Emerging Roles of RNA-Binding Proteins in Seed Development and Performance
Abstract
1. Introduction
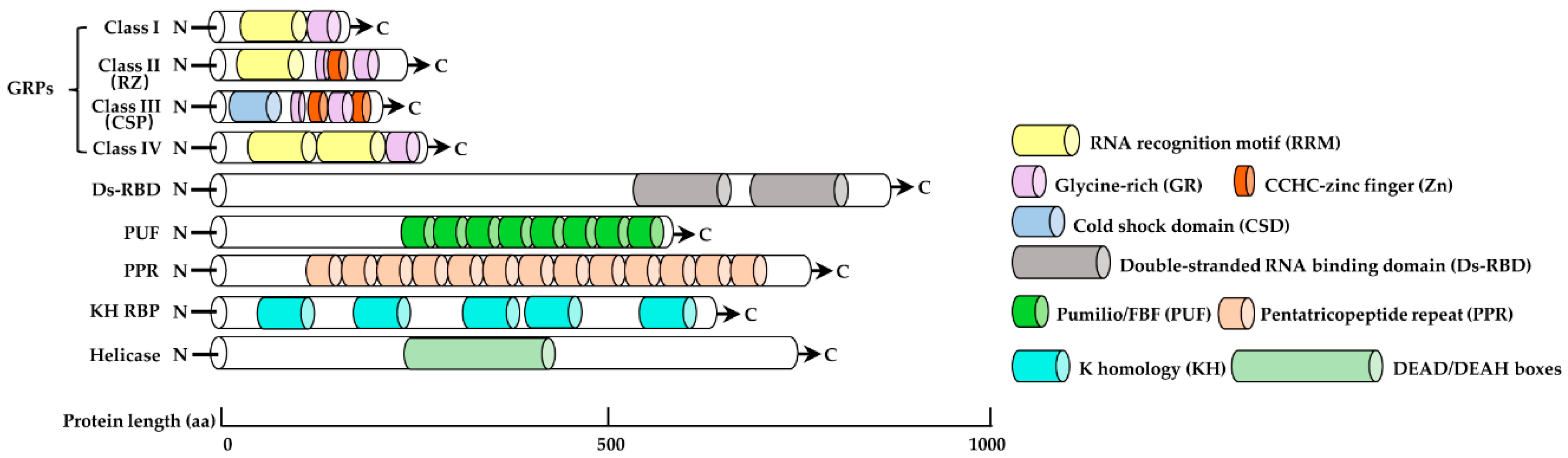
2. Structural Characteristics of RBPs
3. PUF-Type RBPs in Seed Development and Performance
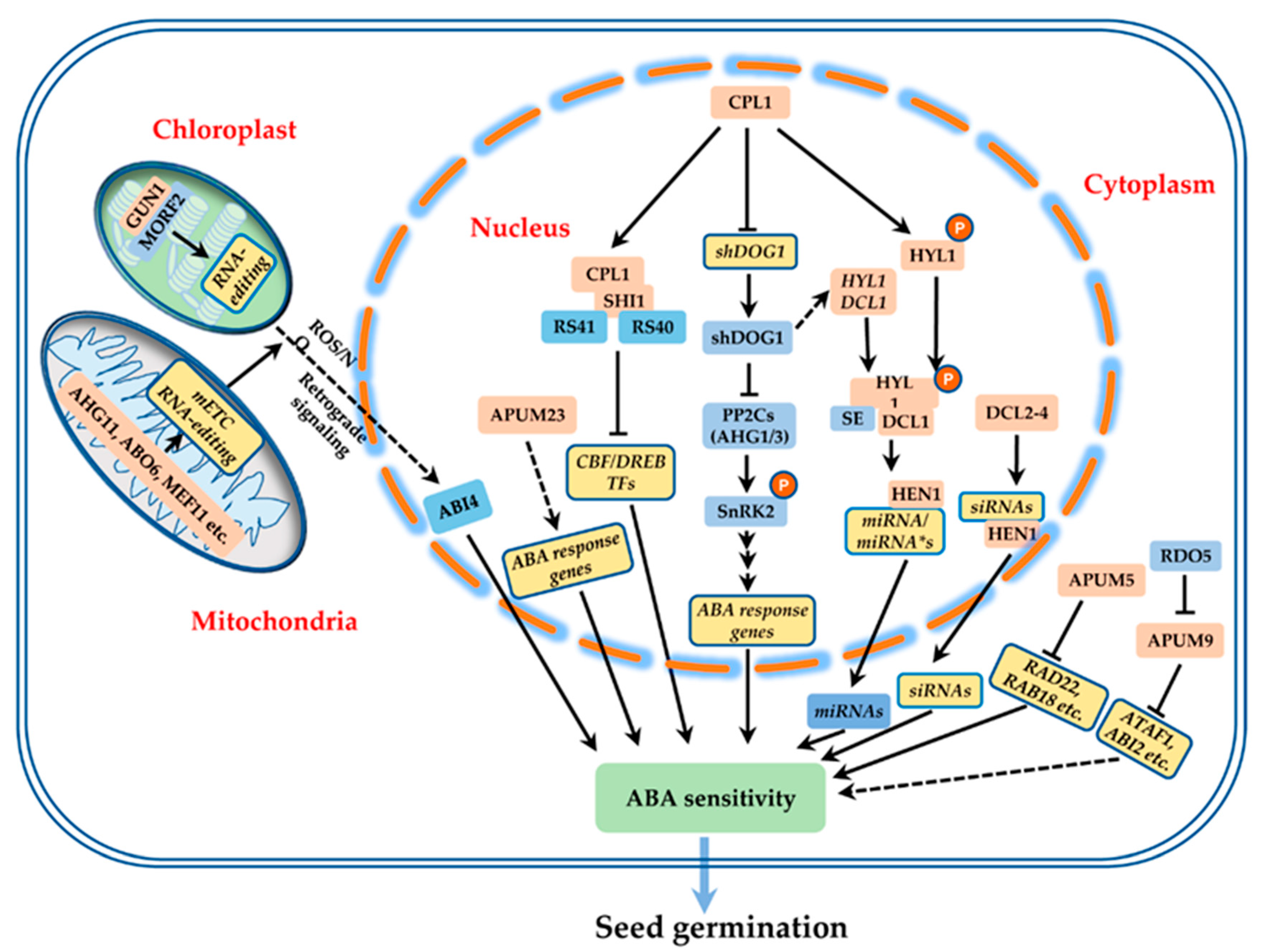
4. DsRBD-Type RBPs in Seed Development and Performance
5. GRPs in Seed Development and Performance
6. PPR-Type RBPs in Seed Development and Performance
| Type | Name | Species | Subcellular Localization a | Seed Development | Seed Germination under Different Conditions | Seed Dormancy | Reference | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Normal | ABA | Salt | Osmotic | Cold | |||||||
| PUF | APUM5 | A. thaliana | Cytoplasm | √ | √ | √ | [38,39] | ||||
| APUM23 | A. thaliana | Nucleus | √ | √ | √ | [42,43] | |||||
| APUM24 | A. thaliana | Nucleus | √ | √ | [16,40] | ||||||
| APUM9-11 | A. thaliana | Cytoplasm/Nuclear/periphery | √ | √ | [17] | ||||||
| dsRBD | CPL1 | A. thaliana | Nucleus | √ | √ | √ | √ | √ | [46,47,52,61] | ||
| CPL2 | A. thaliana | Cytoplasm/Nucleus | √ | √ | [46,51,62] | ||||||
| HYL1 | A. thaliana | Nucleus | √ | √ | √ | √ | [67,71,73,80,81] | ||||
| DCL1 | A. thaliana | Nucleus | √ | √ | √ | √ | [65,71,79,81] | ||||
| HEN1 | A. thaliana | Nucleus | √ | √ | √ | √ | [66,71,80] | ||||
| DCL2-4 | A. thaliana | Nucleus | √ | [66,71] | |||||||
| GRP | CSP1 | A. thaliana | Nucleus/Cytoplasm | √ | √ | [85,89] | |||||
| CSP2 | A. thaliana | Nucleus/Cytoplasm | √ | √ | [84,89,91] | ||||||
| CSP4 | A. thaliana | Nucleus/Cytoplasm | √ | [87] | |||||||
| RZ-1A | A. thaliana | Nucleus/Cytoplasm | √ | √ | √ | √ | [95,97,98] | ||||
| AtRZ-1B | A. thaliana | Nucleus | √ | [95,96] | |||||||
| AtRZ-1C | A. thaliana | Nucleus | √ | [95,96] | |||||||
| GRP4 | A. thaliana | Unknown | √ | √ | [92] | ||||||
| GRP7 | A. thaliana | Nucleus/Cytoplasm | √ | √ | √ | √ | [93,94] | ||||
| OsGRP1 | O. Sativa L. | Unknown | √ | [99] | |||||||
| OsGRP4 | O. sativa L. | Unknown | √ | [99] | |||||||
| OsGRP6 | O. sativa L. | Unknown | [99] | ||||||||
| OsRZ2 | O. sativa L. | Unknown | √ | [100] | |||||||
| TaRZ1 | T. aestivum L. | Nucleus | √ | √ | [101] | ||||||
| TaRZ2 | T. aestivum L. | Nucleus/cytoplasm/ER | √ | √ | [101] | ||||||
| TaRZ3 | T. aestivum L. | Nucleus | √ | √ | [101] | ||||||
| PPR | EMBs | A. thaliana | Chloroplast | √ | [103] | ||||||
| GUN1 | A. thaliana | Chloroplast | √ | [110,112] | |||||||
| AHG11 | A. thaliana | Mitochondria | √ | √ | √ | [113] | |||||
| SLG1 | A. thaliana | Mitochondria | √ | √ | √ | √ | [114] | ||||
| SLO2 | A. thaliana | Mitochondria | √ | √ | √ | [116] | |||||
| ABO5 | A. thaliana | Mitochondria | √ | [118] | |||||||
| ABO6 | A. thaliana | Mitochondria | √ | √ | √ | [119] | |||||
| PPR19 | A. thaliana | Mitochondria | √ | [117] | |||||||
| PNG | A. thaliana | Mitochondria | √ | √ | [120] | ||||||
| PPR40 | A. thaliana | Mitochondria | √ | √ | [121] | ||||||
| PPR96 | A. thaliana | Mitochondria | √ | √ | [122] | ||||||
| GRP23 | A. thaliana | Nucleus | √ | [108] | |||||||
| MEF11 | A. thaliana | Mitochondria | √ | √ | √ | [123,124] | |||||
| WLS | O. sativa L. | Chloroplast | √ | √ | [129] | ||||||
| OGR1 | O. sativa L. | Mitochondria | √ | √ | [107] | ||||||
| EMP4 | Z mays | Mitochondria | √ | √ | [104] | ||||||
| PPR8522 | Z. mays | Chloroplast | √ | √ | [106] | ||||||
7. Conclusions and Open Questions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Moore, M.J. From birth to death: The complex lives of eukaryotic mRNAs. Science 2005, 309, 1514–1518. [Google Scholar] [CrossRef] [PubMed]
- Owttrim, G.W. RNA helicases and abiotic stress. Nucleic Acids Res. 2006, 34, 3220–3230. [Google Scholar] [CrossRef] [PubMed]
- Tam, P.P.; Barrette-Ng, I.H.; Simon, D.M.; Tam, M.W.; Ang, A.L.; Muench, D.G. The Puf family of RNA-binding proteins in plants: Phylogeny, structural modeling, activity and subcellular localization. BMC Plant Biol. 2010, 10, 44. [Google Scholar] [CrossRef] [PubMed]
- Lorkovic, Z.J.; Barta, A. Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Res. 2002, 30, 623–635. [Google Scholar] [CrossRef] [PubMed]
- Sachetto-Martins, G.; Franco, L.O.; De Oliveira, D.E. Plant glycine-rich proteins: a family or just proteins with a common motif? Biochim. Biophys. Acta 2000, 1492, 1–14. [Google Scholar] [CrossRef]
