Pyridoxal 5′-Phosphate-Dependent Enzymes at the Crossroads of Host–Microbe Tryptophan Metabolism
Abstract
1. Introduction
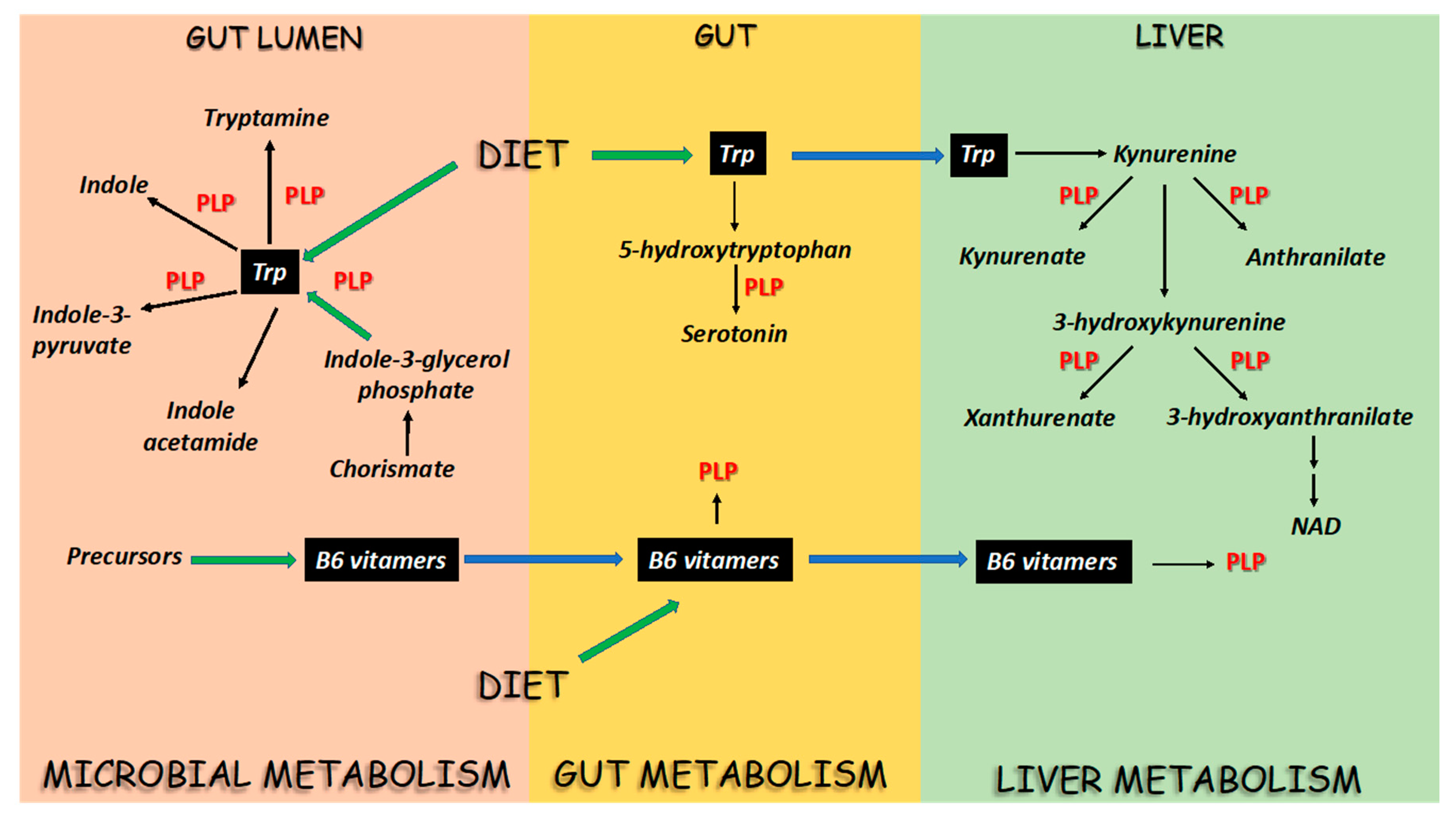
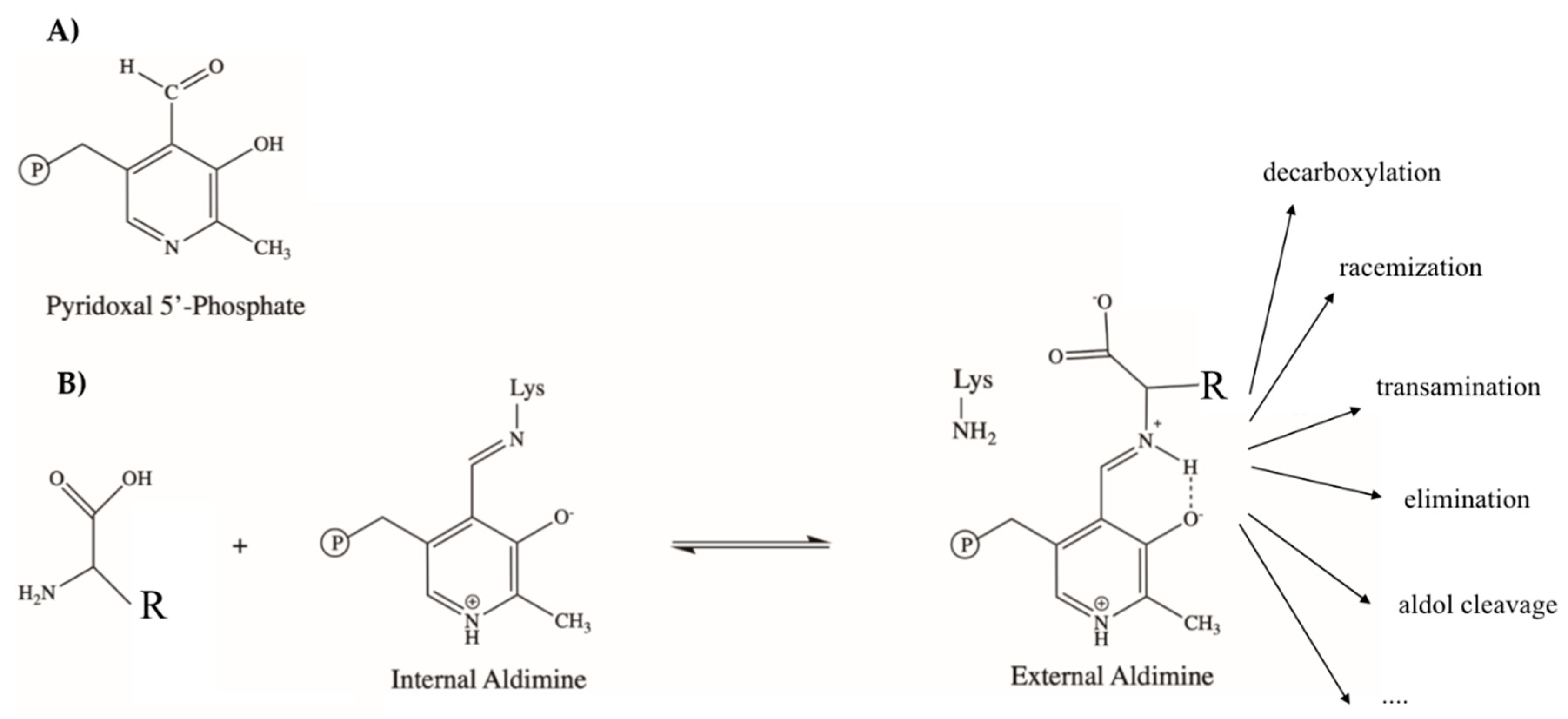
2. PLP-Dependent Enzymes in the Flux of Trp in Mammals and Microbes
3. A Biochemical Overview of Microbial and Host Trp-Metabolizing Enzymes
3.1. Tryptophan Synthase (TS)
3.2. Tryptophan Indole Lyase (Trpase)
3.3. Aromatic Amino Acid Aminotransferase (AroAT)
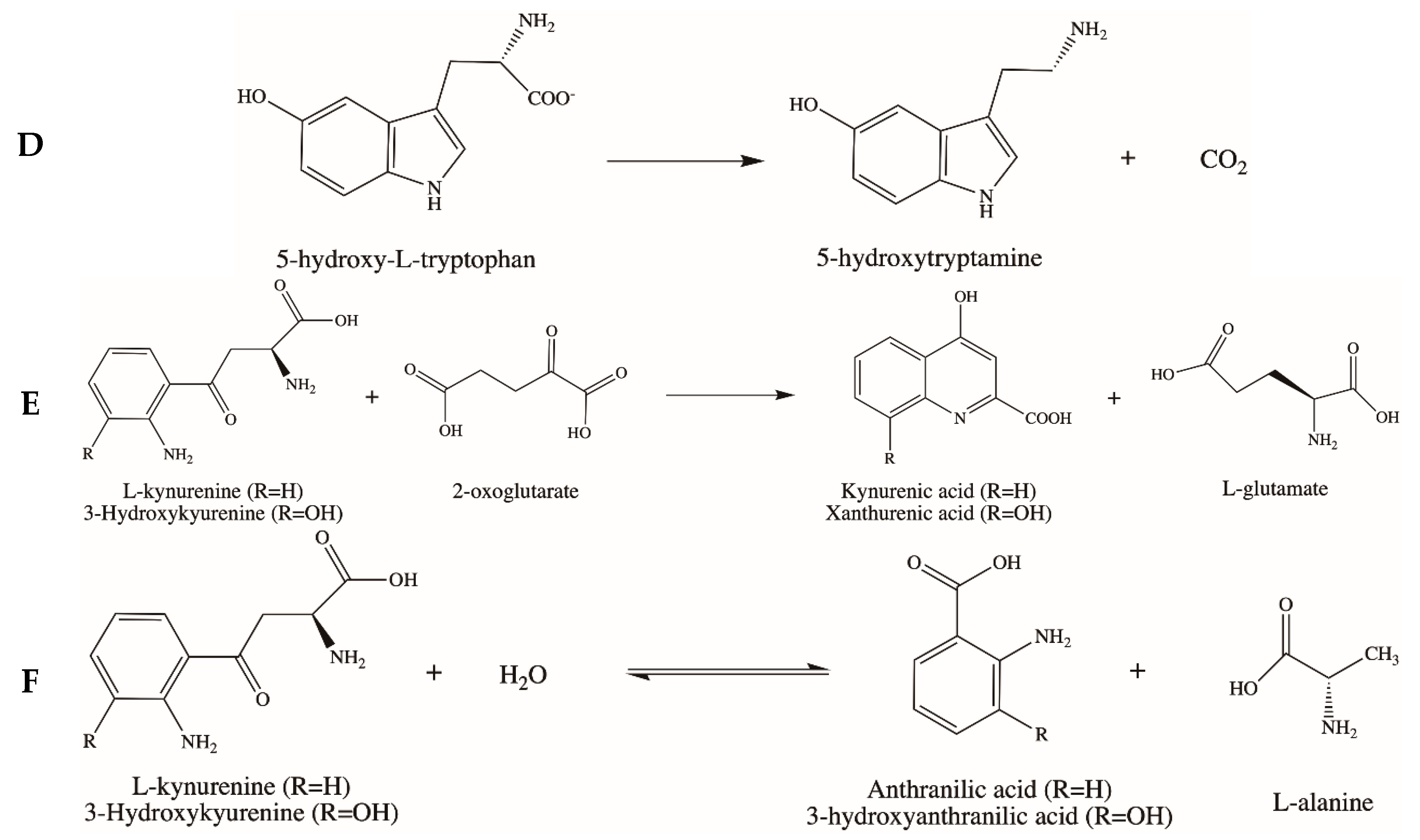
3.4. Aromatic Amino Acid Decarboxylase (AADC)
3.5. Kynurenine Aminotransferase (KAT) and Kynureninase (KYNU)
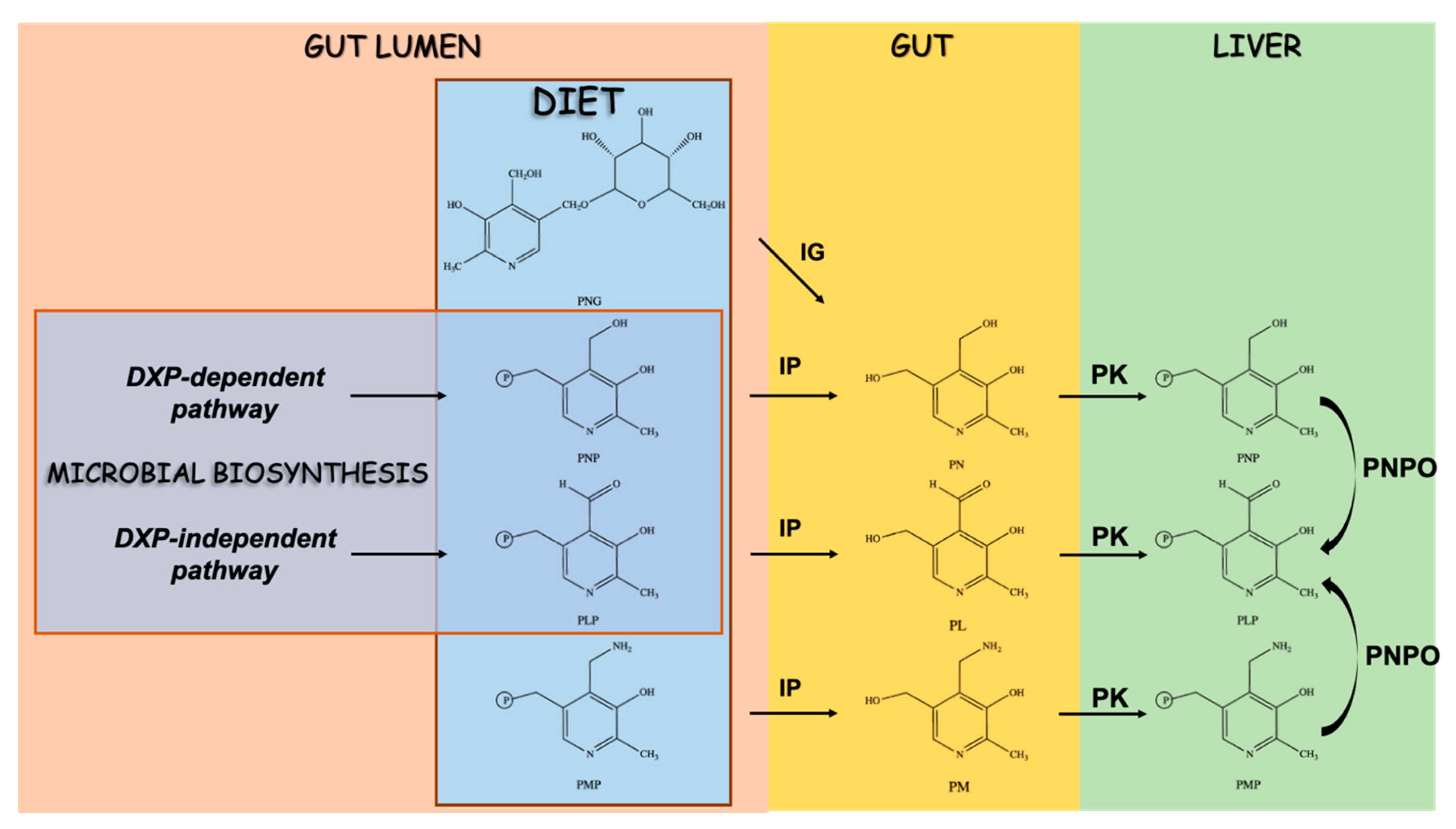
4. Vitamin B6 and the Regulation of Trp Flux in the Host and Microbes
5. Vitamin B6 Dysregulation in Diseases of the Gastrointestinal Tract
6. Vitamin B6 Dysregulation in Immunology
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| 3-hydroxykyn | 3-hydroxykynurenine |
