Structure, Folding and Stability of Nucleoside Diphosphate Kinases
Abstract
:1. Introduction
2. NDPK Monomer Structure
2.1. The α/β Domain
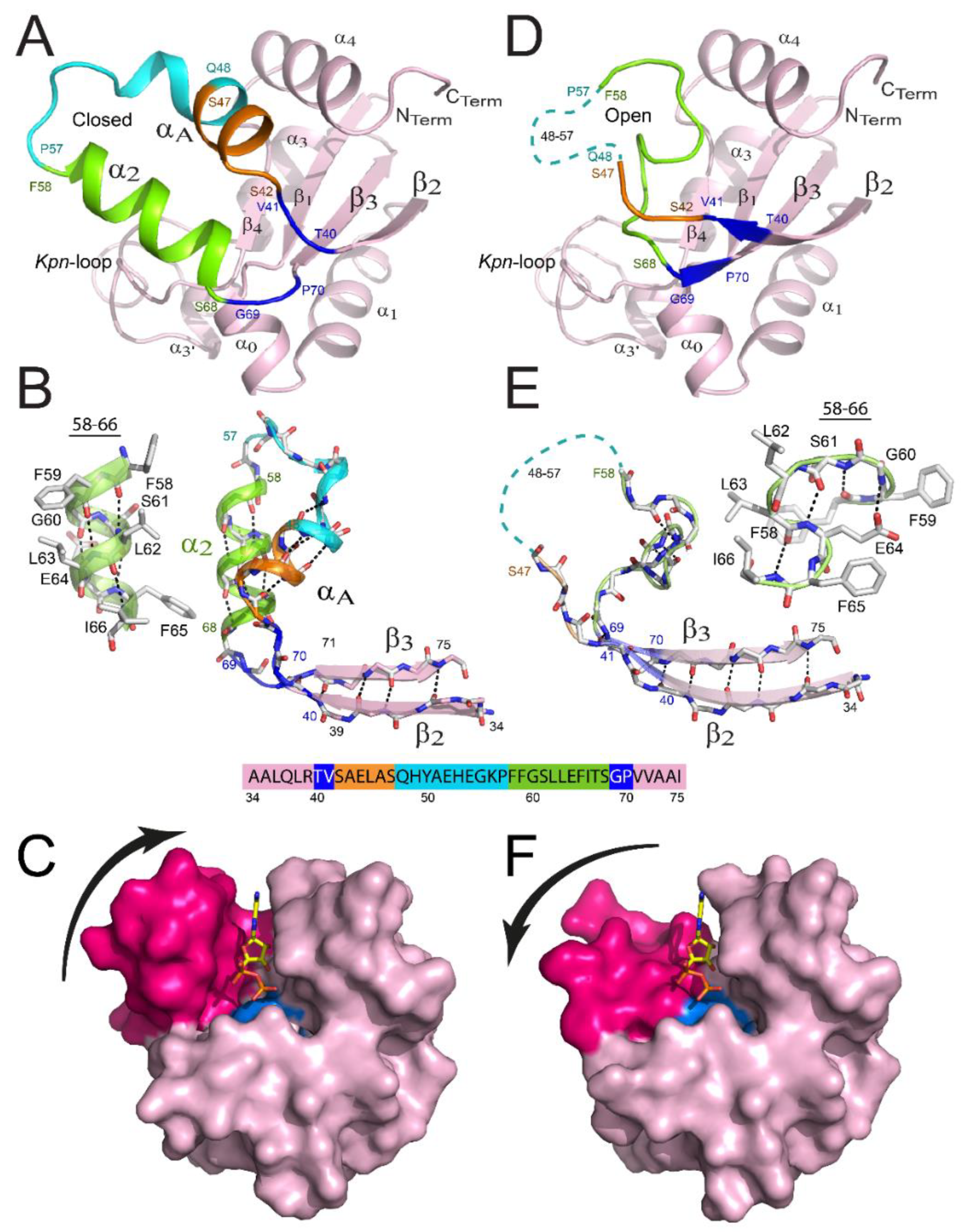
2.2. Structural Specificities of NDPK
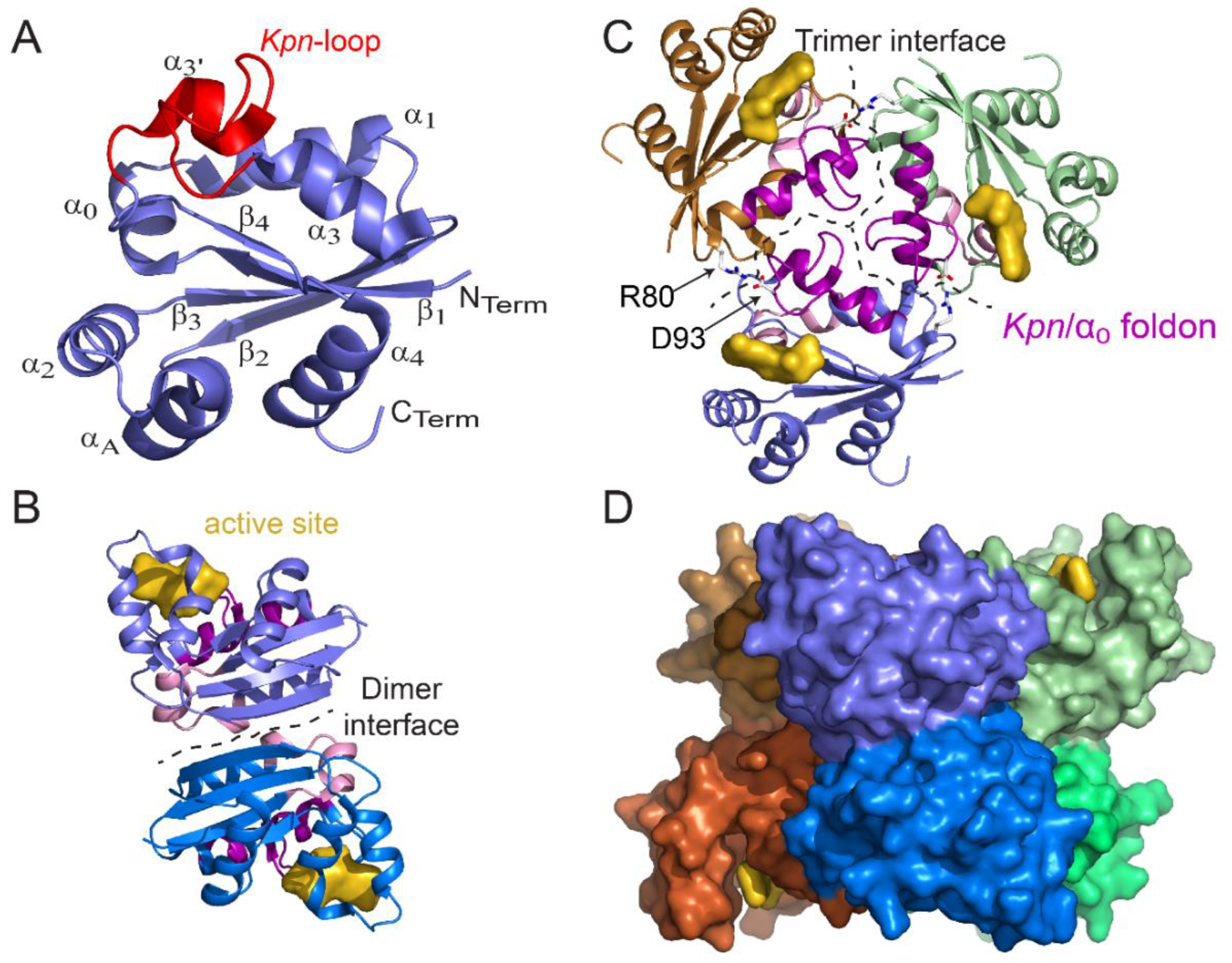
2.3. Active Site and Substrate Fixation
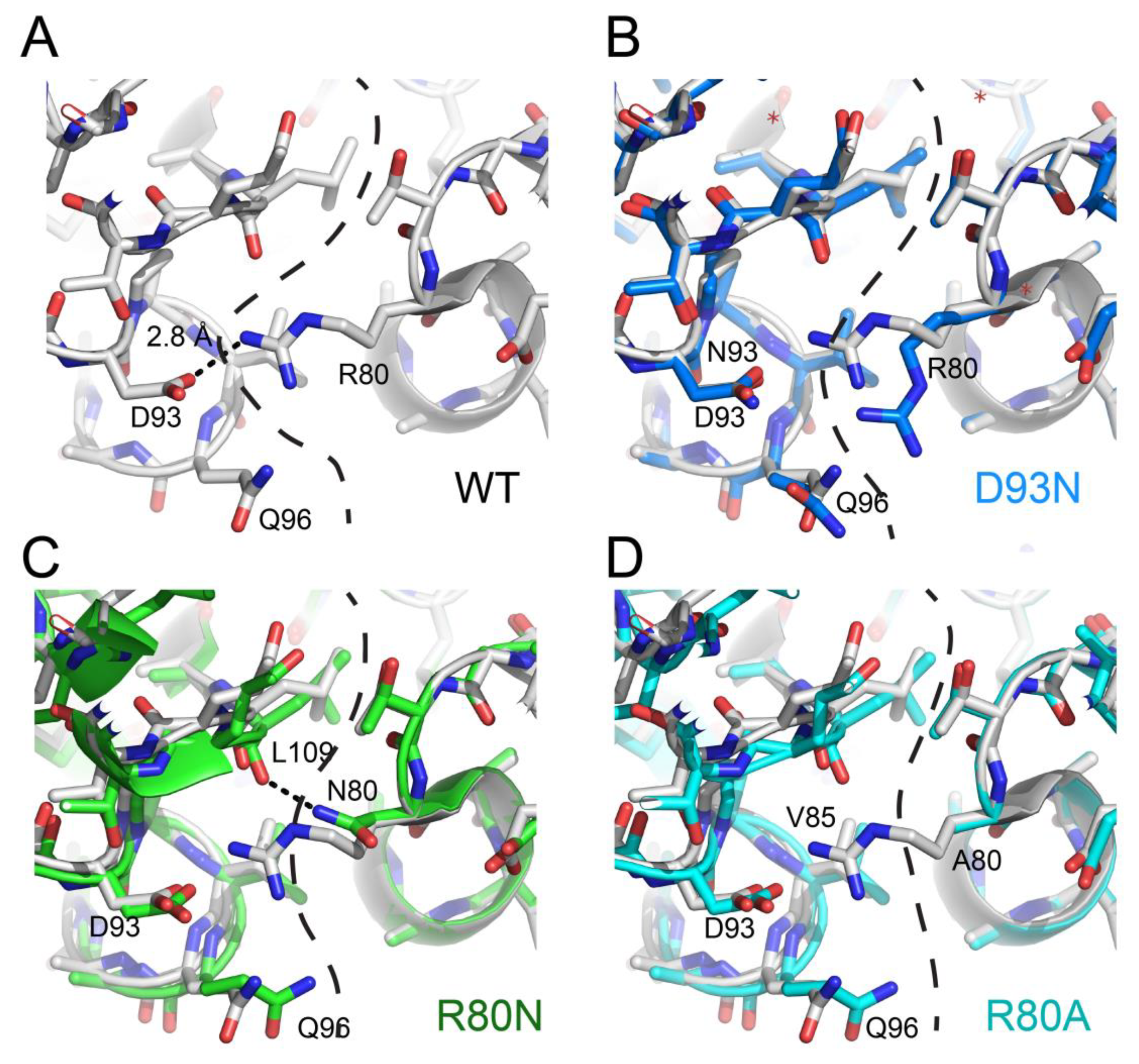
3. NDPK Quaternary Structure
3.1. Dimer: The Basic Subunit of NDPK Oligomer
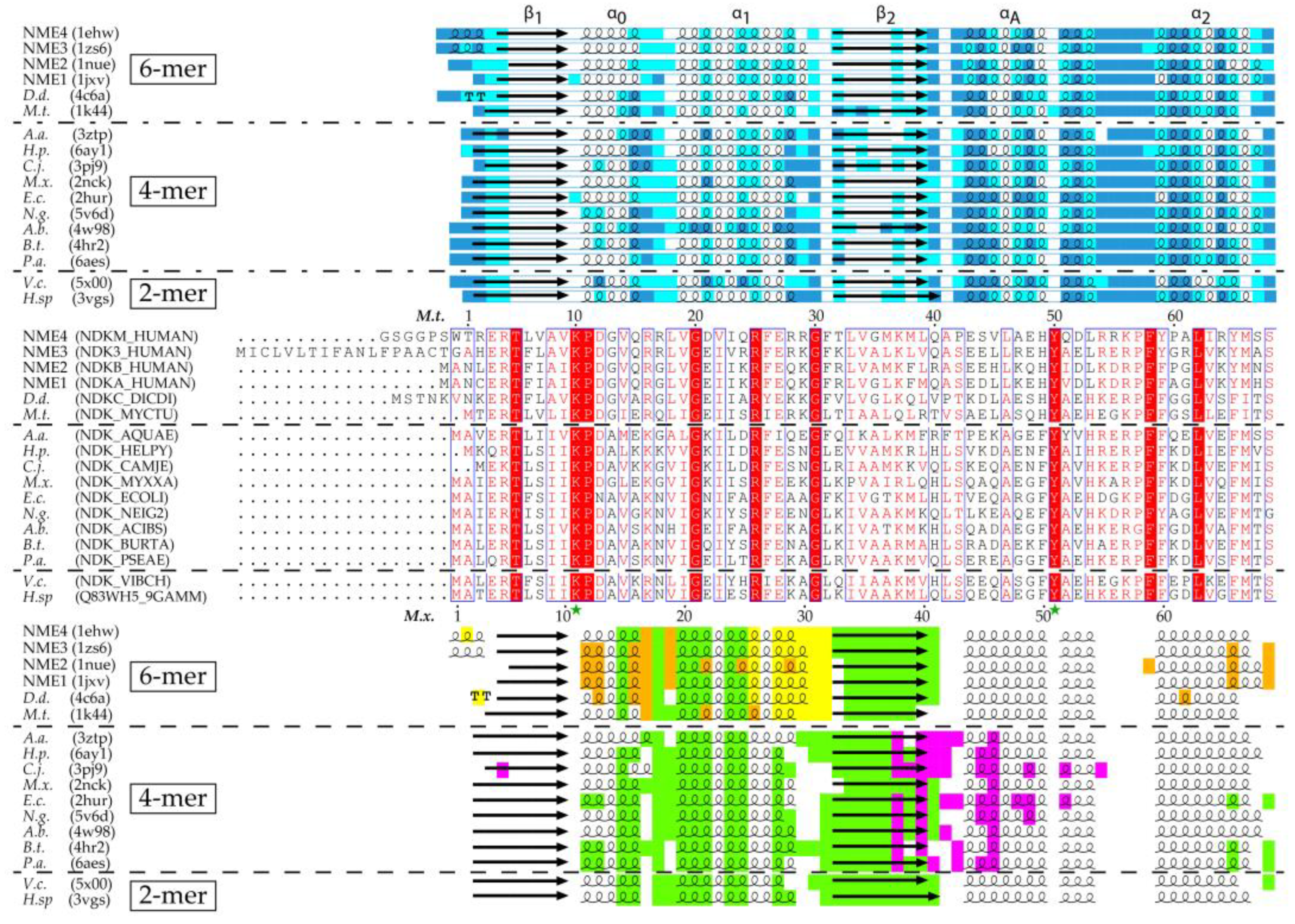
3.2. Hexamer and Tetramer of NDPK
3.3. The NME Family
4. NDPK Folding
5. NDPK Stability
6. Concluding Remarks and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| asa | accessible surface area |
| bsa | buried surface area |
| cryo-EM | cryo-electron microscopy |
| DSSP | Database of Secondary Structure of Proteins |
| FAP67 | Flagellar associated protein 67 |
| HDX-MS | Hydrogen-Deuterium eXchange—Mass Spectrometry |
| Kpn | Killer of prune |
| NDPK | Nucleoside diphosphate kinase |
| NDPK-A | Nucleoside diphosphate kinase isoform A |
| NME | Nucleotide metabolism enzyme |
| PISA | Proteins, Interfaces, Structures and Assemblies |
| rmsd | root mean square deviation |
| SEC-MALLS | Multi-angle scattering of laser light coupled with size exclusion chromatography |
| TMAO | Trimethylamine-N-oxide |
| WT | Wild type |
References
- Boissan, M.; Schlattner, U.; Lacombe, M.L. The NDPK/NME superfamily: State of the art. Lab. Investig. 2018, 98, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Desvignes, T.; Pontarotti, P.; Fauvel, C.; Bobe, J. Nme protein family evolutionary history, a vertebrate perspective. BMC Evol. Biol. 2009, 9, 256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lacombe, M.L.; Milon, L.; Munier, A.; Mehus, J.G.; Lambeth, D.O. The human Nm23/nucleoside diphosphate kinases. J. Bioenerg. Biomembr. 2000, 32, 247–258. [Google Scholar] [CrossRef]
- Lacombe, M.L.; Tokarska-Schlattner, M.; Boissan, M.; Schlattner, U. The mitochondrial nucleoside diphosphate kinase (NDPK-D/NME4), a moonlighting protein for cell homeostasis. Lab. Investig. 2018, 98, 582–588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lascu, I.; Gonin, P. The catalytic mechanism of nucleoside diphosphate kinases. J. Bioenerg. Biomembr. 2000, 32, 237–246. [Google Scholar] [CrossRef]
- Janin, J.; Dumas, C.; Moréra, S.; Xu, Y.; Meyer, P.; Chiadmi, M.; Cherfils, J. Three-dimensional structure of nucleoside diphosphate kinase. J. Bioenerg. Biomembr. 2000, 32, 215–225. [Google Scholar] [CrossRef]
- Bernard, M.A.; Ray, N.B.; Olcott, M.C.; Hendricks, S.P.; Mathews, C.K. Metabolic functions of microbial nucleoside diphosphate kinases. J. Bioenerg. Biomembr. 2000, 32, 259–267. [Google Scholar] [CrossRef]
- Dorion, S.; Rivoal, J. Plant nucleoside diphosphate kinase 1: A housekeeping enzyme with moonlighting activity. Plant Signal. Behav. 2018, 13, e1475804. [Google Scholar] [CrossRef] [Green Version]
- Lu, Z.; Hunter, T. Metabolic kinases moonlighting as protein kinases. Trends Biochem. Sci. 2018, 43, 301–310. [Google Scholar] [CrossRef]
- Wolfe, K.; Kofuji, S.; Yoshino, H.; Sasaki, M.; Okumura, K.; Sasaki, A.T. Dynamic compartmentalization of purine nucleotide metabolic enzymes at leading edge in highly motile renal cell carcinoma. Biochem. Biophys. Res. Commun. 2019, 516, 50–56. [Google Scholar] [CrossRef]
