Abstract
ZIP (zinc-regulated transporters, iron-regulated transporter-like protein) family plays an important role in organism Zn balance. This research identified the promoter regions of ZIP3 and ZIP8, two members of ZIP family, from a freshwater teleost yellow catfish Pelteobagrus fulvidraco, characterized the binding sequences of the metal-responsive transcription factor-1 (MTF-1) and Ras responsive element binding protein 1 (RREB1) on their promoter regions. The present study cloned and obtained the 2027 bp of ZIP3 promoter and 1664 bp of ZIP8 promoter, and predicted several key elements on their promoters, such as the binding sites of CREB (cAMP-response element binding protein), KLF4 (Kruppel like factor 4), MTF-1 and RREB1. The sequence deletion from −361 bp to −895 bp down-regulated the luciferase activity of ZIP3 promoter, and the deletion from −897 bp to −1664 bp down-regulated the luciferase activity of ZIP8 promoter. Within different deletion plasmids, the relative luciferase activities of ZIP3 and ZIP8 promoters changes to Zn incubation in a Zn concentration-dependent manner. The site mutagenesis and EMSA (electrophoretic mobility shift assay) found that the −1327 bp/−1343 bp MTF-1 binding site and the −248 bp/−267 bp RREB1 binding site on the ZIP3 promoter, and the −1543 bp/−1557 bp MTF-1 binding site on the ZIP8 promoter are functional sites. Low Zn increased the binding capability between MTF-1 and its responsive site on the ZIP3 promoter, and high Zn increased the transcriptional activation ZIP3 by RREB1; Zn also promoted the binding ability between MTF-1 and its responsive element on the ZIP8 promoter. This study provides the first direct evidence for the response elements of MTF-1 and RREB1 on ZIP3 and MTF-1 on ZIP8 to Zn, which are very important for the evaluation of Zn nutrition and toxicity in vertebrates.
1. Introduction
Zn is a trace element essential for all the organisms because of its important roles in many important physiological processes [1,2]. Zn deficiency will cause adverse physiological effects, particularly during the period of rapid growth [2]. Therefore, organisms possess effective systems to absorb Zn from the environment. Zn uptake is mediated by Zn transporters of the ZIP (zinc-regulated transporters, iron-regulated transporter-like protein) family, which increase the cytoplasmic Zn concentration by transporting Zn from the extracellular space or organelles into cytoplasma [3,4]. Among these members, ZIP3 and ZIP8 were widely expressed in many tissues and involved in Zn acquisition by many cells of the body [4,5]. However, ZIP3 and ZIP8 possess many differences in the tissue distribution, expression, subcellular localization and regulation. These differences have important implications for their functions in maintaining Zn homeostasis. However, little is known about the regulation and function of ZIP3 and ZIP8 in fish.
In eukaryotic organisms, transcription initiation regulates the gene expression. Promoters have many cis-acting elements and can be bound by transcriptional factors, which regulate the expression of genes at the transcriptional level. Therefore, as the first step for demonstrating the regulatory mechanism of Zn transporters, it is very important to investigate the structure and function of the promoters. In addition, metal-regulatory transcription factor-1 (MTF1) and ras-responsive element-binding protein 1 (RREB1) are two important Zn-finger (Cys2His2) transcription factors and function as sensors of intracellular Zn by binding to their corresponding responsive elements, and thereby regulates cellular adaptation to Zn [6,7,8]. At present, the responsive elements of MTF-1 and RREB1 have been identified in the promoter regions of several Zn transporters [6,8], but not in ZIP3 and ZIP8. Moreover, the role of Zn in regulating ZIP3 and ZIP8 remains unclear.
The yellow catfish (Pelteobagrus fulvidraco) is an omnivorous freshwater teleost that is widely cultured in several Asian countries for their excellent fillets and high economic value. In this study, the promoter regions of ZIP3 and ZIP8 were identified in yellow catfish, and the binding sites of MTF-1 and RREB1 in the promoter regions were studied. This study provides an innovative insight into the mechanism by which Zn regulates the transcriptional activities of ZIP3 and ZIP8, which are very important for the evaluation of Zn nutrition and toxicity in vertebrates.
2. Results
2.1. Cloning and Sequence Analysis of the ZIP3 ((Zinc-Regulated Transporters, Iron-Regulated Transporter-Like Protein)) and ZIP8 Promoters
The 2027 bp of ZIP3 promoter and 1664 bp of ZIP8 promoter were successfully cloned (Figure 1A), and the first nucleotide of 5′ cDNA of ZIP3 and ZIP8 were designated as +1. On the ZIP3 promoter, several core promoter elements, such as a TATA-box (TBP, −60 bp/−67 bp) and a CCAAT-box (nuclear transcription factor Y, NF-Y, −124 bp/−131 bp) were predicted (Figure 1B); a cluster of transcription factor binding sites were also predicted, including CREB (−1922 bp/−1935 bp), STAT2 (signal transducer and activator of transcription 2) (−780 bp/−793 bp), SREBP1 (sterol-regulatory element binding proteins 1) (−574 bp/−584 bp), KLF4 (−294 bp/−304 bp) and PPARγ (peroxisome proliferators-activated receptors γ) (−489 bp/−505 bp and −960 bp/−975 bp), MTF-1 (−1327 bp/−1343 bp) and RREB1 (−248 bp to −267 bp) (Figure 1B). On the ZIP8 promoter, this study discovered two GC-boxes (Sp1, −8 bp/−14 bp and −34 bp/−44 bp), the binding sites of two MTF-1 (−483 bp/−497 bp and −1543 bp/−1557 bp) on its core region, CREB (−1237 bp/−1245 bp), STAT3 (signal transducer and activator of transcription 3) (−408 bp/−418 bp), STAT4 (signal transducer and activator of transcription 4) (−992 bp/−1006 bp), KLF4 (−87 bp/−98 bp), PPARα (peroxisome proliferators-activated receptors α) (at −1421 bp/−1435 bp) and PPARδ (peroxisome proliferators-activated receptors δ) (at −586 bp/−602 bp) on its core region (Figure 1C).

Figure 1.
(A) The schematic diagram of ZIP3 and ZIP8 gene structures. Positive: the region that positively regulated the promoter activity. TSS: transcription start site. ATG: translation initiation site; (B) Nucleotide sequence of yellow catfish ZIP3 promoter. Numbers are relative to the transcription start site (+1). The putative transcription factor binding sites are underlined. The highlighted sequences show putative transcription factor binding sites of MTF-1 and RREB1; (C) Nucleotide sequence of yellow catfish ZIP8 promoter. Numbers are relative to the transcription start site (+1). The putative transcription factor binding sites are underlined. The highlighted sequences show putative transcription factor binding sites of MTF-1.
