSC-based model that we generated to epoxomicin or MG132, both of which interfere with αSyn clearance via the proteasome, we observed a significant increase in cleaved caspase-3 immunoreactivity consistent with the levels of LDH release in mutant neurons. This was accompanied by a pronounced disruption of the MAP2+ neuronal network, confirming once again their susceptibility to proteotoxic stress [38]. The observed stress-induced vulnerability could also be reversed by the three small molecules targeting αSyn [82,83], resulting in restoration of the MAP2+ network [38].
Finally, a recent study has revealed that increased oxidative stress and inflammation are associated with induction of the necroptotic pathway in hiPSC-derived neural cells from patients with a mutation in the OPA1 gene encoding a key player in mitochondrial fusion and structure [58] that has been associated with an inherited form of PD and dementia [85]. Mutant cultures exhibited severe mitochondrial dysfunctions, impaired oxidative phosphorylation, and high oxidative stress levels, leading to neuronal cell loss. Pharmacological treatment of necroptosis with the specific inhibitor necrostatin-1 protected neurons from cell death [58].
The above paradigms provide evidence that, despite the initial concerns in using human iPSC-based models for modeling age-related neurodegenerative pathologies, such systems show multiple disease-associated phenotypes with high relevance to PD pathogenesis and progression and can be of great value in drug discovery.
4. Phenotypic Screens using hiPSC-Derived Models of PD: Empowering Drug Discovery
4.1. Target-Based Versus Phenotype-Based Drug Screening
The two main high-throughput screening approaches for discovering new disease-modifying therapeutics are either target-based or phenotype-based. Historically, phenotypic-based screening strategies shaped the foundations of pharmaceutical drug discovery long before molecular target-based approaches were applied [86]. However, in the past 25 years, molecular target-based drug screening has become the main route to drug discovery in both the academia and the pharmaceutical industry. This change was mainly due to an accelerated progress in molecular biology and genomics that resulted in efficient mining of genes associated with various diseases [87]. The starting point in this approach is a well-defined molecular target with a predicted role in disease allowing the hypothesis that modulation of its activity would have beneficial effects. Screening of chemical libraries of small molecules is then used to identify lead compounds that interact with high affinity and specificity with the target [88]. Hits from such screens are then used for pharmacological target validation and lead compound optimization. The main advantage of the target-based approach is that the mechanism of action is known right from the start, which can accelerate preclinical assessment. Other advantages include the ability to facilitate optimization of the lead compound as there is a clear structure–activity relationship enabling improvement of its physicochemical properties, and the potential to predict target-associated safety liabilities and toxicity. However, knowledge of the molecular targets has not translated into identification of disease-modifying agents for PD or other neurodegenerative diseases [89]. One reason could be that the underlying mechanism is not clear for PD, as for most neurodegenerative diseases, resulting in a universal lack of well-defined targets. Besides, neurodegenerative diseases, including PD, are highly complex disorders and manipulating a single target may not be sufficient to restore the dysfunctional cellular network.
In phenotypic screens, on the other hand, disease-driving phenotypes can be used to determine compounds that change the outcome of multiple biological pathways without prior knowledge of the molecular mechanisms of the disease. Such screens are unbiased and may identify compounds targeting completely unexpected proteins or pathways. Consequently, phenotypic screens hold promise for the identification of previously unrecognized disease pathways and the discovery of new therapeutic targets [90]. It is notable that, during the past 20 years, phenotypic screening has contributed to most of the first-in-class small-molecule drugs approved by the FDA. Among all the new molecular entities approved from 1999 to 2008, 28 were identified through phenotypic screens, whereas target-based approaches contributed to the discovery of 17 compounds [91]. In particular, in the central nervous system field, phenotypic screening has yielded seven out of nine first-in-class drugs. Consequently, there is renewed interest in reinventing phenotypic screens as a means of drug discovery.
4.2. Phenotypic-Based Drug Screening in hiPSC-Derived Models of PD
Although the first PD patient-derived hiPSCs were generated in 2009 [30], surprisingly only two phenotypic screens have been reported so far in hiPSC-derived PD neurons. To identify disease-modifying agents, Yamaguchi et al. established an imaging-based, semi-automatic, high-throughput assay for quantitative detection of mitochondrial clearance and cell viability in dopaminergic neurons from patients with familial PD having Parkin or PINK1 mutations. After screening 320 pharmacologically active inhibitor compounds, the researchers identified four hits, MRS1220, tranylcypromine, flunarizine and bromocriptine, that improved the pathological clearance of mitochondria possibly by promoting mitochondrial degradation through the lysosomal system, without further investigating the underlying mechanism [92]. In another study, Tabata et al. [93] performed a phenotypic screen in Parkin (PARK2) patient-derived dopaminergic neurons displaying increased susceptibility to rotenone-induced mitochondrial stress, to identify neuroprotective compounds. From phenotypic screening of an FDA-approved drug library, one voltage-gated calcium channel antagonist, benidipine, was found to suppress rotenone-induced apoptosis [93]. The selective vulnerability of dopaminergic neurons was further attributed in this study to the dysregulation of intracellular calcium homeostasis via T-type calcium channels, revealing a previously unidentified pathway in PD and offering a potential treatment opportunity. More recently, using A53T-αSyn patient-derived neurons, we performed a small-scale phenotypic screen to identify neuroprotective compounds and identified the multikinase inhibitor BX795 as a candidate therapeutic that rescued the pathological features of PD neurons [94].
5. Looking into the Future: Optimization of hiPSC-Based Models for Understanding and Treating PD
The development of phenotypic screens in PD hiPSC-based neuronal cultures is still underway with the field awaiting the identification of novel disease targets for therapeutic interventions. In the meantime, new scientific discoveries and technical advances underscore the limitations of using two-dimensional (2D) neuronal platforms lacking a more physiological environment, such as extracellular matrix and the presence of glial cells. Emerging developments highlight the enhanced potential of using neuronal and glial co-cultures as well as three-dimensional (3D) systems of various kinds, including brain organoids that simulate more closely the human pathophysiology. Current progress in these approaches complemented by advances in microfluidics, state-of-the-art imaging methodologies and electrophysiological analyses to evaluate the functionality of neuronal networks, is discussed below (Figure 1).
5.1. Glial Cell Involvement in PD Pathogenesis: Mimicking the CNS Microenvironment in hiPSC-Based Co-Culture Systems
As for most neurodegenerative diseases, the majority of PD studies have been performed in neuronal cultures, preferably consisting of midbrain dopaminergic neurons in various degrees of purity, and assessing neuronal degeneration signs after exposure, or not, to different types of stress. However, neuronal death may also be induced by a microenvironment that does not sufficiently support neuronal survival and/or function, while newer concepts suggest that other neural cell types, such as astrocytes [95] and microglia [96], may contribute to PD pathogenesis and progression.
Studies in post-mortem human brains and in animal models of PD, including Parkinsonian macaques [97,98,99,100,101], have demonstrated astroglial activation, suggesting that astrogliosis in combination with the secretion of pro-inflammatory cytokines may contribute to PD progression [102,103,104]. Moreover, abnormal αSyn accumulation has been observed in post-mortem astrocytes indicative of pathological alterations [101]. One explanation that has been provided is that neurons transfer pathological αSyn to astrocytes, which in turn could have an active role in its clearance [105]. It is therefore possible that astrocytes could have either a detrimental or a beneficial role, or both depending on the disease stage. It is plausible to assume that, at early stages of PD, astrocytes act to protect neurons from an overload of pathological protein cargo, while, at later stages, they become dysfunctional and contribute to the deterioration of neuronal health.
As evidenced by positron emission tomography (PET), microgliosis is an early and sustained response in PD [106,107], while reactive microglia have also been detected in toxin-induced or transgenic mouse models of PD [108,109,110,111]. Interestingly, various studies support that microglia-mediated inflammation can have both beneficial and detrimental effects [112,113,114,115,116,117]. For example, the interaction of pathogenic αSyn with different microglial receptors promotes microglial clearance of αSyn and phagocytosis of apoptotic neurons, which may be beneficial in controlling pathology [118,119,120,121]. On the other hand, it has been shown that neurotoxic microglia can induce the generation of A1-type astrocytes exhibiting a neurotoxic phenotype, thus revealing a deleterious effect of microglial activation [122]. Interestingly, blocking the microglial-induced A1 astrocyte conversion has been found to be neuroprotective in models of PD offering new therapeutic prospects [123].
Oligodendrocytes appear to be the least affected cell type in PD, although they too present αSyn depositions [101] and show intrinsic formation of pathological αSyn assemblies [124]. Intriguingly, a reduction in the myelination of neuron projections seen in the earliest stages of the disease provides a possible association between oligodendrocyte function and PD pathogenesis [125].
Human cell-derived in vitro models can provide more specific information on the positive and/or negative involvement of glial cells in PD pathogenesis and progression. So far, only a few studies have been reported that explore hiPSC-derived systems for investigating the astrocytic involvement in PD. Human midbrain astrocytes generated from PD patients carrying the LRRK2-G2019S mutation showed transcriptomic dysregulation associated with compromised ability to degrade αSyn [126]. A more distinctive involvement of hiPSC-derived astrocytes transpired when they were co-cultured with dopaminergic neurons. In particular, when co-cultured with LRRK2-G2019S astrocytes, control hiPSC-derived dopaminergic neurons acquired morphological signs of neurodegeneration and abnormal, astrocyte-derived αSyn accumulation. Conversely, control astrocytes partially prevented the appearance of disease-related phenotypes in PD neurons [127]. In a recent study, astrocytes derived from patients with GBA-associated Parkinsonism displayed a cytokine and chemokine profile indicative of an inflammatory response [128]. Moreover, when these cells were co-cultured with dopaminergic neurons generated from the same hiPSC lines, excessive αSyn released from neurons was endocytosed by the GBA-derived astrocytes, translocating into lysosomes. It therefore seems that in GBA-associated Parkinsonism, astrocytes play a role in αSyn accumulation and processing, contributing to neuroinflammation. Similarly, Sonninen et al. have just published that LRRK2-G2019S hiPSC-derived astrocytes exhibit pathological hallmarks of the disease, including increased αSyn expression, which results in altered metabolism, disturbed Ca2+ homeostasis and increased release of cytokines upon inflammatory stimulation [129]. These findings are in line with the manifestation of αSyn inclusions in activated astrocytes in the post-mortem human PD brain as well as in animal models and suggest that pathogenic astrocytes may contribute to non-cell autonomous build-up of toxic αSyn species and the initiation of neuronal deterioration.
