The Role of ABA in Plant Immunity is Mediated through the PYR1 Receptor
Abstract
1. Introduction
2. Results
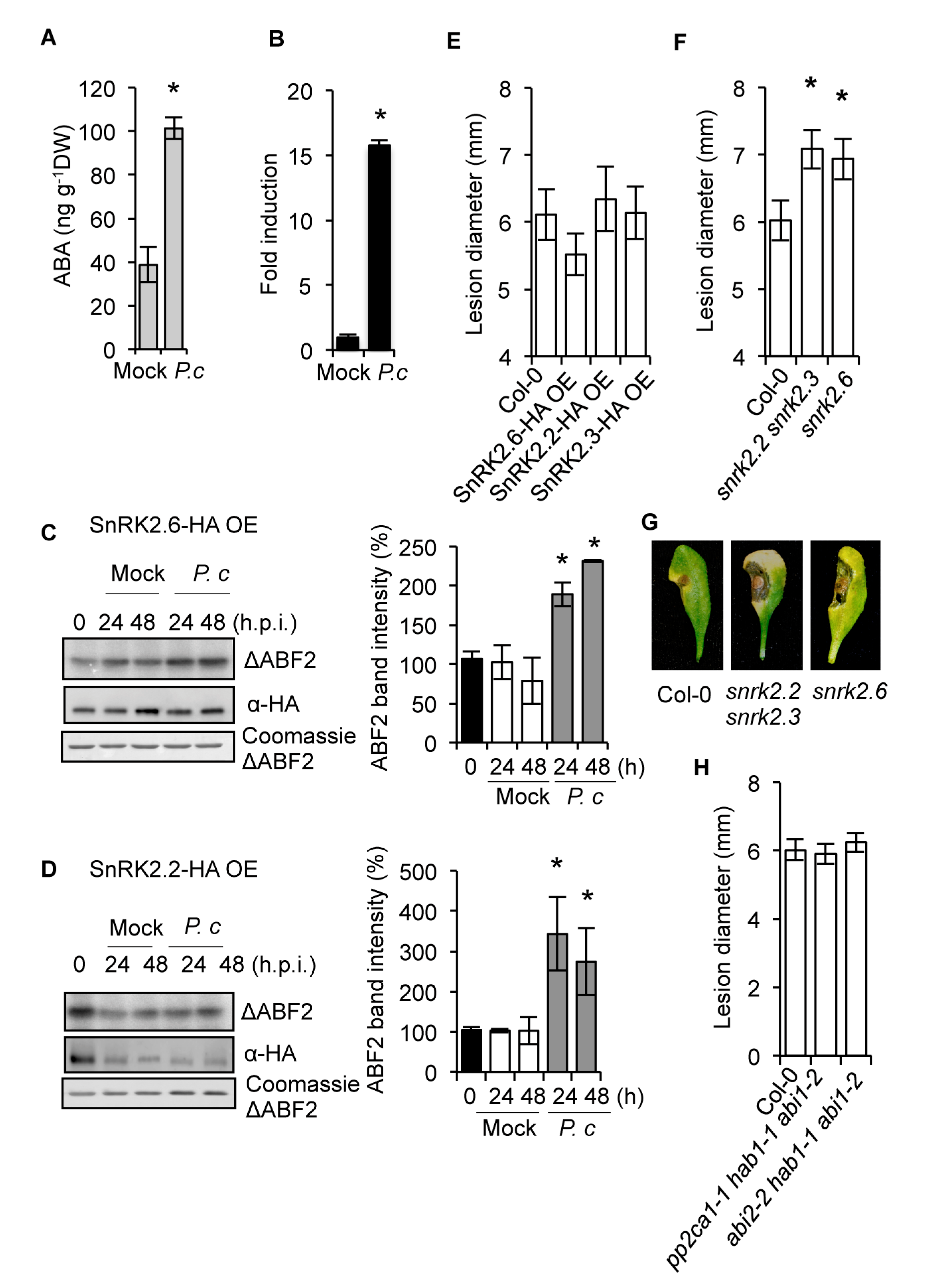
2.1. The SnRK2s Protein Kinases are Engaged in Disease Resistance to Fungal Infection
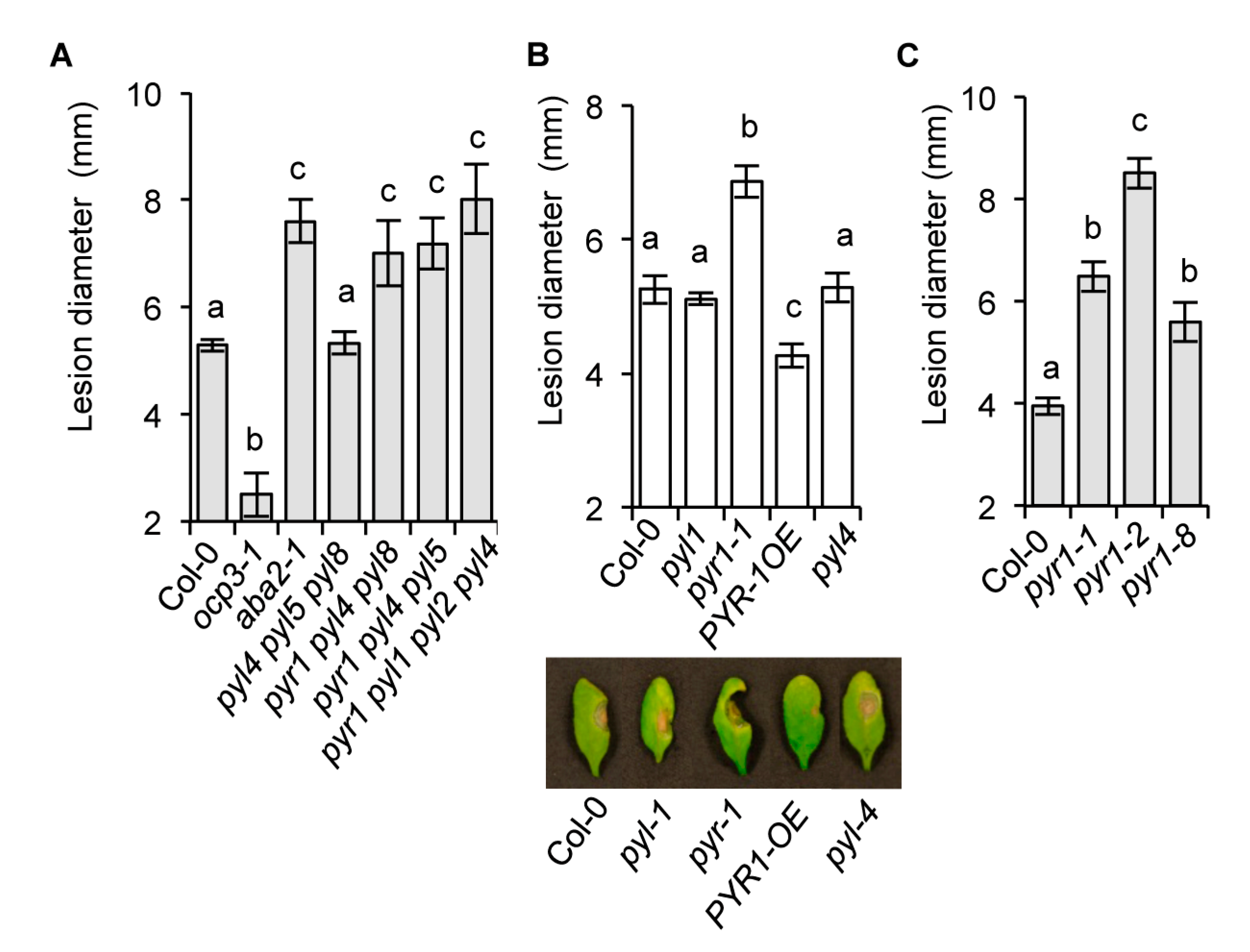
2.2. The Requirement of the PYR1 Receptor for Antifungal Resistance
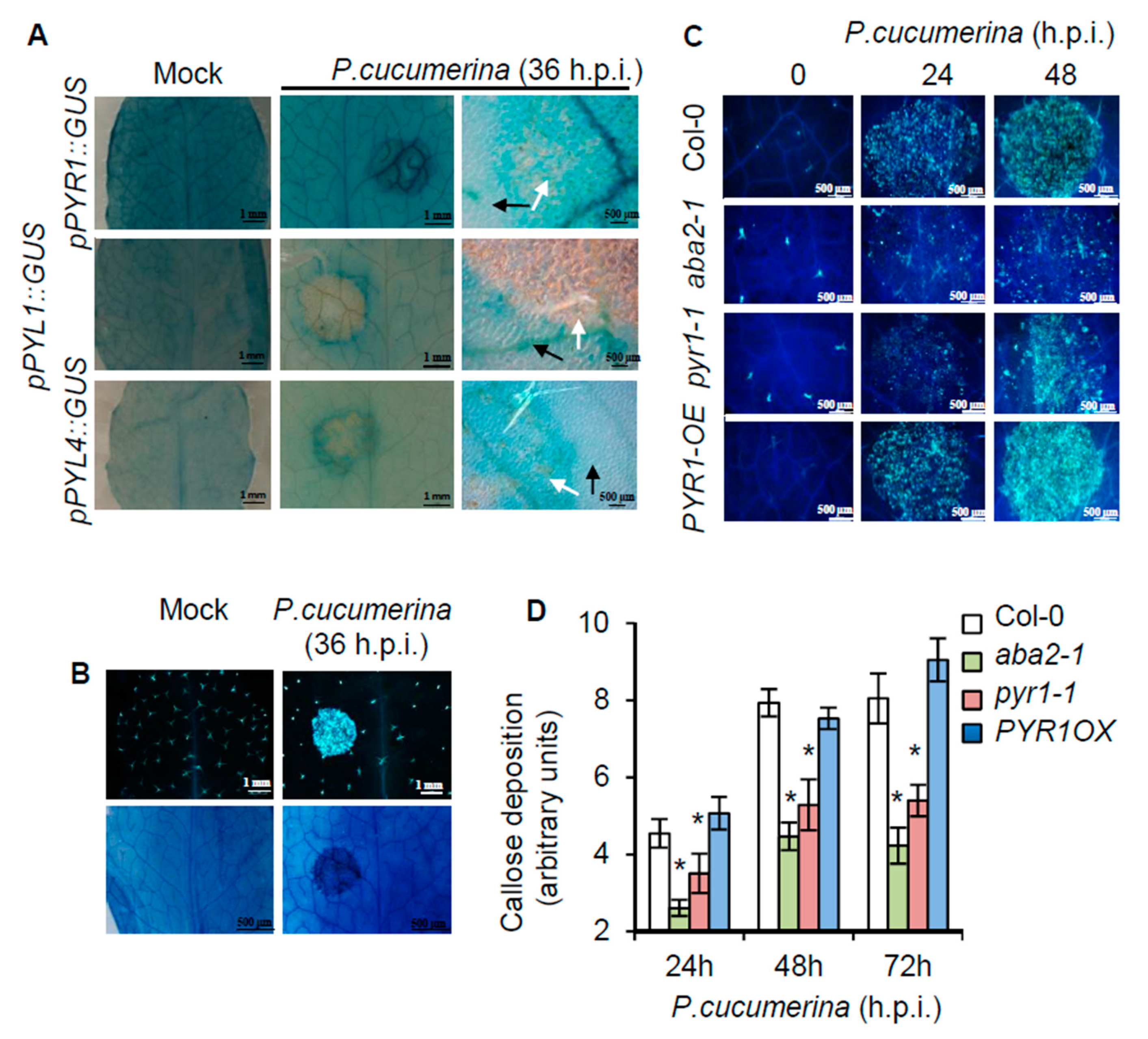
2.3. Local Induction of PYR1 Gene Expression by P. cucumerina
2.4. Resistance Enhancement of pyr1 Plants to Pseudomonas syringae DC3000
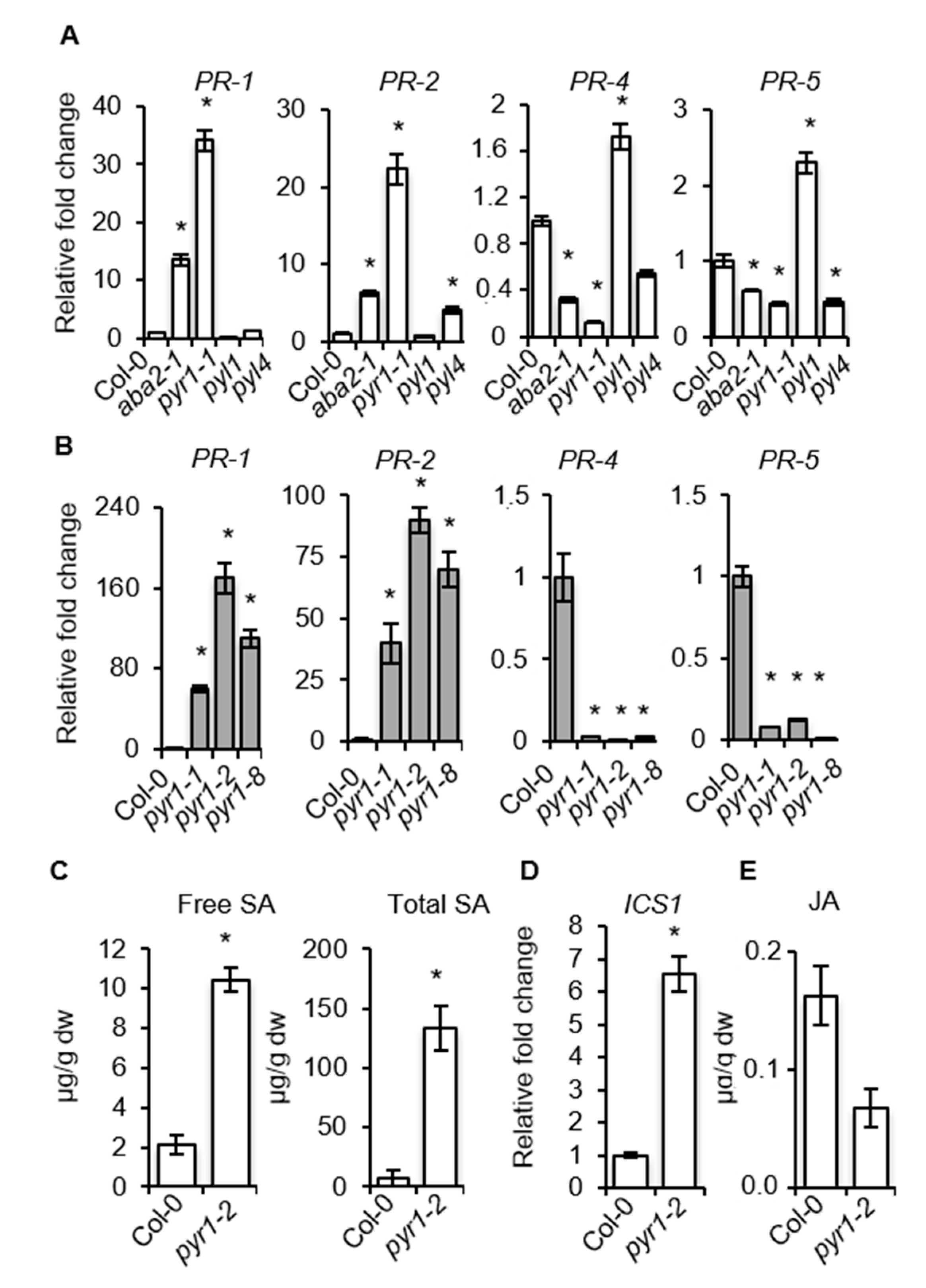
2.5. SA–Responsive Defense Genes are Activated in PYR1 Defective Mutants
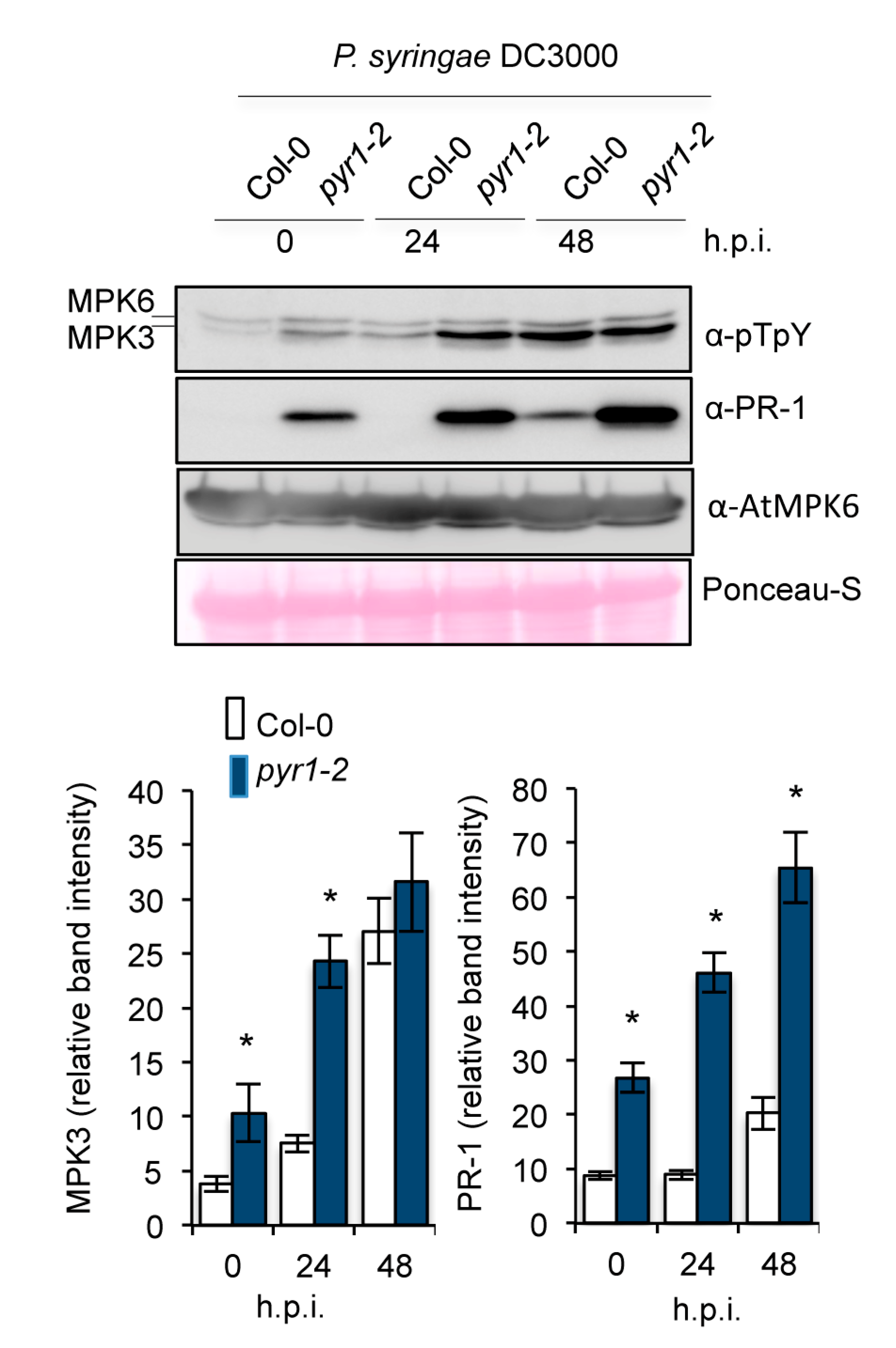
2.6. Enhanced Activation of MAPK Kinases in pyr1 Plants
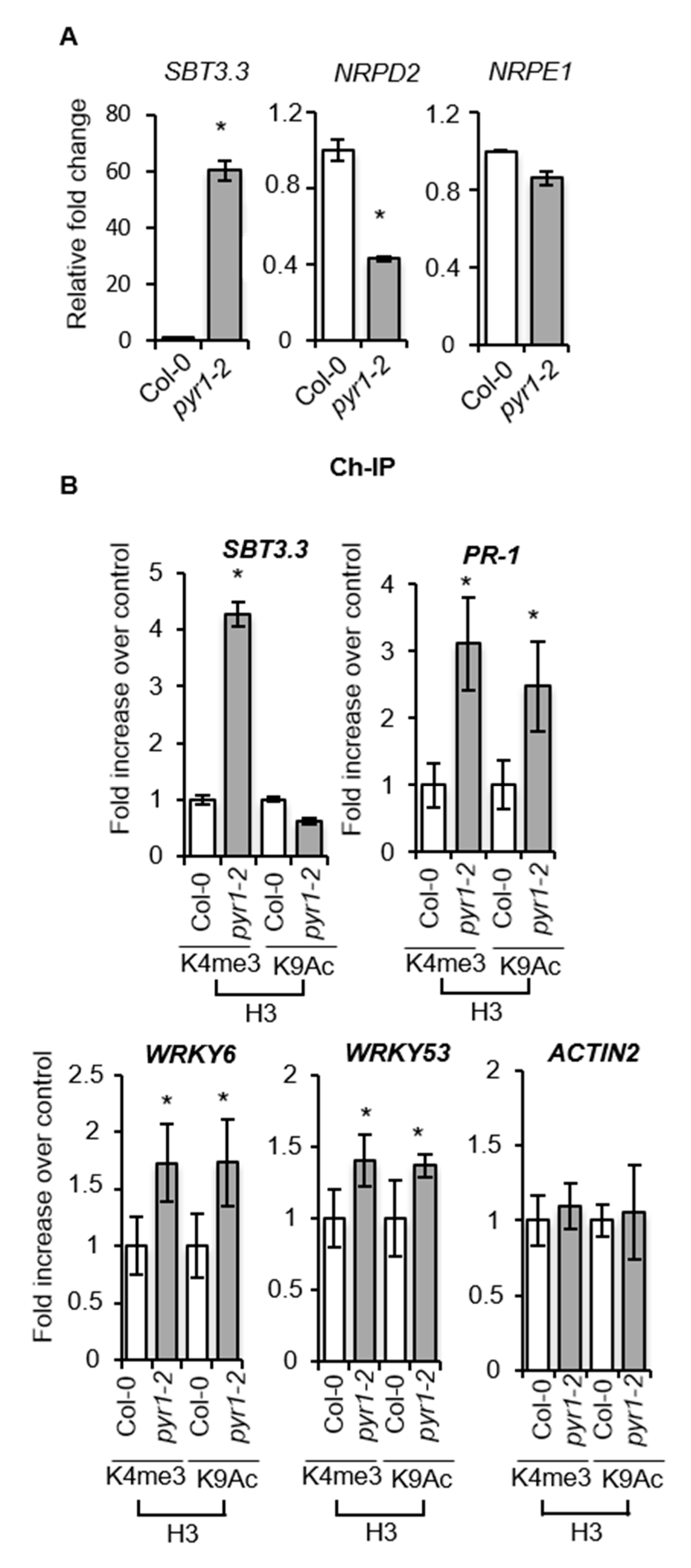
2.7. SA-Mediated Defense Genes are Poised for Enhanced Activation through Chromatin Remodeling in pyr1 Plants
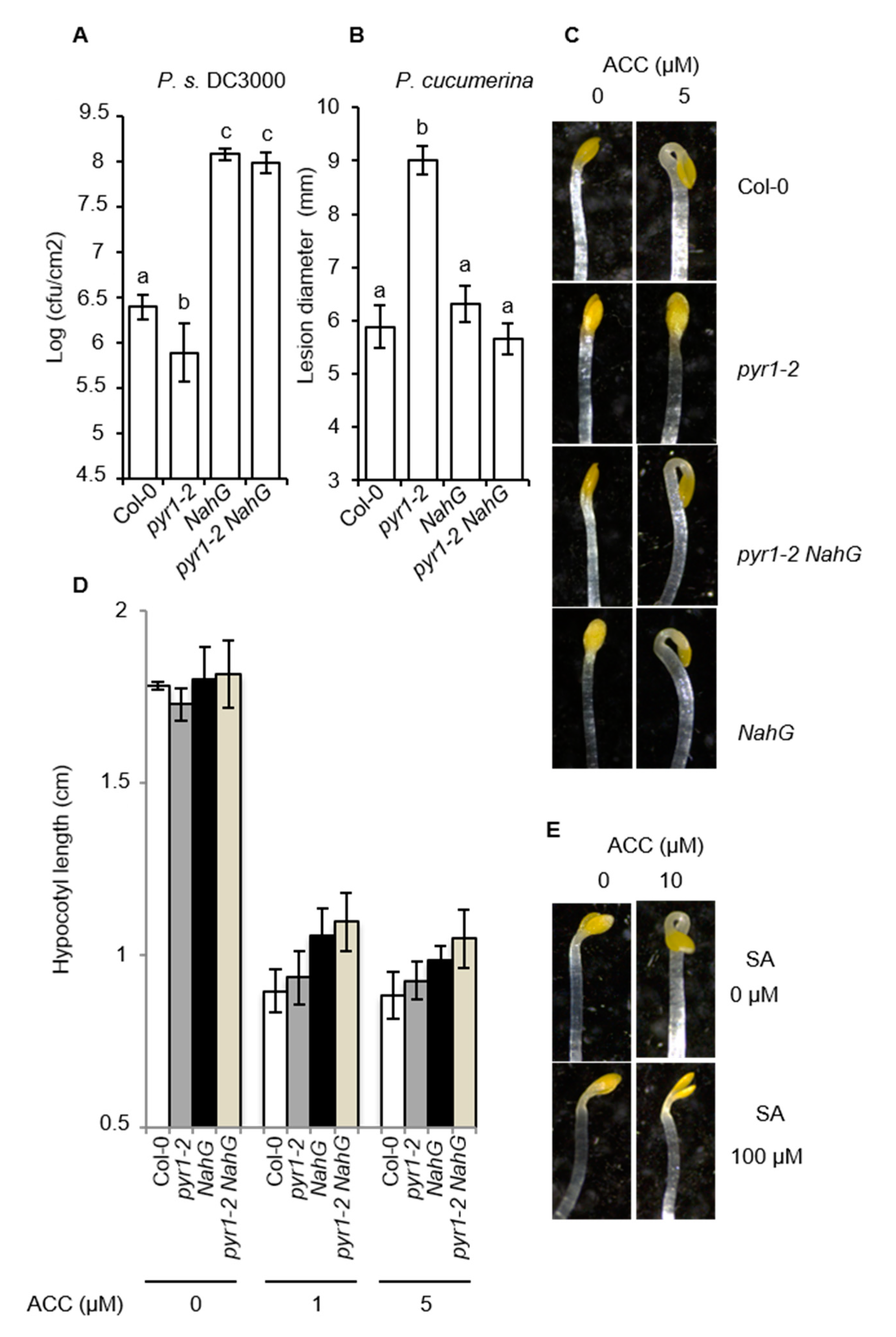
2.8. NahG Plants Abrogate the Altered Disease Resistance Response of pyr1 Plants
2.9. pyr1-Mediated Enhanced SA Content Blocks ET Perception
3. Discussion
4. Materials and Methods
4.1. Plants Growth Conditions
4.2. Gene Expression Analysis
4.3. Immunoprecipitation of HA–SnRKs and In Vitro Phosphorylation
4.4. Chromatin Immunoprecipitation
4.5. Western Blot
4.6. Pathogen Assays
4.7. Determination of Plant Hormones and Metabolites
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Malamy, J.; Carr, J.P.; Klessig, D.F.; Raskin, I. Salicylic acid: A likely endogenous signal in the resistance response of tobacco to viral infection. Science 1990, 250, 1002–1004. [Google Scholar] [CrossRef] [PubMed]
- Métraux, J.P.; Signer, H.; Ryals, J.; Ward, E.; WyssBenz, M.; Gaudin, J.; Raschdorf, K.; Schmid, E.; Blum, W.; Inverardi, B. Increase in salicylic acid at the onset of systemic acquired resistance in cucumber. Science 1990, 250, 1004–1006. [Google Scholar] [CrossRef] [PubMed]
- Glazebrook, J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef]
- Verhage, A.; van Wees, S.C.M.; Pieterse, C.M.J. Plant immunity: It’s the hormones talking, but what do they say? Plant Physiol. 2010, 154, 536–540. [Google Scholar] [CrossRef] [PubMed]
- Broekaert, W.F.; Delaure, S.L.; De Bolle, M.F.; Cammue, B.P. The role of ethylene in host-pathogen interactions. Annu. Rev. Phytopathol. 2006, 44, 393–416. [Google Scholar] [CrossRef]
- Grant, M.R.; Jones, J.D. Hormone (dis)harmony moulds plant health and disease. Science 2009, 324, 750–752. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; Van der Does, D.; Zamioudis, C.; Leon-Reyes, A.; Van Wees, S.C. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 2012, 28, 489–521. [Google Scholar] [CrossRef]
