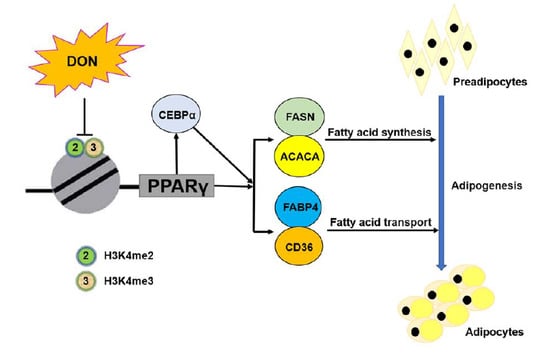
Deoxynivalenol Exposure Suppresses Adipogenesis by Inhibiting the Expression of Peroxisome Proliferator-Activated Receptor Gamma 2 (PPARγ2) in 3T3-L1 Cells
Abstract
1. Introduction
2. Results
2.1. DON Suppresses Intracellular Lipid Accumulation in 3T3-L1 Cells
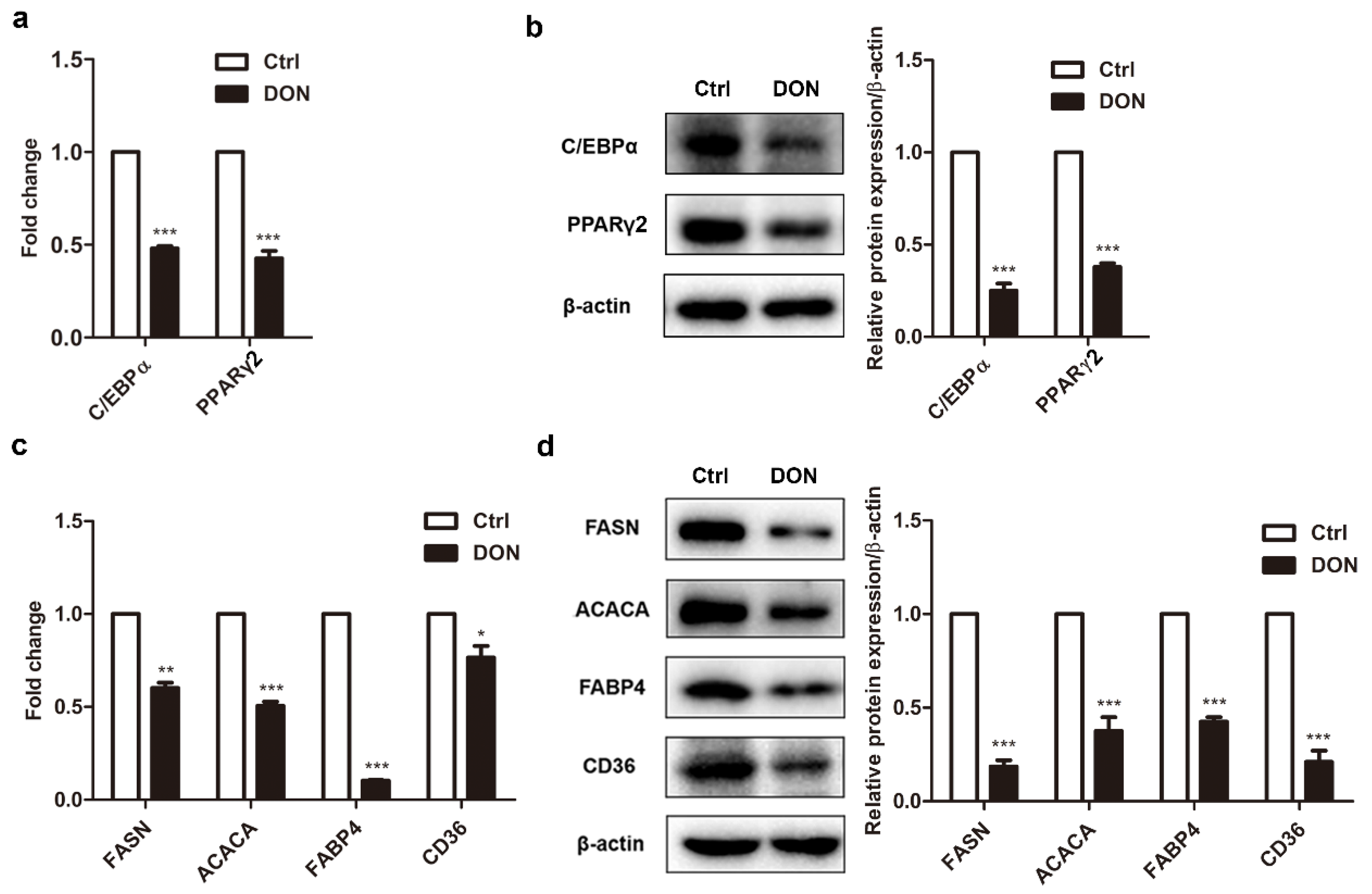
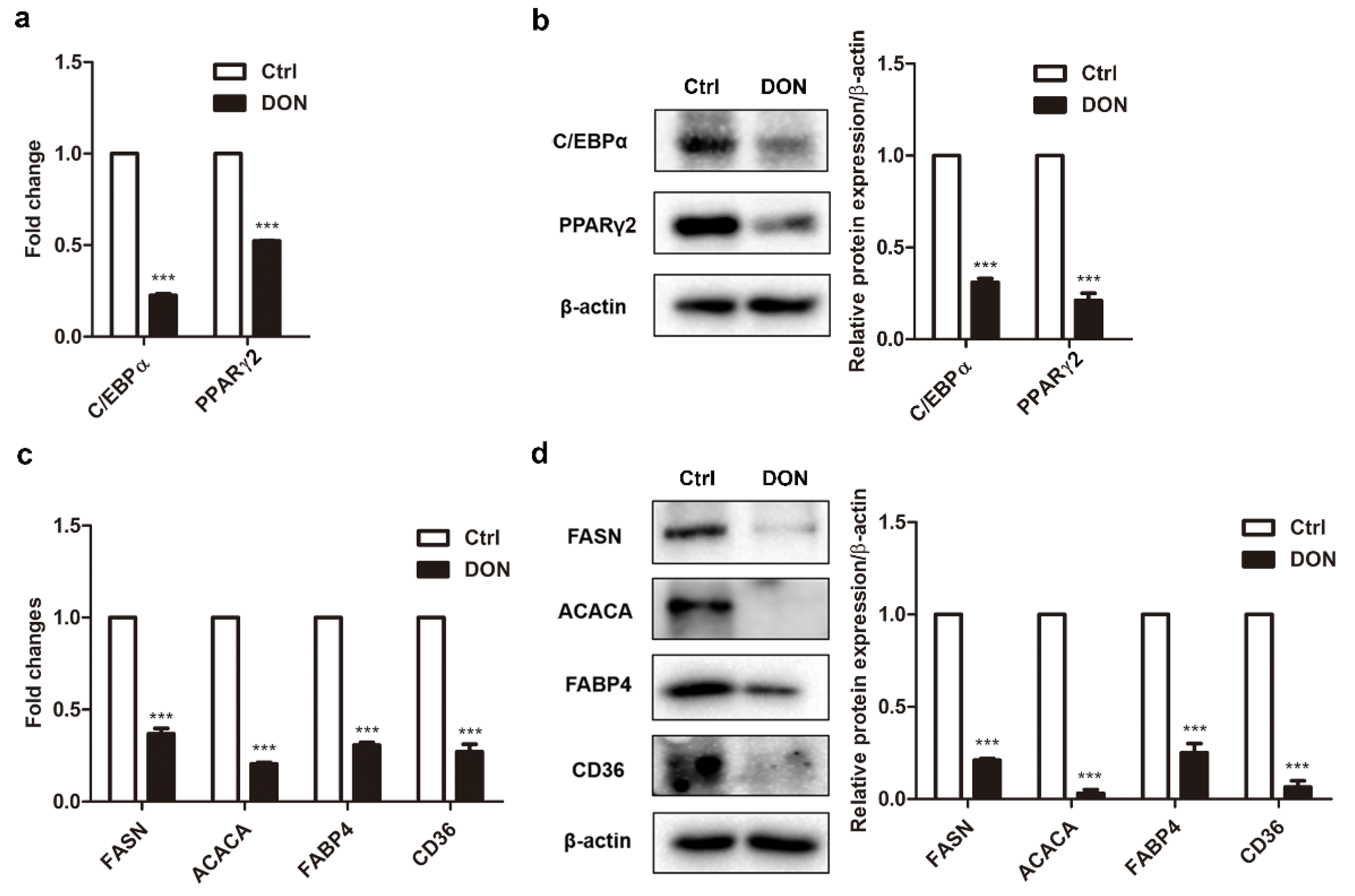
2.2. Low Expression Levels of Adipogenic Marker Genes in Differentiated 3T3-L1 Cells Treated with DON
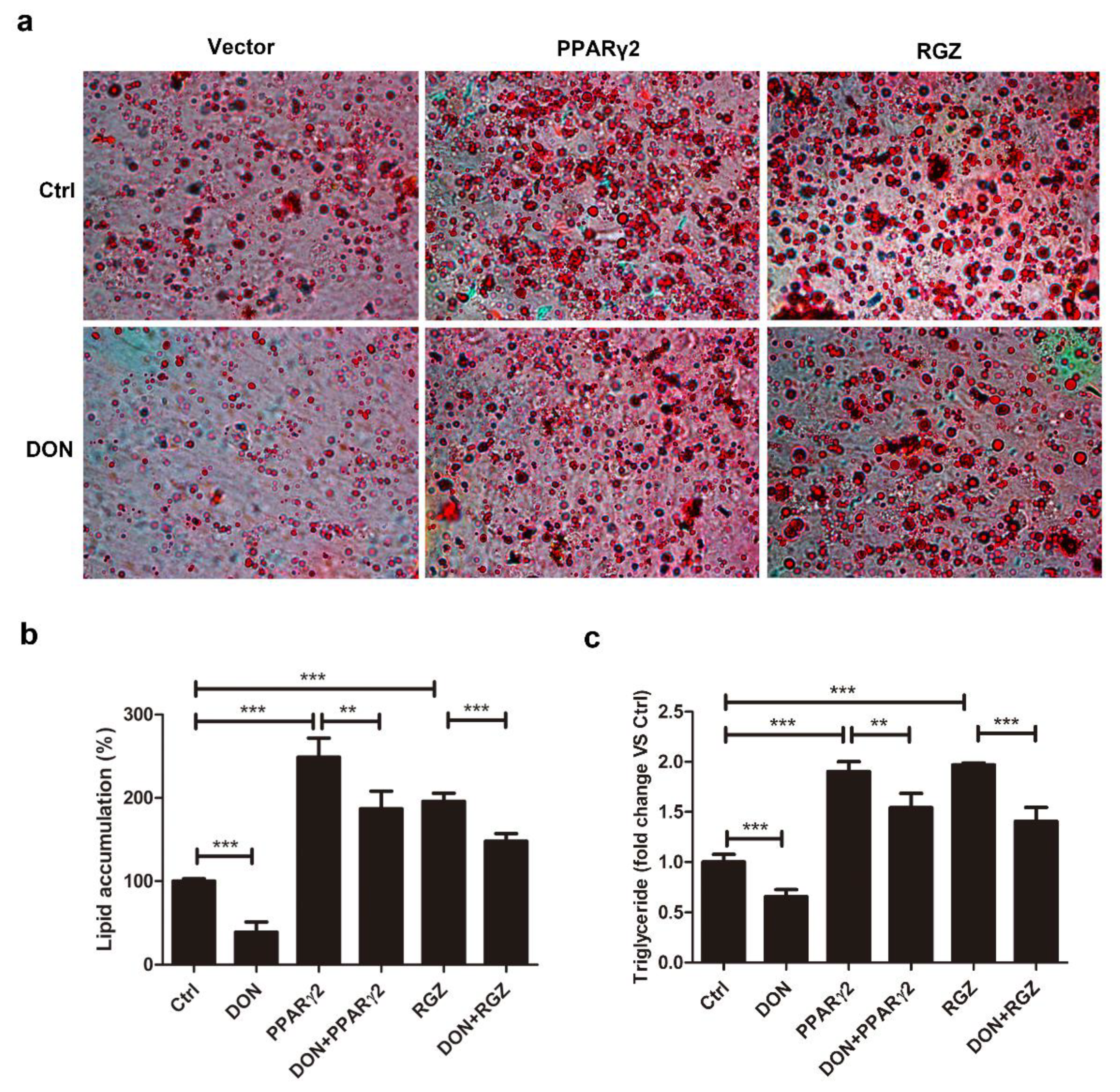
2.3. Overexpression of PPARγ2 Attenuated DON-Induced Inhibition of Adipogenesis
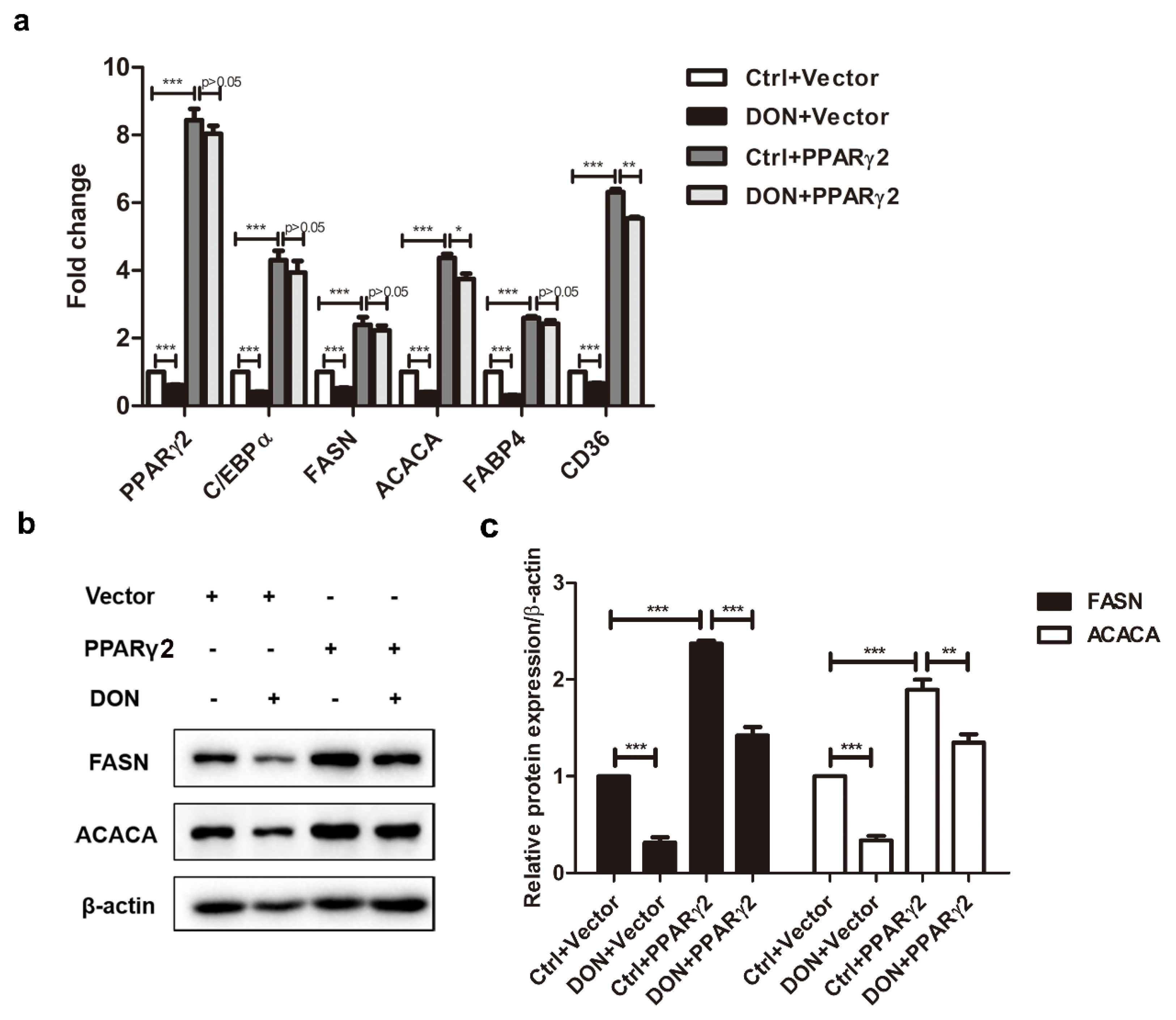
2.4. Overexpression of PPARγ2 Attenuated DON-Induced Downregulation of Adipogenic Marker Genes
2.5. DON Decreases WAT Size and TG Levels in BALB/c Mice
2.6. Low Expression Levels of Adipogenic Marker Genes in Mice Treated with DON
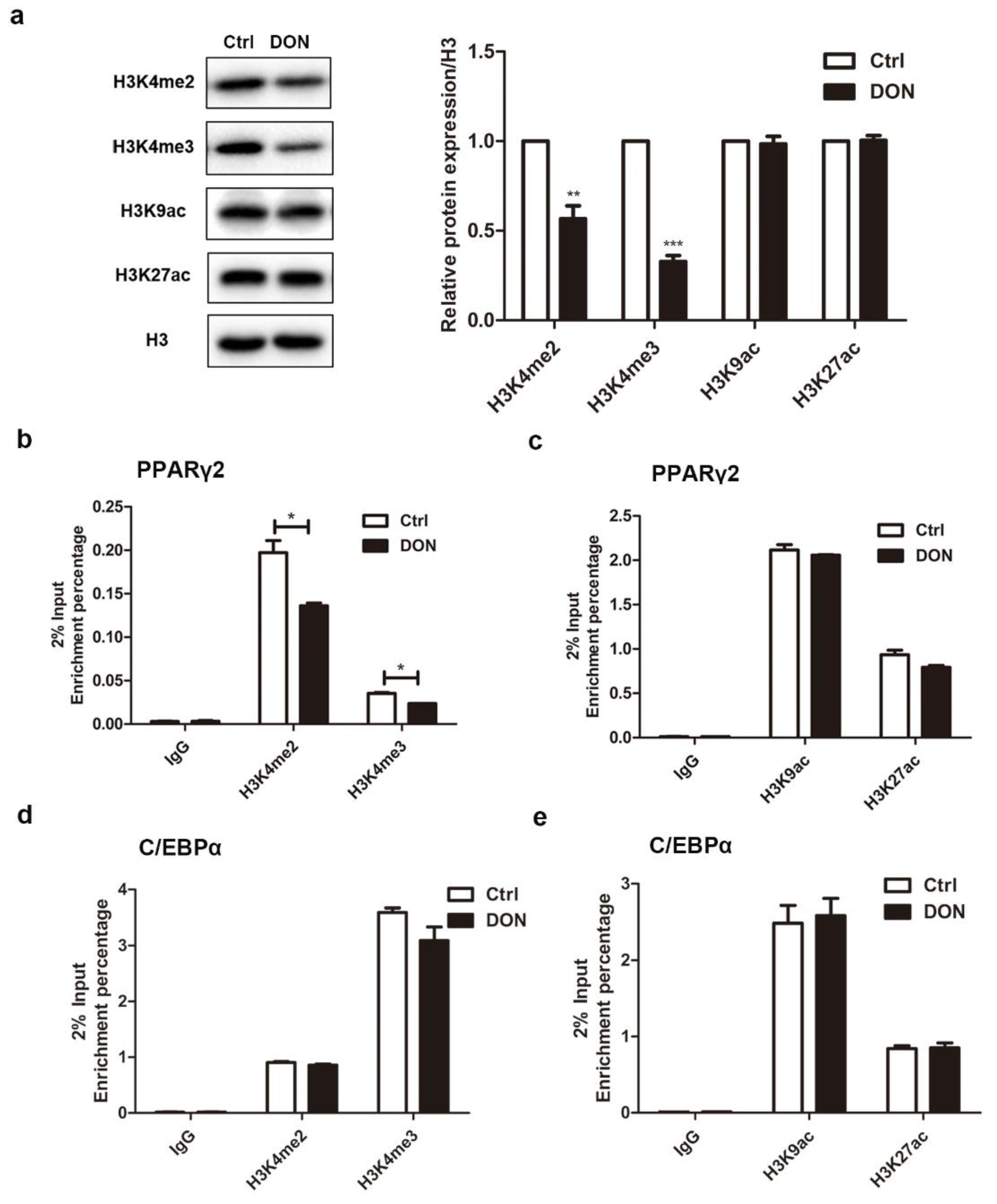
2.7. DON Suppressed the Expression of PPARγ2 by Inhibiting the Enrichment of H3K4me2 and H3K4me3 in 3T3-L1 Cells
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Construction of Plasmids, Adenovirus Transfection, and Luciferase Activity Detection
4.3. Cell Viability Assay
4.4. Oil Red O Staining
4.5. TG Colorimetric Assay
4.6. Real-Time PCR (qPCR)
4.7. Western Blotting
4.8. Chromatin Immunoprecipitation (ChIP) Assays
4.9. Experimental Animals
4.10. Histological Analysis
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ACACA | Fatty acid synthesis acetyl-CoA carboxylase alpha |
| bZIP | Basic region leucine zipper |
| CD36/FAT | Fatty acid translocase |
| C/EBPα | CCAAT/enhancer-binding proteins alpha |
| CHOP | CCAAT/enhancer-binding protein homologous protein |
| DON | Deoxynivalenol |
| eWAT | Epididymal white adipose tissue |
| FABP4 | Fatty acid binding protein 4 |
| FASN | Fatty acid synthase |
| HE | Hematoxylin and eosin |
| MAPK | Mitogen activated protein kinases |
| MLL3/MLL4 | Lysine (K)-specific methyltransferase 2C/2D |
| PPARγ | Peroxisome proliferator-activated receptors gamma |
| PTMs | Post-translational modifications |
| pWAT | Perirenal white adipose tissue |
| RGZ | Rosiglitazone |
| SPF | Specific-pathogen-free |