- Walker, N.S.; Stiffler, N.; Barkan, A. POGs/PlantRBP: A resource for comparative genomics in plants. Nucleic Acids Res. 2007, 35, D852–D856. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, Y.; Xiao, H.; Zheng, Y.; Yue, B. Genome-wide identification, evolution, and expression analysis of RNA-binding glycine-rich protein family in maize. J. Integr. Plant Biol. 2014, 56, 1020–1031. [Google Scholar] [CrossRef]
- Koster, T.; Marondedze, C.; Meyer, K.; Staiger, D. RNA-binding proteins revisited-the emerging Arabidopsis mRNA interactome. Trends Plant Sci. 2017, 22, 512–526. [Google Scholar] [CrossRef]
- Reichel, M.; Liao, Y.; Rettel, M.; Ragan, C.; Evers, M.; Alleaume, A.M.; Horos, R.; Hentze, M.W.; Preiss, T.; Millar, A.A. In planta determination of the mRNA-binding proteome of Arabidopsis etiolated seedlings. Plant Cell 2016, 28, 2435–2452. [Google Scholar] [CrossRef]
- Zhang, Z.; Boonen, K.; Ferrari, P.; Schoofs, L.; Janssens, E.; van Noort, V.; Rolland, F.; Geuten, K. UV crosslinked mRNA-binding proteins captured from leaf mesophyll protoplasts. Plant Methods 2016, 12, 42. [Google Scholar] [CrossRef]
- Howell, K.A.; Narsai, R.; Carroll, A.; Ivanova, A.; Lohse, M.; Usadel, B.; Millar, A.H.; Whelan, J. Mapping metabolic and transcript temporal switches during germination in rice highlights specific transcription factors and the role of RNA instability in the germination process. Plant Physiol. 2009, 149, 961–980. [Google Scholar] [CrossRef] [PubMed]
- Nakabayashi, K.; Okamoto, M.; Koshiba, T.; Kamiya, Y.; Nambara, E. Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: Epigenetic and genetic regulation of transcription in seed. Plant J. 2005, 41, 697–709. [Google Scholar] [CrossRef] [PubMed]
- Rajjou, L.; Gallardo, K.; Debeaujon, I.; Vandekerckhove, J.; Job, C.; Job, D. The effect of alpha-amanitin on the Arabidopsis seed proteome highlights the distinct roles of stored and neosynthesized mRNAs during germination. Plant Physiol. 2004, 134, 1598–1613. [Google Scholar] [CrossRef] [PubMed]
- Masaki, S.; Yamada, T.; Hirasawa, T.; Todaka, D.; Kanekatsu, M. Proteomic analysis of RNA-binding proteins in dry seeds of rice after fractionation by ssDNA affinity column chromatography. Biotechnol. Lett. 2008, 30, 955–960. [Google Scholar] [CrossRef]
- Sano, N.; Masaki, S.; Tanabata, T.; Yamada, T.; Hirasawa, T.; Kashiwagi, M.; Kanekatsu, M. RNA-binding proteins associated with desiccation during seed development in rice. Biotechno. Lett. 2013, 35, 1945–1952. [Google Scholar] [CrossRef]
- Shanmugam, T.; Abbasi, N.; Kim, H.S.; Kim, H.B.; Park, N.I.; Park, G.T.; Oh, S.A.; Park, S.K.; Muench, D.G.; Choi, Y.; et al. An Arabidopsis divergent pumilio protein, APUM24, is essential for embryogenesis and required for faithful pre-rRNA processing. Plant J. 2017, 92, 1092–1105. [Google Scholar] [CrossRef]
- Xiang, Y.; Nakabayashi, K.; Ding, J.; He, F.; Bentsink, L.; Soppe, W.J.J. Reduced Dormancy5 encodes a protein phosphatase 2C that is required for seed dormancy in Arabidopsis. Plant Cell 2014, 26, 4362–4375. [Google Scholar] [CrossRef]
- Lorković, Z.J. Role of plant RNA-binding proteins in development, stress response and genome organization. Trends Plant Sci. 2009, 14, 229–236. [Google Scholar] [CrossRef]
- Hanano, S.; Sugita, M.; Sugiura, M. Isolation of a novel RNA-binding protein and its association with a large ribonucleoprotein particle present in the nucleoplasm of tobacco cells. Plant Mol. Biol. 1996, 31, 57–68. [Google Scholar] [CrossRef]
- Czolpinska, M.; Rurek, M. Plant glycine-rich proteins in stress response: An emerging, still prospective story. Front. Plant Sci. 2018, 9, 302. [Google Scholar] [CrossRef]
- Graumann, P.L.; Marahiel, M.A. A superfamily of proteins that contain the cold-shock domain. Trends Biochem. Sci. 1998, 23, 286–290. [Google Scholar] [CrossRef]
- Manival, X.; Ghisolfi-Nieto, L.; Joseph, G.; Bouvet, P.; Erard, M. RNA-binding strategies common to cold-shock domain- and RNA recognition motif-containing proteins. Nucleic Acids Res. 2001, 29, 2223–2233. [Google Scholar] [CrossRef] [PubMed]
- Maris, C.; Dominguez, C.; Allain, F.H. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005, 272, 2118–2131. [Google Scholar] [CrossRef]
- Tian, B.; Bevilacqua, P.C.; Diegelman-Parente, A.; Mathews, M.B. The double-stranded-RNA-binding motif: Interference and much more. Nat. Rev. Mol. Cell. Biol. 2004, 5, 1013–1023. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Varani, G. Engineering RNA-binding proteins for biology. FEBS J. 2013, 280, 3734–3754. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Gallegos, M.; Puoti, A.; Durkin, E.; Fields, S.; Kimble, J.; Wickens, M.P. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature 1997, 390, 477–484. [Google Scholar] [CrossRef]