- Kapoor, I.; Varshney, U. Diverse roles of nucleoside diphosphate kinase in genome stability and growth fitness. Curr. Genet. 2020, 66, 571–582. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; McCorkle, J.R.; Novak, M.; Yang, M.; Kaetzel, D.M. Metastasis suppressor function of NM23-H1 requires its 3’-5’ exonuclease activity. Int. J. Cancer 2011, 128, 40–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, C.Y.; Chang, C.L. NDPKA is not just a metastasis suppressor—Be aware of its metastasis-promoting role in neuroblastoma. Lab. Investig. 2018, 98, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Steeg, P.S. Metastasis suppressors: Functional pathways. Lab. Investig. 2018, 98, 198–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boissan, M.; Montagnac, G.; Shen, Q.; Griparic, L.; Guitton, J.; Romao, M.; Sauvonnet, N.; Lagache, T.; Lascu, I.; Raposo, G.; et al. Membrane trafficking. Nucleoside diphosphate kinases fuel dynamin superfamily proteins with GTP for membrane remodeling. Science 2014, 344, 1510–1515. [Google Scholar] [CrossRef] [Green Version]
- Snider, N.T.; Altshuler, P.J.; Omary, M.B. Modulation of cytoskeletal dynamics by mammalian nucleoside diphosphate kinase (NDPK) proteins. Naunyn Schmiedebergs Arch. Pharmacol. 2015, 388, 189–197. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Poghosyan, E.; Gopal, R.; Liu, Y.; Ciruelas, K.S.; Maizy, Y.; Diener, D.R.; King, S.M.; Ishikawa, T.; Yang, P. General and specific promotion of flagellar assembly by a flagellar nucleoside diphosphate kinase. Mol. Biol. Cell 2017, 28, 3029–3042. [Google Scholar] [CrossRef]
- Vlatkovic, N.; Chang, S.H.; Boyd, M.T. Janus-faces of NME-oncoprotein interactions. Naunyn Schmiedebergs Arch. Pharmacol. 2015, 388, 175–187. [Google Scholar] [CrossRef]
- Steeg, P.S.; Zollo, M.; Wieland, T. A critical evaluation of biochemical activities reported for the nucleoside diphosphate kinase/Nm23/Awd family proteins: Opportunities and missteps in understanding their biological functions. Naunyn Schmiedebergs Arch. Pharmacol. 2011, 384, 331–339. [Google Scholar] [CrossRef] [Green Version]
- Dumas, C.; Lascu, I.; Moréra, S.; Glaser, P.; Fourme, R.; Wallet, V.; Lacombe, M.L.; Véron, M.; Janin, J. X-ray structure of nucleoside diphosphate kinase. EMBO J. 1992, 11, 3203–3208. [Google Scholar] [CrossRef]
- Williams, R.L.; Oren, D.A.; Munoz-Dorado, J.; Inouye, S.; Inouye, M.; Arnold, E. Crystal structure of Myxococcus xanthus nucleoside diphosphate kinase and its interaction with a nucleotide substrate at 2.0 Å resolution. J. Mol. Biol. 1993, 234, 1230–1247. [Google Scholar] [CrossRef] [PubMed]
- Moynié, L.; Giraud, M.F.; Georgescauld, F.; Lascu, I.; Dautant, A. The structure of the Escherichia coli nucleoside diphosphate kinase reveals a new quaternary architecture for this enzyme family. Proteins 2007, 67, 755–765. [Google Scholar] [CrossRef] [PubMed]
