Current Progress in Understanding and Recovering the Wheat Genes Lost in Evolution and Domestication
Abstract
1. Introduction
2. Evolution of Wheat
3. Changes in the Wheat Genome during Evolution
3.1. Genomic Changes through Domestication
3.2. Genomic Changes through Polyploidization
3.3. Genomic Changes through Natural Mutation
4. Exploring Wild Progenitor-Like Emmer Wheat
4.1. Geographical Distribution of WEM
4.2. WEM: Genetic Resources with a Great Diversity
4.3. Progresses of Using WEM Wheat to Broaden Modern Wheat Gene Pool
4.4. Identifying QTL/Genes for the Traits of Interest by Genetic Mapping
4.4.1. Agronomic Traits
4.4.2. Biotic Stress-Related Traits
4.4.3. Abiotic Stress-Related Traits
4.4.4. Quality and Nutritional Traits
4.5. Genomic Approach Using WEM Genotypes for Novel Allele Identification
5. Conclusions
Funding
Conflicts of Interest
References
- Peng, J.H.; Sun, D.; Nevo, E. Domestication evolution, genetics and genomics in wheat. Mol. Breed. 2011, 28, 281. [Google Scholar] [CrossRef]
- Matsuoka, Y. Evolution of polyploid Triticum wheats under cultivation: The role of domestication, natural hybridization and allopolyploid speciation in their diversification. Plant Cell Physiol. 2011, 52, 750–764. [Google Scholar] [CrossRef] [PubMed]
- Doebley, J.F.; Gaut, B.S.; Smith, B.D. The molecular genetics of crop domestication. Cell 2006, 127, 1309–1321. [Google Scholar] [CrossRef] [PubMed]
- Feldman, M.; Levy, A.A. Genome evolution in allopolyploid wheat—A revolutionary reprogramming followed by gradual changes. J. Genet. Genom. 2009, 36, 511–518. [Google Scholar] [CrossRef]
- Zhao, N.; Xu, L.; Zhu, B.; Li, M.; Zhang, H.; Qi, B.; Xu, C.; Han, F.; Liu, B. Chromosomal and genome-wide molecular changes associated with initial stages of allohexaploidization in wheat can be transit and incidental. Genome 2011, 54, 692–699. [Google Scholar] [CrossRef] [PubMed]
- Feldman, M.; Levy, A. Allopolyploidy–A shaping force in the evolution of wheat genomes. Cytogenet. Genome Res. 2005, 109, 250–258. [Google Scholar] [CrossRef]
- Guo, X.; Han, F. Asymmetric epigenetic modification and elimination of rDNA sequences by polyploidization in wheat. Plant Cell 2014, 26, 4311–4327. [Google Scholar] [CrossRef]
- Peleg, Z.; Fahima, T.; Korol, A.B.; Abbo, S.; Saranga, Y. Genetic analysis of wheat domestication and evolution under domestication. J. Exp. Bot. 2011, 62, 5051–5061. [Google Scholar] [CrossRef]
- Eilam, T.; Anikster, Y.; Millet, E.; Manisterski, J.; Feldman, M. Nuclear DNA amount and genome downsizing in natural and synthetic allopolyploids of the genera Aegilops and Triticum. Genome 2008, 51, 616–627. [Google Scholar] [CrossRef]
- Feldman, M.; Levy, A.A. Genome evolution due to allopolyploidization in wheat. Genetics 2012, 192, 763–774. [Google Scholar] [CrossRef]
- Xie, W.; Nevo, E. Wild emmer: Genetic resources, gene mapping and potential for wheat improvement. Euphytica 2008, 164, 603–614. [Google Scholar] [CrossRef]
- Zaharieva, M.; Ayana, N.G.; Hakimi, A.A.; Misra, S.C.; Monneveux, P. Cultivated emmer wheat (Triticum dicoccon Schrank), an old crop with promising future: A review. Genet. Resour. Crop Evol. 2010, 57, 937–962. [Google Scholar] [CrossRef]
- Zohary, D. Unconscious selection and the evolution of domesticated plants. Econ. Bot. 2004, 58, 5–10. [Google Scholar] [CrossRef]
- Chatzav, M.; Peleg, Z.; Ozturk, L.; Yazici, A.; Fahima, T.; Cakmak, I.; Saranga, Y. Genetic diversity for grain nutrients in wild emmer wheat: Potential for wheat improvement. Ann. Bot. 2010, 105, 1211–1220. [Google Scholar] [CrossRef]
- Nevo, E.; Korol, A.; Beiles, A.; Fahima, T. Evolution of Wild Emmer and Wheat Improvement: Population Genetics, Genetic Resources, and Genome Organization of Wheat’s Progenitor, Triticum Dicoccoides; Springer Science & Business Media: Berlin, Germany, 2013. [Google Scholar]
- Peleg, Z.; Fahima, T.; Abbo, S.; Krugman, T.; Nevo, E.; Yakir, D.; Saranga, Y. Genetic diversity for drought resistance in wild emmer wheat and its ecogeographical associations. Plant Cell Environ. 2005, 28, 176–191. [Google Scholar] [CrossRef]
- Peng, J.; Sun, D.; Nevo, E. Wild emmer wheat,‘Triticum dicoccoides’, occupies a pivotal position in wheat domestication process. Aust. J. Crop Sci. 2011, 5, 1127. [Google Scholar]