- Thaler, J.S.; Bostock, R.M. Interactions between abscisic-acid-mediated responses and plant resistance to pathogens and insects. Ecology 2004, 85, 48–58. [Google Scholar] [CrossRef]
- Spoel, S.H.; Koornneef, A.; Claessens, S.M.; Korzelius, J.P.; Van Pelt, J.A.; Mueller, M.J.; Buchala, A.J.; Metraux, J.P.; Brown, R.; Kazan, K.; et al. NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 2003, 15, 760–770. [Google Scholar] [CrossRef]
- Thomma, B.P.H.J.; Eggermont, K.; Tierens, K.F.M.; Broekaert, W.F. Requirement of functional ethylene-insensitive 2 gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea. Plant Physiol. 1999, 121, 1093–1102. [Google Scholar] [CrossRef]
- Gu, Y.Q.; Yang, C.; Thara, V.K.; Zhou, J.; Martin, G.B. Pti4 is induced by ethylene and salicylic acid, and its product is phosphorylated by the Pto kinase. Plant Cell 2000, 12, 771–786. [Google Scholar] [CrossRef] [PubMed]
- Berrocal-Lobo, M.; Molina, A.; Solano, R. Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 2002, 29, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Díaz, J.; ten Have, A.; van Kan, J.A.L. The role of ethylene and wound signaling in resistance of tomato to Botrytis cinerea. Plant Physiol. 2002, 129, 1341–1351. [Google Scholar] [CrossRef] [PubMed]
- Leslie, C.A.; Romani, R.J. Inhibition of ethylene biosynthesis by salicylic acid. Plant Physiol. 1988, 88, 833–837. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Xue, L.; Chintamanani, S.; Germain, H.; Lin, H.; Cui, H.; Cai, R.; Zuo, J.; Tang, X.; Li, X.; et al. ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 repress SALICYLIC ACID INDUCTION DEFICIENT2 expression to negatively regulate plant innate immunity in Arabidopsis. Plant Cell 2009, 21, 2527–2540. [Google Scholar] [CrossRef]
- Huang, P.; Dong, Z.; Guo, P.; Zhang, X.; Qiu, Y.; Li, B.; Wang, Y.; Guo, H. Salicylic Acid Suppresses Apical Hook Formation via NPR1-Mediated Repression of EIN3 and EIL1 in Arabidopsis. Plant Cell 2020, 32, 612–629. [Google Scholar] [CrossRef] [PubMed]
- Spoel, S.H.; Johnson, J.S.; Dong, X. Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc. Natl. Acad. Sci. USA 2007, 104, 18842–18847. [Google Scholar] [CrossRef]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef]
- Mauch-Mani, B.; Mauch, F. The role of abscisic acid in plant-pathogen interactions. Curr. Opin. Plant Biol. 2005, 8, 409–414. [Google Scholar] [CrossRef]