- Arai, S.; Yonezawa, Y.; Okazaki, N.; Matsumoto, F.; Tamada, T.; Tokunaga, H.; Ishibashi, M.; Blaber, M.; Tokunaga, M.; Kuroki, R. A structural mechanism for dimeric to tetrameric oligomer conversion in Halomonas sp. nucleoside diphosphate kinase. Protein Sci. 2012, 21, 498–510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baugh, L.; Gallagher, L.A.; Patrapuvich, R.; Clifton, M.C.; Gardberg, A.S.; Edwards, T.E.; Armour, B.; Begley, D.W.; Dieterich, S.H.; Dranow, D.M.; et al. Combining functional and structural genomics to sample the essential Burkholderia structome. PLoS ONE 2013, 8, e53851. [Google Scholar] [CrossRef]
- Hu, Y.S.; Feng, F.; Liu, Y.F. Structural and functional characterization of Acinetobacter baumannii nucleoside diphosphate kinase. Prog. Biochem. Biophys. 2015, 42, 260–267. [Google Scholar]
- Priet, S.; Roux, L.; Saez-Ayala, M.; Ferron, F.; Canard, B.; Alvarez, K. Enzymatic synthesis of acyclic nucleoside thiophosphonate diphosphates: Effect of the alpha-phosphorus configuration on HIV-1 RT activity. Antiviral. Res. 2015, 117, 122–131. [Google Scholar] [CrossRef]
- Boissier, F.; Georgescauld, F.; Moynié, L.; Dupuy, J.W.; Sarger, C.; Podar, M.; Lascu, I.; Giraud, M.F.; Dautant, A. An intersubunit disulfide bridge stabilizes the tetrameric nucleoside diphosphate kinase of Aquifex aeolicus. Proteins 2012, 80, 1658–1668. [Google Scholar] [CrossRef]
- Georgescauld, F.; Moynié, L.; Habersetzer, J.; Dautant, A. Structure of Mycobacterium tuberculosis nucleoside diphosphate kinase R80N mutant in complex with citrate. Acta Crystallogr. F Struct. Biol. Commun. 2014, 70, 40–43. [Google Scholar] [CrossRef]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.M.; Krissinel, E.B.; Leslie, A.G.; McCoy, A.; et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Ma, M.; Stoyanova, M.; Rademacher, G.; Dutcher, S.K.; Brown, A.; Zhang, R. Structure of the decorated ciliary doublet microtubule. Cell 2019, 179, 909–922.e12. [Google Scholar] [CrossRef]
- Kabsch, W.; Sander, C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef] [PubMed]
- Krissinel, E.; Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [Green Version]
- Carotenuto, M.; Pedone, E.; Diana, D.; de Antonellis, P.; Dzeroski, S.; Marino, N.; Navas, L.; Di Dato, V.; Scoppettuolo, M.N.; Cimmino, F.; et al. Neuroblastoma tumorigenesis is regulated through the Nm23-H1/h-Prune C-terminal interaction. Sci. Rep. 2013, 3, 1351. [Google Scholar] [CrossRef] [Green Version]
- D’Angelo, A.; Garzia, L.; Andre, A.; Carotenuto, P.; Aglio, V.; Guardiola, O.; Arrigoni, G.; Cossu, A.; Palmieri, G.; Aravind, L.; et al. Prune cAMP phosphodiesterase binds nm23-H1 and promotes cancer metastasis. Cancer Cell 2004, 5, 137–149. [Google Scholar] [CrossRef] [Green Version]
- Dautant, A.; Meyer, P.; Georgescauld, F. Hydrogen/Deuterium Exchange Mass Spectrometry reveals mechanistic details of activation of nucleoside diphosphate kinases by oligomerization. Biochemistry 2017, 56, 2886–2896. [Google Scholar] [CrossRef]
- Georgescauld, F.; Moynié, L.; Habersetzer, J.; Cervoni, L.; Mocan, I.; Borza, T.; Harris, P.; Dautant, A.; Lascu, I. Intersubunit ionic interactions stabilize the nucleoside diphosphate kinase of Mycobacterium tuberculosis. PLoS ONE 2013, 8, e57867. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. PyMOL Molecular Graphics System; Schrodinger: New York, NY, USA, 2002. [Google Scholar]