| 5-hydroxyTrp | 5-hydroxytryptophan |
| AroAT | aromatic amino acid aminotransferases |
| AADC | aromatic-L-amino acid decarboxylase |
| AhR | aryl hydrocarbon receptor |
| CD | Crohn’s disease |
| CRISPR | clustered regularly interspaced short palindromic repeats |
| Cas9 | CRISPR-associated protein-9 nuclease |
| RT-PCR | real-time reverse transcription-polymerase chain reaction |
| ELISA | enzyme-linked immunosorbent assay |
| NF-κB | nuclear factor kappa-light-chain-enhancer of activated B cells |
| PAK1 | p21-activated kinase 1 |
| FAK | focal adhesion kinase |
| SDS-PAGE | sodium dodecyl sulfate polyacrylamide gel electrophoresis |
| HRP | horseradish peroxidase |
| EC | enterochromaffin cells |
| 3-IAld | indole-3-aldehyde |
| IDO | indoleamine-2,3-dioxygenase |
| IBS | irritable bowel syndrome |
| KYNU | kynureninase |
| KA | kynurenic acid |
| Kyn | kynurenine |
| KAT | kynurenine aminotransferase |
| KP | kynurenine pathway |
| PL | pyridoxal |
| PM | pyridoxamine |
| PMP | pyridoxamine phosphate |
| PN | pyridoxine |
| PNG | pyridoxine glucoside |
| PNP | pyridoxine phosphate |
| TDO | tryptophan-2,3-dioxygenase |
| Tph | tryptophan hydroxylase |
| Trpase | tryptophan indole lyase |
| TS | tryptophan synthase |
| UC | ulcerative colitis |
| XA | xanthurenic acid |
References
- Palego, L.; Betti, L.; Rossi, A.; Giannaccini, G. Tryptophan biochemistry: Structural, nutritional, metabolic, and medical aspects in humans. J. Amino Acids. 2016, 2016, 8952520. [Google Scholar] [CrossRef] [PubMed]
- Platten, M.; Nollen, E.A.A.; Rohrig, U.F.; Fallarino, F.; Opitz, C.A. Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat. Rev. Drug. Discov. 2019, 18, 379–401. [Google Scholar] [CrossRef] [PubMed]