- Dvorak, J.; Akhunov, E.D. Tempos of gene locus deletions and duplications and their relationship to recombination rate during diploid and polyploid evolution in the Aegilops-Triticum alliance. Genetics 2005, 171, 323–332. [Google Scholar] [CrossRef]
- Huang, X.; Börner, A.; Röder, M.; Ganal, M. Assessing genetic diversity of wheat (Triticum aestivum L.) germplasm using microsatellite markers. Theor. Appl. Genet. 2002, 105, 699–707. [Google Scholar] [CrossRef]
- Ramsey, J.; Schemske, D.W. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst. 1998, 29, 467–501. [Google Scholar] [CrossRef]
- Luo, M.-C.; Yang, Z.-L.; You, F.; Kawahara, T.; Waines, J.; Dvorak, J. The structure of wild and domesticated emmer wheat populations, gene flow between them, and the site of emmer domestication. Theor. Appl. Genet. 2007, 114, 947–959. [Google Scholar] [CrossRef]
- Feldman, M.; Kislev, M.E. Domestication of emmer wheat and evolution of free-threshing tetraploid wheat. Isr. J. Plant Sci. 2007, 55, 207–221. [Google Scholar] [CrossRef]
- Dvorak, J.; Luo, M.; Yang, Z. Genetic evidence on the origin of Triticum aestivum L. In The Origins of Agriculture and Crop Domestication, Proceedings of the Harlan Symposium, Aleppo, Syria, 10–14 May 1997; ICARDA: Aleppo, Syria, 1998. [Google Scholar]
- Matsuoka, Y.; Nasuda, S. Durum wheat as a candidate for the unknown female progenitor of bread wheat: An empirical study with a highly fertile F 1 hybrid with Aegilops tauschii Coss. Theor. Appl. Genet. 2004, 109, 1710–1717. [Google Scholar] [CrossRef] [PubMed]
- Giles, R.J.; Brown, T.A. GluDy allele variations in Aegilops tauschii and Triticum aestivum: Implications for the origins of hexaploid wheats. Theor. Appl. Genet. 2006, 112, 1563–1572. [Google Scholar] [CrossRef] [PubMed]
- Salamini, F.; Özkan, H.; Brandolini, A.; Schäfer-Pregl, R.; Martin, W. Genetics and geography of wild cereal domestication in the Near East. Nat. Rev. Genet. 2002, 3, 429. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.H. Variation under domestication in plants: 1859 and today. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010, 365, 2523–2530. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zohary, D.; Hopf, M. Domestication of Plants in the Old World: The Origin and Spread of Cultivated Plants in West Asia; Europe, and the Nile Valley Oxford University Press: New York, NY, USA, 2000. [Google Scholar]
- Ladizinsky, G. Plant Evolution under Domestication; Springer: Dordrecht, The Netherlands, 1998. [Google Scholar]
- Hammer, K. Das domestikationssyndrom. Die Kulturpflanze 1984, 32, 11–34. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, X.; Islam, S.; She, M.; Peng, Y.; Yu, Z.; Wylie, S.; Juhasz, A.; Dowla, M.; Yang, R. New insights into the evolution of wheat avenin-like proteins in wild emmer wheat (Triticum dicoccoides). Proc. Natl. Acad. Sci. USA 2018, 115, 13312–13317. [Google Scholar] [CrossRef]
- Fu, Y.-B.; Somers, D.J. Genome-wide reduction of genetic diversity in wheat breeding. Crop Sci. 2009, 49, 161–168. [Google Scholar] [CrossRef]
- Nevo, E. Ecological genomics of natural plant populations: The Israeli perspective. In Plant Genomics; Springer: Berlin, Germany, 2009; pp. 321–344. [Google Scholar]
- Nevo, E. Triticum. In Wild Crop Relatives: Genomic and Breeding Resources; Springer: Berlin, Germany, 2011; pp. 407–456. [Google Scholar]
- Haudry, A.; Cenci, A.; Ravel, C.; Bataillon, T.; Brunel, D.; Poncet, C.; Hochu, I.; Poirier, S.; Santoni, S.; Glémin, S. Grinding up wheat: A massive loss of nucleotide diversity since domestication. Mol. Biol. Evol. 2007, 24, 1506–1517. [Google Scholar] [CrossRef]
- Reif, J.C.; Zhang, P.; Dreisigacker, S.; Warburton, M.L.; van Ginkel, M.; Hoisington, D.; Bohn, M.; Melchinger, A.E. Wheat genetic diversity trends during domestication and breeding. Theor. Appl. Genet. 2005, 110, 859–864. [Google Scholar] [CrossRef]
- Smale, M.; Reynolds, M.; Warburton, M.; Skovmand, B.; Trethowan, R.; Singh, R.; Ortiz-Monasterio, I.; Crossa, J. Dimensions of diversity in modern spring bread wheat in developing countries from 1965. Crop Sci. 2002, 42, 1766–1779. [Google Scholar] [CrossRef]
- Martínez, S.I.; Sanabria, A.; Fleitas, M.C.; Consolo, V.F.; Perelló, A. Wheat blast: Aggressiveness of isolates of Pyricularia oryzae and effect on grain quality. J. King Saud Univ. Sci. 2019, 31, 150–157. [Google Scholar] [CrossRef]