- Edwards, T.A.; Pyle, S.E.; Wharton, R.P.; Aggarwal, A.K. Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell 2001, 105, 281–289. [Google Scholar] [CrossRef]
- Wang, X.; Zamore, P.D.; Hall, T.M. Crystal structure of a pumilio homology domain. Mol. Cell 2001, 7, 855–865. [Google Scholar] [CrossRef]
- Wang, X.; McLachlan, J.; Zamore, P.D.; Hall, T.M. Modular recognition of RNA by a human pumilio-homology domain. Cell 2002, 110, 501–512. [Google Scholar] [CrossRef]
- Francischini, C.W.; Quaggio, R.B. Molecular characterization of Arabidopsis thaliana PUF proteins-binding specificity and target candidates. FEBS J. 2009, 276, 5456–5470. [Google Scholar] [CrossRef]
- Wang, M.; Oge, L.; Perez-Garcia, M.D.; Hamama, L.; Sakr, S. The PUF Protein Family: Overview on PUF RNA targets, biological functions, and post transcriptional regulation. Int. J. Mol. Sci. 2018, 19, 410. [Google Scholar] [CrossRef]
- Takenaka, M.; Zehrmann, A.; Verbitskiy, D.; Hartel, B.; Brennicke, A. RNA editing in plants and its evolution. Annu. Rev. Genet. 2013, 47, 335–352. [Google Scholar] [CrossRef]
- Lurin, C.; Andrés, C.; Aubourg, S.; Bellaoui, M.; Bitton, F.; Bruyère, C.; Caboche, M.; Debast, C.; Gualberto, J.; Hoffmann, B.; et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 2004, 16, 2089–2103. [Google Scholar] [CrossRef]
- Grishin, N.V. KH domain: One motif, two folds. Nucleic Acids Res. 2001, 29, 638–643. [Google Scholar] [CrossRef]
- Makeyev, A.V.; Liebhaber, S.A. The poly(C)-binding proteins: A multiplicity of functions and a search for mechanisms. RNA 2002, 8, 265–278. [Google Scholar] [CrossRef] [PubMed]
- Mingam, A.; Toffano-Nioche, C.; Brunaud, V.; Boudet, N.; Kreis, M.; Lecharny, A. DEAD-box RNA helicases in Arabidopsis thaliana: Establishing a link between quantitative expression, gene structure and evolution of a family of genes. Plant Biotechnol. J. 2004, 2, 401–415. [Google Scholar] [CrossRef]
- Huh, S.U.; Kim, M.J.; Paek, K.H. Arabidopsis Pumilio protein APUM5 suppresses cucumber mosaic virus infection via direct binding of viral RNAs. Proc. Natl. Acad. Sci. USA 2013, 110, 779–784. [Google Scholar] [CrossRef]
- Huh, S.U.; Paek, K.H. APUM5, encoding a Pumilio RNA binding protein, negatively regulates abiotic stress responsive gene expression. BMC Plant Biol. 2014, 14, 75. [Google Scholar] [CrossRef] [PubMed]
- Arae, T.; Morita, K.; Imahori, R.; Suzuki, Y.; Yasuda, S.; Sato, T.; Yamaguchi, J.; Chiba, Y. Identification of Arabidopsis CCR4-NOT complexes with pumilio RNA-binding proteins, APUM5 and APUM2. Plant Cell Physiol. 2019, 60, 2015–2025. [Google Scholar] [CrossRef]
- Maekawa, S.; Ishida, T.; Yanagisawa, S. Reduced expression of APUM24, encoding a novel rRNA processing factor, induces sugar-dependent nucleolar stress and altered sugar responses in Arabidopsis thaliana. Plant Cell 2018, 30, 209–227. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Muench, D.G. A Nucleolar PUF RNA-binding protein with specificity for a unique RNA sequence. J. Biol. Chem. 2015, 290, 30108–30118. [Google Scholar] [CrossRef] [PubMed]
- Abbasi, N.; Kim, H.B.; Park, N.I.; Kim, H.S.; Kim, Y.K.; Park, Y.I.; Choi, S.B. APUM23, a nucleolar Puf domain protein, is involved in pre-ribosomal RNA processing and normal growth patterning in Arabidopsis. Plant J. 2010, 64, 960–976. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.C.; Lin, W.C.; Cheng, W.H. Salt hypersensitive mutant 9, a nucleolar APUM23 protein, is essential for salt sensitivity in association with the ABA signaling pathway in Arabidopsis. BMC Plant Biol. 2018, 18, 40. [Google Scholar] [CrossRef]
- Nyikó, T.; Auber, A.; Bucher, E. Functional and molecular characterization of the conserved Arabidopsis PUMILIO protein, APUM9. Plant Mol. Biol. 2019, 100, 199–214. [Google Scholar] [CrossRef] [PubMed]
- Hiraguri, A.; Itoh, R.; Kondo, N.; Nomura, Y.; Aizawa, D.; Murai, Y.; Koiwa, H.; Seki, M.; Shinozaki, K.; Fukuhara, T. Specific interactions between Dicer-like proteins and HYL1/DRB-family dsRNA-binding proteins in Arabidopsis thaliana. Plant Mol. Biol. 2005, 57, 173–188. [Google Scholar] [CrossRef]
- Koiwa, H.; Hausmann, S.; Bang, W.Y.; Ueda, A.; Kondo, N.; Hiraguri, A.; Fukuhara, T.; Bahk, J.D.; Yun, D.J.; Bressan, R.A.; et al. Arabidopsis C-terminal domain phosphatase-like 1 and 2 are essential Ser-5-specific C-terminal domain phosphatases. Proc. Natl. Acad. Sci. USA 2004, 101, 14539–14544. [Google Scholar] [CrossRef]