| TG | Triglycerides |
References
- Wang, Z.; Wu, Q.; Kuca, K.; Dohnal, V.; Tian, Z. Deoxynivalenol: Signaling pathways and human exposure risk assessment–An update. Arch. Toxicol. 2014, 88, 1915–1928. [Google Scholar] [CrossRef]
- Wu, Q.; Lohrey, L.; Cramer, B.; Yuan, Z.; Humpf, H.U. Impact of physicochemical parameters on the decomposition of deoxynivalenol during extrusion cooking of wheat grits. J. Agric. Food Chem. 2011, 59, 12480–12485. [Google Scholar] [CrossRef] [PubMed]
- Vesonder, R.F.; Ciegler, A.; Jensen, A.H. Isolation of the emetic principle from Fusarium-infected corn. Appl. Microbiol. 1973, 26, 1008–1010. [Google Scholar] [CrossRef] [PubMed]
- Pestka, J.J. Deoxynivalenol-induced proinflammatory gene expression: Mechanisms and pathological sequelae. Toxins 2010, 2, 1300–1317. [Google Scholar] [CrossRef] [PubMed]
- Pestka, J.J. Deoxynivalenol: Mechanisms of action, human exposure, and toxicological relevance. Arch. Toxicol. 2010, 84, 663–679. [Google Scholar] [CrossRef]
- Frankic, T.; Pajk, T.; Rezar, V.; Levart, A.; Salobir, J. The role of dietary nucleotides in reduction of DNA damage induced by T-2 toxin and deoxynivalenol in chicken leukocytes. Food Chem. Toxicol. 2006, 44, 1838–1844. [Google Scholar] [CrossRef]
- Payros, D.; Alassane-Kpembi, I.; Pierron, A.; Loiseau, N.; Pinton, P.; Oswald, I.P. Toxicology of deoxynivalenol and its acetylated and modified forms. Arch. Toxicol. 2016, 90, 2931–2957. [Google Scholar] [CrossRef]
- Pierron, A.; Mimoun, S.; Murate, L.S.; Loiseau, N.; Lippi, Y.; Bracarense, A.P.; Liaubet, L.; Schatzmayr, G.; Berthiller, F.; Moll, W.D.; et al. Intestinal toxicity of the masked mycotoxin deoxynivalenol-3-beta-D-glucoside. Arch. Toxicol. 2016, 90, 2037–2046. [Google Scholar] [CrossRef]
- Lowell, B.B. PPARgamma: An essential regulator of adipogenesis and modulator of fat cell function. Cell 1999, 99, 239–242. [Google Scholar] [CrossRef]
- Lehrke, M.; Lazar, M.A. The many faces of PPARgamma. Cell 2005, 123, 993–999. [Google Scholar] [CrossRef]