- Harlan, J.R.; De Wet, J.; Price, E.G. Comparative evolution of cereals. Evolution 1973, 27, 311–325. [Google Scholar] [CrossRef] [PubMed]
- Gegas, V.C.; Nazari, A.; Griffiths, S.; Simmonds, J.; Fish, L.; Orford, S.; Sayers, L.; Doonan, J.H.; Snape, J.W. A genetic framework for grain size and shape variation in wheat. Plant Cell 2010, 22, 1046–1056. [Google Scholar] [CrossRef] [PubMed]
- Berkman, P.J.; Visendi, P.; Lee, H.C.; Stiller, J.; Manoli, S.; Lorenc, M.T.; Lai, K.; Batley, J.; Fleury, D.; Šimková, H. Dispersion and domestication shaped the genome of bread wheat. Plant Biotechnol. J. 2013, 11, 564–571. [Google Scholar] [CrossRef] [PubMed]
- Gill, B.S.; Friebe, B.; Raupp, W.J.; Wilson, D.L.; Cox, T.S.; Sears, R.G.; Brown-Guedira, G.L.; Fritz, A.K. Wheat genetics resource center: The first 25 years. Adv. Agron. 2006, 89, 73–136. [Google Scholar]
- Nalam, V.J.; Vales, M.I.; Watson, C.J.; Johnson, E.B.; Riera-Lizarazu, O. Map-based analysis of genetic loci on chromosome 2D that affect glume tenacity and threshability, components of the free-threshing habit in common wheat (Triticum aestivum L.). Theor. Appl. Genet. 2007, 116, 135–145. [Google Scholar] [CrossRef]
- Jantasuriyarat, C.; Vales, M.; Watson, C.; Riera-Lizarazu, O. Identification and mapping of genetic loci affecting the free-threshing habit and spike compactness in wheat (Triticum aestivum L.). Theor. Appl. Genet. 2004, 108, 261–273. [Google Scholar] [CrossRef]
- Elias, E.; Steiger, D.; Cantrell, R. Evaluation of lines derived from wild emmer chromosome substitutions: II. Agronomic traits. Crop Sci. 1996, 36, 228–233. [Google Scholar] [CrossRef]
- Peng, J.; Ronin, Y.; Fahima, T.; Röder, M.S.; Li, Y.; Nevo, E.; Korol, A. Domestication quantitative trait loci in Triticum dicoccoides, the progenitor of wheat. Proc. Natl. Acad. Sci. USA 2003, 100, 2489–2494. [Google Scholar] [CrossRef]
- Campbell, B.T.; Baenziger, P.S.; Gill, K.; Eskridge, K.M.; Budak, H.; Erayman, M.; Dweikat, I.; Yen, Y. Identification of QTLs and environmental interactions associated with agronomic traits on chromosome 3A of wheat. Crop Sci. 2003, 43, 1493–1505. [Google Scholar] [CrossRef]
- Kato, K.; Miura, H.; Sawada, S. Mapping QTLs controlling grain yield and its components on chromosome 5A of wheat. Theor. Appl. Genet. 2000, 101, 1114–1121. [Google Scholar] [CrossRef]
- Feldman, M.; Sears, E.R. The wild gene resources of wheat. Sci. Am. 1981, 244, 102–113. [Google Scholar] [CrossRef]
- Nevo, E.; Beiles, A. Genetic diversity of wild emmer wheat in Israel and Turkey. Theor. Appl. Genet. 1989, 77, 421–455. [Google Scholar] [CrossRef] [PubMed]
- Feldman, M.; Levy, A.A.; Fahima, T.; Korol, A. Genomic asymmetry in allopolyploid plants: Wheat as a model. J. Exp. Bot. 2012, 63, 5045–5059. [Google Scholar] [CrossRef]
- Shaked, H.; Kashkush, K.; Ozkan, H.; Feldman, M.; Levy, A.A. Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell 2001, 13, 1749–1759. [Google Scholar] [CrossRef]
- Frankel, O.; Gerlach, W.; Peacock, W. The ribosomal RNA genes in synthetic tetraploids of wheat. Theor. Appl. Genet. 1987, 75, 138–143. [Google Scholar] [CrossRef]
- Akhunova, A.R.; Matniyazov, R.T.; Liang, H.; Akhunov, E.D. Homoeolog-specific transcriptional bias in allopolyploid wheat. BMC Genom. 2010, 11, 505. [Google Scholar] [CrossRef]
- Silva, M.; Pereira, H.S.; Bento, M.; Santos, A.P.; Shaw, P.; Delgado, M.; Neves, N.; Viegas, W. Interplay of ribosomal DNA loci in nucleolar dominance: Dominant NORs are up-regulated by chromatin dynamics in the wheat-rye system. PLoS ONE 2008, 3, e3824. [Google Scholar] [CrossRef]
- Qi, B.; Huang, W.; Zhu, B.; Zhong, X.; Guo, J.; Zhao, N.; Xu, C.; Zhang, H.; Pang, J.; Han, F. Global transgenerational gene expression dynamics in two newly synthesized allohexaploid wheat (Triticum aestivum) lines. BMC Biol. 2012, 10, 3. [Google Scholar] [CrossRef]
- Feldman, M.; Liu, B.; Segal, G.; Abbo, S.; Levy, A.A.; Vega, J.M. Rapid elimination of low-copy DNA sequences in polyploid wheat: A possible mechanism for differentiation of homoeologous chromosomes. Genetics 1997, 147, 1381–1387. [Google Scholar] [PubMed]
- Han, F.; Fedak, G.; Guo, W.; Liu, B. Rapid and repeatable elimination of a parental genome-specific DNA repeat (pGc1R-1a) in newly synthesized wheat allopolyploids. Genetics 2005, 170, 1239–1245. [Google Scholar] [CrossRef] [PubMed]