- Adam, K.; Hunter, T. Histidine kinases and the missing phosphoproteome from prokaryotes to eukaryotes. Lab. Investig. 2018, 98, 233–247. [Google Scholar] [CrossRef] [Green Version]
- Attwood, P.V.; Wieland, T. Nucleoside diphosphate kinase as protein histidine kinase. Naunyn Schmiedebergs Arch Pharmacol. 2015, 388, 153–160. [Google Scholar] [CrossRef]
- Dautant, A.; Henri, J.; Wales, T.E.; Meyer, P.; Engen, J.R.; Georgescauld, F. Remodeling of the binding site of nucleoside diphosphate kinase revealed by X-ray structure and H/D exchange. Biochemistry 2019, 58, 1440–1449. [Google Scholar] [CrossRef]
- Tepper, A.D.; Dammann, H.; Bominaar, A.A.; Véron, M. Investigation of the active site and the conformational stability of nucleoside diphosphate kinase by site-directed mutagenesis. J. Biol. Chem. 1994, 269, 32175–32180. [Google Scholar] [PubMed]
- Schneider, B.; Sarfati, R.; Deville-Bonne, D.; Véron, M. Role of nucleoside diphosphate kinase in the activation of anti-HIV nucleoside analogs. J. Bioenerg. Biomembr. 2000, 32, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Sellam, O.; Moréra, S.; Sarfati, S.; Biondi, R.; Véron, M.; Janin, J. X-ray analysis of azido-thymidine diphosphate binding to nucleoside diphosphate kinase. Proc. Natl. Acad. Sci. USA 1997, 94, 7162–7165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeudy, S.; Lartigue, A.; Claverie, J.M.; Abergel, C. Dissecting the unique nucleotide specificity of mimivirus nucleoside diphosphate kinase. J. Virol. 2009, 83, 7142–7150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lascu, L.; Giartosio, A.; Ransac, S.; Erent, M. Quaternary structure of nucleoside diphosphate kinases. J. Bioenerg. Biomembr. 2000, 32, 227–236. [Google Scholar] [CrossRef]
- Yonezawa, Y.; Izutsu, K.; Tokunaga, H.; Maeda, H.; Arakawa, T.; Tokunaga, M. Dimeric structure of nucleoside diphosphate kinase from moderately halophilic bacterium: Contrast to the tetrameric Pseudomonas counterpart. FEMS Microbiol. Lett. 2007, 268, 52–58. [Google Scholar] [CrossRef] [Green Version]
- Mesnildrey, S.; Agou, F.; Karlsson, A.; Bonne, D.D.; Véron, M. Coupling between catalysis and oligomeric structure in nucleoside diphosphate kinase. J. Biol. Chem. 1998, 273, 4436–4442. [Google Scholar] [CrossRef] [Green Version]
- Karlsson, A.; Mesnildrey, S.; Xu, Y.; Moréra, S.; Janin, J.; Véron, M. Nucleoside diphosphate kinase. Investigation of the intersubunit contacts by site-directed mutagenesis and crystallography. J. Biol. Chem. 1996, 271, 19928–19934. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Moréra, S.; Mocan, J.; Lascu, I.; Janin, J. X-ray structure of Mycobacterium tuberculosis nucleoside diphosphate kinase. Proteins 2002, 47, 556–557. [Google Scholar] [CrossRef]
- Chen, C.J.; Liu, M.Y.; Chang, T.; Chang, W.C.; Wang, B.C.; Le Gall, J. Crystal structure of a nucleoside diphosphate kinase from Bacillus halodenitrificans: Coexpression of its activity with a Mn-superoxide dismutase. J. Struct. Biol. 2003, 142, 247–255. [Google Scholar] [CrossRef]