2.2. Analysis of the 5′-Sequence Deletion of the ZIP3 and ZIP8 Promoters
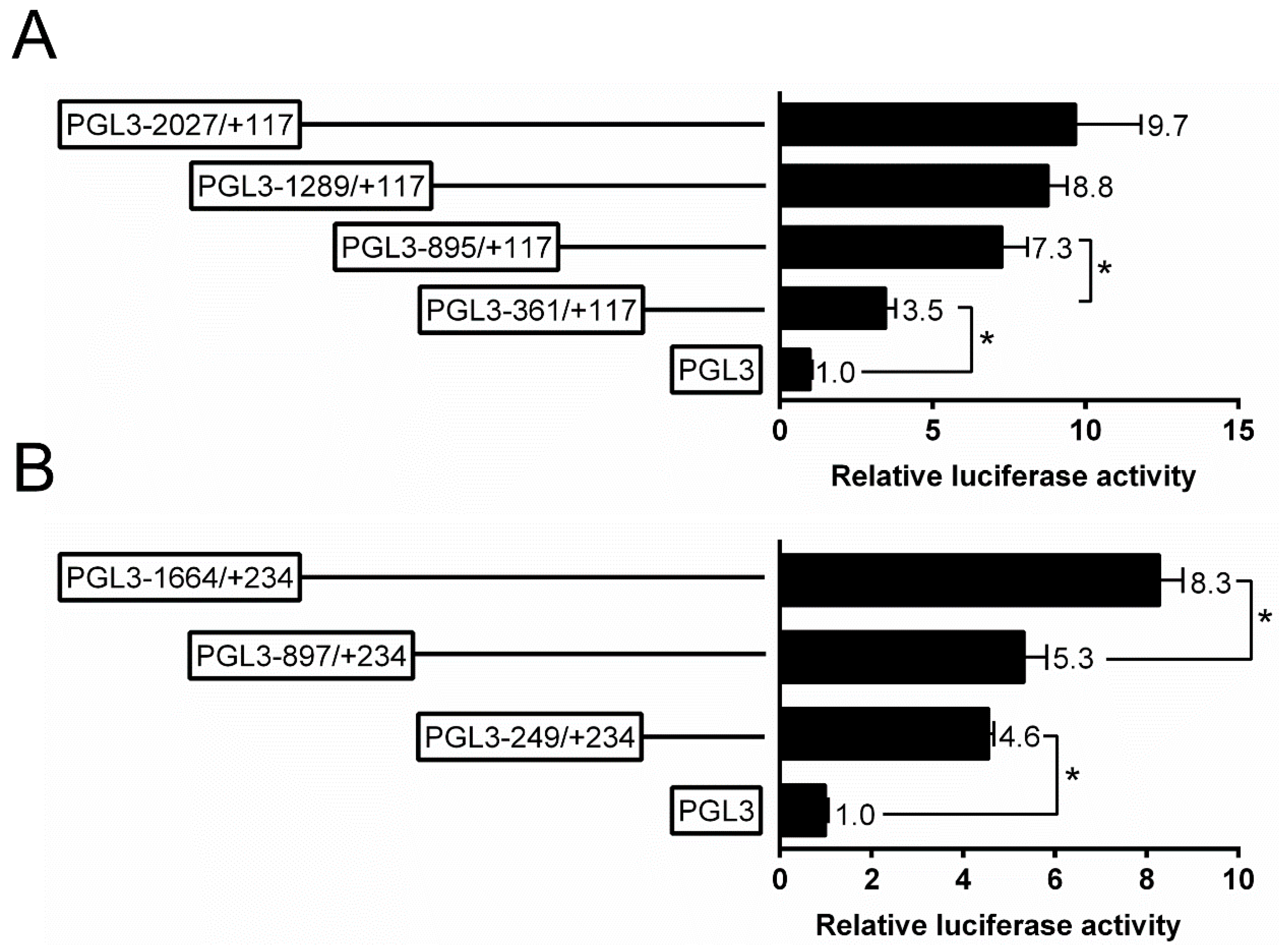
The present study randomly generated the plasmids of different sizes and selected four plasmids for the analysis of 5′-sequence deletion. For ZIP3 promoter, the sequence deletion from −361 bp to −895 bp down-regulated the luciferase activity significantly, and the sequence deletion from −895 bp to −2027 bp did not significantly affect the luciferase activity (Figure 2A). For the ZIP8 promoter, the subsequent absence from +234 to −897 bp did not affect the luciferase activity, whereas the deletion from −897 bp to −1664 bp down-regulated the luciferase activity (Figure 2B).
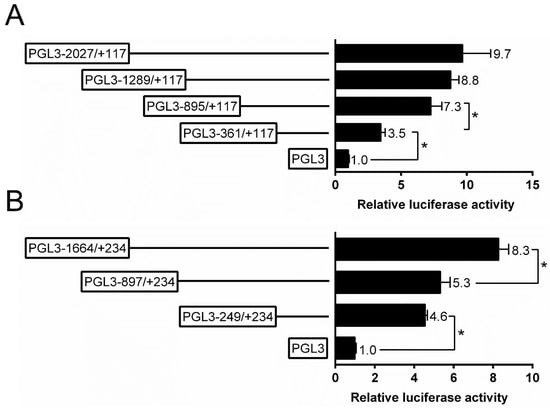
Figure 2.
5′ unidirectional deletion assays of the promoter regions of ZIP3 (A) and ZIP8 (B) of yellow catfish. Values mean the ratio of activities of Firefly to Renilla luciferase, normalized to the control plasmid, and are shown as mean ± standard error of mean (SEM) (n = 3). Asterisk (*) means significant differences between two groups (p < 0.05).