- Kerber, E.; Green, G. Suppression of stem rust resistance in the hexaploid wheat cv. Canthatch by chromosome 7DL. Can. J. Bot. 1980, 58, 1347–1350. [Google Scholar] [CrossRef]
- Akhunov, E.D.; Akhunova, A.R.; Anderson, O.D.; Anderson, J.A.; Blake, N.; Clegg, M.T.; Coleman-Derr, D.; Conley, E.J.; Crossman, C.C.; Deal, K.R. Nucleotide diversity maps reveal variation in diversity among wheat genomes and chromosomes. BMC Genom. 2010, 11, 702. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, N.; Sugiyama, K.; Yamagishi, Y.; Sakata, Y. Comparative telosomic mapping of homoeologous genes for brittle rachis in tetraploid and hexaploid wheats. Hereditas 2002, 137, 180–185. [Google Scholar] [CrossRef]
- Simons, K.J.; Fellers, J.P.; Trick, H.N.; Zhang, Z.; Tai, Y.-S.; Gill, B.S.; Faris, J.D. Molecular characterization of the major wheat domestication gene Q. Genetics 2006, 172, 547–555. [Google Scholar] [CrossRef]
- Merchuk-Ovnat, L.; Fahima, T.; Krugman, T.; Saranga, Y. Ancestral QTL alleles from wild emmer wheat improve grain yield, biomass and photosynthesis across enviroinments in modern wheat. Plant Sci. 2016, 251, 23–34. [Google Scholar] [CrossRef]
- Körnicke, F. Wilde Stammformen unserer Kulturweizen. Niederrheiner Gesellsch. f. Natur-und Heilkunde in Bonn, Sitzungsber 1889, 46, 21. [Google Scholar]
- Saranga, Y. Foreword by the Guest Editor. Isr. J. Plant Sci. 2007, 55, i–xii. [Google Scholar] [CrossRef]
- Kato, K.; Mori, Y.; Beiles, A.; Nevo, E. Geographical variation in heading traits in wild emmer wheat, Triticum dicoccoides. I. Variation in vernalization response and ecological differentiation. Theor. Appl. Genet. 1997, 95, 546–552. [Google Scholar] [CrossRef]
- Fahima, T.; Röder, M.; Wendehake, K.; Kirzhner, V.; Nevo, E. Microsatellite polymorphism in natural populations of wild emmer wheat, Triticum dicoccoides, in Israel. Theor. Appl. Genet. 2002, 104, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Nevo, E. “Evolution Canyon,” a potential microscale monitor of global warming across life. Proc. Natl. Acad. Sci. USA 2012, 109, 2960–2965. [Google Scholar] [CrossRef] [PubMed]
- Ben-Abu, Y.; Beiles, A.; Flom, D.; Nevo, E. Adaptive evolution of benzoxazinoids in wild emmer wheat, Triticum dicoccoides, at" Evolution Canyon", Mount Carmel, Israel. PLoS ONE 2018, 13, e0190424. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Ben-Abu, Y.; Wang, H.; Li, A.; Nevo, E.; Kong, L. Natural selection causes adaptive genetic resistance in wild emmer wheat against powdery mildew at “evolution canyon” microsite, mt. Carmel, Israel. PLoS ONE 2015, 10, e0122344. [Google Scholar] [CrossRef]
- Nevo, E. Evolution of genome–phenome diversity under environmental stress. Proc. Natl. Acad. Sci. USA 2001, 98, 6233–6240. [Google Scholar] [CrossRef]
- Li, X.; Wang, A.; Xiao, Y.; Yan, Y.; He, Z.; Appels, R.; Ma, W.; Hsam, S.; Zeller, F. Cloning and characterization of a novel low molecular weight glutenin subunit gene at the Glu-A3 locus from wild emmer wheat (Triticum turgidum L. var. dicoccoides). Euphytica 2008, 159, 181–190. [Google Scholar] [CrossRef]
- Uauy, C.; Distelfeld, A.; Fahima, T.; Blechl, A.; Dubcovsky, J. A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 2006, 314, 1298–1301. [Google Scholar] [CrossRef]
- Nevo, E.; Beiles, A. Amino-Acid Resources in the Wild Progenitor of Wheats, Triticum dicoccoides, in Israel—Polymorphisms and Predictability by Ecology and Isozymes. Plant Breed. 1992, 108, 190–201. [Google Scholar] [CrossRef]
- Çakmak, İ.; Torun, A.; Millet, E.; Feldman, M.; Fahima, T.; Korol, A.; Nevo, E.; Braun, H.; Özkan, H. Triticum dicoccoides: An important genetic resource for increasing zinc and iron concentration in modern cultivated wheat. Soil Sci. Plant Nutr. 2004, 50, 1047–1054. [Google Scholar] [CrossRef]
- Nevo, E.; Moseman, J.; Beiles, A.; Zohary, D. Patterns of resistance of Israeli wild emmer wheat to pathogens I. Predictive method by ecology and allozyme genotypes for powdery mildew and leaf rust. Genetica 1985, 67, 209–222. [Google Scholar] [CrossRef]
- Oliver, R.; Stack, R.; Miller, J.; Cai, X. Reaction of wild emmer wheat accessions to Fusarium head blight. Crop Sci. 2007, 47, 893–897. [Google Scholar] [CrossRef]