- Potel, C.M.; Fasci, D.; Heck, A.J.R. Mix and match of the tumor metastasis suppressor Nm23 protein isoforms in vitro and in vivo. FEBS J. 2018, 285, 2856–2868. [Google Scholar] [CrossRef] [Green Version]
- Shen, R.; Wheeler, L.J.; Mathews, C.K. Molecular interactions involving Escherichia coli nucleoside diphosphate kinase. J. Bioenerg. Biomembr. 2006, 38, 255–259. [Google Scholar] [CrossRef] [PubMed]
- Puts, G.S.; Leonard, M.K.; Pamidimukkala, N.V.; Snyder, D.E.; Kaetzel, D.M. Nuclear functions of NME proteins. Lab. Investig. 2018, 98, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Schlattner, U.; Tokarska-Schlattner, M.; Epand, R.M.; Boissan, M.; Lacombe, M.L.; Kagan, V.E. NME4/nucleoside diphosphate kinase D in cardiolipin signaling and mitophagy. Lab. Investig. 2018, 98, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Steeg, P.S.; Bevilacqua, G.; Kopper, L.; Thorgeirsson, U.P.; Talmadge, J.E.; Liotta, L.A.; Sobel, M.E. Evidence for a novel gene associated with low tumor metastatic potential. J. Natl. Cancer Inst. 1988, 80, 200–204. [Google Scholar] [CrossRef]
- Chang, C.L.; Zhu, X.X.; Thoraval, D.H.; Ungar, D.; Rawwas, J.; Hora, N.; Strahler, J.R.; Hanash, S.M.; Radany, E. Nm23-H1 mutation in neuroblastoma. Nature 1994, 370, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.L.; Strahler, J.R.; Thoraval, D.H.; Qian, M.G.; Hinderer, R.; Hanash, S.M. A nucleoside diphosphate kinase A (nm23-H1) serine 120-->glycine substitution in advanced stage neuroblastoma affects enzyme stability and alters protein-protein interaction. Oncogene 1996, 12, 659–667. [Google Scholar]
- Georgescauld, F.; Mocan, I.; Lacombe, M.L.; Lascu, I. Rescue of the neuroblastoma mutant of the human nucleoside diphosphate kinase A/nm23-H1 by the natural osmolyte trimethylamine-N-oxide. FEBS Lett. 2009, 583, 820–824. [Google Scholar] [CrossRef] [Green Version]
- Giraud, M.F.; Georgescauld, F.; Lascu, I.; Dautant, A. Crystal structures of S120G mutant and wild type of human nucleoside diphosphate kinase A in complex with ADP. J. Bioenerg. Biomembr. 2006, 38, 261–264. [Google Scholar] [CrossRef]
- Lascu, I.; Schaertl, S.; Wang, C.; Sarger, C.; Giartosio, A.; Briand, G.; Lacombe, M.L.; Konrad, M. A point mutation of human nucleoside diphosphate kinase A found in aggressive neuroblastoma affects protein folding. J. Biol. Chem. 1997, 272, 15599–15602. [Google Scholar] [CrossRef] [Green Version]
- Mocan, I.; Georgescauld, F.; Gonin, P.; Thoraval, D.; Cervoni, L.; Giartosio, A.; Dabernat-Arnaud, S.; Crouzet, M.; Lacombe, M.L.; Lascu, I. Protein phosphorylation corrects the folding defect of the neuroblastoma (S120G) mutant of human nucleoside diphosphate kinase A/Nm23-H1. Biochem. J. 2007, 403, 149–156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Georgescauld, F.; Sabaté, R.; Espargarόo, A.; Ventura, S.; Chaignepain, S.; Lacombe, M.L.; Lascu, I. Aggregation of the neuroblastoma-associated mutant (S120G) of the human nucleoside diphosphate kinase-A/NM23-H1 into amyloid fibrils. Naunyn Schmiedebergs Arch. Pharmacol. 2011, 384, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Lascu, I. Nm23-H1/NDP kinase folding intermediates and cancer: A hypothesis. J. Bioenerg. Biomembr. 2006, 38, 265–268. [Google Scholar] [CrossRef] [PubMed]