Recent developments in more efficient methodologies have allowed for the generation of hiPSC-derived macrophages that can be induced towards a microglial phenotype by co-culture with neurons [130]. These derived microglial cells acquired a highly dynamic ramified morphology and exhibited neuronal surveillance activity mimicking the in vivo situation. Building on this advancement, a subsequent study demonstrated that LRRK2 expression in macrophages and microglia plays an important role in phagosome maturation and in the regulation of recycling pathways, implying that LRRK2 mutations in PD patients may disrupt microglial clearance mechanisms [131]. Additionally, it has been shown that the LRRK2-G2019S mutation influences fate decision in hiPSC-derived human monocytes, further endorsing the involvement of the immune system in the development of PD [132].
Overall, the limited research performed so far on more complex human experimental set-ups has emphasized that considering bilateral or non-cell autonomous interactions between neurons, astrocytes and microglia is critical for elucidating the pathogenic processes occurring in PD and uncovering the molecular mechanisms triggering disease appearance and progression. Furthermore, the few studies performed indicated the need for using more complex systems as physiologically relevant disease models for drug discovery.
5.2. Emergence of Three-Dimensional (3D) hiPSC- Based Platforms for PD
The appearance of 3D systems of increasing complexity mimicking the brain microenvironment meets the requirement for more efficient modeling of disease phenotypes not only towards elucidating context-dependent human pathologies, but also for the development of novel platforms for high-throughput drug screening. Yet, there are still serious hurdles to overcome, including the reduced long-term viability of such systems due to lack of sufficient oxygenation within the 3D core and the large variability observed [133]. In a first attempt within the PD field, the culture of patient neurons derived from hiPSCs carrying the LRRK2-G2019S mutation was optimized in 3D microfluidics [134]. Automated high-content imaging revealed decreased dopaminergic differentiation and branching complexity, altered mitochondrial morphology, and increased cell death in the absence of external stressors. In two subsequent efforts, midbrain-like organoids were produced from sporadic or familial PD patients carrying the LRRK2-G2019S mutation that, respectively, displayed αSyn accumulation and dopaminergic neuron degeneration [135,136].
Clearly, further optimization and analysis are needed for the production of advanced patient-specific platforms that can be useful in modelling and treating PD. In this respect, research is progressing fast to generate more complex 3D cultures yielding organoids or spheroids that incorporate neurons and different glial cell types [137,138,139,140,141] offering promising tools to simulate PD pathology with higher fidelity to the in vivo scenario. Enriched 3D systems can be more informative on the cellular and molecular basis of neuron-glia cross-talk, monitoring glial activation and inflammation, also in correlation with toxic αSyn accumulation and neuronal decline. Of relevance, an optimized protocol was devised recently for the generation of midbrain-like organoids containing mature midbrain dopaminergic and GABAergic neurons, functional astrocytes and oligodendrocytes, exhibiting electrophysiological activity and producing dopamine and neuromelanin-like granules [142]. A PD-mimicking neurotoxin-based protocol was then applied to assess cell-to-cell interactions in neurodegeneration, demonstrating glia-mediated massive cell death of dopaminergic neurons. Interestingly, Ormel et al. showed that microglia can also develop innately within cerebral organoids and display their characteristic ramified morphology [141]. This was an unexpected finding since the consensus was that cerebral organoids consist of cells derived from the neuroectodermal lineage and should therefore lack mesodermal-derived microglia. Yet, this study exemplified a model where the interplay between microglia, macroglia and neurons can be studied in human brain development and disease.
Even though 3D organoids have not been used yet for high-throughput drug screening in PD, the technology for adaptation is developing and has been used successfully in other neurodegenerative diseases, indicating the feasibility of such strategies [143]. Developments include a number of microtissue and nanoculture products to support 3D architecture in 96- or 384-well format, introduction of enriched extracellular matrix (ECM) products and inducers of endogenous ECM proteins and specialized synthetic biomaterials [144]. Nevertheless, the high cost of such products, the disruption of viscosity and temperature during automated handling, the sample processing for high-content analysis and the poor penetration of tested compounds present serious limitations for performing preclinical testing in a large scale and within a reasonable timescale. New approaches that target the issues of high-throughput scale and cost while offering amenable systems with a high level of biological complexity and clinical relevance, such as formulation and optimization of engineered microenvironments, are in great demand. Such brain-on-a-chip platforms comprising miniaturized microfluidic perfusion systems that permit long-term growth in a format that is financially viable and has the potential of scaling up for launching high-throughput discovery campaigns, might pave the way for future fundamental discoveries and the development of more effective drugs [145]. Microfluidic devices have already been used to study the interaction of microglia and neurons in PD and to demonstrate neuronal internalization of αSyn fibrils before their propagation along the axons [146,147,148]. However, today, the most relevant formats are restricted to low-throughput applications awaiting their adaptation for automated screening on a large scale.
6. Functional Assays for PD Studies and Drug Screening in hiPSC-Derived Systems
As the ultimate success of drug discovery depends on the functional recovery it can produce, a pipeline based solely on assessment of morphological rescue of human PD neurons will have a high chance of failing. PD is characterized by severe changes in neuronal connectivity and data derived from electrophysiological analyses of patient-derived neurons highlight the importance of recording changes in synaptogenesis, network formation and neurotransmitter balance when new drugs are tested or during unbiased phenotypic screens. Two functional assays may be used in drug screening based on their potential for adaptation and scalability: calcium imaging and the multi-electrode array system (Figure 1).
6.1. Calcium Imaging
Calcium homeostasis is fundamental to neuronal survival and function and, when deregulated, can lead to neurodegeneration via complex and diverse mechanisms resulting in selective neuronal impairment and death. Fluorescent calcium imaging is a well-established method, which enables the visualization of free intracellular Ca2+ in populations of cells. Calcium indicators are sensitive to calcium changes and can be loaded in a non-invasive manner to neuronal cells, although prolonged exposure to the dye is toxic. Fluorescent dyes can either be single or multiple wavelengths. Calcium imaging allows to explore calcium-mediated processes happening over different time scales. For example, calcium-mediated neurotransmitter release occurs much more rapidly than calcium-mediated gene expression in the nucleus [149]. The development of genetically encoded calcium indicators offers a better alternative to the use of fluorescent dyes, since they are non-invasive and can be targeted to specific neurons and/or astrocytes, allowing for a longer duration of imaging without the risk of phototoxicity [150]. Additionally, other issues observed with fluorescent dyes such as background fluorescence and non-specific dye loading can be overcome with genetic indicators [150].
In relation to PD, increasing evidence suggests that defective calcium signaling plays an important role in disease pathogenesis. Schwab and Ebert showed that LRRK2-G2019S hiPSC-derived sensory neurons display altered calcium dynamics and treatment with LRRK2 kinase inhibitors resulted in significant rescue [151]. Moreover, Tabata et al. demonstrated that there is a dysregulation of calcium homeostasis in Parkin and PINK1 hiPSC-derived dopamine neurons which is prevented by T-type calcium channel knockdown or antagonists [93]. We have also demonstrated aberrant Ca+ fluxes in A53T-αSyn neurons [152] that could be linked to the decreased spontaneous synaptic activity recorded by patch-clamp electrophysiology [38]. Currently, high-content imaging or drug screening based on recording Ca+ dynamics has not yet been performed in PD hiPSC-derived systems.
6.2. Multi-Electrode Arrays (MEAs)
High-resolution MEA systems enable one to assess novel electrophysiological parameters of hiPSC-derived neurons, which can be potentially used as biomarkers for phenotype screening and drug testing. MEAs comprise a platform for monitoring prolonged, non-destructive recordings of spontaneously firing neurons in vitro with applications in neurodegenerative diseases. A MEA system combines a cell culture plate with an embedded array of high- or low-impedance electrodes allowing for parallel detection of local field potentials generated by spontaneous or evoked firing of neurons [153,154,155]. The generation of synchronously active neuronal networks depends on sequential developmental processes. Neuronal network formation starts with excitable and spontaneously active neurons that are asynchronously active due to lack of functional connectivity between neurons. With the formation of functional synapses, two or more neurons become functionally interconnected and capable to generate synchronous bursting. In a population of neurons, the connectivity between neurons is increasing over time and finally results in synchronous bursting activity of hundreds or thousands of interconnected cells. So, calcium imaging and MEA recordings could reveal the inability of pathological cells to make the physiological transition of asynchronously active neurons into few synchronous bursting neurons and finally into a population of neurons that are highly synchronously active. One characteristic example is described by Woodard et al., in which hiPSC-derived dopamine neurons were examined from monozygotic male twins of Ashkenazi Jewish background that were discordant for PD [156]. The investigators noticed that neurons from the unaffected twin developed robust synchronous bursting patterns indicative of maturing neuronal networks in contrast to neurons from the affected twin that did not produce synchronous bursting patterns with their spontaneous activity also being significantly lower [156]. In another study, however, on hiPSC-derived dopaminergic neurons from patients with young-onset PD, MEA recordings did not show differences between disease and control cultures [157].
Nonetheless, the value of MEA recordings in uncovering disease-related phenotypes in an in vitro setting is highlighted in primary cultures derived from animal models of neurodegenerative diseases. Amin et al. applied MEA recordings to characterize the early activity-dependent changes induced by toxic Aβ-oligomers in neuronal networks using a simple in vitro model based on a rat hippocampal cell culture system. It is interesting to note that, in this study, a clinically applied N-methyl D-aspartate antagonist used for Alzheimer’s disease treatment, could reverse Aβ-neurotoxicity and rescue network-wide firing [158]. The importance of monitoring coordinated neuronal activity and its disruption in the development of neurodegeneration has also been emphasized by Iaccarino et al. in a mouse model of Alzheimer’s disease [159].