- Anikster, Y.; Manisterski, J.; Long, D.; Leonard, K. Leaf rust and stem rust resistance in Triticum dicoccoides populations in Israel. Plant Dis. 2005, 89, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Zhang, Z.; Peng, Y.; Nevo, E.; Peng, J. Resistance in wild emmer wheat (Triticum dicoccoides) from the fertile crescent to yellow rust in China. J. Food Agric. Environ. 2013, 11, 1395–1403. [Google Scholar]
- Wang, Y.-M.; Long, X.-Y.; Yan, Z.-H.; Nevo, E.; Baum, B.R.; Zheng, Y.-L. Molecular evolution of dimeric α-amylase inhibitor genes in wild emmer wheat and its ecological association. BMC Evol. Biol. 2008, 8, 91. [Google Scholar] [CrossRef]
- Avni, R.; Nave, M.; Barad, O.; Baruch, K.; Twardziok, S.O.; Gundlach, H.; Hale, I.; Mascher, M.; Spannagl, M.; Wiebe, K. Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 2017, 357, 93–97. [Google Scholar] [CrossRef]
- Maccaferri, M.; Harris, N.S.; Twardziok, S.O.; Pasam, R.K.; Gundlach, H.; Spannagl, M.; Ormanbekova, D.; Lux, T.; Prade, V.M.; Milner, S.G. Durum wheat genome highlights past domestication signatures and future improvement targets. Nat. Genet. 2019, 51, 885. [Google Scholar] [CrossRef]
- Peng, J.; Sun, D.; Peng, Y.; Nevo, E. Gene discovery inTriticum dicoccoides, the direct progenitor of cultivated wheats. Cereal. Res. Commun. 2013, 41, 1–22. [Google Scholar] [CrossRef]
- Pasam, R.K.; Sharma, R.; Malosetti, M.; van Eeuwijk, F.A.; Haseneyer, G.; Kilian, B.; Graner, A. Genome-wide association studies for agronomical traits in a world wide spring barley collection. BMC Plant Biol. 2012, 12, 16. [Google Scholar] [CrossRef]
- Zhu, C.; Gore, M.; Buckler, E.S.; Yu, J. Status and prospects of association mapping in plants. Plant Genome 2008, 1, 5–20. [Google Scholar] [CrossRef]
- Nevo, E.; Fu, Y.-B.; Pavlicek, T.; Khalifa, S.; Tavasi, M.; Beiles, A. Evolution of wild cereals during 28 years of global warming in Israel. Proc. Natl. Acad. Sci. USA 2012, 109, 3412–3415. [Google Scholar] [CrossRef]
- Spielmeyer, W.; Richards, R. Comparative mapping of wheat chromosome 1AS which contains the tiller inhibition gene (tin) with rice chromosome 5S. Theor. Appl. Genet. 2004, 109, 1303–1310. [Google Scholar] [CrossRef] [PubMed]
- Bennett, F.G. Resistance to powdery mildew in wheat: A review of its use in agriculture and breeding programmes. Plant Pathol. 1984, 33, 279–300. [Google Scholar] [CrossRef]
- Reader, S.; Miller, T. The introduction into bread wheat of a major gene for resistance to powdery mildew from wild emmer wheat. Euphytica 1991, 53, 57–60. [Google Scholar] [CrossRef]
- Chen, X.; Luo, Y.; Xia, X.; Xia, L.; Chen, X.; Ren, Z.; He, Z.; Jia, J. Chromosomal location of powdery mildew resistance gene Pm16 in wheat using SSR marker analysis. Plant Breed. 2005, 124, 225–228. [Google Scholar] [CrossRef]
- Rong, J.; Millet, E.; Manisterski, J.; Feldman, M. A new powdery mildew resistance gene: Introgression from wild emmer into common wheat and RFLP-based mapping. Euphytica 2000, 115, 121–126. [Google Scholar] [CrossRef]
- Liu, Z.; Sun, Q.; Ni, Z.; Nevo, E.; Yang, T. Molecular characterization of a novel powdery mildew resistance gene Pm30 in wheat originating from wild emmer. Euphytica 2002, 123, 21–29. [Google Scholar] [CrossRef]
- McIntosh, R.; Dubcovsky, J.; Rogers, J.W.; Morris, C.; Appels, R.; Xia, X. Catalogue of gene symbols for wheat: 2011 Supplement. Annu. Wheat Newsl. 2010, 57, 1–29. [Google Scholar]
- Li, G.; Fang, T.; Zhang, H.; Xie, C.; Li, H.; Yang, T.; Nevo, E.; Fahima, T.; Sun, Q.; Liu, Z. Molecular identification of a new powdery mildew resistance gene Pm41 on chromosome 3BL derived from wild emmer (Triticum turgidum var. dicoccoides). Theor. Appl. Genet. 2009, 119, 531–539. [Google Scholar] [CrossRef]
- Hua, W.; Liu, Z.; Zhu, J.; Xie, C.; Yang, T.; Zhou, Y.; Duan, X.; Sun, Q.; Liu, Z. Identification and genetic mapping of pm42, a new recessive wheat powdery mildew resistance gene derived from wild emmer (Triticum turgidum var. dicoccoides). Theor. Appl. Genet. 2009, 119, 223–230. [Google Scholar] [CrossRef]
- Mohler, V.; Zeller, F.J.; Wenzel, G.; Hsam, S.L. Chromosomal location of genes for resistance to powdery mildew in common wheat (Triticum aestivum L. em Thell.). 9. Gene MlZec1 from the Triticum dicoccoides-derived wheat line Zecoi-1. Euphytica 2005, 142, 161–167. [Google Scholar] [CrossRef]