- Xiong, L.; Lee, H.; Ishitani, M.; Tanaka, Y.; Stevenson, B.; Koiwa, H.; Bressan, R.A.; Hasegawa, P.M.; Zhu, J.K. Repression of stress-responsive genes by FIERY2, a novel transcriptional regulator in Arabidopsis. Proc. Natl. Acad. Sci. USA 2002, 99, 10899–10904. [Google Scholar] [CrossRef]
- Chen, T.; Cui, P.; Chen, H.; Ali, S.; Zhang, S.; Xiong, L. A KH-domain RNA-binding protein interacts with FIERY2/CTD phosphatase-like 1 and splicing factors and is important for pre-mRNA splicing in Arabidopsis. PLoS Genet. 2013, 9, e1003875. [Google Scholar] [CrossRef]
- Jeong, I.S.; Fukudome, A.; Aksoy, E.; Bang, W.Y.; Kim, S.; Guan, Q.; Bahk, J.D.; May, K.A.; Russell, W.K.; Zhu, J.; et al. Regulation of abiotic stress signalling by Arabidopsis C-terminal domain phosphatase-like 1 requires interaction with a k-homology domain-containing protein. PLoS ONE 2013, 8, e80509. [Google Scholar] [CrossRef]
- Jiang, J.; Wang, B.; Shen, Y.; Wang, H.; Feng, Q.; Shi, H. The Arabidopsis RNA binding protein with K homology motifs, SHINY1, interacts with the C-terminal domain phosphatase-like 1 (CPL1) to repress stress-inducible gene expression. PLoS Genet. 2013, 9, e1003625. [Google Scholar] [CrossRef]
- Ueda, A.; Li, P.; Feng, Y.; Vikram, M.; Kim, S.; Kang, C.H.; Kang, J.S.; Bahk, J.D.; Lee, S.Y.; Fukuhara, T.; et al. The Arabidopsis thaliana carboxyl-terminal domain phosphatase-like 2 regulates plant growth, stress and auxin responses. Plant Mol. Biol. 2008, 67, 683–697. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Cazorla, E.; Ortuño-Miquel, S.; Candela, H.; Bailey-Steinitz, L.J.; Yanofsky, M.F.; Martínez-Laborda, A.; Ripoll, J.J.; Vera, A. Ovule identity mediated by pre-mRNA processing in Arabidopsis. PLoS Genet. 2018, 14, e1007182. [Google Scholar] [CrossRef] [PubMed]
- Yoine, M.; Ohto, M.A.; Onai, K.; Mita, S.; Nakamura, K. The lba1 mutation of UPF1 RNA helicase involved in nonsense-mediated mRNA decay causes pleiotropic phenotypic changes and altered sugar signalling in Arabidopsis. Plant J. 2006, 47, 49–62. [Google Scholar] [CrossRef]
- Cui, P.; Chen, T.; Qin, T.; Ding, F.; Wang, Z.; Chen, H.; Xiong, L. The RNA Polymerase II C-terminal domain phosphatase-like protein FIERY2/CPL1 interacts with eIF4AIII and is essential for nonsense-mediated mRNA decay in Arabidopsis. Plant Cell 2016, 28, 770–785. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Li, W.; An, L. NMD mechanism and the functions of Upf proteins in plant. Plant Cell Rep. 2016, 35, 5–15. [Google Scholar] [CrossRef]
- Bentsink, L.; Jowett, J.; Hanhart, C.J.; Koornneef, M. Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 17042–17047. [Google Scholar] [CrossRef]
- Soppe, W.J.J.; Bentsink, L. Seed dormancy back on track, its definition and regulation by DOG1. New Phytol. 2020. [Google Scholar] [CrossRef]
- Li, X.; Chen, T.; Li, Y.; Wang, Z.; Cao, H.; Chen, F.; Li, Y.; Soppe, W.J.J.; Li, W.; Liu, Y. ETR1/RDO3 regulates seed dormancy by relieving the inhibitory effect of the ERF12-TPL complex on DELAY OF GERMINATION1 expression. Plant Cell 2019, 31, 832–847. [Google Scholar] [CrossRef]
- Zheng, J.; Chen, F.; Wang, Z.; Cao, H.; Li, X.; Deng, X.; Soppe, W.J.J.; Li, Y.; Liu, Y. A novel role for histone methyltransferase KYP/SUVH4 in the control of Arabidopsis primary seed dormancy. New Phytol. 2012, 193, 605–616. [Google Scholar] [CrossRef]
- Cyrek, M.; Fedak, H.; Ciesielski, A.; Guo, Y.; Sliwa, A.; Brzezniak, L.; Krzyczmonik, K.; Pietras, Z.; Kaczanowski, S.; Liu, F.; et al. Seed dormancy in Arabidopsis is controlled by alternative polyadenylation of DOG1. Plant Physiol. 2016, 170, 947–955. [Google Scholar] [CrossRef]
- Kowalczyk, J.; Palusinska, M.; Wroblewska-Swiniarska, A.; Pietras, Z.; Szewc, L.; Dolata, J.; Jarmolowski, A.; Swiezewski, S. Alternative polyadenylation of the sense transcript controls antisense transcription of DELAY OF GERMINATION 1 in Arabidopsis. Mol. Plant 2017, 10, 1349–1352. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Dean, G.H. ECERIFERUM11/C-TERMINAL DOMAIN PHOSPHATASE-LIKE2 affects secretory trafficking. Plant cell 2019, 181, 901–915. [Google Scholar] [CrossRef] [PubMed]
- Penfield, S.; Meissner, R.C.; Shoue, D.A.; Carpita, N.C.; Bevan, M.W. MYB61 is required for mucilage deposition and extrusion in the Arabidopsis seed coat. Plant Cell 2001, 13, 2777–2791. [Google Scholar] [CrossRef]
- Debeaujon, I.; Léon-Kloosterziel, K.M.; Koornneef, M. Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol. 2000, 122, 403–414. [Google Scholar] [CrossRef] [PubMed]