- Cervoni, L.; Egistelli, L.; Mocan, I.; Giartosio, A.; Lascu, I. Quaternary structure of Dictyostelium discoideum nucleoside diphosphate kinase counteracts the tendency of monomers to form a molten globule. Biochemistry 2003, 42, 14599–14605. [Google Scholar] [CrossRef] [PubMed]
- Lascu, I.; Deville-Bonne, D.; Glaser, P.; Véron, M. Equilibrium dissociation and unfolding of nucleoside diphosphate kinase from Dictyostelium discoideum. Role of proline 100 in the stability of the hexameric enzyme. J. Biol. Chem. 1993, 268, 20268–20275. [Google Scholar]
- Arakawa, T.; Tokunaga, M. Electrostatic and hydrophobic interactions play a major role in the stability and refolding of halophilic proteins. Protein Pept. Lett. 2004, 11, 125–132. [Google Scholar] [CrossRef]
- Ishibashi, M.; Ida, K.; Tatsuda, S.; Arakawa, T.; Tokunaga, M. Interaction of hexa-His tag with acidic amino acids results in facilitated refolding of halophilic nucleoside diphosphate kinase. Int. J. Biol. Macromol. 2011, 49, 778–783. [Google Scholar] [CrossRef]
- Lascu, I.; Chaffotte, A.; Limbourg-Bouchon, B.; Véron, M. A Pro/Ser substitution in nucleoside diphosphate kinase of Drosophila melanogaster (mutation killer of prune) affects stability but not catalytic efficiency of the enzyme. J. Biol. Chem. 1992, 267, 12775–12781. [Google Scholar]
- Giartosio, A.; Erent, M.; Cervoni, L.; Moréra, S.; Janin, J.; Konrad, M.; Lascu, I. Thermal stability of hexameric and tetrameric nucleoside diphosphate kinases. Effect of subunit interaction. J. Biol. Chem. 1996, 271, 17845–17851. [Google Scholar] [CrossRef] [Green Version]
- Ishibashi, M.; Hayashi, T.; Yoshida, C.; Tokunaga, M. Increase of salt dependence of halophilic nucleoside diphosphate kinase caused by a single amino acid substitution. Extremophiles 2013, 17, 585–591. [Google Scholar] [CrossRef]
- Ishibashi, M.; Uchino, M.; Arai, S.; Kuroki, R.; Arakawa, T.; Tokunaga, M. Reduction of salt-requirement of halophilic nucleoside diphosphate kinase by engineering S-S bond. Arch. Biochem. Biophys. 2012, 525, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Tokunaga, H.; Ishibashi, M.; Arisaka, F.; Arai, S.; Kuroki, R.; Arakawa, T.; Tokunaga, M. Residue 134 determines the dimer-tetramer assembly of nucleoside diphosphate kinase from moderately halophilic bacteria. FEBS Lett. 2008, 582, 1049–1054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamura, A.; Ichimura, T.; Kamekura, M.; Mizuki, T.; Usami, R.; Makino, T.; Ohtsuka, J.; Miyazono, K.; Okai, M.; Nagata, K.; et al. Molecular mechanism of distinct salt-dependent enzyme activity of two halophilic nucleoside diphosphate kinases. Biophys. J. 2009, 96, 4692–4700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pedelacq, J.D.; Waldo, G.S.; Cabantous, S.; Liong, E.C.; Terwilliger, T.C. Structural and functional features of an NDP kinase from the hyperthermophile crenarchaeon Pyrobaculum aerophilum. Protein Sci. 2005, 14, 2562–2573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yonezawa, Y.; Nagayama, A.; Tokunaga, H.; Ishibashi, M.; Arai, S.; Kuroki, R.; Watanabe, K.; Arakawa, T.; Tokunaga, M. Nucleoside diphosphate kinase from psychrophilic Pseudoalteromonas sp. AS-131 Isolated from Antarctic Ocean. Protein J. 2015, 34, 275–283. [Google Scholar] [CrossRef]