- Moseti, D.; Regassa, A.; Kim, W.K. Molecular Regulation of Adipogenesis and Potential Anti-Adipogenic Bioactive Molecules. Int. J. Mol. Sci. 2016, 17, 124. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Lee, S.J.; Jang, S.H.; Kim, T.H.; Kim, H.D.; Kim, S.W.; Won, C.K.; Cho, J.H. Annual Wormwood Leaf Inhibits the Adipogenesis of 3T3-L1 and Obesity in High-Fat Diet-Induced Obese Rats. Nutrients 2017, 9, 554. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Umek, R.M.; McKnight, S.L. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 1991, 5, 1538–1552. [Google Scholar] [CrossRef] [PubMed]
- Farmer, S.R. Transcriptional control of adipocyte formation. Cell Metab. 2006, 4, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Rosen, E.D.; Spiegelman, B.M. Adipocytes as regulators of energy balance and glucose homeostasis. Nature 2006, 444, 847–853. [Google Scholar] [CrossRef] [PubMed]
- Rosen, E.D.; Walkey, C.J.; Puigserver, P.; Spiegelman, B.M. Transcriptional regulation of adipogenesis. Genes Dev. 2000, 14, 1293–1307. [Google Scholar] [PubMed]
- Landschulz, W.H.; Johnson, P.F.; Adashi, E.Y.; Graves, B.J.; McKnight, S.L. Isolation of a recombinant copy of the gene encoding C/EBP. Genes Dev. 1988, 2, 786–800. [Google Scholar] [CrossRef]
- Koschmieder, S.; Halmos, B.; Levantini, E.; Tenen, D.G. Dysregulation of the C/EBPα differentiation pathway in human cancer. J. Clin. Oncol. 2009, 27, 619–628. [Google Scholar] [CrossRef]
- Tontonoz, P.; Hu, E.; Spiegelman, B.M. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell 1994, 79, 1147–1156. [Google Scholar] [CrossRef]
- Ren, D.; Collingwood, T.N.; Rebar, E.J.; Wolffe, A.P.; Camp, H.S.; Ren, D.; Collingwood, T.N.; Rebar, E.J.; Wolffe, A.P.; Camp, H.S. PPARγ knockdown by engineered transcription factors: Exogenous PPARγ2 but not PPARγ1 reactivates adipogenesis. Genes Dev. 2002, 16, 27–32. [Google Scholar] [CrossRef]
- Rosen, E.D.; Spiegelman, B.M. PPARgamma: A nuclear regulator of metabolism, differentiation, and cell growth. J. Biol. Chem. 2001, 276, 37731–37734. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.; Wang, C.; Kuang, X.; Feng, X.; Sartorelli, V.; Ying, H.; Ge, K.; Dent, S.Y. Gcn5 and PCAF regulate PPARgamma and Prdm16 expression to facilitate brown adipogenesis. Mol. Cell. Biol. 2014, 34, 3746–3753. [Google Scholar] [CrossRef] [PubMed]