- Papp, I.; Mette, M.F.; Aufsatz, W.; Daxinger, L.; Schauer, S.E.; Ray, A.; van der Winden, J.; Matzke, M.; Matzke, A.J. Evidence for nuclear processing of plant micro RNA and short interfering RNA precursors. Plant Physiol. 2003, 132, 1382–1390. [Google Scholar] [CrossRef]
- Pontes, O.; Vitins, A.; Ream, T.S.; Hong, E.; Pikaard, C.S.; Costa-Nunes, P. Intersection of small RNA pathways in Arabidopsis thaliana sub-nuclear domains. PLoS ONE 2013, 8, e65652. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Fedoroff, N. A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell 2000, 12, 2351–2366. [Google Scholar] [CrossRef]
- Voinnet, O. Origin, biogenesis, and activity of plant microRNAs. Cell 2009, 136, 669–687. [Google Scholar] [CrossRef]
- Yang, S.W.; Chen, H.Y.; Yang, J.; Machida, S.; Chua, N.H.; Yuan, Y.A. Structure of Arabidopsis HYPONASTIC LEAVES1 and its molecular implications for miRNA processing. Structure 2010, 18, 594–605. [Google Scholar] [CrossRef]
- Curtin, S.J.; Watson, J.M.; Smith, N.A.; Eamens, A.L.; Blanchard, C.L.; Waterhouse, P.M. The roles of plant dsRNA-binding proteins in RNAi-like pathways. FEBS Lett. 2008, 582, 2753–2760. [Google Scholar] [CrossRef]
- Zhang, J.F.; Yuan, L.J.; Shao, Y.; Du, W.; Yan, D.W.; Lu, Y.T. The disturbance of small RNA pathways enhanced abscisic acid response and multiple stress responses in Arabidopsis. Plant Cell Environ. 2008, 31, 562–574. [Google Scholar] [CrossRef]
- Li, J.; Yang, Z.; Yu, B.; Liu, J.; Chen, X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr. Biol. 2005, 15, 1501–1507. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Han, M.H.; Guevara-Garcia, A.; Fedoroff, N.V. Mitogen-activated protein kinase signaling in postgermination arrest of development by abscisic acid. Proc. Natl. Acad. Sci. USA 2002, 99, 15812–15817. [Google Scholar] [CrossRef]
- Duarte, G.T.; Matiolli, C.C.; Pant, B.D.; Schlereth, A.; Scheible, W.R.; Stitt, M.; Vicentini, R.; Vincentz, M. Involvement of microRNA-related regulatory pathways in the glucose-mediated control of Arabidopsis early seedling development. J. Exp. Bot. 2013, 64, 4301–4312. [Google Scholar] [CrossRef] [PubMed]
- Manavella, P.A.; Hagmann, J.; Ott, F.; Laubinger, S.; Franz, M.; Macek, B.; Weigel, D. Fast-forward genetics identifies plant CPL phosphatases as regulators of miRNA processing factor HYL1. Cell 2012, 151, 859–870. [Google Scholar] [CrossRef]
- Raghuram, B.; Sheikh, A.H.; Rustagi, Y.; Sinha, A.K. MicroRNA biogenesis factor DRB1 is a phosphorylation target of mitogen activated protein kinase MPK3 in both rice and Arabidopsis. FEBS J. 2015, 282, 521–536. [Google Scholar] [CrossRef]
- Yan, J.; Wang, P. The SnRK2 kinases modulate miRNA accumulation in Arabidopsis. PLoS Genet. 2017, 13, e1006753. [Google Scholar] [CrossRef]
- Huo, H.; Wei, S.; Bradford, K.J. Delay of Germination1 (DOG1) regulates both seed dormancy and flowering time through microRNA pathways. Proc. Natl. Acad. Sci. USA 2016, 113, E2199–E2206. [Google Scholar] [CrossRef]
- Robinson-Beers, K.; Pruitt, R.E.; Gasser, C.S. Ovule development in wild-type Arabidopsis and two female-sterile mutants. Plant Cell 1992, 4, 1237–1249. [Google Scholar] [CrossRef]
- Wei, S.J.; Chai, S.; Zhu, R.M.; Duan, C.Y.; Zhang, Y.; Li, S. HUA ENHANCER1 mediates ovule development. Front. Plant Sci. 2020, 11, 397. [Google Scholar] [CrossRef]
- Armenta-Medina, A.; Lepe-Soltero, D.; Xiang, D.; Datla, R.; Abreu-Goodger, C.; Gillmor, C.S. Arabidopsis thaliana miRNAs promote embryo pattern formation beginning in the zygote. Dev. Biol. 2017, 431, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Guo, G.; Liu, X. Wheat miR9678 affects seed germination by generating phased siRNAs and modulating abscisic acid/gibberellin signaling. Plant Cell 2018, 30, 796–814. [Google Scholar] [CrossRef] [PubMed]
- Miao, C.; Wang, Z.; Zhang, L.; Yao, J.; Hua, K.; Liu, X.; Shi, H. The grain yield modulator miR156 regulates seed dormancy through the gibberellin pathway in rice. Nat. Commun. 2019, 10, 3822. [Google Scholar] [CrossRef]
- Fusaro, A.F.; Bocca, S.N.; Ramos, R.L.B.; Barrôco, R.M.; Magioli, C.; Jorge, V.C.; Coutinho, T.C.; Rangel-Lima, C.M.; De Rycke, R.; Inzé, D.; et al. AtGRP2, a cold-induced nucleo-cytoplasmic RNA-binding protein, has a role in flower and seed development. Planta 2007, 225, 1339–1351. [Google Scholar] [CrossRef] [PubMed]