- Costantini, C.; Bellet, M.M.; Renga, G.; Stincardini, C.; Borghi, M.; Pariano, M.; Cellini, B.; Keller, N.; Romani, L.; Zelante, T. Tryptophan Co-Metabolism at the Host-Pathogen interface. Front. Immunol. 2020, 11, 67. [Google Scholar] [CrossRef] [PubMed]
- Agus, A.; Planchais, J.; Sokol, H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell. Host. Microbe. 2018, 23, 716–724. [Google Scholar] [CrossRef]
- Wilson, M.P.; Plecko, B.; Mills, P.B.; Clayton, P.T. Disorders affecting vitamin B6 metabolism. J. Inherit. Metab. Dis. 2019, 42, 629–646. [Google Scholar]
- Percudani, R.; Peracchi, A. The B6 database: A tool for the description and classification of vitamin B6-dependent enzymatic activities and of the corresponding protein families. BMC Bioinform. 2009, 10, 273. [Google Scholar] [CrossRef]
- Mir, R.; Jallu, S.; Singh, T.P. The shikimate pathway: Review of amino acid sequence, function and three-dimensional structures of the enzymes. Crit. Rev. Microbiol. 2015, 41, 172–189. [Google Scholar] [CrossRef]
- Dunn, M.F.; Niks, D.; Ngo, H.; Barends, T.R.; Schlichting, I. Tryptophan synthase: The workings of a channeling nanomachine. Trends Biochem. Sci. 2008, 33, 254–264. [Google Scholar] [CrossRef]
- Van der Leek, A.P.; Yanishevsky, Y.; Kozyrskyj, A.L. The kynurenine pathway as a novel link between allergy and the gut microbiome. Front. Immunol. 2017, 8, 1374. [Google Scholar] [CrossRef]
- Svetlana, A.D. Epilepsy as a Pyridoxine-Dependent Condition: Quantitative Urinary Biomarkers of Epilepsy. Family Disorders of Pyridoxine Metabolism. 2016. Available online: https://www.intechopen.com/books/epileptology-the-modern-state-of-science/epilepsy-as-a-pyridoxine-dependent-condition-quantitative-urinary-biomarkers-of-epilepsy-family-diso (accessed on 13 August 2020).