Regarding the adaptation of the methodology for use in high-throughput screening, Durens et al. have described a method for multiplexing MEA recordings and Ca2+ imaging to examine local microcircuits in 3D brain organoids and assess inter-experimental consistency, necessary for drug screening [160]. Examination of such circuit-based mechanisms in hiPSC-derived systems would be of great interest and importance for the development of new and more efficient drugs. However, while it seems a very attractive readout for the development of high-throughput drug screening platforms, one should be careful with the interpretation of MEA and calcium imaging results as chemically induced effects on neuronal network activity, such as spontaneous firing and bursting behavior, depend on the ratio of inhibitory to excitatory neurons that are present in the culture system and may vary either due to the differentiation and enrichment protocols applied or to intrinsic defects of the lines used. Therefore, hiPSC-derived neuronal models must be carefully characterized prior to large-scale functional applications in drug screening.
7. Artificial Intelligence Technologies
Concurrent with advances in imaging technologies and the morphological, biochemical or functional analyses of neuronal populations has been the increase in volume and complexity of data sets generated. To exploit the ever-growing amount of data that become available, computational techniques are constantly evolving. As a result, artificial intelligence technologies such as deep learning and machine learning methods have emerged to identify meaningful patterns and cellular features that may be interpreted into novel biological insights. Still, these approaches have yet to be developed and adapted for high-throughput analyses in the medical and biological sciences. The creation of automated platforms that can perform phenotypic screens, process raw data and analyze them using deep neural network algorithms will increase both the capacity of the screens as well as the quality and size of the data collected. Several groups are trying to develop new methods that would have the potential to be used in large-scale phenotypic screens of hiPSC-based models. For example, Schwartz et al. developed a 3D model of hiPSC-derived neural tissue constructs comprising diverse neuronal and glial populations, interconnected vascular networks, and ramified microglia by seeding neural precursor cells on synthetic hydrogels [161]. Machine learning was used to build a predictive model from the changes occurring in global gene expression resulting from exposure to toxic and nontoxic training chemicals. This combined strategy reveals the value of human cell-based assays for predictive toxicology. Similarly, Monzel et al. developed a pipeline for a machine learning method, allowing for detailed image-based cell profiling and toxicity prediction in brain organoids treated with the neurotoxic compound 6-hydroxydopamine (6-OHDA) [162].
The analysis of large biological data sets derived from high-throughput screens assessing simultaneously multiple morphological and functional parameters in hiPSC-based platforms is time-consuming and will certainly benefit from artificial intelligence methodologies. Machine learning has the potential to shrink drug discovery timelines, helping researchers accelerate drug discovery and ultimately patients obtain disease-modifying therapies. Because these methodologies are expected to make the quest for new pharmaceuticals not only quicker, but also cheaper and more effective, leading biopharmaceutical companies to begin to embrace artificial intelligence platforms in their pursuit for new therapies. However, in the short term, these technologies have a number of challenges to overcome, especially when combined with automation.
8. Concluding Remarks
Many hiPSC lines have been generated from patients with familial or sporadic PD uncovering known or previously unrecognized disease-relevant phenotypes that, in many cases, could be effectively restored using small molecules. These investigations have laid the foreground for developing bioassays for screening small or larger chemical libraries in the search of lead compounds that may evolve into PD disease-modifying therapeutics. However, robust assays still need to be established before hiPSC-based systems become amenable to high-throughput technologies. In the meantime, more advanced co-culture systems encompassing neurons and glial cells or 3D brain organoids mimicking more closely the in vivo human situation are being developed to assist in PD studies and drug discovery. The necessity for functional assays to predict drug efficacy is also being recognized while technological advancements render complicated screens more feasible. Last but not least, the emergence of artificial intelligence over the past few years may prove to be a game-changing technology in drug discovery. Nevertheless, opportunities and challenges still remain ahead before these young technologies come to fruition and fulfill their promise for understanding and treating neurodegeneration in PD.
Author Contributions
All authors wrote and corrected the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Hellenic Foundation for Research and Innovation 899-PARKINSynapse grant to G.K.; a Stavros Niarchos Foundation grant to the Hellenic Pasteur Institute as part of the Foundation’s initiative to support the Greek Research Center ecosystem; the Greek General Secretariat for Research and Technology grant BIOIMAGING-GR MIS 5002755 implemented under the Action “Reinforcement of Research and Innovation Infrastructure”, funded by the Operational Programme “Competitiveness, Entrepreneurship and Innovation” (NSRF 2014–2020) and co-financed by Greece and the European Union (European Regional Development Fund).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Abbreviations
| PD | Parkinson’s disease |
| SNCA (PARK1/4) | α-synuclein gene |
| LRRK2 (PARK8) | leucine-rich repeat kinase 2 gene |
| PINK1 (PARK6) | PTEN-induced putative kinase 1 |
| DJ-1 (PARK7) | protein deglycase |
| ATP13A2 (PARK9) | endo-/lysosomal-associated P5 type transport ATPase |
| GBA | β-glucocerebrosidase gene |
| αSyn | α-synuclein protein |
| hiPSC | human induced pluripotent stem cell |
| GMP | good manufacturing practices |
| ROS | reactive oxygen species |
| ER | endoplasmic reticulum |
| GCase | β-glucocerebrosidase |
| ASO | antisense oligonucleotide |
| 6-OHDA | 6-hydroxydopamine |
| LDH | lactate dehydrogenase |
| OPA1 | mitochondrial dynamin-like GTPase |
| PET | positron emission tomography |
| ECM | extracellular matrix |
| MEA | multi-electrode array |
References
- Lees, A.J.; Hardy, J.; Revesz, T. Parkinson’s disease. Lancet 2009, 373, 2055–2066. [Google Scholar] [CrossRef]
- Kalia, L.V.; Lang, A.E. Parkinson’s disease. Lancet 2015, 386, 896–912. [Google Scholar] [CrossRef]
- Pfeiffer, R.F. Non-motor symptoms in Parkinson’s disease. Parkinsonism Relat. Disord. 2016, 22 (Suppl. 1), S119–S122. [Google Scholar] [CrossRef] [PubMed]
- Klein, C.; Westenberger, A. Genetics of Parkinson’s disease. Cold Spring Harb. Perspect. Med. 2012, 2, a008888. [Google Scholar] [CrossRef] [PubMed]
- Dettmer, U.; Newman, A.J.; Soldner, F.; Luth, E.S.; Kim, N.C.; von Saucken, V.E.; Sanderson, J.B.; Jaenisch, R.; Bartels, T.; Selkoe, D. Parkinson-causing alpha-synuclein missense mutations shift native tetramers to monomers as a mechanism for disease initiation. Nat. Commun. 2015, 6, 7314. [Google Scholar] [CrossRef]
- Zimprich, A.; Biskup, S.; Leitner, P.; Lichtner, P.; Farrer, M.; Lincoln, S.; Kachergus, J.; Hulihan, M.; Uitti, R.J.; Calne, D.B.; et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron 2004, 44, 601–607. [Google Scholar] [CrossRef]
- Valente, E.M.; Abou-Sleiman, P.M.; Caputo, V.; Muqit, M.M.; Harvey, K.; Gispert, S.; Ali, Z.; Del Turco, D.; Bentivoglio, A.R.; Healy, D.G.; et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science 2004, 304, 1158–1160. [Google Scholar] [CrossRef]
- Kitada, T.; Asakawa, S.; Hattori, N.; Matsumine, H.; Yamamura, Y.; Minoshima, S.; Yokochi, M.; Mizuno, Y.; Shimizu, N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 1998, 392, 605–608. [Google Scholar] [CrossRef]