- Seale, P.; Bjork, B.; Yang, W.; Kajimura, S.; Chin, S.; Kuang, S.; Scime, A.; Devarakonda, S.; Conroe, H.M.; Erdjument-Bromage, H.; et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature 2008, 454, 961–967. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, T.S.; Xu, Z.; Zhang, X.; Wang, L.; Gimble, J.M.; Lander, E.S.; Rosen, E.D. Comparative epigenomic analysis of murine and human adipogenesis. Cell 2010, 143, 156–169. [Google Scholar] [CrossRef] [PubMed]
- Ali, D.; Hamam, R.; Alfayez, M.; Kassem, M.; Aldahmash, A.; Alajez, N.M. Epigenetic Library Screen Identifies Abexinostat as Novel Regulator of Adipocytic and Osteoblastic Differentiation of Human Skeletal (Mesenchymal) Stem Cells. Stem Cells Transl. Med. 2016, 5, 1036–1047. [Google Scholar] [CrossRef] [PubMed]
- Nolte, R.T.; Wisely, G.B.; Westin, S.; Cobb, J.E.; Lambert, M.H.; Kurokawa, R.; Rosenfeld, M.G.; Willson, T.M.; Glass, C.K.; Milburn, M.V. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature 1998, 395, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Hadrich, F.; Sayadi, S. Apigetrin inhibits adipogenesis in 3T3-L1 cells by downregulating PPARγ and CEBP-α. Lipids Health Dis. 2018, 17, 95. [Google Scholar] [CrossRef]
- Levert, K.L.; Waldrop, G.L.; Stephens, J.M. A biotin analog inhibits acetyl-CoA carboxylase activity and adipogenesis. J. Biol. Chem. 2002, 277, 16347–16350. [Google Scholar] [CrossRef]
- Kralisch, S.; Fasshauer, M. Adipocyte fatty acid binding protein: A novel adipokine involved in the pathogenesis of metabolic and vascular disease? Diabetologia 2013, 56, 10–21. [Google Scholar] [CrossRef]
- Terra, X.; Quintero, Y.; Auguet, T.; Porras, J.A.; Hernandez, M.; Sabench, F.; Aguilar, C.; Luna, A.M.; Del Castillo, D.; Richart, C. FABP 4 is associated with inflammatory markers and metabolic syndrome in morbidly obese women. Eur. J. Endocrinol. 2011, 164, 539–547. [Google Scholar] [CrossRef]
- Wolf Greenstein, A.; Majumdar, N.; Yang, P.; Subbaiah, P.V.; Kineman, R.D.; Cordoba-Chacon, J. Hepatocyte-specific, PPARgamma-regulated mechanisms to promote steatosis in adult mice. J. Endocrinol. 2017, 232, 107–121. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Q.; Zhao, S.; Yu, B.; Wang, X.; Matyal, R.; Li, Y.; Jiang, Z. High-density lipoprotein increases the uptake of oxidized low density lipoprotein via PPARγ/CD36 pathway in inflammatory adipocytes. Int. J. Biol. Sci. 2015, 11, 256–265. [Google Scholar] [CrossRef] [PubMed]