- Thomas, S.R.; Salahifar, H.; Mashima, R.; Hunt, N.H.; Richardson, D.R.; Stocker, R. Antioxidants inhibit indoleamine 2,3-dioxygenase in IFN-gamma-activated human macrophages: Posttranslational regulation by pyrrolidine dithiocarbamate. J. Immunol. 2001, 166, 6332–6340. [Google Scholar] [CrossRef]
- Nelp, M.T.; Kates, P.A.; Hunt, J.T.; Newitt, J.A.; Balog, A.; Maley, D.; Zhu, X.; Abell, L.; Allentoff, A.; Borzilleri, R.; et al. Immune-modulating enzyme indoleamine 2,3-dioxygenase is effectively inhibited by targeting its apo-form. Proc. Natl. Acad. Sci. USA 2018, 115, 3249–3254. [Google Scholar] [CrossRef] [PubMed]
- Ueland, P.M.; Ulvik, A.; Rios-Avila, L.; Midttun, O.; Gregory, J.F. Direct and functional biomarkers of Vitamin B6 Status. Annu. Rev. Nutr. 2015, 35, 33–70. [Google Scholar] [CrossRef] [PubMed]
- Vitamin B6 deficiency and tryptophan metabolism. Nutr. Rev. 1963, 21, 89–91. Available online: https://academic.oup.com/nutritionreviews/article-abstract/21/3/89/1877909? (accessed on 13 August 2020).
- Yeh, J.K.; Brown, R.R. Effects of vitamin B-6 deficiency and tryptophan loading on urinary excretion of tryptophan metabolites in mammals. J. Nutr. 1977, 107, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Rios-Avila, L.; Nijhout, H.F.; Reed, M.C.; Sitren, H.S.; Gregory, J.F., 3rd. A mathematical model of tryptophan metabolism via the kynurenine pathway provides insights into the effects of vitamin B-6 deficiency, tryptophan loading, and induction of tryptophan 2,3-dioxygenase on tryptophan metabolites. J. Nutr. 2013, 143, 1509–1519. [Google Scholar] [CrossRef]
- Ulvik, A.; Theofylaktopoulou, D.; Midttun, O.; Nygard, O.; Eussen, S.J.; Ueland, P.M. Substrate product ratios of enzymes in the kynurenine pathway measured in plasma as indicators of functional vitamin B-6 status. Am. J. Clin. Nutr. 2013, 98, 934–940. [Google Scholar] [CrossRef]
- Ulvik, A.; Midttun, O.; McCann, A.; Meyer, K.; Tell, G.; Nygard, O.; Ueland, P.M. Tryptophan catabolites as metabolic markers of vitamin B-6 status evaluated in cohorts of healthy adults and cardiovascular patients. Am. J. Clin. Nutr. 2020, 111, 178–186. [Google Scholar] [CrossRef]
- Hvas, A.M.; Juul, S.; Bech, P.; Nexo, E. Vitamin B6 level is associated with symptoms of depression. Psychother. Psychosom. 2004, 73, 340–343. [Google Scholar] [CrossRef]
- Roager, H.M.; Licht, T.R. Microbial tryptophan catabolites in health and disease. Nat. Commun. 2018, 9, 3294. [Google Scholar] [CrossRef]
- Stover, P.J.; Field, M.S. Vitamin B-6. Adv. Nutr. 2015, 6, 132–133. [Google Scholar] [CrossRef]