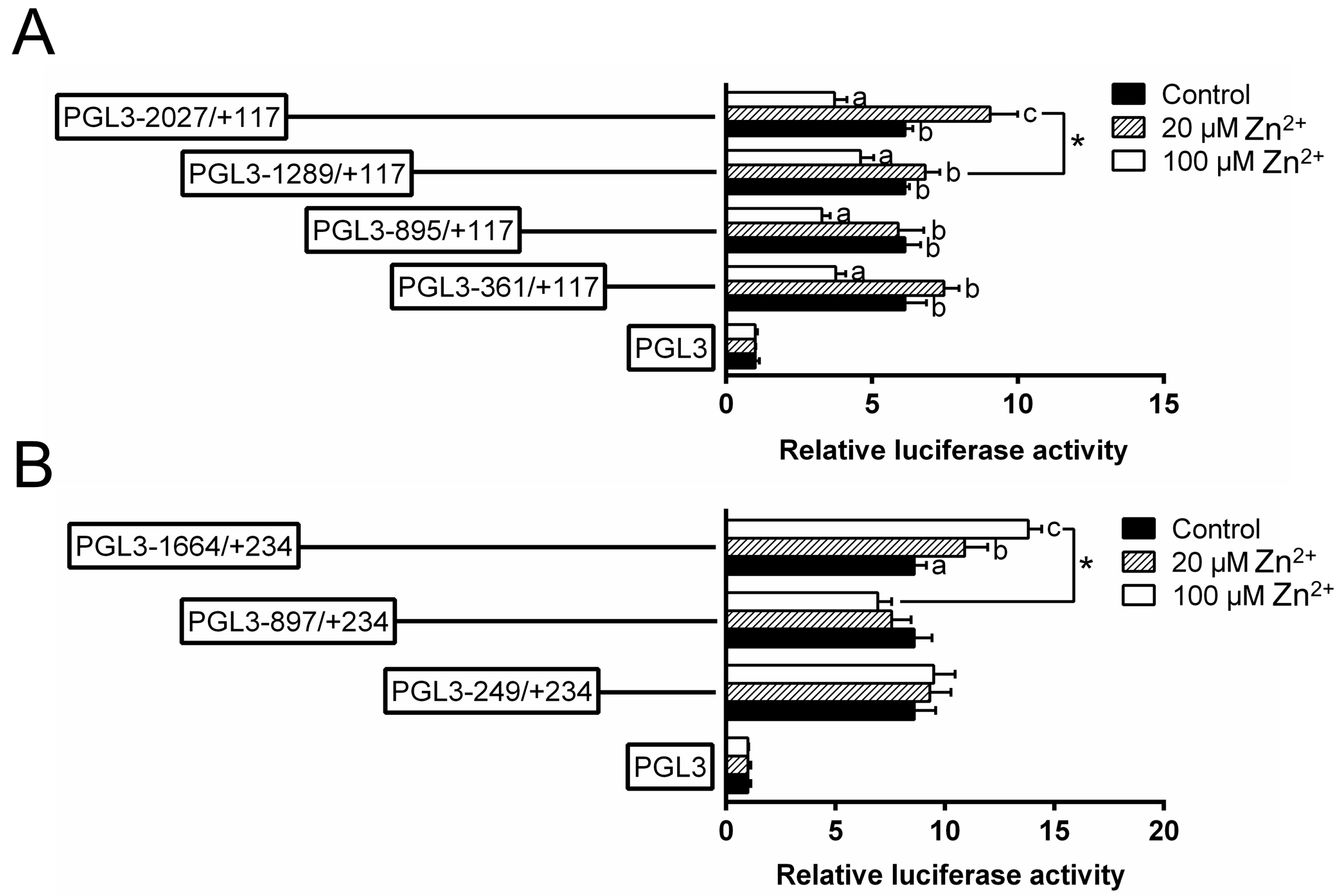
To investigate the responses of these promoters to Zn, 20 μmol/L (low Zn) and 100 μmol/L Zn2+ (high Zn) were used to incubate the HEK293T cells and conducted the 5′-sequence deletion assay (Figure 3). For the ZIP3 promoter, within the deletion plasmids of −2027 bp/+117 bp, the relative luciferase activities were the lowest in the high Zn group and the highest in the low Zn group; for other plasmids, the luciferase activities in the high Zn group were significantly lower than those in other two groups. For the ZIP8 promoter, within the plasmid of −1664 bp/+234 bp, the luciferase activities increased with increasing Zn concentrations; however, for other plasmids, Zn incubation did not significantly affect the luciferase activities. Meantime, when the sequence was deleted from −1664 bp to −897 bp, high Zn significantly reduced the luciferase activities.
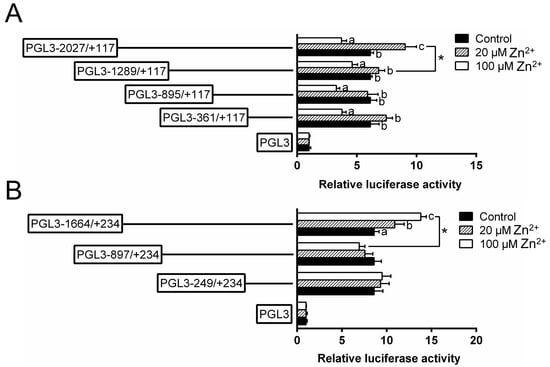
Figure 3.
5′ unidirectional deletion assays for promoter regions of ZIP3 (A) and ZIP8 (B) after Zn incubation. Values showed the ratio of activities of Firefly to Renilla luciferase, normalized to the control, and were presented as mean ± SEM (n = 3). Asterisk (*) indicates significant differences between different 5′ unidirectional deletion plasmids under the same treatment (p < 0.05). Different letters (a, b and c) indicate significant differences among different treatments in the same plasmid (p < 0.05).