- Bonifati, V.; Rizzu, P.; van Baren, M.J.; Schaap, O.; Breedveld, G.J.; Krieger, E.; Dekker, M.C.; Squitieri, F.; Ibanez, P.; Joosse, M.; et al. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science 2003, 299, 256–259. [Google Scholar] [CrossRef]
- Ramirez, A.; Heimbach, A.; Grundemann, J.; Stiller, B.; Hampshire, D.; Cid, L.P.; Goebel, I.; Mubaidin, A.F.; Wriekat, A.L.; Roeper, J.; et al. Hereditary parkinsonism with dementia is caused by mutations in ATP13A2, encoding a lysosomal type 5 P-type ATPase. Nat. Genet. 2006, 38, 1184–1191. [Google Scholar] [CrossRef]
- Goker-Alpan, O.; Schiffmann, R.; LaMarca, M.E.; Nussbaum, R.L.; McInerney-Leo, A.; Sidransky, E. Parkinsonism among Gaucher disease carriers. J. Med. Genet. 2004, 41, 937–940. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Jeon, B.S.; Jenner, P. Hallmarks of Treatment Aspects: Parkinson’s Disease Throughout Centuries Including l-Dopa. Int. Rev. Neurobiol. 2017, 132, 295–343. [Google Scholar] [CrossRef] [PubMed]
- Kulisevsky, J.; Oliveira, L.; Fox, S.H. Update in therapeutic strategies for Parkinson’s disease. Curr. Opin. Neurol. 2018, 31, 439–447. [Google Scholar] [CrossRef] [PubMed]
- Dijk, J.M.; Espay, A.J.; Katzenschlager, R.; de Bie, R.M.A. The Choice Between Advanced Therapies for Parkinson’s Disease Patients: Why, What, and When? J. Parkinsons Dis. 2020. [Google Scholar] [CrossRef] [PubMed]
- Fields, C.R.; Bengoa-Vergniory, N.; Wade-Martins, R. Targeting Alpha-Synuclein as a Therapy for Parkinson’s Disease. Front. Mol. Neurosci. 2019, 12, 299. [Google Scholar] [CrossRef] [PubMed]
- Polymeropoulos, M.H.; Lavedan, C.; Leroy, E.; Ide, S.E.; Dehejia, A.; Dutra, A.; Pike, B.; Root, H.; Rubenstein, J.; Boyer, R.; et al. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science 1997, 276, 2045–2047. [Google Scholar] [CrossRef]
- Spillantini, M.G.; Schmidt, M.L.; Lee, V.M.; Trojanowski, J.Q.; Jakes, R.; Goedert, M. Alpha-synuclein in Lewy bodies. Nature 1997, 388, 839–840. [Google Scholar] [CrossRef]
- Goedert, M.; Jakes, R.; Spillantini, M.G. The Synucleinopathies: Twenty Years On. J. Parkinsons Dis. 2017, 7, S51–S69. [Google Scholar] [CrossRef]
- Simon-Sanchez, J.; Schulte, C.; Bras, J.M.; Sharma, M.; Gibbs, J.R.; Berg, D.; Paisan-Ruiz, C.; Lichtner, P.; Scholz, S.W.; Hernandez, D.G.; et al. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat. Genet. 2009, 41, 1308–1312. [Google Scholar] [CrossRef]
- Petrucci, S.; Ginevrino, M.; Valente, E.M. Phenotypic spectrum of alpha-synuclein mutations: New insights from patients and cellular models. Parkinsonism Relat. Disord. 2016, 22 (Suppl. 1), S16–S20. [Google Scholar] [CrossRef]
- Lashuel, H.A.; Overk, C.R.; Oueslati, A.; Masliah, E. The many faces of alpha-synuclein: From structure and toxicity to therapeutic target. Nat. Rev. Neurosci. 2013, 14, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Savitt, D.; Jankovic, J. Targeting alpha-Synuclein in Parkinson’s Disease: Progress Towards the Development of Disease-Modifying Therapeutics. Drugs 2019, 79, 797–810. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.; Wong, Y.C.; Ysselstein, D.; Severino, A.; Krainc, D. Synaptic, Mitochondrial, and Lysosomal Dysfunction in Parkinson’s Disease. Trends Neurosci 2019, 42, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Shahmoradian, S.H.; Lewis, A.J.; Genoud, C.; Hench, J.; Moors, T.E.; Navarro, P.P.; Castano-Diez, D.; Schweighauser, G.; Graff-Meyer, A.; Goldie, K.N.; et al. Lewy pathology in Parkinson’s disease consists of crowded organelles and lipid membranes. Nat. Neurosci. 2019, 22, 1099–1109. [Google Scholar] [CrossRef] [PubMed]
- Caligiore, D.; Helmich, R.C.; Hallett, M.; Moustafa, A.A.; Timmermann, L.; Toni, I.; Baldassarre, G. Parkinson’s disease as a system-level disorder. NPJ Parkinsons Dis. 2016, 2, 16025. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef]
- Parmar, M.; Grealish, S.; Henchcliffe, C. The future of stem cell therapies for Parkinson disease. Nat. Rev. Neurosci. 2020, 21, 103–115. [Google Scholar] [CrossRef]
- Kim, T.W.; Koo, S.Y.; Studer, L. Pluripotent Stem Cell Therapies for Parkinson Disease: Present Challenges and Future Opportunities. Front. Cell Dev. Biol. 2020, 8, 729. [Google Scholar] [CrossRef]
- Park, I.H.; Arora, N.; Huo, H.; Maherali, N.; Ahfeldt, T.; Shimamura, A.; Lensch, M.W.; Cowan, C.; Hochedlinger, K.; Daley, G.Q. Disease-specific induced pluripotent stem cells. Cell 2008, 134, 877–886. [Google Scholar] [CrossRef]
- Soldner, F.; Hockemeyer, D.; Beard, C.; Gao, Q.; Bell, G.W.; Cook, E.G.; Hargus, G.; Blak, A.; Cooper, O.; Mitalipova, M.; et al. Parkinson’s disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell 2009, 136, 964–977. [Google Scholar] [CrossRef]
- Marotta, N.; Kim, S.; Krainc, D. Organoid and pluripotent stem cells in Parkinson’s disease modeling: An expert view on their value to drug discovery. Expert Opin. Drug Discov. 2020, 15, 427–441. [Google Scholar] [CrossRef] [PubMed]
- Weykopf, B.; Haupt, S.; Jungverdorben, J.; Flitsch, L.J.; Hebisch, M.; Liu, G.H.; Suzuki, K.; Belmonte, J.C.I.; Peitz, M.; Blaess, S.; et al. Induced pluripotent stem cell-based modeling of mutant LRRK2-associated Parkinson’s disease. Eur. J. Neurosci. 2019, 49, 561–589. [Google Scholar] [CrossRef] [PubMed]
- Sison, S.L.; Vermilyea, S.C.; Emborg, M.E.; Ebert, A.D. Using Patient-Derived Induced Pluripotent Stem Cells to Identify Parkinson’s Disease-Relevant Phenotypes. Curr. Neurol. Neurosci. Rep. 2018, 18, 84. [Google Scholar] [CrossRef] [PubMed]
- Bendor, J.T.; Logan, T.P.; Edwards, R.H. The function of alpha-synuclein. Neuron 2013, 79, 1044–1066. [Google Scholar] [CrossRef] [PubMed]
- Wong, Y.C.; Krainc, D. alpha-synuclein toxicity in neurodegeneration: Mechanism and therapeutic strategies. Nat. Med. 2017, 23, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ryan, S.D.; Dolatabadi, N.; Chan, S.F.; Zhang, X.; Akhtar, M.W.; Parker, J.; Soldner, F.; Sunico, C.R.; Nagar, S.; Talantova, M.; et al. Isogenic human iPSC Parkinson’s model shows nitrosative stress-induced dysfunction in MEF2-PGC1alpha transcription. Cell 2013, 155, 1351–1364. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.Y.; Khurana, V.; Auluck, P.K.; Tardiff, D.F.; Mazzulli, J.R.; Soldner, F.; Baru, V.; Lou, Y.; Freyzon, Y.; Cho, S.; et al. Identification and rescue of alpha-synuclein toxicity in Parkinson patient-derived neurons. Science 2013, 342, 983–987. [Google Scholar] [CrossRef]
- Kouroupi, G.; Taoufik, E.; Vlachos, I.S.; Tsioras, K.; Antoniou, N.; Papastefanaki, F.; Chroni-Tzartou, D.; Wrasidlo, W.; Bohl, D.; Stellas, D.; et al. Defective synaptic connectivity and axonal neuropathology in a human iPSC-based model of familial Parkinson’s disease. Proc. Natl. Acad. Sci. USA 2017, 114, E3679–E3688. [Google Scholar] [CrossRef]
- Little, D.; Luft, C.; Mosaku, O.; Lorvellec, M.; Yao, Z.; Paillusson, S.; Kriston-Vizi, J.; Gandhi, S.; Abramov, A.Y.; Ketteler, R.; et al. A single cell high content assay detects mitochondrial dysfunction in iPSC-derived neurons with mutations in SNCA. Sci. Rep. 2018, 8, 9033. [Google Scholar] [CrossRef]
- Prots, I.; Grosch, J.; Brazdis, R.M.; Simmnacher, K.; Veber, V.; Havlicek, S.; Hannappel, C.; Krach, F.; Krumbiegel, M.; Schutz, O.; et al. alpha-Synuclein oligomers induce early axonal dysfunction in human iPSC-based models of synucleinopathies. Proc. Natl. Acad. Sci. USA 2018, 115, 7813–7818. [Google Scholar] [CrossRef]
- Byers, B.; Cord, B.; Nguyen, H.N.; Schule, B.; Fenno, L.; Lee, P.C.; Deisseroth, K.; Langston, J.W.; Pera, R.R.; Palmer, T.D. SNCA triplication Parkinson’s patient’s iPSC-derived DA neurons accumulate alpha-synuclein and are susceptible to oxidative stress. PLoS ONE 2011, 6, e26159. [Google Scholar] [CrossRef] [PubMed]
- Devine, M.J.; Ryten, M.; Vodicka, P.; Thomson, A.J.; Burdon, T.; Houlden, H.; Cavaleri, F.; Nagano, M.; Drummond, N.J.; Taanman, J.W.; et al. Parkinson’s disease induced pluripotent stem cells with triplication of the alpha-synuclein locus. Nat. Commun. 2011, 2, 440. [Google Scholar] [CrossRef] [PubMed]