- Xie, W. Identification and Molecular Mapping of Powdery Mildew Resistance Genes Derived from Wild Relatives of Wheat; University of Haifa; The Graduate Studies Authority; The Committee of Doctorate Studies: Haifa, Israel, 2006. [Google Scholar]
- Ji, X.; Xie, C.; Ni, Z.; Yang, T.; Nevo, E.; Fahima, T.; Liu, Z.; Sun, Q. Identification and genetic mapping of a powdery mildew resistance gene in wild emmer (Triticum dicoccoides) accession IW72 from Israel. Euphytica 2008, 159, 385–390. [Google Scholar] [CrossRef]
- Zhang, H.; Guan, H.; Li, J.; Zhu, J.; Xie, C.; Zhou, Y.; Duan, X.; Yang, T.; Sun, Q.; Liu, Z. Genetic and comparative genomics mapping reveals that a powdery mildew resistance gene Ml3D232 originating from wild emmer co-segregates with an NBS-LRR analog in common wheat (Triticum aestivum L.). Theor. Appl. Genet. 2010, 121, 1613–1621. [Google Scholar] [CrossRef] [PubMed]
- Roelfs, A.P. Rust Diseases of Wheat: Concepts and Methods of Disease Management; Cimmyt: Mexico City, Mexico, 1992. [Google Scholar]
- McIntosh, R.; Silk, J. Cytogenetic studies in wheat XVII. Monosomic analysis and linkage relationships of gene Yr15 for resistance to stripe rust. Euphytica 1996, 89, 395–399. [Google Scholar]
- Zakari, A.; McIntosh, R.; Wellings, C.; Shariflou, M.; Hayden, M.; Bariana, H.; Hovmøller, M.S. Recombination of Yr15 and Yr24 in chromosome 1BS. In Recombination of Yr15 and Yr24 in Chromosome 1BS; International Wheat Genetics Symposium: Paestum, Italy, 2003. [Google Scholar]
- Peng, J.; Fahima, T.; Röder, M.; Li, Y.; Grama, A.; Nevo, E. Microsatellite high-density mapping of the stripe rust resistance gene YrH52 region on chromosome 1B and evaluation of its marker-assisted selection in the F 2 generation in wild emmer wheat. New Phytol. 2000, 146, 141–154. [Google Scholar] [CrossRef]
- Marais, G.; Pretorius, Z.; Wellings, C.; McCallum, B.; Marais, A. Leaf rust and stripe rust resistance genes transferred to common wheat from Triticum dicoccoides. Euphytica 2005, 143, 115–123. [Google Scholar] [CrossRef]
- Uauy, C.; Brevis, J.C.; Chen, X.; Khan, I.; Jackson, L.; Chicaiza, O.; Distelfeld, A.; Fahima, T.; Dubcovsky, J. High-temperature adult-plant (HTAP) stripe rust resistance gene Yr36 from Triticum turgidum ssp. dicoccoides is closely linked to the grain protein content locus Gpc-B1. Theor. Appl. Genet. 2005, 112, 97. [Google Scholar] [CrossRef]
- Dadkhodaie, N.; Karaoglou, H.; Wellings, C.; Park, R. Mapping genes Lr53 and Yr35 on the short arm of chromosome 6B of common wheat with microsatellite markers and studies of their association with Lr36. Theor. Appl. Genet. 2011, 122, 479–487. [Google Scholar] [CrossRef]
- Saccomanno, A.; Matny, O.; Marone, D.; Laidò, G.; Petruzzino, G.; Mazzucotelli, E.; Desiderio, F.; Blanco, A.; Gadaleta, A.; Pecchioni, N. Genetic mapping of loci for resistance to stem rust in a tetraploid wheat collection. Int. J. Mol. Sci. 2018, 19, 3907. [Google Scholar] [CrossRef]
- Otto, C.; Kianian, S.; Elias, E.; Stack, R.; Joppa, L. Genetic dissection of a major Fusarium head blight QTL in tetraploid wheat. Plant Mol. Biol. 2002, 48, 625–632. [Google Scholar] [CrossRef]
- Kumar, S.; Stack, R.; Friesen, T.; Faris, J. Identification of a novel Fusarium head blight resistance quantitative trait locus on chromosome 7A in tetraploid wheat. Phytopathology 2007, 97, 592–597. [Google Scholar] [CrossRef]
- Garvin, D.F.; Stack, R.W.; Hansen, J.M. Quantitative trait locus mapping of increased Fusarium head blight susceptibility associated with a wild emmer wheat chromosome. Phytopathology 2009, 99, 447–452. [Google Scholar] [CrossRef] [PubMed]
- Nevo, E.; Krugman, T.; Beiles, A. Genetic resources for salt tolerance in the wild progenitors of wheat (Triticum dicoccoides) and barley (Hordeum spontaneum) in Israel. Plant Breed. 1993, 110, 338–341. [Google Scholar] [CrossRef]
- Husain, S.; Munns, R.; Condon, A.T. Effect of sodium exclusion trait on chlorophyll retention and growth of durum wheat in saline soil. Aust. J. Agric. Res. 2003, 54, 589–597. [Google Scholar] [CrossRef]
- Peleg, Z.; Fahima, T.; Krugman, T.; Abbo, S.; Yakir, D.; Korol, A.B.; Saranga, Y. Genomic dissection of drought resistance in durum wheat× wild emmer wheat recombinant inbreed line population. Plant Cell Environ. 2009, 32, 758–779. [Google Scholar] [CrossRef] [PubMed]