- Green, H.; Meuth, M. An established pre-adipose cell line and its differentiation in culture. Cell 1974, 3, 127–133. [Google Scholar] [CrossRef]
- Park, B.O.; Ahrends, R.; Teruel, M.N. Consecutive positive feedback loops create a bistable switch that controls preadipocyte-to-adipocyte conversion. Cell Rep. 2012, 2, 976–990. [Google Scholar] [CrossRef]
- Wang, M.; Wang, J.J.; Li, J.; Park, K.; Qian, X.; Ma, J.X.; Zhang, S.X. Pigment epithelium-derived factor suppresses adipogenesis via inhibition of the MAPK/ERK pathway in 3T3-L1 preadipocytes. Am. J. Physiol. Endocrinol. Metab. 2009, 297, E1378–E1387. [Google Scholar] [CrossRef]
- Li, X.; Mu, P.; Qiao, H.; Wen, J.; Deng, Y. JNK-AKT-NF-kappaB controls P-glycoprotein expression to attenuate the cytotoxicity of deoxynivalenol in mammalian cells. Biochem. Pharmacol. 2018, 156, 120–134. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.R.; Islam, Z.; Pestka, J.J. Rapid, sequential activation of mitogen-activated protein kinases and transcription factors precedes proinflammatory cytokine mRNA expression in spleens of mice exposed to the trichothecene vomitoxin. Toxicol. Sci. 2003, 72, 130–142. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xu, S.; Lee, J.E.; Baldridge, A.; Grullon, S.; Peng, W.; Ge, K. Histone H3K9 methyltransferase G9a represses PPARγ expression and adipogenesis. Embo. J. 2013, 32, 45–59. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Pestka, J.J. Comparative induction of 28S ribosomal RNA cleavage by ricin and the trichothecenes deoxynivalenol and T-2 toxin in the macrophage. Toxicol. Sci. 2008, 105, 67–78. [Google Scholar] [CrossRef]
- Mishra, S.; Dwivedi, P.D.; Pandey, H.P.; Das, M. Role of oxidative stress in Deoxynivalenol induced toxicity. Food Chem. Toxicol. 2014, 72, 20–29. [Google Scholar] [CrossRef]