2.3. Site-Mutation Analysis of MTF-1 (Metal-Responsive Transcription Factor-1) and RREB1 (Ras Responsive Element Binding Protein 1) Binding Sites on the ZIP3 and ZIP8 Promoters
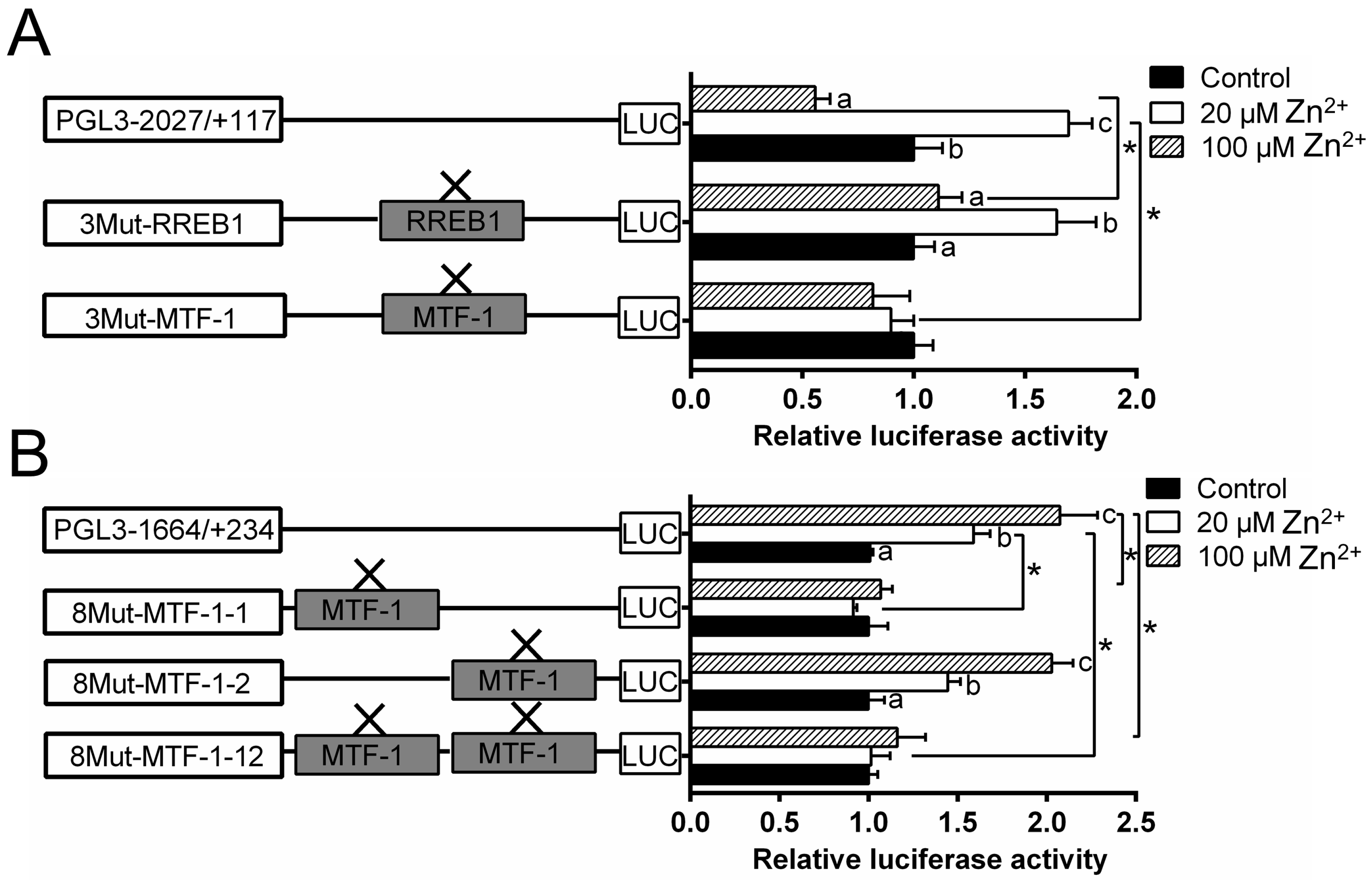
Since MTF-1 and RREB1 mediated the regulation of Zn homeostasis, it is important to determine if the observed changes in gene expression are directly related to their regulation. Thus, this study performed the analysis of site mutation in the ZIP3 and ZIP8 promoters that may possess MTF-1 and RREB1 binding sites. For the ZIP3 promoter, the mutation of the −1327/−1343 MTF-1 binding site (3Mut-MTF-1) influenced the Zn-induced variation of the luciferase activity, indicating that this site acted as a functional one for the ZIP3 transcriptional response to Zn incubation; the mutation of the −248/−267 RREB1 binding site (3Mut-RREB1) increased the luciferase activity of the high Zn group; for the same RREB1-mutated group, the luciferase activity was significantly higher in the low Zn group than those in the control and the high Zn group (Figure 4A).
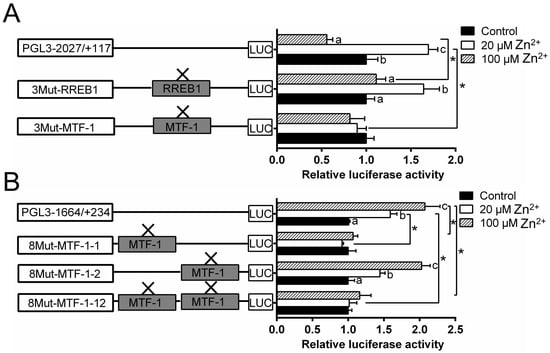
Figure 4.
Assays of predicted MTF-1 and RREB1 binding sites of yellow catfish ZIP3 and ZIP8 promoters after site-directed mutagenesis. (A) Site mutagenesis of MTF-1 on pGl3-ZIP3-2027 vector and RREB1 on pGl3-ZIP3-2027 vector; (B) site mutagenesis of MTF-1 on pGl3-ZIP8-1664 vector. Values mean the ratio of activities of Firefly to Renilla luciferase, normalized to the control. Results were presented as mean ± SEM (n = 3). Asterisk (*) indicates significant differences between different 5′ unidirectional deletion plasmids under the same treatment (p < 0.05). Different letters (a, b and c) indicate significant differences between different treatments in the same plasmid (p < 0.05).
For ZIP8 promoter, the mutation of the −1543/−1557 MTF-1 binding site (8Mut-MTF-1-1), but not −483/−497 binding site (8Mut-MTF-1-2), eliminated the Zn-induced increase of promoter activity (Figure 4B).