- Krugman, T.; Chagué, V.; Peleg, Z.; Balzergue, S.; Just, J.; Korol, A.B.; Nevo, E.; Saranga, Y.; Chalhoub, B.; Fahima, T. Multilevel regulation and signalling processes associated with adaptation to terminal drought in wild emmer wheat. Funct. Integr. Genom. 2010, 10, 167–186. [Google Scholar] [CrossRef] [PubMed]
- Lucas, S.; Dogan, E.; Budak, H. TMPIT1 from wild emmer wheat: First characterisation of a stress-inducible integral membrane protein. Gene 2011, 483, 22–28. [Google Scholar] [CrossRef]
- Gerechter-Amitai, Z.; Grama, A. Use of alien genes in wheat breeding. Ann. Wheat Newsl. 1977, 23, 57–58. [Google Scholar]
- Joppa, L.; Du, C.; Hart, G.; Hareland, G. Mapping gene(s) for grain protein in tetraploid wheat (Triticum turgidum L.) using a population of recombinant inbred chromosome lines. Crop Sci. 1997, 37, 1586–1589. [Google Scholar] [CrossRef]
- Olmos, S.; Distelfeld, A.; Chicaiza, O.; Schlatter, A.; Fahima, T.; Echenique, V.; Dubcovsky, J. Precise mapping of a locus affecting grain protein content in durum wheat. Theor. Appl. Genet. 2003, 107, 1243–1251. [Google Scholar] [CrossRef]
- Gonzalez-Hernandez, J.; Elias, E.; Kianian, S. Mapping genes for grain protein concentration and grain yield on chromosome 5B of Triticum turgidum (L.) var. dicoccoides. Euphytica 2004, 139, 217–225. [Google Scholar] [CrossRef]
- Blanco, A.; Gadaleta, A.; Cenci, A.; Carluccio, A.; Abdelbacki, A.; Simeone, R. Molecular mapping of the novel powdery mildew resistance gene Pm36 introgressed from Triticum turgidum var. dicoccoides in durum wheat. Theor. Appl. Genet. 2008, 117, 135. [Google Scholar] [CrossRef] [PubMed]
- Klindworth, D.; Hareland, G.; Elias, E.; Faris, J.; Chao, S.; Xu, S. Agronomic and quality characteristics of two new sets of Langdon durum–wild emmer wheat chromosome substitution lines. J. Cereal Sci. 2009, 50, 29–35. [Google Scholar] [CrossRef]
- Chandrasekhar, K.; Nashef, K.; Ben-David, R. Agronomic and genetic characterization of wild emmer wheat (Triticum turgidum subsp. dicoccoides) introgression lines in a bread wheat genetic background. Genet. Resour. Crop Evol. 2017, 64, 1917–1926. [Google Scholar] [CrossRef]
- Ciaffi, M.; Lafiandra, D.; Porceddu, E.; Benedettelli, S. Storage-protein variation in wild emmer wheat (Triticum turgidum ssp. dicoccoides) from Jordan and Turkey. I. Electrophoretic characterization of genotypes. Theor. Appl. Genet. 1993, 86, 474–480. [Google Scholar] [CrossRef]
- Jiang, C.; Pei, Y.; Zhang, Y.; Li, X.; Yao, D.; Yan, Y.; Ma, W.; Hsam, S.; Zeller, F. Molecular cloning and characterization of four novel LMW glutenin subunit genes from Aegilops longissima, Triticum dicoccoides and T. zhukovskyi. Hereditas 2008, 145, 92–98. [Google Scholar] [CrossRef]
- Liang, X.; Zhen, S.; Han, C.; Wang, C.; Li, X.; Ma, W.; Yan, Y. Molecular characterization and marker development for hexaploid wheat-specific HMW glutenin subunit 1By18 gene. Mol. Breed. 2015, 35, 221. [Google Scholar] [CrossRef]
- Wang, J.-R.; Wei, Y.-M.; Deng, M.; Nevo, E.; Yan, Z.-H.; Zheng, Y.-L. The impact of single nucleotide polymorphism in monomeric alpha-amylase inhibitor genes from wild emmer wheat, primarily from Israel and Golan. BMC Evol. Biol. 2010, 10, 170. [Google Scholar] [CrossRef]
- Wang, Z.; Huang, L.; Wu, B.; Hu, J.; Jiang, Z.; Qi, P.; Zheng, Y.; Liu, D. Characterization of an integrated active Glu-1Ay allele in common wheat from wild emmer and its potential role in flour improvement. Int. J. Mol. Sci. 2018, 19, 923. [Google Scholar] [CrossRef]
- Yu, J.; Zhao, Y.; Ding, M.; Yu, Z.; Jiang, Y.; Ma, W.; Rong, J. Wild emmer chromosome arm substitution lines: Useful resources for wheat genetic study and breeding. Crop Sci. 2019. [Google Scholar] [CrossRef]
- Peleg, Z.; Cakmak, I.; Ozturk, L.; Yazici, A.; Jun, Y.; Budak, H.; Korol, A.B.; Fahima, T.; Saranga, Y. Quantitative trait loci conferring grain mineral nutrient concentrations in durum wheat× wild emmer wheat RIL population. Theor. Appl. Genet. 2009, 119, 353–369. [Google Scholar] [CrossRef]
- Schreiber, M.; Stein, N.; Mascher, M. Genomic approaches for studying crop evolution. Genome Biol. 2018, 19, 140. [Google Scholar] [CrossRef] [PubMed]