- Vieira, P.S.; de Giuseppe, P.O.; de Oliveira, A.H.C.; Murakami, M.T. The role of the C-terminus and Kpn loop in the quaternary structure stability of nucleoside diphosphate kinase from Leishmania parasites. J. Struct. Biol. 2015, 192, 336–341. [Google Scholar] [CrossRef]
- Vieira, P.S.; de Giuseppe, P.O.; Murakami, M.T.; de Oliveira, A.H. Crystal structure and biophysical characterization of the nucleoside diphosphate kinase from Leishmania braziliensis. BMC Struct. Biol. 2015, 15, 2. [Google Scholar] [CrossRef] [Green Version]
- Vieira, P.S.; de Jesus Santos, A.P.; de Oliveira, A.H. Biophysical characterization of the nucleoside diphosphate kinase of Leishmania major and Effect of the P95S Mutation. Protein Pept. Lett. 2016, 23, 99–106. [Google Scholar] [CrossRef]
- Souza, T.A.; Trindade, D.M.; Tonoli, C.C.; Santos, C.R.; Ward, R.J.; Arni, R.K.; Oliveira, A.H.; Murakami, M.T. Molecular adaptability of nucleoside diphosphate kinase b from trypanosomatid parasites: Stability, oligomerization and structural determinants of nucleotide binding. Mol. Biosyst. 2011, 7, 2189–2195. [Google Scholar] [CrossRef]
- Akanuma, S.; Yokobori, S.; Nakajima, Y.; Bessho, M.; Yamagishi, A. Robustness of predictions of extremely thermally stable proteins in ancient organisms. Evolution 2015, 69, 2954–2962. [Google Scholar] [CrossRef]
- Garcia, A.K.; Schopf, J.W.; Yokobori, S.I.; Akanuma, S.; Yamagishi, A. Reconstructed ancestral enzymes suggest long-term cooling of Earth’s photic zone since the Archean. Proc. Natl. Acad. Sci. USA 2017, 114, 4619–4624. [Google Scholar] [CrossRef] [Green Version]
- Kimura, M.; Akanuma, S. Reconstruction and characterization of thermally stable and catalytically active proteins comprising an alphabet of ~13 amino acids. J. Mol. Evol. 2020, 88, 372–381. [Google Scholar] [CrossRef] [PubMed]
- Shibue, R.; Sasamoto, T.; Shimada, M.; Zhang, B.; Yamagishi, A.; Akanuma, S. Comprehensive reduction of amino acid set in a protein suggests the importance of prebiotic amino acids for stable proteins. Sci. Rep. 2018, 8, 1227. [Google Scholar] [CrossRef] [PubMed]
- Chopra, P.; Singh, A.; Koul, A.; Ramachandran, S.; Drlica, K.; Tyagi, A.K.; Singh, Y. Cytotoxic activity of nucleoside diphosphate kinase secreted from Mycobacterium tuberculosis. Eur. J. Biochem. 2003, 270, 625–634. [Google Scholar] [CrossRef] [Green Version]
- Ganaie, A.A.; Lella, R.K.; Solanki, R.; Sharma, C. Thermostable hexameric form of Eis (Rv2416c) protein of M. tuberculosis plays an important role for enhanced intracellular survival within macrophages. PLoS ONE 2011, 6, e27590. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Singh, V.; Lau, A.; Stokes, R.W.; Obregon-Henao, A.; Orme, I.M.; Wong, D.; Av-Gay, Y.; Hmama, Z. Mycobacterium tuberculosis nucleoside diphosphate kinase inactivates small GTPases leading to evasion of innate immunity. PLoS Pathog. 2013, 9, e1003499. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, T. NDP kinase 7 is a conserved microtubule-binding protein preferentially expressed in ciliated cells. Cell Struct. Funct. 2010, 35, 23–30. [Google Scholar] [CrossRef] [Green Version]