- Juntawong, P.; Sorenson, R.; Bailey-Serres, J. Cold shock protein 1 chaperones mRNAs during translation in Arabidopsis thaliana. Plant J. 2013, 74, 1016–1028. [Google Scholar] [CrossRef]
- Karlson, D.; Imai, R. Conservation of the cold shock domain protein family in plants. Plant Physiol. 2003, 131, 12–15. [Google Scholar] [CrossRef]
- Yang, Y.; Karlson, D.T. Overexpression of AtCSP4 affects late stages of embryo development in Arabidopsis. J. Exp. Bot. 2011, 62, 2079–2091. [Google Scholar] [CrossRef]
- Nakaminami, K.; Hill, K.; Perry, S.E.; Sentoku, N.; Long, J.A.; Karlson, D.T. Arabidopsis cold shock domain proteins: Relationships to floral and silique development. J. Exp. Bot. 2009, 60, 1047–1062. [Google Scholar] [CrossRef]
- Park, S.J.; Kwak, K.J.; Oh, T.R.; Kim, Y.O.; Kang, H. Cold shock domain proteins affect seed germination and growth of Arabidopsis thaliana under abiotic stress conditions. Plant Cell Physiol. 2009, 50, 869–878. [Google Scholar] [CrossRef]
- Sasaki, K.; Kim, M.H.; Imai, R. Arabidopsis COLD SHOCK DOMAIN PROTEIN2 is a RNA chaperone that is regulated by cold and developmental signals. Biochem. Biophys. Res. Commun. 2007, 364, 633–638. [Google Scholar] [CrossRef]
- Kim, J.Y.; Park, S.J.; Jang, B.; Jung, C.H.; Ahn, S.J.; Goh, C.H.; Cho, K.; Han, O.; Kang, H. Functional characterization of a glycine-rich RNA-binding protein 2 in Arabidopsis thaliana under abiotic stress conditions. Plant J. 2007, 50, 439–451. [Google Scholar] [CrossRef] [PubMed]
- Kwak, K.J.; Kim, Y.O.; Kang, H. Characterization of transgenic Arabidopsis plants overexpressing GR-RBP4 under high salinity, dehydration, or cold stress. J. Exp. Bot. 2005, 56, 3007–3016. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Jiang, L.; Song, S.; Jing, R.; Xu, G. AtGRP7 is involved in the regulation of abscisic acid and stress responses in Arabidopsis. Cell. Mol. Biol. Lett. 2006, 11, 526–535. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Jung, H.J.; Lee, H.J.; Kim, K.A.; Goh, C.-H.; Woo, Y.; Oh, S.H.; Han, Y.S.; Kang, H. Glycine-rich RNA-binding protein 7 affects abiotic stress responses by regulating stomata opening and closing in Arabidopsis thaliana. Plant J. 2008, 55, 455–466. [Google Scholar] [CrossRef]
- Kim, W.Y.; Kim, J.Y.; Jung, H.J.; Oh, S.H.; Han, Y.S.; Kang, H. Comparative analysis of Arabidopsis zinc finger-containing glycine-rich RNA-binding proteins during cold adaptation. Plant Physiol. Biochem. 2010, 48, 866–872. [Google Scholar] [CrossRef]
- Wu, Z.; Zhu, D.; Lin, X.; Miao, J.; Gu, L.; Deng, X.; Yang, Q.; Sun, K.; Zhu, D.; Cao, X.; et al. RNA Binding Proteins RZ-1B and RZ-1C play critical roles in regulating pre-mRNA splicing and gene expression during development in Arabidopsis. Plant Cell 2016, 28, 55–73. [Google Scholar] [CrossRef]
- Kim, Y.O.; Kim, J.S.; Kang, H. Cold-inducible zinc finger-containing glycine-rich RNA-binding protein contributes to the enhancement of freezing tolerance in Arabidopsis thaliana. Plant J. 2005, 42, 890–900. [Google Scholar] [CrossRef]
- Kim, Y.O.; Pan, S.; Jung, C.H.; Kang, H. A zinc finger-containing glycine-rich RNA-binding protein, atRZ-1a, has a negative impact on seed germination and seedling growth of Arabidopsis thaliana under salt or drought stress conditions. Plant Cell Physiol. 2007, 48, 1170–1181. [Google Scholar] [CrossRef]
- Kim, J.Y.; Kim, W.Y.; Kwak, K.J.; Oh, S.H.; Han, Y.S.; Kang, H. Glycine-rich RNA-binding proteins are functionally conserved in Arabidopsis thaliana and Oryza sativa during cold adaptation process. J. Exp. Bot. 2010, 61, 2317–2325. [Google Scholar] [CrossRef]
- Kim, J.Y.; Kim, W.Y.; Kwak, K.J.; Oh, S.H.; Han, Y.S.; Kang, H. Zinc finger-containing glycine-rich RNA-binding protein in Oryza sativa has an RNA chaperone activity under cold stress conditions. Plant Cell Environ. 2010, 33, 759–768. [Google Scholar]
- Xu, T.; Gu, L.; Choi, M.J.; Kim, R.J.; Suh, M.C.; Kang, H. Comparative functional analysis of wheat (Triticum aestivum) zinc finger-containing glycine-rich RNA-binding proteins in response to abiotic stresses. PLoS ONE 2014, 9, e96877. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Kang, H. Emerging roles of RNA-Binding proteins in plant growth, development, and stress responses. Mol. Cells 2016, 39, 179–185. [Google Scholar] [PubMed]
- Bryant, N.; Lloyd, J.; Sweeney, C.; Myouga, F.; Meinke, D. Identification of nuclear genes encoding chloroplast-localized proteins required for embryo development in Arabidopsis. Plant Physiol. 2011, 155, 1678–1689. [Google Scholar] [CrossRef] [PubMed]