- Tang, S.; Chen, S.; Huang, B.; Jiang, J.; Wen, J.; Deng, Y. Deoxynivalenol induces inhibition of cell proliferation via the Wnt/β-catenin signaling pathway. Biochem. Pharmacol. 2019, 166, 12–22. [Google Scholar] [CrossRef] [PubMed]
- Shan, T.; Liu, J.; Wu, W.; Xu, Z.; Wang, Y. Roles of Notch Signaling in Adipocyte Progenitor Cells and Mature Adipocytes. J. Cell. Physiol. 2017, 232, 1258–1261. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.B.; Moon, H.M.; Kim, W.S.; Lee, Y.S.; Jeong, H.W.; Yoo, E.J.; Ham, J.; Kang, H.; Park, M.G.; Steffensen, K.R.; et al. Activated liver X receptors stimulate adipocyte differentiation through induction of peroxisome proliferator-activated receptor γ expression. Mol. Cell. Biol. 2004, 24, 3430–3444. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.T.; Hochfeld, W.E.; Myburgh, R.; Pepper, M.S. Adipocyte and adipogenesis. Eur. J. Cell. Biol. 2013, 92, 229–236. [Google Scholar] [CrossRef]
- Rosen, E.D.; Hsu, C.H.; Wang, X.; Sakai, S.; Freeman, M.W.; Gonzalez, F.J.; Spiegelman, B.M. C/EBPα induces adipogenesis through PPARγ: A unified pathway. Genes Dev. 2002, 16, 22–26. [Google Scholar] [CrossRef]
- Alassane-Kpembi, I.; Gerez, J.R.; Cossalter, A.M.; Neves, M.; Laffitte, J.; Naylies, C.; Lippi, Y.; Kolf-Clauw, M.; Bracarense, A.P.L.; Pinton, P.; et al. Intestinal toxicity of the type B trichothecene mycotoxin fusarenon-X: Whole transcriptome profiling reveals new signaling pathways. Sci. Rep. 2017, 7, 7530. [Google Scholar] [CrossRef]
- Park, S.H.; Choi, H.J.; Yang, H.; Do, K.H.; Kim, J.; Moon, Y. Repression of peroxisome proliferator-activated receptor γ by mucosal ribotoxic insult-activated CCAAT/enhancer-binding protein homologous protein. J. Immunol. 2010, 185, 5522–5530. [Google Scholar] [CrossRef]
- Mashima, T.; Seimiya, H.; Tsuruo, T. De novo fatty-acid synthesis and related pathways as molecular targets for cancer therapy. Br. J. Cancer 2009, 100, 1369–1372. [Google Scholar] [CrossRef]
- Jang, M.K.; Yun, Y.R.; Kim, J.H.; Park, M.H.; Jung, M.H. Gomisin N inhibits adipogenesis and prevents high-fat diet-induced obesity. Sci. Rep. 2017, 7, 40345. [Google Scholar] [CrossRef]
- Lazra, Y.; Falach, A.; Frenkel, L.; Rozenberg, K.; Sampson, S.; Rosenzweig, T. Autocrine/paracrine function of globular adiponectin: Inhibition of lipid metabolism and inflammatory response in 3T3-L1 adipocytes. J. Cell. Biochem. 2015, 116, 754–766. [Google Scholar] [CrossRef]