- Koenig, D.; Jiménez-Gómez, J.M.; Kimura, S.; Fulop, D.; Chitwood, D.H.; Headland, L.R.; Kumar, R.; Covington, M.F.; Devisetty, U.K.; Tat, A.V. Comparative transcriptomics reveals patterns of selection in domesticated and wild tomato. Proc. Natl. Acad. Sci. USA 2013, 110, E2655–E2662. [Google Scholar] [CrossRef] [PubMed]
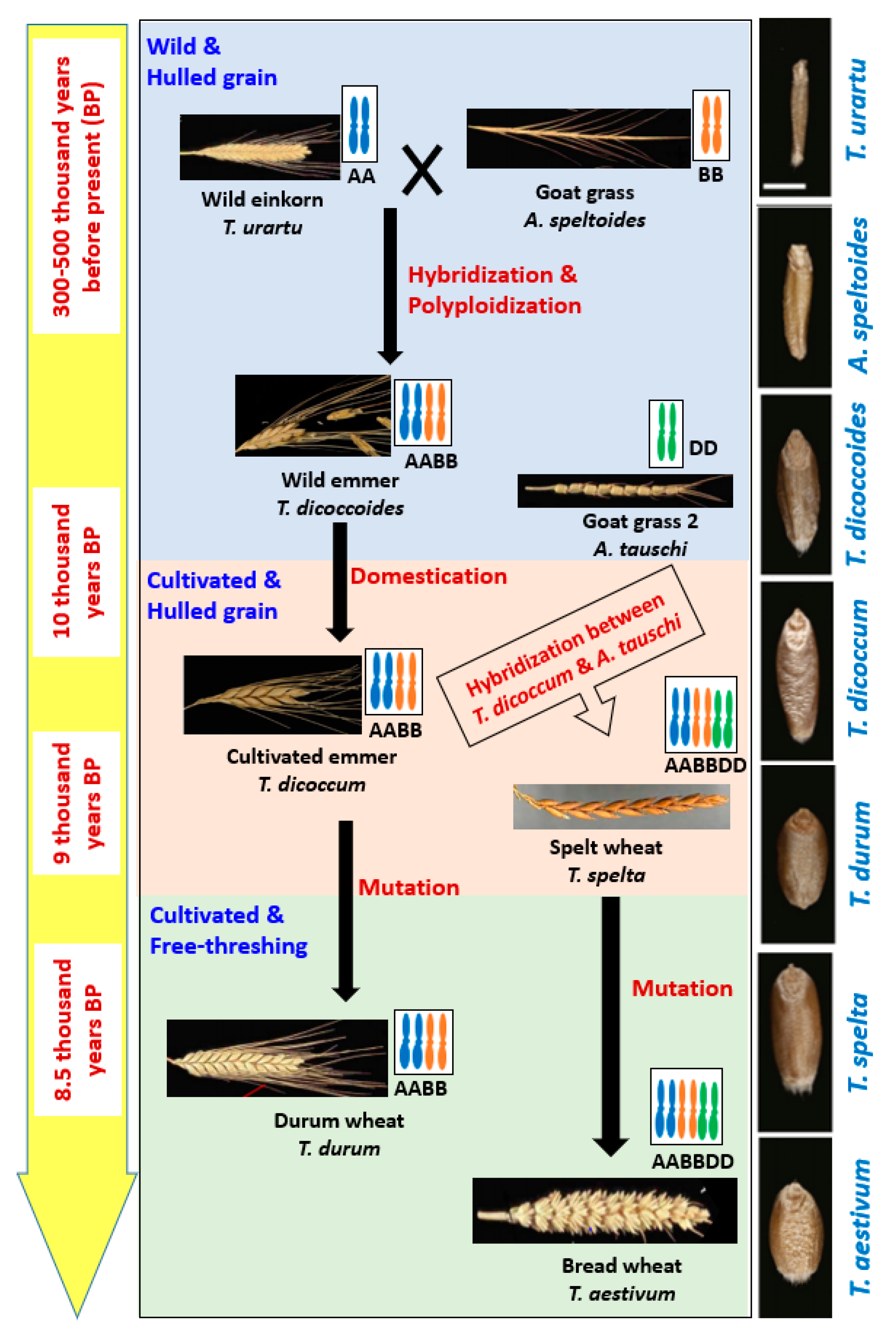
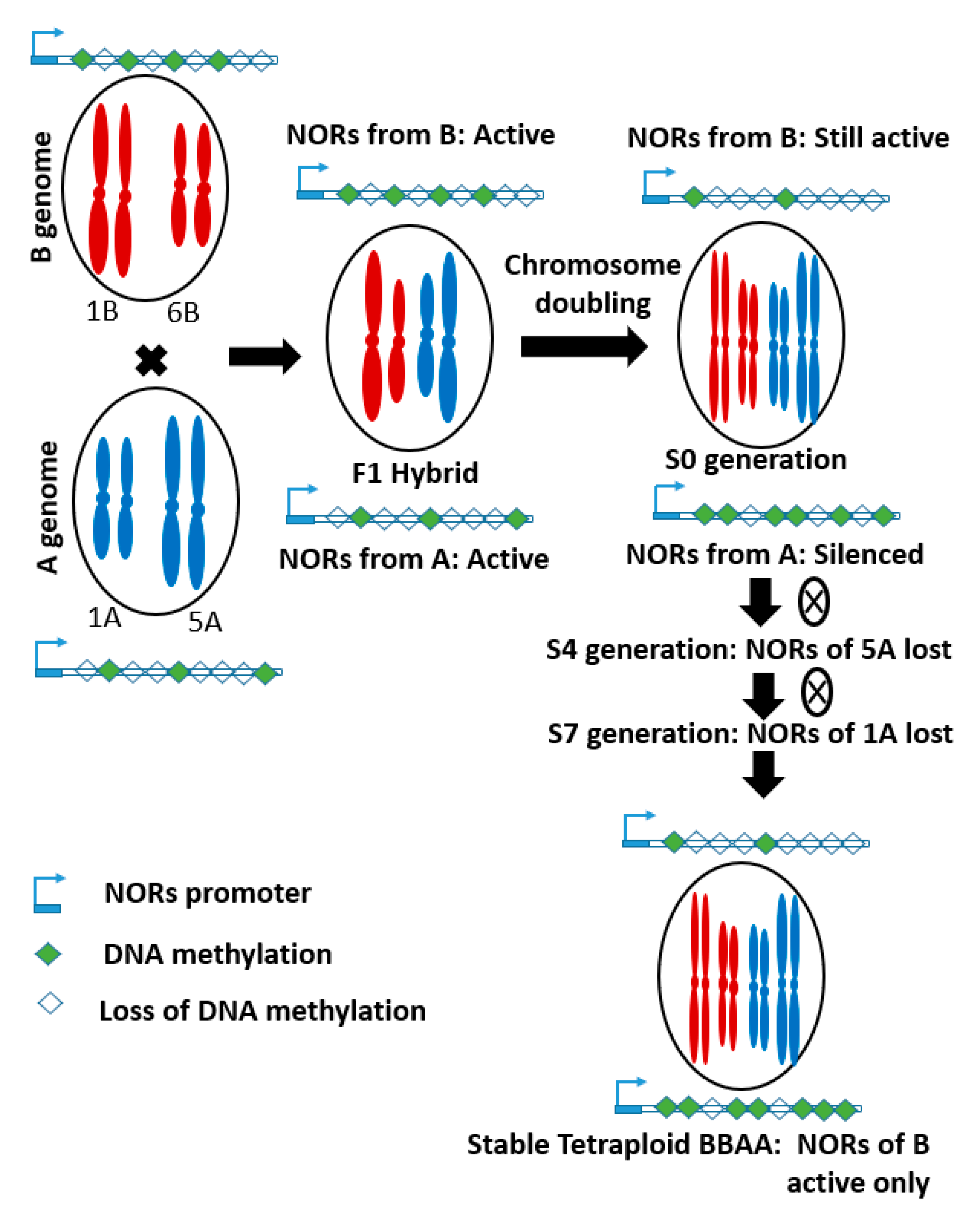

| Trait | Description of the Traits in Relation to Domestication | Gene Name | QTL Position |
|---|---|---|---|
| Brittle rachis [42,43] | This trait is agriculturally deleterious, and thus, the transformation of brittle rachis to non-Br is perhaps the first symbol of domestication in wheat. | Brittle rachis (Br1, Br2 and Br3) | 3DS, 3AS and 3BS |
| Glume tenacity [43,44] | The wild wheat floret is wrapped by tough glumes that make spikes difficult to thresh, whereas cultivated wheats have soft glumes and are free-threshing. | Tough glumed (Tg1) soft glumes (sog) | 2A, 2B, 2D, 5A, 6A, 6D and 7B |
| Free-threshing [1] | The Q gene is a major domestication gene conferring spike shape and threshability in wheat. Increased transcription of Q was associated with spike compactness and reduced plant height. | Threshability gene (Q gene) | 5AL |
| Seed size/weight [40,45,46] | Increase in seed size or weight took place before the evolution of non-shattering ears. The trait is under complex polygenic control for all domesticated cereals. | - | 1A, 1B, 2A, 3A, 3B, 4A, 4B, 5A, 5B, 6A, 6B, 7A, 7B |
| Seed shape [40] | Grain shape is an important attribute for ensuring market quality. Domestication has transformed long and thin primitive grains to wider and shorter modern grain. | Grain size (GS3) grain weight (GW2) seed width (SW5) | 1A, 3A, 4B, 5A, 6A |
| Flowering time [1,40,46] | Domestication involved selection of spring wheat that lack of vernalization and specific photoperiod requirement. The wild allele on 5A of T. dicoccoides, responsible for late-flowering, is similar to the VRN1 gene and also present in a collinear position with Ppd genes. | - | 2A, 4B, 5A, 6B |
| Grain yield [1,47,48] | Yield was considered to be one of the important traits for domestication which minimize the labor input and land needs. Yield QTL is overlapped with QTL for other traits. | - | 1B, 2A, 3A, 4A, 5A, 5B, |
| Plant height [1,46] | Though reduced plant height is desired for modern wheat breeding, tall mutants with higher biomass and yielding potential were historically selected. | - | 5A, 7B |
| Spike number/plant [1,46] | Spike number is strongly correlated with tillering capacity. A single recessive gene (tin) on 5A, controlling tiller number is assumed to be homologous with QTL for spike number on 1B of T. dicoccoides. | - | 1B, 2A, 2B, 5A, 7A |
| Spike weight/plant [1,46] | These all traits are highly correlated with each other and also with grain yield. QTL for these yield-related traits were found in different chromosomes, among them 5A, 2A and 1B had the most significant role in domestication. | - | 1B, 2A, 3A, 5A, 5B, 7A |
| Single spike weigh [1,46] | - | 1B, 2A, 3A, 5A | |
| Kernel number/plant [1,46] | - | 1B, 2A, 3A, 5A, 5B, 7A | |
| Kernel number/spike [1,46] | - | 1B, 2A, 3A, 5A, 6B | |
| Kernel number/spikelet [1,46] | - | 1B, 2A, 3A, 5A, 5B, 7B | |
| Spikelet number/spike [1,46] | - | 1B, 2A, 5A, 6B |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahman, S.; Islam, S.; Yu, Z.; She, M.; Nevo, E.; Ma, W. Current Progress in Understanding and Recovering the Wheat Genes Lost in Evolution and Domestication. Int. J. Mol. Sci. 2020, 21, 5836. https://doi.org/10.3390/ijms21165836
Rahman S, Islam S, Yu Z, She M, Nevo E, Ma W. Current Progress in Understanding and Recovering the Wheat Genes Lost in Evolution and Domestication. International Journal of Molecular Sciences. 2020; 21(16):5836. https://doi.org/10.3390/ijms21165836
Chicago/Turabian StyleRahman, Shanjida, Shahidul Islam, Zitong Yu, Maoyun She, Eviatar Nevo, and Wujun Ma. 2020. "Current Progress in Understanding and Recovering the Wheat Genes Lost in Evolution and Domestication" International Journal of Molecular Sciences 21, no. 16: 5836. https://doi.org/10.3390/ijms21165836
APA StyleRahman, S., Islam, S., Yu, Z., She, M., Nevo, E., & Ma, W. (2020). Current Progress in Understanding and Recovering the Wheat Genes Lost in Evolution and Domestication. International Journal of Molecular Sciences, 21(16), 5836. https://doi.org/10.3390/ijms21165836