2.4. EMSA (Electrophoretic Mobility Shift Assay) for the Confirmation of the Functional Binding of MTF-1 and RREB1 on the ZIP3 and ZIP8 Promoters
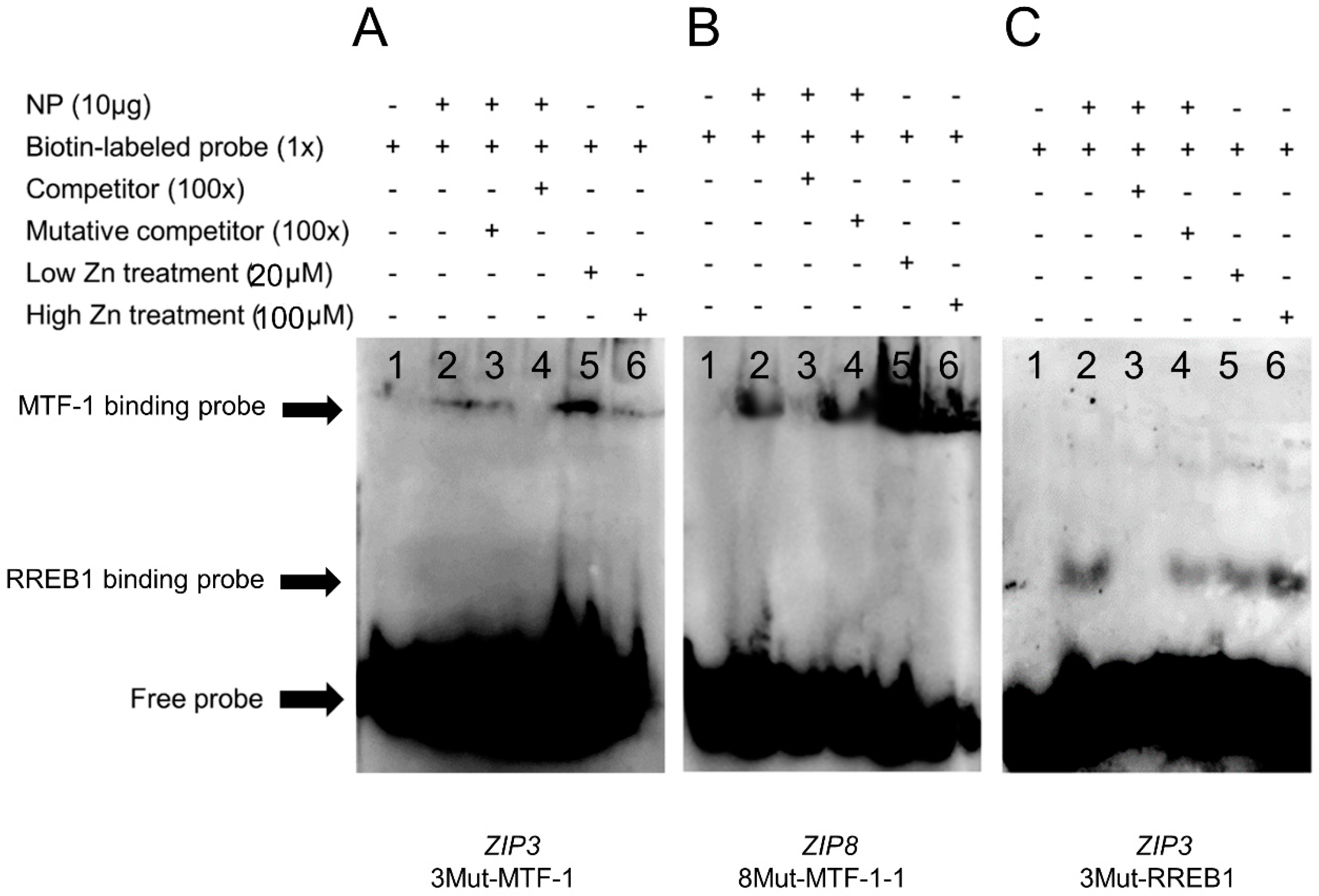
Based on the site mutation assays, the −1327 bp/−1343 bp for MTF-1 binding and −248 bp/−267 bp for RREB1 binding on the ZIP3 promoter, and −1543 bp/−1557 bp for MTF-1 bindings on the ZIP8 promoter were considered to be functional. In order to confirm whether these sites can interact with MTF-1 and RREB1, the EMSA was conducted. For the ZIP3 promoter, when the MTF-1 binding sequence was used as the probe, the 100-fold unlabeled MTF-1 binding sites (−1327 bp/−1343 bp) competed for the binding, and the 100-fold unlabeled Mut-MTF-1 binding sites reduced this competition, suggesting that MTF-1 could bind with this region (Figure 5A). Meantime, results showed that low Zn promoted the binding of MTF-1 to its corresponding binding site on the ZIP3 promoter. When the RREB1 binding sequence was used as the probe, the 100-fold unlabeled RREB1 binding site (−248 bp/−267 bp) competed for the binding, and the 100-fold unlabeled Mut-RREB1 binding region reduced this competition, suggesting that RREB1 could bind with this region (Figure 5B). Additionally, compared with the band in the control, high Zn increased the brightness of the band, indicating that high Zn mediated the transcriptional regulation of ZIP3 by RREB1.
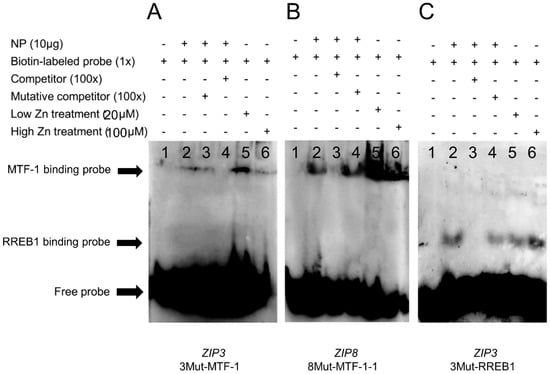
Figure 5.
EMSA (electrophoretic mobility shift assay) of predicted MTF-1 and RREB1 binding sequences on the yellow catfish ZIP3 and ZIP8 promoters. (A) MTF-1 binding sequences sited between −1327 bp and −1343 bp of ZIP3 promoter; (B) MTF-1 binding sequences sited between −1543 bp and −1557 bp of ZIP8 promoter; (C) RREB1 binding sequences sited between −248 bp and −267 bp of ZIP3 promoter. NP, nuclear protein. The numbers 1–6 represent the six different lanes.
For ZIP8 promoter, when the MTF-1 binding sequence was used as the probe, the 100-fold unlabeled MTF-1 binding site (−1543 bp/−1557 bp) competed for the binding, and the 100-fold unlabeled Mut-MTF-1 binding region reduced this competition, suggesting that MTF-1 could bind this region (Figure 5C). Besides, Zn increased the brightness of bands compared with the bands in the control, indicating that Zn promoted the binding of MTF-1 to its binding site on the ZIP8 promoter.
3. Discussion
The investigation of the transcriptional regulation of Zn transporter proteins is an important aspect of Zn homeostasis in organisms. This study, for the first time, cloned and characterized the ZIP3 and ZIP8 promoters from a teleost fish, yellow catfish.
Studies suggested that the core promoter region was located most proximal to the start codon and contained the RNA polymerase binding sites [9]. In the present study, the core region of ZIP promoter had one TATA-box and one CAAT-box (NF-Y), similar to those in mouse ZIP3 promoter (Ensembl ENSMUSG00000046822). TATA-box and CAAT-box are considered to dock the RNA polymerase transcriptional complex [10,11]. However, the core region of yellow catfish ZIP had neither TATA-box nor CAAT-box. Roy and Singer [9] reported that only about 5–7% of eukaryotic promoters had the TATA-box. Smale and Kadonaga [12] found that usually TATA-less promoters possessed Sp1 binding sites, which was also observed in the present study since two Sp1 binding sites were found in the core region of yellow catfish ZIP8 promoter. Aiba et al. [13] pointed out that suppression of binding site Sp1 down-regulated ZIP8, suggesting the role of Sp1 in the control of ZIP8 gene expression.