- Sugii, S.; Evans, R.M. Epigenetic codes of PPARγ in metabolic disease. FEBS Lett. 2011, 585, 2121–2128. [Google Scholar] [CrossRef] [PubMed]
- Noh, J.R.; Kim, Y.H.; Hwang, J.H.; Gang, G.T.; Yeo, S.H.; Kim, K.S.; Oh, W.K.; Ly, S.Y.; Lee, I.K.; Lee, C.H. Scoparone inhibits adipocyte differentiation through down-regulation of peroxisome proliferators-activated receptor γ in 3T3-L1 preadipocytes. Food Chem. 2013, 141, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, A.; Tsujinaka, T.; Yano, M.; Morita, S.; Taniguchi, M.; Kaneko, K.; Doki, Y.; Shiozaki, H.; Monden, M. Changes of liver-enriched nuclear transcription factors for albumin gene in starvation in rats. Nutrition 1999, 15, 213–216. [Google Scholar] [CrossRef]
- Chapin, R.B.; Brady, P.S.; Barke, R.A.; Brady, L.J. Hepatic CCAAT/enhancer binding protein (C/EBP-α and C/EBP-β) expression changes with riboflavin deficiency, diet restriction and starvation in rats. J. Nutr. 1994, 124, 2365–2375. [Google Scholar] [CrossRef] [PubMed]
- Floyd, Z.E.; Stephens, J.M. Controlling a master switch of adipocyte development and insulin sensitivity: Covalent modifications of PPARγ. Biochim. Biophys. Acta 2012, 1822, 1090–1095. [Google Scholar] [CrossRef]
- Baek, S.J.; Wilson, L.C.; His, L.C.; Eling, T.E. Troglitazone, a Peroxisome Proliferator-activated Receptor γ (PPARγ) Ligand, Selectively Induces the Early Growth Response-1 Gene Independently of PPARγ: A novel mechanism for its anti-tumorigenic activity. J. Biol. Chem. 2003, 278, 5845–5853. [Google Scholar] [CrossRef]
- Yuan, L.; Mu, P.; Huang, B.; Li, H.; Mu, H.; Deng, Y. EGR1 is essential for deoxynivalenol-induced G2/M cell cycle arrest in HepG2 cells via the ATF3DeltaZip2a/2b-EGR1-p21 pathway. Toxicol. Lett. 2018, 299, 95–103. [Google Scholar] [CrossRef]
- Jozwik, K.M.; Chernukhin, I.; Serandour, A.A.; Nagarajan, S.; Carroll, J.S. FOXA1 Directs H3K4 Monomethylation at Enhancers via Recruitment of the Methyltransferase MLL3. Cell Rep. 2016, 17, 2715–2723. [Google Scholar] [CrossRef]