- Gabotti, D.; Caporali, E.; Manzotti, P.; Persico, M.; Vigani, G.; Consonni, G. The maize pentatricopeptide repeat gene empty pericarp4 (emp4) is required for proper cellular development in vegetative tissues. Plant Sci. 2014, 223, 25–35. [Google Scholar] [CrossRef]
- Liu, Y.J.; Xiu, Z.H.; Meeley, R.; Tan, B.C. Empty pericarp5 encodes a pentatricopeptide repeat protein that is required for mitochondrial RNA editing and seed development in maize. Plant Cell 2013, 25, 868–883. [Google Scholar] [CrossRef]
- Sosso, D.; Canut, M.; Gendrot, G.; Dedieu, A.; Chambrier, P.; Barkan, A.; Consonni, G.; Rogowsky, P.M. PPR8522 encodes a chloroplast-targeted pentatricopeptide repeat protein necessary for maize embryogenesis and vegetative development. J. Exp. Bot. 2012, 63, 5843–5857. [Google Scholar] [CrossRef]
- Kim, S.R.; Yang, J.I.; Moon, S.; Ryu, C.H.; An, K.; Kim, K.M.; Yim, J.; An, G. Rice OGR1 encodes a pentatricopeptide repeat-DYW protein and is essential for RNA editing in mitochondria. Plant J. 2009, 59, 738–749. [Google Scholar] [CrossRef]
- Ding, Y.H.; Liu, N.Y.; Tang, Z.S.; Liu, J.; Yang, W.C. Arabidopsis GLUTAMINE-RICH PROTEIN23 is essential for early embryogenesis and encodes a novel nuclear PPR motif protein that interacts with RNA polymerase II subunit III. Plant Cell 2006, 18, 815–830. [Google Scholar] [CrossRef]
- Barkan, A.; Small, I. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 2014, 65, 415–442. [Google Scholar] [CrossRef]
- Koussevitzky, S.; Nott, A.; Mockler, T.C.; Hong, F.; Sachetto-Martins, G.; Surpin, M.; Lim, J.; Mittler, R.; Chory, J. Signals from chloroplasts converge to regulate nuclear gene expression. Science 2007, 316, 715–719. [Google Scholar] [CrossRef]
- Zhao, X.; Huang, J.; Chory, J. GUN1 interacts with MORF2 to regulate plastid RNA editing during retrograde signaling. Proc. Natl. Acad. Sci. USA 2019, 116, 10162–10167. [Google Scholar] [CrossRef] [PubMed]
- Cottage, A.; Mott, E.K.; Kempster, J.A.; Gray, J.C. The Arabidopsis plastid-signalling mutant gun1 (genomes uncoupled1) shows altered sensitivity to sucrose and abscisic acid and alterations in early seedling development. J. Exp. Bot. 2010, 61, 3773–3786. [Google Scholar] [CrossRef] [PubMed]
- Murayama, M.; Hayashi, S.; Nishimura, N.; Ishide, M.; Kobayashi, K.; Yagi, Y.; Asami, T.; Nakamura, T.; Shinozaki, K.; Hirayama, T. Isolation of Arabidopsis ahg11, a weak ABA hypersensitive mutant defective in nad4 RNA editing. J. Exp. Bot. 2012, 63, 5301–5310. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Liu, D. Functional disruption of the pentatricopeptide protein SLG1 affects mitochondrial RNA editing, plant development, and responses to abiotic stresses in Arabidopsis. Plant J. 2012, 70, 432–444. [Google Scholar] [CrossRef]
- Zhu, Q.; Dugardeyn, J.; Zhang, C.; Takenaka, M.; Kuhn, K.; Craddock, C.; Smalle, J.; Karampelias, M.; Denecke, J.; Peters, J.; et al. SLO2, a mitochondrial pentatricopeptide repeat protein affecting several RNA editing sites, is required for energy metabolism. Plant J. 2012, 71, 836–849. [Google Scholar] [CrossRef]
- Zhu, Q.; Dugardeyn, J.; Zhang, C.; Mühlenbock, P.; Eastmond, P.J.; Valcke, R.; De Coninck, B.; Öden, S.; Karampelias, M.; Cammue, B.P.A.; et al. The Arabidopsis thaliana RNA editing factor SLO2, which affects the mitochondrial electron transport chain, participates in multiple stress and hormone responses. Mol. Plant 2014, 7, 290–310. [Google Scholar] [CrossRef]
- Lee, K.; Han, J.H.; Park, Y.I.; Colas des Francs-Small, C.; Small, I.; Kang, H. The mitochondrial pentatricopeptide repeat protein PPR19 is involved in the stabilization of NADH dehydrogenase 1 transcripts and is crucial for mitochondrial function and Arabidopsis thaliana development. New Phytol. 2017, 215, 202–216. [Google Scholar] [CrossRef]
- Liu, Y.; He, J.; Chen, Z.; Ren, X.; Hong, X.; Gong, Z. ABA overly-sensitive 5 (ABO5), encoding a pentatricopeptide repeat protein required for cis-splicing of mitochondrial nad2 intron 3, is involved in the abscisic acid response in Arabidopsis. Plant J. 2010, 63, 749–765. [Google Scholar] [CrossRef]
- He, J.; Duan, Y.; Hua, D.; Fan, G.; Wang, L.; Liu, Y.; Chen, Z.; Han, L.; Qu, L.J.; Gong, Z. DEXH box RNA helicase-mediated mitochondrial reactive oxygen species production in Arabidopsis mediates crosstalk between abscisic acid and auxin signaling. Plant Cell 2012, 24, 1815–1833. [Google Scholar] [CrossRef]