- Asselbergh, B.; De Vleesschauwer, D.; Hofte, M. Global switches and fine-tuning-aba modulates plant pathogen defense. Mol. Plant Microbe Interact. 2008, 21, 709–719. [Google Scholar] [CrossRef]
- Fan, J.; Hill, L.; Crooks, C.; Doerner, P.; Lamb, C. Abscisic acid has a key role in modulating diverse plant-pathogen interactions. Plant Physiol. 2009, 150, 1750–1761. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Vallet, A.; Lopez, G.; Ramos, B.; Delgado-Cerezo, M.; Riviere, M.P.; Llorente, F.; Fernandez, P.V.; Miedes, E.; Estevez, J.M.; Grant, M.; et al. Disruption of abscisic acid signaling constitutively activates arabidopsis resistance to the necrotrophic fungus plectosphaerella cucumerina. Plant Physiol. 2012, 160, 2109–2124. [Google Scholar] [CrossRef] [PubMed]
- Ton, J.; Mauch-Mani, B. Beta-amino-butyric acid-induced resistance against necrotrophic pathogens is based on aba-dependent priming for callose. Plant J. 2004, 38, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Adie, B.A.; Perez-Perez, J.; Perez-Perez, M.M.; Godoy, M.; Sanchez-Serrano, J.J.; Schmelz, E.A.; Solano, R. ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 2007, 19, 1665–1681. [Google Scholar] [CrossRef]
- García-Andrade, J.; Ramírez, V.; Flors, V.; Vera, P. Arabidopsis ocp3 mutant reveals a mechanism linking aba and ja to pathogen-induced callose deposition. Plant J. 2011, 67, 783–794. [Google Scholar]
- Mohr, P.G.; Cahill, D.M. Suppression by ABA of salicylic acid and lignin accumulation and the expression of multiple genes, in arabidopsis infected with Pseudomonas syringae pv. tomato. Funct. Integr. Genomics 2007, 7, 181–191. [Google Scholar] [CrossRef]
- Jensen, M.K.; Hagedorn, P.H.; de Torres-Zabala, M.; Grant, M.R.; Rung, J.H.; Collinge, D.B.; Lyngkjaer, M.F. Transcriptional regulation by an nac (nam-ataf1,2-cuc2) transcription factor attenuates aba signalling for efficient basal defence towards Blumeria graminis f. sp. Hordei in Arabidopsis. Plant J. 2008, 56, 867–880. [Google Scholar] [CrossRef]
- Torres-Zabala, M.; Truman, W.; Bennett, M.H.; Lafforgue, G.; Mansfield, J.W.; Rodriguez Egea, P.; Bögre, L.; Grant, M. Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. EMBO J. 2007, 26, 1434–1443. [Google Scholar] [CrossRef]
- Mine, A.; Berens, M.L.; Nobori, T.; Anver, S.; Fukumoto, K.; Winkelmüller, T.M.; Takeda, A.; Becker, D.; Tsuda, K. Pathogen exploitation of an abscisic acid- and jasmonate-inducible MAPK phosphatase and its interception by Arabidopsis immunity. Proc. Natl. Acad. Sci. USA 2017, 114, 7456–7461. [Google Scholar] [CrossRef]