- Flierl, A.; Oliveira, L.M.; Falomir-Lockhart, L.J.; Mak, S.K.; Hesley, J.; Soldner, F.; Arndt-Jovin, D.J.; Jaenisch, R.; Langston, J.W.; Jovin, T.M.; et al. Higher vulnerability and stress sensitivity of neuronal precursor cells carrying an alpha-synuclein gene triplication. PLoS ONE 2014, 9, e112413. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, L.M.; Falomir-Lockhart, L.J.; Botelho, M.G.; Lin, K.H.; Wales, P.; Koch, J.C.; Gerhardt, E.; Taschenberger, H.; Outeiro, T.F.; Lingor, P.; et al. Elevated alpha-synuclein caused by SNCA gene triplication impairs neuronal differentiation and maturation in Parkinson’s patient-derived induced pluripotent stem cells. Cell Death Dis. 2015, 6, e1994. [Google Scholar] [CrossRef] [PubMed]
- Heman-Ackah, S.M.; Manzano, R.; Hoozemans, J.J.M.; Scheper, W.; Flynn, R.; Haerty, W.; Cowley, S.A.; Bassett, A.R.; Wood, M.J.A. Alpha-synuclein induces the unfolded protein response in Parkinson’s disease SNCA triplication iPSC-derived neurons. Hum. Mol. Genet. 2017, 26, 4441–4450. [Google Scholar] [CrossRef]
- Ludtmann, M.H.R.; Angelova, P.R.; Horrocks, M.H.; Choi, M.L.; Rodrigues, M.; Baev, A.Y.; Berezhnov, A.V.; Yao, Z.; Little, D.; Banushi, B.; et al. alpha-synuclein oligomers interact with ATP synthase and open the permeability transition pore in Parkinson’s disease. Nat. Commun. 2018, 9, 2293. [Google Scholar] [CrossRef]
- Chen, C.; Turnbull, D.M.; Reeve, A.K. Mitochondrial Dysfunction in Parkinson’s Disease-Cause or Consequence? Biology (Basel) 2019, 8, 38. [Google Scholar] [CrossRef]
- Su, Y.C.; Qi, X. Inhibition of excessive mitochondrial fission reduced aberrant autophagy and neuronal damage caused by LRRK2 G2019S mutation. Hum. Mol. Genet. 2013, 22, 4545–4561. [Google Scholar] [CrossRef]
- Imaizumi, Y.; Okada, Y.; Akamatsu, W.; Koike, M.; Kuzumaki, N.; Hayakawa, H.; Nihira, T.; Kobayashi, T.; Ohyama, M.; Sato, S.; et al. Mitochondrial dysfunction associated with increased oxidative stress and alpha-synuclein accumulation in PARK2 iPSC-derived neurons and postmortem brain tissue. Mol. Brain 2012, 5, 35. [Google Scholar] [CrossRef]
- Aboud, A.A.; Tidball, A.M.; Kumar, K.K.; Neely, M.D.; Han, B.; Ess, K.C.; Hong, C.C.; Erikson, K.M.; Hedera, P.; Bowman, A.B. PARK2 patient neuroprogenitors show increased mitochondrial sensitivity to copper. Neurobiol. Dis. 2015, 73, 204–212. [Google Scholar] [CrossRef]
- Chung, S.Y.; Kishinevsky, S.; Mazzulli, J.R.; Graziotto, J.; Mrejeru, A.; Mosharov, E.V.; Puspita, L.; Valiulahi, P.; Sulzer, D.; Milner, T.A.; et al. Parkin and PINK1 Patient iPSC-Derived Midbrain Dopamine Neurons Exhibit Mitochondrial Dysfunction and alpha-Synuclein Accumulation. Stem Cell Rep. 2016, 7, 664–677. [Google Scholar] [CrossRef] [PubMed]
- Zanon, A.; Kalvakuri, S.; Rakovic, A.; Foco, L.; Guida, M.; Schwienbacher, C.; Serafin, A.; Rudolph, F.; Trilck, M.; Grunewald, A.; et al. SLP-2 interacts with Parkin in mitochondria and prevents mitochondrial dysfunction in Parkin-deficient human iPSC-derived neurons and Drosophila. Hum. Mol. Genet. 2017, 26, 2412–2425. [Google Scholar] [CrossRef] [PubMed]
- Cartelli, D.; Amadeo, A.; Calogero, A.M.; Casagrande, F.V.M.; De Gregorio, C.; Gioria, M.; Kuzumaki, N.; Costa, I.; Sassone, J.; Ciammola, A.; et al. Parkin absence accelerates microtubule aging in dopaminergic neurons. Neurobiol. Aging 2018, 61, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Azkona, G.; Lopez de Maturana, R.; Del Rio, P.; Sousa, A.; Vazquez, N.; Zubiarrain, A.; Jimenez-Blasco, D.; Bolanos, J.P.; Morales, B.; Auburger, G.; et al. LRRK2 Expression Is Deregulated in Fibroblasts and Neurons from Parkinson Patients with Mutations in PINK1. Mol. Neurobiol. 2018, 55, 506–516. [Google Scholar] [CrossRef]
- Schondorf, D.C.; Ivanyuk, D.; Baden, P.; Sanchez-Martinez, A.; De Cicco, S.; Yu, C.; Giunta, I.; Schwarz, L.K.; Di Napoli, G.; Panagiotakopoulou, V.; et al. The NAD+ Precursor Nicotinamide Riboside Rescues Mitochondrial Defects and Neuronal Loss in iPSC and Fly Models of Parkinson’s Disease. Cell Rep. 2018, 23, 2976–2988. [Google Scholar] [CrossRef]
- Shaltouki, A.; Sivapatham, R.; Pei, Y.; Gerencser, A.A.; Momcilovic, O.; Rao, M.S.; Zeng, X. Mitochondrial alterations by PARKIN in dopaminergic neurons using PARK2 patient-specific and PARK2 knockout isogenic iPSC lines. Stem Cell Rep. 2015, 4, 847–859. [Google Scholar] [CrossRef]
- Schwab, A.J.; Sison, S.L.; Meade, M.R.; Broniowska, K.A.; Corbett, J.A.; Ebert, A.D. Decreased Sirtuin Deacetylase Activity in LRRK2 G2019S iPSC-Derived Dopaminergic Neurons. Stem Cell Rep. 2017, 9, 1839–1852. [Google Scholar] [CrossRef]
- Iannielli, A.; Bido, S.; Folladori, L.; Segnali, A.; Cancellieri, C.; Maresca, A.; Massimino, L.; Rubio, A.; Morabito, G.; Caporali, L.; et al. Pharmacological Inhibition of Necroptosis Protects from Dopaminergic Neuronal Cell Death in Parkinson’s Disease Models. Cell Rep. 2018, 22, 2066–2079. [Google Scholar] [CrossRef]
- Ryan, T.; Bamm, V.V.; Stykel, M.G.; Coackley, C.L.; Humphries, K.M.; Jamieson-Williams, R.; Ambasudhan, R.; Mosser, D.D.; Lipton, S.A.; Harauz, G.; et al. Cardiolipin exposure on the outer mitochondrial membrane modulates alpha-synuclein. Nat. Commun. 2018, 9, 817. [Google Scholar] [CrossRef]
- Cooper, O.; Seo, H.; Andrabi, S.; Guardia-Laguarta, C.; Graziotto, J.; Sundberg, M.; McLean, J.R.; Carrillo-Reid, L.; Xie, Z.; Osborn, T.; et al. Pharmacological rescue of mitochondrial deficits in iPSC-derived neural cells from patients with familial Parkinson’s disease. Sci. Transl. Med. 2012, 4, 141ra190. [Google Scholar] [CrossRef]
- Toulorge, D.; Schapira, A.H.; Hajj, R. Molecular changes in the postmortem parkinsonian brain. J. Neurochem. 2016, 139 (Suppl. 1), 27–58. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.N.; Byers, B.; Cord, B.; Shcheglovitov, A.; Byrne, J.; Gujar, P.; Kee, K.; Schule, B.; Dolmetsch, R.E.; Langston, W.; et al. LRRK2 mutant iPSC-derived DA neurons demonstrate increased susceptibility to oxidative stress. Cell Stem Cell 2011, 8, 267–280. [Google Scholar] [CrossRef] [PubMed]
- Ohta, E.; Nihira, T.; Uchino, A.; Imaizumi, Y.; Okada, Y.; Akamatsu, W.; Takahashi, K.; Hayakawa, H.; Nagai, M.; Ohyama, M.; et al. I2020T mutant LRRK2 iPSC-derived neurons in the Sagamihara family exhibit increased Tau phosphorylation through the AKT/GSK-3beta signaling pathway. Hum. Mol. Genet. 2015, 24, 4879–4900. [Google Scholar] [CrossRef]
- Jiang, H.; Ren, Y.; Yuen, E.Y.; Zhong, P.; Ghaedi, M.; Hu, Z.; Azabdaftari, G.; Nakaso, K.; Yan, Z.; Feng, J. Parkin controls dopamine utilization in human midbrain dopaminergic neurons derived from induced pluripotent stem cells. Nat. Commun. 2012, 3, 668. [Google Scholar] [CrossRef]
- Vera, E.; Bosco, N.; Studer, L. Generating Late-Onset Human iPSC-Based Disease Models by Inducing Neuronal Age-Related Phenotypes through Telomerase Manipulation. Cell Rep. 2016, 17, 1184–1192. [Google Scholar] [CrossRef]
- Khurana, V.; Peng, J.; Chung, C.Y.; Auluck, P.K.; Fanning, S.; Tardiff, D.F.; Bartels, T.; Koeva, M.; Eichhorn, S.W.; Benyamini, H.; et al. Genome-Scale Networks Link Neurodegenerative Disease Genes to alpha-Synuclein through Specific Molecular Pathways. Cell Syst. 2017, 4, 157–170 e114. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Danes, A.; Richaud-Patin, Y.; Carballo-Carbajal, I.; Jimenez-Delgado, S.; Caig, C.; Mora, S.; Di Guglielmo, C.; Ezquerra, M.; Patel, B.; Giralt, A.; et al. Disease-specific phenotypes in dopamine neurons from human iPS-based models of genetic and sporadic Parkinson’s disease. EMBO Mol. Med. 2012, 4, 380–395. [Google Scholar] [CrossRef] [PubMed]
- Schondorf, D.C.; Aureli, M.; McAllister, F.E.; Hindley, C.J.; Mayer, F.; Schmid, B.; Sardi, S.P.; Valsecchi, M.; Hoffmann, S.; Schwarz, L.K.; et al. iPSC-derived neurons from GBA1-associated Parkinson’s disease patients show autophagic defects and impaired calcium homeostasis. Nat. Commun. 2014, 5, 4028. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, H.J.; Hartfield, E.M.; Christian, H.C.; Emmanoulidou, E.; Zheng, Y.; Booth, H.; Bogetofte, H.; Lang, C.; Ryan, B.J.; Sardi, S.P.; et al. ER Stress and Autophagic Perturbations Lead to Elevated Extracellular alpha-Synuclein in GBA-N370S Parkinson’s iPSC-Derived Dopamine Neurons. Stem Cell Rep. 2016, 6, 342–356. [Google Scholar] [CrossRef]