- Laluk, K.; Abuqamar, S.; Mengiste, T. The Arabidopsis mitochondria-localized pentatricopeptide repeat protein PGN functions in defense against necrotrophic fungi and abiotic stress tolerance. Plant Physiol. 2011, 156, 2053–2068. [Google Scholar] [CrossRef]
- Zsigmond, L.; Rigó, G.; Szarka, A.; Székely, G.; Ötvös, K.; Darula, Z.; Medzihradszky, K.F.; Koncz, C.; Koncz, Z.; Szabados, L. Arabidopsis PPR40 connects abiotic stress responses to mitochondrial electron transport. Plant Physiol. 2008, 146, 1721–1737. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.M.; Zhao, J.Y.; Lu, P.P.; Chen, M.; Guo, C.H.; Xu, Z.S.; Ma, Y.Z. The E-subgroup pentatricopeptide repeat protein family in Arabidopsis thaliana and confirmation of the responsiveness PPR96 to abiotic stresses. Front. Plant Sci. 2016, 7, 1825. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Suzuki, M.; Tang, J.; Nagata, N.; Ohyama, K.; Seki, H.; Kiuchi, R.; Kaneko, Y.; Nakazawa, M.; Matsui, M.; et al. Lovastatin insensitive 1, a novel pentatricopeptide repeat protein, is a potential regulatory factor of isoprenoid biosynthesis in Arabidopsis. Plant Cell Physiol. 2007, 48, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Sechet, J.; Roux, C.; Plessis, A.; Effroy, D.; Frey, A.; Perreau, F.; Biniek, C.; Krieger-Liszkay, A.; Macherel, D.; North, H.M.; et al. The ABA-deficiency suppressor locus HAS2 encodes the PPR protein LOI1/MEF11 involved in mitochondrial RNA editing. Mol. Plant 2015, 8, 644–656. [Google Scholar] [CrossRef]
- Bailly, C.; El-Maarouf-Bouteau, H.; Corbineau, F. From intracellular signaling networks to cell death: The dual role of reactive oxygen species in seed physiology. C. R. Biol. 2008, 331, 806–814. [Google Scholar] [CrossRef]
- Nonogaki, H. Seed biology updates-highlights and new discoveries in seed dormancy and germination research. Front. Plant Sci. 2017, 8, 524. [Google Scholar] [CrossRef]
- Locato, V.; Cimini, S.; De Gara, L. ROS and redox balance as multifaceted players of cross-tolerance: Epigenetic and retrograde control of gene expression. J. Exp. Bot. 2018, 69, 3373–3391. [Google Scholar] [CrossRef]
- Nonogaki, H. The long-standing paradox of seed dormancy unfolded? Trends Plant Sci. 2019, 24, 989–998. [Google Scholar] [CrossRef]
- Tan, J.; Tan, Z.; Wu, F.; Sheng, P.; Heng, Y.; Wang, X.; Ren, Y.; Wang, J.; Guo, X.; Zhang, X.; et al. A novel chloroplast-localized pentatricopeptide repeat protein involved in splicing affects chloroplast development and abiotic stress response in rice. Mol. Plant 2014, 7, 1329–1349. [Google Scholar] [CrossRef]
- Lapointe, C.P.; Wilinski, D.; Saunders, H.A.J.; Wickens, M. Protein-RNA networks revealed through covalent RNA marks. Nat. Methods 2015, 12, 1163–1170. [Google Scholar] [CrossRef]
- Rahman, R.; Xu, W.; Jin, H.; Rosbash, M. Identification of RNA-binding protein targets with HyperTRIBE. Nat. Protoc. 2018, 13, 1829–1849. [Google Scholar] [CrossRef] [PubMed]
- Laloum, T.; Martín, G.; Duque, P. Alternative splicing control of abiotic stress responses. Trends Plant Sci. 2018, 23, 140–150. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.; Qian, B.; Cao, F.; Wu, W.; Yang, L.; Guan, Q.; Gu, X.; Wang, P.; Okusolubo, T.A.; Dunn, S.L.; et al. An Arabidopsis PWI and RRM motif-containing protein is critical for pre-mRNA splicing and ABA responses. Nat. Commun. 2015, 6, 8139. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Kalinowska, K.; Muller, B.; Mergner, J.; Deutzmann, R.; Schwechheimer, C.; Hammes, U.Z.; Dresselhaus, T. DiSUMO-LIKE interacts with RNA-binding proteins and affects cell-cycle progression during maize embryogenesis. Curr. Biol. 2018, 28, 1548–1560. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lou, L.; Ding, L.; Wang, T.; Xiang, Y. Emerging Roles of RNA-Binding Proteins in Seed Development and Performance. Int. J. Mol. Sci. 2020, 21, 6822. https://doi.org/10.3390/ijms21186822
Lou L, Ding L, Wang T, Xiang Y. Emerging Roles of RNA-Binding Proteins in Seed Development and Performance. International Journal of Molecular Sciences. 2020; 21(18):6822. https://doi.org/10.3390/ijms21186822
Chicago/Turabian StyleLou, Lijuan, Ling Ding, Tao Wang, and Yong Xiang. 2020. "Emerging Roles of RNA-Binding Proteins in Seed Development and Performance" International Journal of Molecular Sciences 21, no. 18: 6822. https://doi.org/10.3390/ijms21186822
APA StyleLou, L., Ding, L., Wang, T., & Xiang, Y. (2020). Emerging Roles of RNA-Binding Proteins in Seed Development and Performance. International Journal of Molecular Sciences, 21(18), 6822. https://doi.org/10.3390/ijms21186822