- Peng, Z.; Hu, Y.; Zhang, J.; Huguet-Tapia, J.; Block, A.K.; Park, S.; Sapkota, S.; Liu, Z.; Liu, S.; White, F.F. Xanthomonas translucens commandeers the host rate-limiting step in ABA biosynthesis for disease susceptibility. Proc. Natl Acad. Sci. USA 2019, 116, 20938–20946. [Google Scholar] [CrossRef]
- Fujimoto, S.Y.; Ohta, M.; Usui, A.; Shinshi, H.; Ohme-Takagi, M. Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of gcc box-mediated gene expression. Plant Cell 2000, 12, 393–404. [Google Scholar] [PubMed]
- Chen, W.; Provart, N.J.; Glazebrook, J.; Katagiri, F.; Chang, H.S.; Eulgem, T.; Mauch, F.; Luan, S.; Zou, G.; Whitham, S.A.; et al. Expression profile matrix of arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 2002, 14, 559–574. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.P.; Badruzsaufari, E.; Schenk, P.M.; Manners, J.M.; Desmond, O.J.; Ehlert, C.; Maclean, D.J.; Ebert, P.R.; Kazan, K. Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in arabidopsis. Plant Cell 2004, 16, 3460–3479. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Tian, L.; Latoszek-Green, M.; Brown, D.; Wu, K. Arabidopsis erf4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol. Biol. 2005, 58, 585–596. [Google Scholar] [CrossRef]
- Dittrich, M.; Mueller, H.M.; Bauer, H.; Peirats-Llobet, M.; Rodriguez, P.L.; Geilfus, C.M.; Carpentier, S.C.; Al Rasheid, K.A.S.; Kollist, H.; Merilo, E.; et al. The role of Arabidopsis ABA receptors from the PYR/PYL/RCAR family in stomatal acclimation and closure signal integration. Nat. Plants 2019, 5, 1002–1011. [Google Scholar] [CrossRef]
- Hubbard, K.E.; Nishimura, N.; Hitomi, K.; Getzoff, E.D.; Schroeder, J.I. Early abscisic acid signal transduction mechanisms: Newly discovered components and newly emerging questions. Genes Dev. 2010, 24, 1695–1708. [Google Scholar] [CrossRef]
- Klingler, J.P.; Batelli, G.; Zhu, J.K. Aba receptors: The start of a new paradigm in phytohormone signalling. J. Exp. Bot. 2010, 61, 3199–3210. [Google Scholar] [CrossRef]
- Raghavendra, A.S.; Gonugunta, V.K.; Christmann, A.; Grill, E. ABA perception and signalling. Trends Plant Sci. 2010, 15, 395–401. [Google Scholar] [CrossRef]
- Umezawa, T.; Sugiyama, N.; Mizoguchi, M.; Hayashi, S.; Myouga, F.; Yamaguchi-Shinozaki, K.; Ishihama, Y.; Hirayama, T.; Shinozaki, K. Type 2c protein phosphatases directly regulate abscisic acid-activated protein kinases in arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 17588–17593. [Google Scholar] [CrossRef]
- Vlad, F.; Rubio, S.; Rodrigues, A.; Sirichandra, C.; Belin, C.; Robert, N.; Leung, J.; Rodriguez, P.L.; Lauriere, C.; Merlot, S. Protein phosphatases 2c regulate the activation of the snf1-related kinase ost1 by abscisic acid in arabidopsis. Plant Cell 2009, 21, 3170–3184. [Google Scholar] [CrossRef]