- Jang, M.K.; Kim, J.H.; Jung, M.H. Histone H3K9 demethylase JMJD2B activates adipogenesis by regulating H3K9 methylation on PPARγ and C/EBPα during adipogenesis. PLoS ONE 2017, 12, e0168185. [Google Scholar] [CrossRef]
- Reinhard, B.; Feng, X. Functional Regulation of PPARs through Post-Translational Modifications. Int. J. Mol. Sci. 2018, 19, 1738–1754. [Google Scholar]
- Grenier, B.; Applegate, T.J. Modulation of intestinal functions following mycotoxin ingestion: Meta-analysis of published experiments in animals. Toxins 2013, 5, 396–430. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.C.; Fang, L.W.; Liou, C.J. Phloretin Attenuates Allergic Airway Inflammation and Oxidative Stress in Asthmatic Mice. Front. Immunol. 2017, 8, 134. [Google Scholar] [CrossRef] [PubMed]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Tang, S.; Lin, R.; Zheng, T.; Li, D.; Chen, X.; Zhu, J.; Wen, J.; Deng, Y. Deoxynivalenol Exposure Suppresses Adipogenesis by Inhibiting the Expression of Peroxisome Proliferator-Activated Receptor Gamma 2 (PPARγ2) in 3T3-L1 Cells. Int. J. Mol. Sci. 2020, 21, 6300. https://doi.org/10.3390/ijms21176300
Zhao Y, Tang S, Lin R, Zheng T, Li D, Chen X, Zhu J, Wen J, Deng Y. Deoxynivalenol Exposure Suppresses Adipogenesis by Inhibiting the Expression of Peroxisome Proliferator-Activated Receptor Gamma 2 (PPARγ2) in 3T3-L1 Cells. International Journal of Molecular Sciences. 2020; 21(17):6300. https://doi.org/10.3390/ijms21176300
Chicago/Turabian StyleZhao, Yurong, Shulin Tang, Ruqin Lin, Ting Zheng, Danyang Li, Xiaoxuan Chen, Jiahui Zhu, Jikai Wen, and Yiqun Deng. 2020. "Deoxynivalenol Exposure Suppresses Adipogenesis by Inhibiting the Expression of Peroxisome Proliferator-Activated Receptor Gamma 2 (PPARγ2) in 3T3-L1 Cells" International Journal of Molecular Sciences 21, no. 17: 6300. https://doi.org/10.3390/ijms21176300
APA StyleZhao, Y., Tang, S., Lin, R., Zheng, T., Li, D., Chen, X., Zhu, J., Wen, J., & Deng, Y. (2020). Deoxynivalenol Exposure Suppresses Adipogenesis by Inhibiting the Expression of Peroxisome Proliferator-Activated Receptor Gamma 2 (PPARγ2) in 3T3-L1 Cells. International Journal of Molecular Sciences, 21(17), 6300. https://doi.org/10.3390/ijms21176300