- Liu, G.H.; Qu, J.; Suzuki, K.; Nivet, E.; Li, M.; Montserrat, N.; Yi, F.; Xu, X.; Ruiz, S.; Zhang, W.; et al. Progressive degeneration of human neural stem cells caused by pathogenic LRRK2. Nature 2012, 491, 603–607. [Google Scholar] [CrossRef]
- Reinhardt, P.; Schmid, B.; Burbulla, L.F.; Schondorf, D.C.; Wagner, L.; Glatza, M.; Hoing, S.; Hargus, G.; Heck, S.A.; Dhingra, A.; et al. Genetic correction of a LRRK2 mutation in human iPSCs links parkinsonian neurodegeneration to ERK-dependent changes in gene expression. Cell Stem Cell 2013, 12, 354–367. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Goke, J.; Cukuroglu, E.; Dranias, M.R.; VanDongen, A.M.; Stanton, L.W. Molecular Features Underlying Neurodegeneration Identified through In Vitro Modeling of Genetically Diverse Parkinson’s Disease Patients. Cell Rep. 2016, 15, 2411–2426. [Google Scholar] [CrossRef] [PubMed]
- Qing, X.; Walter, J.; Jarazo, J.; Arias-Fuenzalida, J.; Hillje, A.L.; Schwamborn, J.C. CRISPR/Cas9 and piggyBac-mediated footprint-free LRRK2-G2019S knock-in reveals neuronal complexity phenotypes and alpha-Synuclein modulation in dopaminergic neurons. Stem Cell Res. 2017, 24, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Korecka, J.A.; Talbot, S.; Osborn, T.M.; de Leeuw, S.M.; Levy, S.A.; Ferrari, E.J.; Moskites, A.; Atkinson, E.; Jodelka, F.M.; Hinrich, A.J.; et al. Neurite Collapse and Altered ER Ca(2+) Control in Human Parkinson Disease Patient iPSC-Derived Neurons with LRRK2 G2019S Mutation. Stem Cell Rep. 2019, 12, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Jiang, H.; Hu, Z.; Fan, K.; Wang, J.; Janoschka, S.; Wang, X.; Ge, S.; Feng, J. Parkin mutations reduce the complexity of neuronal processes in iPSC-derived human neurons. Stem Cells 2015, 33, 68–78. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Aflaki, E.; Borger, D.K.; Moaven, N.; Stubblefield, B.K.; Rogers, S.A.; Patnaik, S.; Schoenen, F.J.; Westbroek, W.; Zheng, W.; Sullivan, P.; et al. A New Glucocerebrosidase Chaperone Reduces alpha-Synuclein and Glycolipid Levels in iPSC-Derived Dopaminergic Neurons from Patients with Gaucher Disease and Parkinsonism. J. Neurosci. 2016, 36, 7441–7452. [Google Scholar] [CrossRef] [PubMed]
- Mazzulli, J.R.; Zunke, F.; Tsunemi, T.; Toker, N.J.; Jeon, S.; Burbulla, L.F.; Patnaik, S.; Sidransky, E.; Marugan, J.J.; Sue, C.M.; et al. Activation of beta-Glucocerebrosidase Reduces Pathological alpha-Synuclein and Restores Lysosomal Function in Parkinson’s Patient Midbrain Neurons. J. Neurosci. 2016, 36, 7693–7706. [Google Scholar] [CrossRef]
- Burbulla, L.F.; Jeon, S.; Zheng, J.; Song, P.; Silverman, R.B.; Krainc, D. A modulator of wild-type glucocerebrosidase improves pathogenic phenotypes in dopaminergic neuronal models of Parkinson’s disease. Sci. Transl. Med. 2019, 11. [Google Scholar] [CrossRef]
- Kim, M.J.; Jeon, S.; Burbulla, L.F.; Krainc, D. Acid ceramidase inhibition ameliorates alpha-synuclein accumulation upon loss of GBA1 function. Hum. Mol. Genet. 2018, 27, 1972–1988. [Google Scholar] [CrossRef]
- Burbulla, L.F.; Song, P.; Mazzulli, J.R.; Zampese, E.; Wong, Y.C.; Jeon, S.; Santos, D.P.; Blanz, J.; Obermaier, C.D.; Strojny, C.; et al. Dopamine oxidation mediates mitochondrial and lysosomal dysfunction in Parkinson’s disease. Science 2017, 357, 1255–1261. [Google Scholar] [CrossRef]
- Plowey, E.D.; Cherra, S.J., 3rd; Liu, Y.J.; Chu, C.T. Role of autophagy in G2019S-LRRK2-associated neurite shortening in differentiated SH-SY5Y cells. J. Neurochem. 2008, 105, 1048–1056. [Google Scholar] [CrossRef] [PubMed]
- Toth, G.; Gardai, S.J.; Zago, W.; Bertoncini, C.W.; Cremades, N.; Roy, S.L.; Tambe, M.A.; Rochet, J.C.; Galvagnion, C.; Skibinski, G.; et al. Targeting the intrinsically disordered structural ensemble of alpha-synuclein by small molecules as a potential therapeutic strategy for Parkinson’s disease. PLoS ONE 2014, 9, e87133. [Google Scholar] [CrossRef] [PubMed]
- Wrasidlo, W.; Tsigelny, I.F.; Price, D.L.; Dutta, G.; Rockenstein, E.; Schwarz, T.C.; Ledolter, K.; Bonhaus, D.; Paulino, A.; Eleuteri, S.; et al. A de novo compound targeting alpha-synuclein improves deficits in models of Parkinson’s disease. Brain 2016, 139, 3217–3236. [Google Scholar] [CrossRef] [PubMed]
- Simmnacher, K.; Lanfer, J.; Rizo, T.; Kaindl, J.; Winner, B. Modeling Cell-Cell Interactions in Parkinson’s Disease Using Human Stem Cell-Based Models. Front. Cell. Neurosci. 2019, 13, 571. [Google Scholar] [CrossRef] [PubMed]
- Carelli, V.; Musumeci, O.; Caporali, L.; Zanna, C.; La Morgia, C.; Del Dotto, V.; Porcelli, A.M.; Rugolo, M.; Valentino, M.L.; Iommarini, L.; et al. Syndromic parkinsonism and dementia associated with OPA1 missense mutations. Ann. Neurol. 2015, 78, 21–38. [Google Scholar] [CrossRef]
- Zheng, W.; Thorne, N.; McKew, J.C. Phenotypic screens as a renewed approach for drug discovery. Drug Discov. Today 2013, 18, 1067–1073. [Google Scholar] [CrossRef]
- Goodfellow, P.N. Genomics and drugs. An interview conducted by Holger Breithaupt and Caroline Hadley. EMBO Rep. 2004, 5, 843–846. [Google Scholar] [CrossRef]
- Burbaum, J.J.; Sigal, N.H. New technologies for high-throughput screening. Curr. Opin. Chem. Biol. 1997, 1, 72–78. [Google Scholar] [CrossRef]
- Gitler, A.D.; Dhillon, P.; Shorter, J. Neurodegenerative disease: Models, mechanisms, and a new hope. Dis. Model. Mech. 2017, 10, 499–502. [Google Scholar] [CrossRef]
- Zhang, M.; Luo, G.; Zhou, Y.; Wang, S.; Zhong, Z. Phenotypic screens targeting neurodegenerative diseases. J. Biomol. Screen. 2014, 19, 1–16. [Google Scholar] [CrossRef][Green Version]
- Swinney, D.C.; Anthony, J. How were new medicines discovered? Nat. Rev. Drug Discov. 2011, 10, 507–519. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, A.; Ishikawa, K.I.; Inoshita, T.; Shiba-Fukushima, K.; Saiki, S.; Hatano, T.; Mori, A.; Oji, Y.; Okuzumi, A.; Li, Y.; et al. Identifying Therapeutic Agents for Amelioration of Mitochondrial Clearance Disorder in Neurons of Familial Parkinson Disease. Stem Cell Rep. 2020, 14, 1060–1075. [Google Scholar] [CrossRef] [PubMed]
- Tabata, Y.; Imaizumi, Y.; Sugawara, M.; Andoh-Noda, T.; Banno, S.; Chai, M.; Sone, T.; Yamazaki, K.; Ito, M.; Tsukahara, K.; et al. T-type Calcium Channels Determine the Vulnerability of Dopaminergic Neurons to Mitochondrial Stress in Familial Parkinson Disease. Stem Cell Rep. 2018, 11, 1171–1184. [Google Scholar] [CrossRef]
- Antoniou, N.; Prodromidou, K.; Kouroupi, G.; Samiotaki, M.; Panayotou, G.; Xilouri, M.; Stefanis, L.; Grailhe, R.; Taoufik, E.; Matsas, R. High Content Screening and Proteomic Analysis Identify the Kinase Inhibitor BX795 as a Potent Neuroprotective Compound in a Patient-Derived Model of Parkinson’s Disease. BioRxiv 2020. [Google Scholar] [CrossRef]
- Oksanen, M.; Lehtonen, S.; Jaronen, M.; Goldsteins, G.; Hamalainen, R.H.; Koistinaho, J. Astrocyte alterations in neurodegenerative pathologies and their modeling in human induced pluripotent stem cell platforms. Cell Mol. Life Sci. 2019, 76, 2739–2760. [Google Scholar] [CrossRef]
- Heneka, M.T. Microglia take centre stage in neurodegenerative disease. Nat. Rev. Immunol. 2019, 19, 79–80. [Google Scholar] [CrossRef]
- Braak, H.; Sastre, M.; Del Tredici, K. Development of alpha-synuclein immunoreactive astrocytes in the forebrain parallels stages of intraneuronal pathology in sporadic Parkinson’s disease. Acta Neuropathol. 2007, 114, 231–241. [Google Scholar] [CrossRef]
- Kovacs, G.G.; Wagner, U.; Dumont, B.; Pikkarainen, M.; Osman, A.A.; Streichenberger, N.; Leisser, I.; Verchere, J.; Baron, T.; Alafuzoff, I.; et al. An antibody with high reactivity for disease-associated alpha-synuclein reveals extensive brain pathology. Acta Neuropathol. 2012, 124, 37–50. [Google Scholar] [CrossRef]
- Takeda, A.; Hashimoto, M.; Mallory, M.; Sundsumo, M.; Hansen, L.; Masliah, E. C-terminal alpha-synuclein immunoreactivity in structures other than Lewy bodies in neurodegenerative disorders. Acta Neuropathol. 2000, 99, 296–304. [Google Scholar] [CrossRef]
- Terada, S.; Ishizu, H.; Yokota, O.; Tsuchiya, K.; Nakashima, H.; Ishihara, T.; Fujita, D.; Ueda, K.; Ikeda, K.; Kuroda, S. Glial involvement in diffuse Lewy body disease. Acta Neuropathol. 2003, 105, 163–169. [Google Scholar] [CrossRef]