- Fujii, H.; Chinnusamy, V.; Rodrigues, A.; Rubio, S.; Antoni, R.; Park, S.-Y.; Cutler, S.R.; Sheen, J.; Rodriguez, P.L.; Zhu, J.-K. In vitro reconstitution of an aba signaling pathway. Nature 2009, 462, 660–664. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of pp2c phosphatase activity function as abscisic acid sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.F.; et al. Abscisic acid inhibits type 2c protein phosphatases via the pyr/pyl family of start proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef] [PubMed]
- Umezawa, T.; Sugiyama, N.; Takahashi, F.; Anderson, J.C.; Ishihama, Y.; Peck, S.C.; Shinozaki, K. Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in arabidopsis thaliana. Sci. Signal 2013, 6, rs8. [Google Scholar] [CrossRef]
- Gonzalez-Guzman, M.; Pizzio, G.A.; Antoni, R.; Vera-Sirera, F.; Merilo, E.; Bassel, G.W.; Fernandez, M.A.; Holdsworth, M.J.; Perez-Amador, M.A.; Kollist, H.; et al. Arabidopsis pyr/pyl/rcar receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 2012, 24, 2483–2496. [Google Scholar] [CrossRef]
- Gonzalez-Guzman, M.; Rodriguez, L.; Lorenzo-Orts, L.; Pons, C.; Sarrion-Perdigones, A.; Fernandez, M.A.; Peirats-Llobet, M.; Forment, J.; Moreno-Alvero, M.; Cutler, S.R.; et al. Tomato pyr/pyl/rcar abscisic acid receptors show high expression in root, differential sensitivity to the abscisic acid agonist quinabactin, and the capability to enhance plant drought resistance. J. Exp. Bot. 2014, 65, 4451–4464. [Google Scholar] [CrossRef]
- Helander, J.D.; Vaidya, A.S.; Cutler, S.R. Chemical manipulation of plant water use. Bioorg. Med. Chem. 2016, 24, 493–500. [Google Scholar] [CrossRef]
- Antoni, R.; Gonzalez-Guzman, M.; Rodriguez, L.; Rodrigues, A.; Pizzio, G.A.; Rodriguez, P.L. Selective inhibition of clade a phosphatases type 2c by pyr/pyl/rcar abscisic acid receptors. Plant Physiol. 2012, 158, 970–980. [Google Scholar] [CrossRef]
- Tischer, S.V.; Wunschel, C.; Papacek, M.; Kleigrewe, K.; Hofmann, T.; Christmann, A.; Grill, E. Combinatorial interaction network of abscisic acid receptors and coreceptors from arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2017, 114, 10280–10285. [Google Scholar] [CrossRef]
- Antoni, R.; Gonzalez-Guzman, M.; Rodriguez, L.; Peirats-Llobet, M.; Pizzio, G.A.; Fernandez, M.A.; De Winne, N.; De Jaeger, G.; Dietrich, D.; Bennett, M.J.; et al. Pyrabactin resistance1-like8 plays an important role for the regulation of abscisic acid signaling in root. Plant Physiol. 2013, 161, 931–941. [Google Scholar] [CrossRef]