- Islam, K.; Burns, R.G. Microtubules and nucleoside diphosphate kinase. Nucleoside diphosphate kinase binds to co-purifying contaminants rather than to microtubule proteins. Biochem. J. 1985, 232, 651–656. [Google Scholar] [CrossRef]
- Liu, P.; Choi, Y.K.; Qi, R.Z. NME7 is a functional component of the gamma-tubulin ring complex. Mol. Biol. Cell 2014, 25, 2017–2025. [Google Scholar] [CrossRef]
- Lombardi, D.; Sacchi, A.; D’Agostino, G.; Tibursi, G. The association of the Nm23-M1 protein and beta-tubulin correlates with cell differentiation. Exp. Cell Res. 1995, 217, 267–271. [Google Scholar] [CrossRef]

| Id | Species (Resolution, Å) | 4D | Authors or References |
|---|---|---|---|
| Betaproteobacteria | |||
| 4dut | Burkholderia thailandensis (2.50 Å) | 4-mer | [24] |
| 4ek2 | Burkholderia thailandensis (2.00 Å) | 4-mer | [24] |
| 4hr2 | Burkholderia thailandensis bound to ADP (1.95 Å) | 4-mer | Clifton, M.C.; Abendroth, J.A. |
| 5v6d | Neisseria gonorrhoeae in complex with citrate (1.85 Å) | 4-mer | Abendroth, J.; Mayclin, S.J.; Lorimer, D.D.; Edwards, T.E. |
| Gammaproteobacteria | |||
| 4s0m | Acinetobacter baumannii (1.92 Å) | 4-mer | Sikarwar, J.; Shukla, P.K.; Kaur, P.; Sharma, S.; Singh, T.P. |
| 4wbf | Acinetobacter baumannii (2.64 Å) | 4-mer | [25] |
| 4w98 | Acinetobacter baumannii (1.43 Å) | 4-mer | [25] |
| 5yih | Acinetobacter baumannii (1.98 Å) | 4-mer | Bairagya, H.R.; Sikarwar, J.; Iqbal, N.; Singh, P.K.; Kaur, P.; Sharma, S.; Singh, T.P. |
| 5yol | Acinetobacter baumannii (2.2 Å) | 4-mer | Singh, P.K.; Sikarwar, J.; Kaur, P.; Sharma, S.; Singh, T.P. |
| 3vgs 3vgt | Halomonas sp. 593 WT (2.30 Å and 2.70 Å) | 2-mer | [23] |
| 3vgu 3vgv | Halomonas sp. 593 E134A mutant (2.30 Å and 2.50 Å) | 4-mer | [23] |
| 6aes | Pseudomonas aeruginosa (3.55 Å) | 4-mer | Sikarwar, J.; Singh, P.K.; Sharma, S.; Singh, T.P. |
| 5x00 | Vibrio cholerae (3.06 Å) | 2-mer | Agnihotri, P., Mishra, A.K.; Pratap, J.V. |
| Epsilonproteobacteria | |||
| 3pj9 | Campylobacter jejuni (2.10 Å) | 4-mer | Filippova, E.V.; Wawrzak, Z.; Onopriyenko, O.; Edwards, A.; Savchenko, A.; Anderson, W.F. |
| 6ay1 | Helicobacter pylori (2.05 Å) | 4-mer | Edwards, T.E.; Dranow, D.M.; Lorimer, D.D. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Georgescauld, F.; Song, Y.; Dautant, A. Structure, Folding and Stability of Nucleoside Diphosphate Kinases. Int. J. Mol. Sci. 2020, 21, 6779. https://doi.org/10.3390/ijms21186779
Georgescauld F, Song Y, Dautant A. Structure, Folding and Stability of Nucleoside Diphosphate Kinases. International Journal of Molecular Sciences. 2020; 21(18):6779. https://doi.org/10.3390/ijms21186779
Chicago/Turabian StyleGeorgescauld, Florian, Yuyu Song, and Alain Dautant. 2020. "Structure, Folding and Stability of Nucleoside Diphosphate Kinases" International Journal of Molecular Sciences 21, no. 18: 6779. https://doi.org/10.3390/ijms21186779
APA StyleGeorgescauld, F., Song, Y., & Dautant, A. (2020). Structure, Folding and Stability of Nucleoside Diphosphate Kinases. International Journal of Molecular Sciences, 21(18), 6779. https://doi.org/10.3390/ijms21186779