- Wakabayashi, K.; Hayashi, S.; Yoshimoto, M.; Kudo, H.; Takahashi, H. NACP/alpha-synuclein-positive filamentous inclusions in astrocytes and oligodendrocytes of Parkinson’s disease brains. Acta Neuropathol. 2000, 99, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Barcia, C.; Ros, C.M.; Annese, V.; Gomez, A.; Ros-Bernal, F.; Aguado-Yera, D.; Martinez-Pagan, M.E.; de Pablos, V.; Fernandez-Villalba, E.; Herrero, M.T. IFN-gamma signaling, with the synergistic contribution of TNF-alpha, mediates cell specific microglial and astroglial activation in experimental models of Parkinson’s disease. Cell Death Dis. 2011, 2, e142. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.L.; Long, C.X.; Sun, L.; Xie, C.; Lin, X.; Cai, H. Astrocytic expression of Parkinson’s disease-related A53T alpha-synuclein causes neurodegeneration in mice. Mol. Brain 2010, 3, 12. [Google Scholar] [CrossRef]
- Halliday, G.M.; Stevens, C.H. Glia: Initiators and progressors of pathology in Parkinson’s disease. Mov. Disord. 2011, 26, 6–17. [Google Scholar] [CrossRef] [PubMed]
- Loria, F.; Vargas, J.Y.; Bousset, L.; Syan, S.; Salles, A.; Melki, R.; Zurzolo, C. alpha-Synuclein transfer between neurons and astrocytes indicates that astrocytes play a role in degradation rather than in spreading. Acta Neuropathol. 2017, 134, 789–808. [Google Scholar] [CrossRef] [PubMed]
- Bartels, A.L.; Willemsen, A.T.; Doorduin, J.; de Vries, E.F.; Dierckx, R.A.; Leenders, K.L. [11C]-PK11195 PET: Quantification of neuroinflammation and a monitor of anti-inflammatory treatment in Parkinson’s disease? Parkinsonism Relat. Disord. 2010, 16, 57–59. [Google Scholar] [CrossRef]
- Gerhard, A.; Pavese, N.; Hotton, G.; Turkheimer, F.; Es, M.; Hammers, A.; Eggert, K.; Oertel, W.; Banati, R.B.; Brooks, D.J. In vivo imaging of microglial activation with [11C](R)-PK11195 PET in idiopathic Parkinson’s disease. Neurobiol. Dis. 2006, 21, 404–412. [Google Scholar] [CrossRef]
- Sanchez-Guajardo, V.; Febbraro, F.; Kirik, D.; Romero-Ramos, M. Microglia acquire distinct activation profiles depending on the degree of alpha-synuclein neuropathology in a rAAV based model of Parkinson’s disease. PLoS ONE 2010, 5, e8784. [Google Scholar] [CrossRef]
- McGeer, P.L.; McGeer, E.G.; Kawamata, T.; Yamada, T.; Akiyama, H. Reactions of the immune system in chronic degenerative neurological diseases. Can. J. Neurol. Sci. 1991, 18, 376–379. [Google Scholar] [CrossRef]
- Hoenen, C.; Gustin, A.; Birck, C.; Kirchmeyer, M.; Beaume, N.; Felten, P.; Grandbarbe, L.; Heuschling, P.; Heurtaux, T. Alpha-Synuclein Proteins Promote Pro-Inflammatory Cascades in Microglia: Stronger Effects of the A53T Mutant. PLoS ONE 2016, 11, e0162717. [Google Scholar] [CrossRef]
- Czlonkowska, A.; Kohutnicka, M.; Kurkowska-Jastrzebska, I.; Czlonkowski, A. Microglial reaction in MPTP (1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine) induced Parkinson’s disease mice model. Neurodegeneration 1996, 5, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Ransohoff, R.M. A polarizing question: Do M1 and M2 microglia exist? Nat. Neurosci. 2016, 19, 987–991. [Google Scholar] [CrossRef] [PubMed]
- Wyss-Coray, T.; Mucke, L. Inflammation in neurodegenerative disease—A double-edged sword. Neuron 2002, 35, 419–432. [Google Scholar] [CrossRef]
- Wu, D.C.; Jackson-Lewis, V.; Vila, M.; Tieu, K.; Teismann, P.; Vadseth, C.; Choi, D.K.; Ischiropoulos, H.; Przedborski, S. Blockade of microglial activation is neuroprotective in the 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine mouse model of Parkinson disease. J. Neurosci. 2002, 22, 1763–1771. [Google Scholar] [CrossRef]
- Manocha, G.D.; Floden, A.M.; Puig, K.L.; Nagamoto-Combs, K.; Scherzer, C.R.; Combs, C.K. Defining the contribution of neuroinflammation to Parkinson’s disease in humanized immune system mice. Mol. Neurodegener. 2017, 12, 17. [Google Scholar] [CrossRef]
- Marinova-Mutafchieva, L.; Sadeghian, M.; Broom, L.; Davis, J.B.; Medhurst, A.D.; Dexter, D.T. Relationship between microglial activation and dopaminergic neuronal loss in the substantia nigra: A time course study in a 6-hydroxydopamine model of Parkinson’s disease. J. Neurochem. 2009, 110, 966–975. [Google Scholar] [CrossRef]
- Walsh, S.; Finn, D.P.; Dowd, E. Time-course of nigrostriatal neurodegeneration and neuroinflammation in the 6-hydroxydopamine-induced axonal and terminal lesion models of Parkinson’s disease in the rat. Neuroscience 2011, 175, 251–261. [Google Scholar] [CrossRef]
- Stefanova, N.; Fellner, L.; Reindl, M.; Masliah, E.; Poewe, W.; Wenning, G.K. Toll-like receptor 4 promotes alpha-synuclein clearance and survival of nigral dopaminergic neurons. Am. J. Pathol. 2011, 179, 954–963. [Google Scholar] [CrossRef]
- Lee, H.J.; Suk, J.E.; Bae, E.J.; Lee, S.J. Clearance and deposition of extracellular alpha-synuclein aggregates in microglia. Biochem. Biophys. Res. Commun. 2008, 372, 423–428. [Google Scholar] [CrossRef]
- Fellner, L.; Irschick, R.; Schanda, K.; Reindl, M.; Klimaschewski, L.; Poewe, W.; Wenning, G.K.; Stefanova, N. Toll-like receptor 4 is required for alpha-synuclein dependent activation of microglia and astroglia. Glia 2013, 61, 349–360. [Google Scholar] [CrossRef]
- Kim, C.; Ho, D.H.; Suk, J.E.; You, S.; Michael, S.; Kang, J.; Joong Lee, S.; Masliah, E.; Hwang, D.; Lee, H.J.; et al. Neuron-released oligomeric alpha-synuclein is an endogenous agonist of TLR2 for paracrine activation of microglia. Nat. Commun. 2013, 4, 1562. [Google Scholar] [CrossRef] [PubMed]
- Liddelow, S.A.; Guttenplan, K.A.; Clarke, L.E.; Bennett, F.C.; Bohlen, C.J.; Schirmer, L.; Bennett, M.L.; Munch, A.E.; Chung, W.S.; Peterson, T.C.; et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature 2017, 541, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Yun, S.P.; Kam, T.I.; Panicker, N.; Kim, S.; Oh, Y.; Park, J.S.; Kwon, S.H.; Park, Y.J.; Karuppagounder, S.S.; Park, H.; et al. Block of A1 astrocyte conversion by microglia is neuroprotective in models of Parkinson’s disease. Nat. Med. 2018, 24, 931–938. [Google Scholar] [CrossRef] [PubMed]
- Mavroeidi, P.; Arvanitaki, F.; Karakitsou, A.K.; Vetsi, M.; Kloukina, I.; Zweckstetter, M.; Giller, K.; Becker, S.; Sorrentino, Z.A.; Giasson, B.I.; et al. Endogenous oligodendroglial alpha-synuclein and TPPP/p25alpha orchestrate alpha-synuclein pathology in experimental multiple system atrophy models. Acta Neuropathol. 2019, 138, 415–441. [Google Scholar] [CrossRef]
- Alieva, A.K.; Zyrin, V.S.; Rudenok, M.M.; Kolacheva, A.A.; Shulskaya, M.V.; Ugryumov, M.V.; Slominsky, P.A.; Shadrina, M.I. Whole-Transcriptome Analysis of Mouse Models with MPTP-Induced Early Stages of Parkinson’s Disease Reveals Stage-Specific Response of Transcriptome and a Possible Role of Myelin-Linked Genes in Neurodegeneration. Mol. Neurobiol. 2018, 55, 7229–7241. [Google Scholar] [CrossRef]
- Booth, H.D.E.; Wessely, F.; Connor-Robson, N.; Rinaldi, F.; Vowles, J.; Browne, C.; Evetts, S.G.; Hu, M.T.; Cowley, S.A.; Webber, C.; et al. RNA sequencing reveals MMP2 and TGFB1 downregulation in LRRK2 G2019S Parkinson’s iPSC-derived astrocytes. Neurobiol.Dis. 2019, 129, 56–66. [Google Scholar] [CrossRef]
- di Domenico, A.; Carola, G.; Calatayud, C.; Pons-Espinal, M.; Munoz, J.P.; Richaud-Patin, Y.; Fernandez-Carasa, I.; Gut, M.; Faella, A.; Parameswaran, J.; et al. Patient-Specific iPSC-Derived Astrocytes Contribute to Non-Cell-Autonomous Neurodegeneration in Parkinson’s Disease. Stem Cell Rep. 2019, 12, 213–229. [Google Scholar] [CrossRef]
- Aflaki, E.; Stubblefield, B.K.; McGlinchey, R.P.; McMahon, B.; Ory, D.S.; Sidransky, E. A characterization of Gaucher iPS-derived astrocytes: Potential implications for Parkinson’s disease. Neurobiol. Dis. 2020, 134, 104647. [Google Scholar] [CrossRef]
- Sonninen, T.M.; Hamalainen, R.H.; Koskuvi, M.; Oksanen, M.; Shakirzyanova, A.; Wojciechowski, S.; Puttonen, K.; Naumenko, N.; Goldsteins, G.; Laham-Karam, N.; et al. Metabolic alterations in Parkinson’s disease astrocytes. Sci. Rep. 2020, 10, 14474. [Google Scholar] [CrossRef]
- Haenseler, W.; Zambon, F.; Lee, H.; Vowles, J.; Rinaldi, F.; Duggal, G.; Houlden, H.; Gwinn, K.; Wray, S.; Luk, K.C.; et al. Excess alpha-synuclein compromises phagocytosis in iPSC-derived macrophages. Sci. Rep. 2017, 7, 9003. [Google Scholar] [CrossRef]