- Fujii, H.; Zhu, J.K. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. USA 2009, 106, 8380–8385. [Google Scholar] [CrossRef] [PubMed]
- Fujii, H.; Verslues, P.E.; Zhu, J.K. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in arabidopsis. Plant Cell 2007, 19, 485–494. [Google Scholar] [CrossRef] [PubMed]
- García-Andrade, J.; Ramirez, V.; Lopez, A.; Vera, P. Mediated plastid rna editing in plant immunity. PLoS Pathog. 2013, 9, e1003713. [Google Scholar] [CrossRef] [PubMed]
- López, A.; Ramírez, V.; García-Andrade, J.; Flors, V.; Vera, P. The RNA silencing enzyme RNA polymerase V is required for plant immunity. PLoS Genet. 2011, 7, e1002434. [Google Scholar] [CrossRef] [PubMed]
- Rubio, S.; Rodrigues, A.; Saez, A.; Dizon, M.B.; Galle, A.; Kim, T.H.; Santiago, J.; Flexas, J.; Schroeder, J.I.; Rodriguez, P.L. Triple loss of function of protein phosphatases type 2c leads to partial constitutive response to endogenous abscisic acid. Plant Physiol. 2009, 150, 1345–1355. [Google Scholar] [CrossRef] [PubMed]
- Schweighofer, A.; Hirt, H.; Meskiene, I. Plant PP2C phosphatases: Emerging functions in stress signaling. Trends Plant Sci. 2004, 9, 236–243. [Google Scholar] [CrossRef]
- Carrasco, J.L.; Ancillo, G.; Mayda, E.; Vera, P. A novel transcription factor involved in plant defense endowed with protein phosphatase activity. EMBO J. 2003, 22, 3376–3384. [Google Scholar] [CrossRef]
- Carrasco, J.L.; Castello, M.J.; Naumann, K.; Lassowskat, I.; Navarrete-Gomez, M.; Scheel, D.; Vera, P. Arabidopsis protein phosphatase dbp1 nucleates a protein network with a role in regulating plant defense. PLoS ONE 2014, 9, e90734. [Google Scholar] [CrossRef]
- Clarke, J.D.; Volko, S.M.; Ledford, H.; Ausubel, F.M.; Dong, X. Roles of salicylic acid, jasmonic acid, and ethylene in cpr-induced resistance in Arabidopsis. Plant Cell 2000, 12, 2175–2190. [Google Scholar] [CrossRef]
- Wildermuth, M.C.; Dewdney, J.; Wu, G.; Ausubel, F.M. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 2001, 414, 562–565. [Google Scholar] [CrossRef]
- Garcion, C.; Lohmann, A.; Lamodière, E.; Catinot, J.; Buchala, A.; Doermann, P.; Métraux, J.-P. Characterization and biological function of the isochorismate synthase2 gene of Arabidopsis. Plant Physiol. 2008, 147, 1279–1287. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, V.; López, A.; Mauch-Mani, B.; Gil, M.J.; Vera, P. An extracellular subtilase switch for immune priming in arabidopsis. PLoS Pathog. 2013, 9, e1003445. [Google Scholar] [CrossRef]