Identification of TFBS (transcription factor binding sites) contributes to decipher the mechanisms of gene regulation. The current study showed that the luciferase activities of the ZIP3 promoter reduced when the sequence between −361 bp and −895 bp was deleted, suggesting that the positive regulator existed in this region. Further investigation found a cluster of binding sites, such as PPARγ, STAT2, SREBP, MTF-1, RREB1, CREB, STAT2 and KLF4 in the promoter region of ZIP3, reflecting that ZIP3 participated in many physiological progress. For ZIP8 promoter, some other binding sites, such as STAT3, STAT4 and CREB, in the ZIP promoter region were found. This implies that the transcriptional regulation of ZIP8 could be complicated and diverse transcription factors were involved in the regulation, and thereby play important roles in organisms. Moreover, the deletion of −897 bp/−1664 bp sequence reduced the luciferase activity, suggesting positive regulatory factors existed in this region. Similarly, one MTF-1 binding site were found in this region. MTF-1 was reported to positively regulate ZIP family [7,8].
Next, this study explored whether Zn incubation affected the activities of ZIP3 and ZIP8 promoters. The present studies indicated that for the ZIP3 promoter, within the deletion plasmids of −2027 bp/+117 bp, the relative luciferase activities were the highest in the low Zn group and the lowest in the high Zn group, indicating that the regulation of ZIP3 by Zn is Zn concentration-dependent. Similarly, several studies reported that ZIP3 transcription and mRNA expression increased in responses to Zn deficiency [14,15], but reduced as Zn concentration increased [16]. In our laboratory, Chen et al. [5] found Zn addition down-regulated mRNA levels of ZIP3 compared to the control in the hepatocytes. These results are consistent with the reported functions of these proteins in maintaining the homeostasis of Zn. Further site mutagenesis and EMSA identified a functional binding site of MTF-1 in these regions. MTF-1 functions as a cellular Zn sensor and binds specifically to MREs (metal response elements) to activate the expression of genes encoding Zn transporters in response to Zn ions [8,17,18]. Ling et al. [19] reported that 0.18 mg/L of waterborne Zn addition could significantly improve the mRNA expression level of MTF-1 of juvenile goby Synechogobius hasta. Interestingly, EMSA showed that low zinc treatment had higher MTF-1 binding activity than high zinc treatment (Figure 5A). One possible explanation was the presence of other transcriptional inhibitors in the promoter region of ZIP3, which can be activated by high zinc concentrations. This research implied that low Zn positively impacted ZIP3 expression probably through the MTF-1 binding site located between −1289 bp and −2027 bp. Meantime, this study predicted a RREB1 binding site from −248 bp to −267 bp and indicated that the mutation of the −248/−267 bp RREB1 binding site in ZIP3 alleviated the high Zn-induced reduction of the relative luciferase activities of ZIP3 promoter. Furthermore, EMSA showed that the −248 bp/−267 bp sequence was a functional binding site, and that high Zn concentration promoted the RREB1 binding to this site. Similarly, Franklin et al. [20] reported the presence of the potential RREB1 binding sites in the ZIP3 promoter and found that RREB1 was a positive regulator for ZIP3 gene expression. RREB1 is a transcription factor which could activate or inhibit the transcription of various target genes, depending on the cell type, promoter features and co-binding proteins [21,22,23,24] of gene expression. Thus, the −248 bp/−267 bp RREB1 site may play important roles in high Zn-induced down-regulation of ZIP3 promoter activity.
For the ZIP8 promoter, this study indicated that within the plasmid of −1664 bp/+234 bp, the luciferase activities increased with increasing Zn concentrations. Generally speaking, increasing ZIP8 expression will increase intracellular Zn concentration, as suggested by Kim et al. [25]. Results also showed that, when the sequence was deleted from −1664 bp to −897 bp, high Zn reduced the luciferase activities, indicating that this region had the binding sequence that regulated ZIP8 transcription. Further EMSA indicated that the −1543 bp/−1557 bp MTF-1 binding site could be bound by MTF-1, and that Zn promoted the binding of MTF-1 to its binding site on the ZIP8 promoter. The indispensable role of Zn-MTF-1 axis in regulating Zn homeostasis has been mentioned in several reports [25,26], and that fish and mammalian cells could respond to Zn2+ by increasing the MTF-1 binding [8,27]. Andrews [17] pointed out that the MTF-1 binding to the MRE was dependent on Zn and was easily disrupted when Zn was depleted.
In conclusion, the promoter regions of yellow catfish ZIP3 and ZIP8 were identified, and the binding sites of MTF-1 and RREB1 in the promoter regions were characterized. This study provides the first direct evidence for the interaction between MTF-1, RREB1 and ZIP3 and ZIP8 genes in fish, and accordingly elucidates an innovative mechanism by which Zn regulates the transcriptional activities of ZIP3 and ZIP8. These relevant studies are very important for the evaluation of Zn nutrition and toxicity in vertebrates.
4. Materials and Methods
4.1. Animals and Reagents
Yellow catfish used for promoter cloning were from a local commercial farm (Wuhan, China). HEK293T cell lines were obtained from the Cell Resource Center of Huazhong Agricultural University. Lipofectamine 2000 were purchased from Invitrogen. Passive Lysis Buffer and Dual-Luciferase were from Promega. Dulbecco’s Modified Eagle’s medium (DMEM), 0.25% trypsin-EDTA and fetal bovine serum (FBS) were bought from Gibco company (ThermoFisher Scientific, Waltham, MA, USA). Other reagents were analytical ones from Shanghai Sinopharm Group Corporation (Shanghai, China). The protocols for all animal and cell experiments followed the ethical guidelines of Huazhong Agricultural University (HZAU) for the care and use of laboratory animals and cells, and were approved by that university’s Ethics Committee (identification code: Fish-2018-0721, Date: 21 July 2018).
4.2. Promoter Cloning and Plasmid Construction
According to the full-length ZIP3 and ZIP8 cDNA sequences obtained in the studies [5], RNA ligase-mediated rapid amplification of 5′ cDNA ends (RLM-5′RACE) method was used to identify their 5′ cDNA sequences and the transcription start sites (TSS). The promoter cloning followed the method described in Xu et al. [28]. Briefly, genomic DNA was extracted from the tail fins of yellow catfish via the commercial kit (Omega, Norcross, GA, USA). The different primers were designed to analyze the sites of the first introns of ZIP3 and ZIP8 (Table S1). The high-efficiency thermal asymmetric interlaced-PCR (hiTAIL-PCR) method was used to clone the promoter sequences by designing the specific primers with overlapping sequences (Table S1). The luciferase reporter constructs were producted via the purified PCR product and pGl3-Basic vectors (Promega, Fitchburg, WI, USA). The ClonExpress II One Step Cloning Kit (Vazyme, Piscataway, NJ, USA) was utilized to ligate the products. Based on the distances from their TSS, this study decided to name the plasmids as the pGl3-2027/+117 of ZIP3 vector and pGl3-1664/+234 of ZIP8 vector, respectively. With the Erase-a-Base system (Promega) using templates of pGl3-2024/+117 of ZIP3 vector, plasmids pGl3-361/+117, pGl3-895/+117 and pGl3-1289/+117 of ZIP3 vector were generated. Utilizing the pGl3-1664/+234 of ZIP8 vector as a template, the pGl3-249/+234 and pGl3-897/+234 of ZIP8 vectors were obtained, respectively. The primers for the plasmid construction are listed in Table S2.