- Lee, H.; Flynn, R.; Sharma, I.; Haberman, E.; Carling, P.J.; Nicholls, F.J.; Stegmann, M.; Vowles, J.; Haenseler, W.; Wade-Martins, R.; et al. LRRK2 Is Recruited to Phagosomes and Co-recruits RAB8 and RAB10 in Human Pluripotent Stem Cell-Derived Macrophages. Stem Cell Rep. 2020, 14, 940–955. [Google Scholar] [CrossRef] [PubMed]
- Speidel, A.; Felk, S.; Reinhardt, P.; Sterneckert, J.; Gillardon, F. Leucine-Rich Repeat Kinase 2 Influences Fate Decision of Human Monocytes Differentiated from Induced Pluripotent Stem Cells. PLoS ONE 2016, 11, e0165949. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Song, H.; Ming, G.L. Brain organoids: Advances, applications and challenges. Development 2019, 146. [Google Scholar] [CrossRef] [PubMed]
- Bolognin, S.; Fossepre, M.; Qing, X.; Jarazo, J.; Scancar, J.; Moreno, E.L.; Nickels, S.L.; Wasner, K.; Ouzren, N.; Walter, J.; et al. 3D Cultures of Parkinson’s Disease-Specific Dopaminergic Neurons for High Content Phenotyping and Drug Testing. Adv. Sci. 2019, 6, 1800927. [Google Scholar] [CrossRef]
- Kim, H.; Park, H.J.; Choi, H.; Chang, Y.; Park, H.; Shin, J.; Kim, J.; Lengner, C.J.; Lee, Y.K. Modeling G2019S-LRRK2 Sporadic Parkinson’s Disease in 3D Midbrain Organoids. Stem Cell Rep. 2019, 12, 518–531. [Google Scholar] [CrossRef]
- Smits, L.M.; Reinhardt, L.; Reinhardt, P.; Glatza, M.; Monzel, A.S.; Stanslowsky, N.; Rosato-Siri, M.D.; Zanon, A.; Antony, P.M.; Bellmann, J.; et al. Modeling Parkinson’s disease in midbrain-like organoids. NPJ Parkinsons Dis. 2019, 5, 5. [Google Scholar] [CrossRef]
- Brull, M.; Spreng, A.S.; Gutbier, S.; Loser, D.; Krebs, A.; Reich, M.; Kraushaar, U.; Britschgi, M.; Patsch, C.; Leist, M. Incorporation of stem cell-derived astrocytes into neuronal organoids to allow neuro-glial interactions in toxicological studies. ALTEX 2020, 37, 409–428. [Google Scholar] [CrossRef]
- Qian, X.; Su, Y.; Adam, C.D.; Deutschmann, A.U.; Pather, S.R.; Goldberg, E.M.; Su, K.; Li, S.; Lu, L.; Jacob, F.; et al. Sliced Human Cortical Organoids for Modeling Distinct Cortical Layer Formation. Cell Stem Cell 2020, 26, 766–781 e769. [Google Scholar] [CrossRef]
- Marton, R.M.; Miura, Y.; Sloan, S.A.; Li, Q.; Revah, O.; Levy, R.J.; Huguenard, J.R.; Pasca, S.P. Differentiation and maturation of oligodendrocytes in human three-dimensional neural cultures. Nat. Neurosci. 2019, 22, 484–491. [Google Scholar] [CrossRef]
- Madhavan, M.; Nevin, Z.S.; Shick, H.E.; Garrison, E.; Clarkson-Paredes, C.; Karl, M.; Clayton, B.L.L.; Factor, D.C.; Allan, K.C.; Barbar, L.; et al. Induction of myelinating oligodendrocytes in human cortical spheroids. Nat. Methods 2018, 15, 700–706. [Google Scholar] [CrossRef]
- Ormel, P.R.; Vieira de Sa, R.; van Bodegraven, E.J.; Karst, H.; Harschnitz, O.; Sneeboer, M.A.M.; Johansen, L.E.; van Dijk, R.E.; Scheefhals, N.; Berdenis van Berlekom, A.; et al. Microglia innately develop within cerebral organoids. Nat. Commun. 2018, 9, 4167. [Google Scholar] [CrossRef]
- Kwak, T.H.; Kang, J.H.; Hali, S.; Kim, J.; Kim, K.P.; Park, C.; Lee, J.H.; Ryu, H.K.; Na, J.E.; Jo, J.; et al. Generation of homogeneous midbrain organoids with in vivo-like cellular composition facilitates neurotoxin-based Parkinson’s disease modeling. Stem Cells 2020, 38, 727–740. [Google Scholar] [CrossRef] [PubMed]
- Yadav, A.; Seth, B.; Chaturvedi, R.K. Brain Organoids: Tiny Mirrors of Human Neurodevelopment and Neurological Disorders. Neuroscientist 2020, 1073858420943192. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, Y.; Cui, K.; Guo, Y.; Zhang, X.; Qin, J. Advances in Hydrogels in Organoids and Organs-on-a-Chip. Adv. Mater. 2019, 31, e1902042. [Google Scholar] [CrossRef] [PubMed]
- Miccoli, B.; Braeken, D.; Li, Y.E. Brain-on-a-chip Devices for Drug Screening and Disease Modeling Applications. Curr. Pharm. Des. 2018, 24, 5419–5436. [Google Scholar] [CrossRef]
- Fernandes, J.T.; Chutna, O.; Chu, V.; Conde, J.P.; Outeiro, T.F. A Novel Microfluidic Cell Co-culture Platform for the Study of the Molecular Mechanisms of Parkinson’s Disease and Other Synucleinopathies. Front. Neurosci. 2016, 10, 511. [Google Scholar] [CrossRef]
- Seidi, A.; Kaji, H.; Annabi, N.; Ostrovidov, S.; Ramalingam, M.; Khademhosseini, A. A microfluidic-based neurotoxin concentration gradient for the generation of an in vitro model of Parkinson’s disease. Biomicrofluidics 2011, 5, 22214. [Google Scholar] [CrossRef]
- Freundt, E.C.; Maynard, N.; Clancy, E.K.; Roy, S.; Bousset, L.; Sourigues, Y.; Covert, M.; Melki, R.; Kirkegaard, K.; Brahic, M. Neuron-to-neuron transmission of alpha-synuclein fibrils through axonal transport. Ann. Neurol. 2012, 72, 517–524. [Google Scholar] [CrossRef]
- Grienberger, C.; Konnerth, A. Imaging calcium in neurons. Neuron 2012, 73, 862–885. [Google Scholar] [CrossRef]
- Mank, M.; Griesbeck, O. Genetically encoded calcium indicators. Chem. Rev. 2008, 108, 1550–1564. [Google Scholar] [CrossRef]
- Schwab, A.J.; Ebert, A.D. Neurite Aggregation and Calcium Dysfunction in iPSC-Derived Sensory Neurons with Parkinson’s Disease-Related LRRK2 G2019S Mutation. Stem Cell Rep. 2015, 5, 1039–1052. [Google Scholar] [CrossRef] [PubMed]
- Zygogianni, O.; Antoniou, N.; Kalomoiri, M.; Kouroupi, G.; Taoufik, E.; Matsas, R. In Vivo Phenotyping of Familial Parkinson’s Disease with Human Induced Pluripotent Stem Cells: A Proof-of-Concept Study. Neurochem. Res. 2019, 44, 1475–1493. [Google Scholar] [CrossRef] [PubMed]
- Negri, J.; Menon, V.; Young-Pearse, T.L. Assessment of Spontaneous Neuronal Activity In Vitro Using Multi-Well Multi-Electrode Arrays: Implications for Assay Development. ENeuro 2020, 7. [Google Scholar] [CrossRef]
- Spira, M.E.; Hai, A. Multi-electrode array technologies for neuroscience and cardiology. Nat. Nanotechnol. 2013, 8, 83–94. [Google Scholar] [CrossRef]
- Obien, M.E.; Deligkaris, K.; Bullmann, T.; Bakkum, D.J.; Frey, U. Revealing neuronal function through microelectrode array recordings. Front. Neurosci. 2014, 8, 423. [Google Scholar] [CrossRef] [PubMed]
- Woodard, C.M.; Campos, B.A.; Kuo, S.H.; Nirenberg, M.J.; Nestor, M.W.; Zimmer, M.; Mosharov, E.V.; Sulzer, D.; Zhou, H.; Paull, D.; et al. iPSC-derived dopamine neurons reveal differences between monozygotic twins discordant for Parkinson’s disease. Cell Rep. 2014, 9, 1173–1182. [Google Scholar] [CrossRef] [PubMed]
- Laperle, A.H.; Sances, S.; Yucer, N.; Dardov, V.J.; Garcia, V.J.; Ho, R.; Fulton, A.N.; Jones, M.R.; Roxas, K.M.; Avalos, P.; et al. iPSC modeling of young-onset Parkinson’s disease reveals a molecular signature of disease and novel therapeutic candidates. Nat. Med. 2020, 26, 289–299. [Google Scholar] [CrossRef] [PubMed]
- Amin, H.; Nieus, T.; Lonardoni, D.; Maccione, A.; Berdondini, L. High-resolution bioelectrical imaging of Abeta-induced network dysfunction on CMOS-MEAs for neurotoxicity and rescue studies. Sci. Rep. 2017, 7, 2460. [Google Scholar] [CrossRef]
- Iaccarino, H.F.; Singer, A.C.; Martorell, A.J.; Rudenko, A.; Gao, F.; Gillingham, T.Z.; Mathys, H.; Seo, J.; Kritskiy, O.; Abdurrob, F.; et al. Gamma frequency entrainment attenuates amyloid load and modifies microglia. Nature 2016, 540, 230–235. [Google Scholar] [CrossRef]
- Durens, M.; Nestor, J.; Williams, M.; Herold, K.; Niescier, R.F.; Lunden, J.W.; Phillips, A.W.; Lin, Y.C.; Dykxhoorn, D.M.; Nestor, M.W. High-throughput screening of human induced pluripotent stem cell-derived brain organoids. J. Neurosci. Methods 2020, 335, 108627. [Google Scholar] [CrossRef]
- Schwartz, M.P.; Hou, Z.; Propson, N.E.; Zhang, J.; Engstrom, C.J.; Santos Costa, V.; Jiang, P.; Nguyen, B.K.; Bolin, J.M.; Daly, W.; et al. Human pluripotent stem cell-derived neural constructs for predicting neural toxicity. Proc. Natl. Acad. Sci. USA 2015, 112, 12516–12521. [Google Scholar] [CrossRef] [PubMed]
- Monzel, A.S.; Hemmer, K.; Kaoma, T.; Smits, L.M.; Bolognin, S.; Lucarelli, P.; Rosety, I.; Zagare, A.; Antony, P.; Nickels, S.L.; et al. Machine learning-assisted neurotoxicity prediction in human midbrain organoids. Parkinsonism Relat. Disord. 2020, 75, 105–109. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).