- Mauch-Mani, B.; Baccelli, I.; Luna, E.; Flors, V. Defense priming: An adaptive part of induced resistance. Annu. Rev. Plant Biol. 2017, 68, 485–512. [Google Scholar] [CrossRef]
- Gaffney, T.; Friedrich, L.; Vernooij, B.; Negrotto, D.; Nye, G.; Uknes, S.; Ward, E.; Kessmann, H.; Ryals, J. Requirement of salicylic acid for the induction of systemic acquired resistance. Science 1993, 261, 754–756. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.H.; Endo, A.; Zhou, L.; Penney, J.; Chen, H.C.; Arroyo, A.; Leon, P.; Nambara, E.; Asami, T.; Seo, M.; et al. A unique short-chain dehydrogenase/reductase in arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 2002, 14, 2723–2743. [Google Scholar] [CrossRef] [PubMed]
- Koiwai, H.; Nakaminami, K.; Seo, M.; Mitsuhashi, W.; Toyomasu, T.; Koshiba, T. Tissue-specific localization of an abscisic acid biosynthetic enzyme, AAO3, in Arabidopsis. Plant Physiol. 2004, 134, 1697–1707. [Google Scholar] [CrossRef]
- Endo, A.; Koshiba, T.; Kamiya, Y.; Nambara, E. Vascular system is a node of systemic stress responses: Competence of the cell to synthesize abscisic acid and its responsiveness to external cues. Plant Signal Behav. 2008, 3, 1138–1140. [Google Scholar] [CrossRef][Green Version]
- Endo, A.; Sawada, Y.; Takahashi, H.; Okamoto, M.; Ikegami, K.; Koiwai, H.; Seo, M.; Toyomasu, T.; Mitsuhashi, W.; Shinozaki, K.; et al. Drought induction of arabidopsis 9-cis-epoxycarotenoid dioxygenase occurs in vascular parenchyma cells. Plant Physiol. 2008, 147, 1984–1993. [Google Scholar] [CrossRef]
- Alvarez, M.E.; Pennell, R.I.; Meijer, P.J.; Ishikawa, A.; Dixon, R.A.; Lamb, C. Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 1998, 92, 773–784. [Google Scholar] [CrossRef]
- Yu, A.; Lepere, G.; Jay, F.; Wang, J.; Bapaume, L.; Wang, Y.; Abraham, A.L.; Penterman, J.; Fischer, R.L.; Voinnet, O.; et al. Dynamics and biological relevance of DNA demethylation in Arabidopsis antibacterial defense. Proc. Natl. Acad. Sci. USA 2013, 110, 2389–2394. [Google Scholar] [CrossRef]
- Flors, V.; Ton, J.; van Doorn, R.; Jakab, G.; Garcia-Agustin, P.; Mauch-Mani, B. Interplay between JA, SA and ABA signalling during basal and induced resistance against pseudomonas syringae and alternaria brassicicola. Plant J. 2008, 54, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Petriacq, P.; Stassen, J.H.; Ton, J. Spore density determines infection strategy by the plant pathogenic fungus plectosphaerella cucumerina. Plant Physiol. 2016, 170, 2325–2339. [Google Scholar] [CrossRef] [PubMed]