4.3. Sequence Analysis
This research predicted the putative transcription factor binding sites (TFBS) by these online tools (http://www.genomatix.de/ and http://jaspar.genereg.net/). The reference binding sequences are shown in Table S3. The Clustal-W multiple alignment algorithm was used to assess the sequence alignments.
4.4. Plasmid Transfections and Assays of Luciferase Activities
The experiment transfected the plasmid into HEK293T cells and assayed the luciferase activities followed the methods of Xu et al. [28]. Briefly, HEK293T cells were cultured in DMEM medium + 10% FBS. They were placed in a SANYO incubator for the cell culture with 5% CO2 at 37 °C. All these reporter plasmids were used in the equimolar amounts. They were co-transfected with 20 ng pRL-TK as the control. After 4 h, the transfection medium was changed to DMEM with 10% FBS. HEK293T cells were incubated with three Zn concentrations, including the control (without extra Zn addition), low Zn (20 μM Zn2+) and high Zn (100 μM Zn2+), respectively. Zn was added in the form of ZnSO4. The incubation continued for 24 h. The Dual-Luciferase Reporter Assay System was used to determine the luciferase activity.
4.5. Site-Mutation Assays of MTF-1 and RREB1 Binding Sites on the ZIP3 and ZIP8 Promoters
To analyze the MTF-1 and RREB1 binding sites on ZIP3 and ZIP8 promoters in yellow catfish, QuickChange II Site-Directed Mutagenesis Kit (Vazyme, Jiangsu, China) was used to perform this site mutagenesis. The pGl3-ZIP3-2144 and pGl3-ZIP8-1940 were used as the templates. The primers for mutagenic analysis are shown in Table S4. These constructs were named the 3Mut-MTF-1, 3Mut-RREB1, 8Mut-MTF-1-1 and 8Mut-MTF-1-2, respectively. Then, the present study utilized the reagent Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) to co-transfect the pRL-TK and constructs into HEK293T cells. After 4h transfection, the transfection medium was changed to DMEM (10% FBS) (Thermo Fisher Scientific, Wilmington, DE, USA). After 24 h incubation, HEK293T cells were collected and the luciferase activities were analyzed.
4.6. Analysis of the Functional Binding Sites of MTF-1 and RREB1 on the ZIP3 and ZIP8 Promoters Based on Electrophoretic Mobility-Shift Assay (EMSA)
This study isolated proteins from HEK293T cells and performed EMSA assays to analyze the functional binding sites of MTF-1 and RREB1 on the regions of ZIP3 and ZIP8 promoters, based on Xu et al. [28]. Cytoplasmic and nuclear extracts were obtained based on the methods of Read et al. [29] and determined the protein contents by the bicinchoninic acid assay (BCA) method. Then, according to the LightShift Chemiluminescent EMSA Kit (Invitrogen, Carlsbad, CA, USA), each oligonucleotide duplex of MTF-1 and RREB1 binding sites were incubated with 10 μg nuclear extracts. Before the biotin-labeled probe was added, each unlabeled probe was pre-incubated for 10 min. The biotin-labeled probe was added at room temperature and the reaction continued for 30 min. Then they were detected via the electrophoresis on 6% native polyacrylamide gels. This research performed competition analyses by using 100-fold unlabeled oligonucleotide duplex with or without the mutation. Table S5 showed these oligonucleotide sequences for EMSA.
4.7. Statistical Analysis
All the data were presented as mean ± standard error of mean (SEM). Before the statistical analysis, the Kolmogorov–Smirnov test was used to determine the normality of the distribution of all the data. The homogeneity of variances was tested by Bartlett’s test. Data were analyzed with one-way ANOVA, Duncan’s multiple comparison and Student’s t-test where appropriate. p < 0.05 means statistically significant differences within the treatments. SPSS 19.0 software (SPSS, Chicago, IL, USA) was used to perform the statistical analyses.
Supplementary Materials
The following are available online at https://www.mdpi.com/1422-0067/21/17/6135/s1, Table S1: Primers used for the cloning of ZIP3 and ZIP8 promoters in yellow catfish, Table S2: Primers used for 5′-deletion plasmids construction of yellow catfish ZIP3 and ZIP8 promoters, Table S3: The reference binding site sequences of multiple transcription factors on the promoter regions, Table S4: Primers used for site-mutation analysis, Table S5: Primers used for electrophoretic mobility-shift assay.
Author Contributions
Z.L. and S.-W.C. designed the experiment. S.-W.C. conducted the experiment with the help of K.W., W.-H.L., C.-C.S. and F.C. S.-W.C. analyzed the data with the help of Z.L. and K.W. S.-W.C. drafted the manuscript and Z.L. revised the manuscript. All the authors read and approved the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was founded by National Natural Science Foundation of China (31872585), and the National Key R&D Program of China (grant no.: 2018YFD0900400).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Maret, W.; Li, Y. Coordination dynamics of zinc in proteins. Chem. Rev. 2009, 109, 4682–4707. [Google Scholar] [CrossRef] [PubMed]
- Kambe, T.; Tsuji, T.; Hashimoto, A.; Itsumura, N. The physiological, biochemical, and molecular roles of zinc transporters in zinc homeostasis and metabolism. Physiol. Rev. 2015, 95, 749–784. [Google Scholar] [CrossRef] [PubMed]
- Taylor, K.M.; Nicholson, R.I. The LZT proteins; the LIV-1 subfamily of zinc transporters. BBA-Biomembr. 2003, 1611, 16–30. [Google Scholar] [CrossRef]
- Lichten, L.A.; Cousins, R.J. Mammalian zinc transporters: Nutritional and physiologic regulation. Annu. Rev. Nutr. 2009, 29, 153–176. [Google Scholar] [CrossRef]