- Koornneef, A.; Pieterse, C.M. Cross talk in defense signaling. Plant Physiol. 2008, 146, 839–844. [Google Scholar] [CrossRef]
- Ton, J.; Flors, V.; Mauch-Mani, B. The multifaceted role of aba in disease resistance. Trends Plant Sci. 2009, 14, 310–317. [Google Scholar] [CrossRef]
- De Vleesschauwer, D.; Yang, Y.; Cruz, C.V.; Hofte, M. Abscisic acid-induced resistance against the brown spot pathogen cochliobolus miyabeanus in rice involves map kinase-mediated repression of ethylene signaling. Plant Physiol. 2010, 152, 2036–2052. [Google Scholar] [CrossRef] [PubMed]
- de Torres Zabala, M.; Bennett, M.H.; Truman, W.H.; Grant, M.R. Antagonism between salicylic and abscisic acid reflects early host-pathogen conflict and moulds plant defence responses. Plant J. 2009, 59, 375–386. [Google Scholar] [CrossRef]
- Belin, C.; de Franco, P.O.; Bourbousse, C.; Chaignepain, S.; Schmitter, J.M.; Vavasseur, A.; Giraudat, J.; Barbier-Brygoo, H.; Thomine, S. Identification of features regulating ost1 kinase activity and ost1 function in guard cells. Plant Physiol. 2006, 141, 1316–1327. [Google Scholar] [CrossRef]
- Planes, M.D.; Ninoles, R.; Rubio, L.; Bissoli, G.; Bueso, E.; Garcia-Sanchez, M.J.; Alejandro, S.; Gonzalez-Guzman, M.; Hedrich, R.; Rodriguez, P.L.; et al. A mechanism of growth inhibition by abscisic acid in germinating seeds of arabidopsis thaliana based on inhibition of plasma membrane H+-atpase and decreased cytosolic pH, K+, and anions. J. Exp. Bot. 2015, 66, 813–825. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Andrade, J.; González, B.; Gonzalez-Guzman, M.; Rodriguez, P.L.; Vera, P. The Role of ABA in Plant Immunity is Mediated through the PYR1 Receptor. Int. J. Mol. Sci. 2020, 21, 5852. https://doi.org/10.3390/ijms21165852
García-Andrade J, González B, Gonzalez-Guzman M, Rodriguez PL, Vera P. The Role of ABA in Plant Immunity is Mediated through the PYR1 Receptor. International Journal of Molecular Sciences. 2020; 21(16):5852. https://doi.org/10.3390/ijms21165852
Chicago/Turabian StyleGarcía-Andrade, Javier, Beatriz González, Miguel Gonzalez-Guzman, Pedro L. Rodriguez, and Pablo Vera. 2020. "The Role of ABA in Plant Immunity is Mediated through the PYR1 Receptor" International Journal of Molecular Sciences 21, no. 16: 5852. https://doi.org/10.3390/ijms21165852
APA StyleGarcía-Andrade, J., González, B., Gonzalez-Guzman, M., Rodriguez, P. L., & Vera, P. (2020). The Role of ABA in Plant Immunity is Mediated through the PYR1 Receptor. International Journal of Molecular Sciences, 21(16), 5852. https://doi.org/10.3390/ijms21165852