- Chen, S.W.; Wu, K.; Lv, W.H.; Song, C.C.; Luo, Z. Molecular characterization of ten zinc (Zn) transporter genes and their regulation to Zn metabolism in freshwater teleost yellow catfish Pelteobagrus fulvidraco. J. Trace Elem. Exp. Med. 2020, 59, 126433. [Google Scholar] [CrossRef]
- Plum, L.M.; Rink, L.; Haase, H. The essential toxin: Impact of zinc on human health. Int. J. Environ. Res. Public Health 2010, 7, 1342–1365. [Google Scholar] [CrossRef]
- Gunther, V.; Lindert, U.; Schaffner, W. The taste of heavy metals: Gene regulation by MTF-1. BBA-Mol. Cell. Res. 2012, 1823, 1416–1425. [Google Scholar] [CrossRef]
- Chen, G.H.; Lv, W.; Xu, Y.H.; Wei, X.L.; Xu, Y.C.; Luo, Z. Functional analysis of MTF-1 and MT promoters and their transcriptional response to zinc (Zn) and copper (Cu) in yellow catfish Pelteobagrus fulvidraco. Chemosphere 2020, 246, 125792. [Google Scholar] [CrossRef]
- Roy, A.L.; Singer, D.S. Core promoters in transcription: Old problem, new insights. Trends Biochem. Sci. 2015, 40, 165–171. [Google Scholar] [CrossRef]
- Basehoar, A.D.; Zanton, S.J.; Pugh, B.F. Identification and distinct regulation of yeast TATA box-containing genes. Cell 2004, 116, 699–709. [Google Scholar] [CrossRef]
- Xu, M.; Gonzalez-Hurtado, E.; Martinez, E. Core promoter-specific gene regulation: TATA box selectivity and Initiator-dependent bi-directionality of serum response factor-activated transcription. BBA-Gene Regul. Mech. 2016, 1859, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Smale, S.T.; Kadonaga, J.T. The RNA polymerase II core promoter. Annu. Rev. Biochem. 2003, 72, 449–479. [Google Scholar] [CrossRef] [PubMed]
- Aiba, I.; Hossain, A.; Kuo, M.T. Elevated GSH level increases cadmium resistance through down-regulation of Sp1-dependent expression of the cadmium transporter ZIP8. Mol. Pharmacol. 2008, 74, 823–833. [Google Scholar] [CrossRef] [PubMed]
- Aydemir, T.B.; Blanchard, R.K.; Cousins, R.J. Zinc supplementation of young men alters metallothionein, zinc transporter, and cytokine gene expression in leukocyte populations. Proc. Natl. Acad. Sci. USA 2006, 103, 1699–1704. [Google Scholar] [CrossRef]
- Zheng, D.; Feeney, G.P.; Kille, P.; Hogstrand, C. Regulation of ZIP and ZnT zinc transporters in zebrafish gill: Zinc repression of ZIP10 transcription by an intronic MRE cluster. Physiol. Genom. 2008, 34, 205–214. [Google Scholar] [CrossRef]
- Kelleher, S.L.; Lonnerdal, B. Zn transporter levels and localization change throughout lactation in rat mammary gland and are regulated by Zn in mammary cells. J. Nutr. 2003, 133, 3378–3385. [Google Scholar] [CrossRef]
- Andrews, G.K. Cellular zinc sensors: MTF-1 regulation of gene expression. Biometals 2001, 14, 223–237. [Google Scholar] [CrossRef]
- Hogstrand, C.; Zheng, D.; Feeney, G.; Cunningham, P.; Kille, P. Zinc-controlled gene expression by metal-regulatory transcription factor 1 (MTF1) in a model vertebrate, the zebrafish. Biochem. Soc. Trans. 2008, 36, 1252–1257. [Google Scholar] [CrossRef]
- Ling, S.C.; Luo, Z.; Chen, G.H.; Zhang, D.G.; Liu, X. Waterborne Zn influenced Zn uptake and lipid metabolism in two intestinal regions of juvenile goby Synechogobius hasta. Ecotoxicol. Environ. Saf. 2018, 148, 578–584. [Google Scholar] [CrossRef]
- Franklin, R.B.; Zou, J.; Costello, L.C. The cytotoxic role of RREB1, ZIP3 zinc transporter, and zinc in human pancreatic adenocarcinoma. Cancer Biol. Ther. 2014, 15, 1431–1437. [Google Scholar] [CrossRef]
- Thiagalingam, A.; De Bustros, A.; Borges, M.; Jasti, R.; Compton, D.; Diamond, L.; Mabry, M.; Ball, D.W.; Baylin, S.B.; Nelkin, B.D. RREB-1, a novel zinc finger protein, is involved in the differentiation response to Ras in human medullary thyoid carcinomas. Mol. Cell. Biol. 1996, 16, 5335–5345. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.K.; Nishitani, J.; Petry, M.W.; Fessing, M.Y.; Leiter, A.B. Novel transcriptional potentiation of BETA2/NeuroD on the secretin gene promoter by the DNA-binding protein Finb/RREB-1. Mol. Cell. Biol. 2003, 23, 259–271. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, N.K.; Cinar, B.; Mukhopadhyay, L.; Lutchman, M.; Ferdinand, A.; Kim, J.; Freeman, M.R. The zinc finger protein ras-responsive element binding protein-1 is a coregulator of the androgen receptor: Implications for the role of the ras pathway in enhancing androgenic signaling in prostate cancer. Mol. Endocrinol. 2007, 21, 2056–2070. [Google Scholar] [CrossRef] [PubMed]
- Flajollet, S.; Poras, I.; Carosella, E.D.; Moreau, P. RREB-1 is a transcriptional repressor of HLA-G. J. Immunol. 2009, 183, 6948–6959. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Jeon, J.; Shin, M.; Won, Y.; Lee, M.; Kwak, J.S.; Chun, J.S. Regulation of the catabolic cascade in osteoarthritis by the zinc-ZIP8-MTF1 axis. Cell 2014, 156, 730–743. [Google Scholar] [CrossRef]
- Lee, M.; Won, Y.; Shin, Y.; Kim, J.H.; Chun, J.S. Reciprocal activation of hypoxia-inducible factor (HIF)-2α and the zinc-ZIP8-MTF1 axis amplifies catabolic signaling in osteoarthritis. Osteoarthr. Cartil. 2016, 24, 134–145. [Google Scholar] [CrossRef] [PubMed]
- Dalton, T.P.; Solis, W.A.; Nebert, D.W.; Carvan, M.J. Characterization of the MTF-1 transcription factor from zebrafish and trout cells. Comp. Biochem. Phys. 2000, 126B, 325–335. [Google Scholar] [CrossRef]
- Xu, Y.H.; Luo, Z.; Wu, K.; Fan, Y.F.; You, W.J.; Zhang, L.H. Structure and functional analysis of promoters from two liver isoforms of CPT I in grass carp Ctenopharyngodon idella. Int. J. Mol. Sci. 2017, 18, 2405. [Google Scholar] [CrossRef]
- Read, M.A.; Cordle, S.R.; Veach, R.A.; Carlisle, C.D.; Hawiger, J. Cell-free pool of CD14 mediates activation of transcription factor NF-kappa B by lipopolysaccharide in human endothelial cells. Proc. Natl. Acad. Sci. USA 1993, 90, 9887–9891. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).