- Liang, J.; Han, Q.; Tan, Y.; Ding, H.; Li, J. Current advances on structure-function relationships of Pyridoxal 5′-Phosphate-Dependent Enzymes. Front. Mol. Biosci. 2019, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Eliot, A.C.; Kirsch, J.F. Pyridoxal phosphate enzymes: Mechanistic, structural, and evolutionary considerations. Annu. Rev. Biochem. 2004, 73, 383–415. [Google Scholar] [CrossRef] [PubMed]
- Toney, M.D. Controlling reaction specificity in pyridoxal phosphate enzymes. Biochim. Biophys. Acta. 2011, 1814, 1407–1418. [Google Scholar] [CrossRef] [PubMed]
- Percudani, R.; Peracchi, A. A genomic overview of pyridoxal-phosphate-dependent enzymes. EMBO Rep. 2003, 4, 850–854. [Google Scholar] [CrossRef]
- Dunathan, H.C. Conformation and reaction specificity in pyridoxal phosphate enzymes. Proc. Natl. Acad. Sci. USA 1966, 55, 712–716. [Google Scholar] [CrossRef]
- Cellini, B.; Montioli, R.; Oppici, E.; Astegno, A.; Voltattorni, C.B. The chaperone role of the pyridoxal 5′-phosphate and its implications for rare diseases involving B6-dependent enzymes. Clin. Biochem. 2014, 47, 158–165. [Google Scholar] [CrossRef]
- Contestabile, R.; di Salvo, M.L.; Bunik, V.; Tramonti, A.; Verni, F. The multifaceted role of vitamin B6 in cancer: Drosophila as a model system to investigate DNA damage. Open. Biol. 2020, 10, 200034. [Google Scholar] [CrossRef]
- Ueland, P.M.; McCann, A.; Midttun, O.; Ulvik, A. Inflammation, vitamin B6 and related pathways. Mol. Aspects. Med. 2017, 53, 10–27. [Google Scholar] [CrossRef]
- Amadasi, A.; Bertoldi, M.; Contestabile, R.; Bettati, S.; Cellini, B.; di Salvo, M.L.; Borri-Voltattorni, C.; Bossa, F.; Mozzarelli, A. Pyridoxal 5′-phosphate enzymes as targets for therapeutic agents. Curr. Med. Chem. 2007, 14, 1291–1324. [Google Scholar] [CrossRef]
- Michalska, K.; Gale, J.; Joachimiak, G.; Chang, C.; Hatzos-Skintges, C.; Nocek, B.; Johnston, S.E.; Bigelow, L.; Bajrami, B.; Jedrzejczak, R.P.; et al. Conservation of the structure and function of bacterial tryptophan synthases. IUCr J. 2019, 6, 649–664. [Google Scholar] [CrossRef]
- Ro, H.S.; Miles, E.W. Structure and function of the tryptophan synthase alpha(2)beta(2) complex. Roles of beta subunit histidine 86. J. Biol. Chem. 1999, 274, 36439–36445. [Google Scholar] [CrossRef]
- Merino, E.; Jensen, R.A.; Yanofsky, C. Evolution of bacterial Trp operons and their regulation. Curr. Opin. Microbiol. 2008, 11, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Dunn, M.F. Allosteric regulation of substrate channeling and catalysis in the tryptophan synthase bienzyme complex. Arch. Biochem. Biophys. 2012, 519, 154–166. [Google Scholar] [CrossRef]
- Peracchi, A.; Bettati, S.; Mozzarelli, A.; Rossi, G.L.; Miles, E.W.; Dunn, M.F. Allosteric regulation of tryptophan synthase: Effects of pH, temperature, and alpha-subunit ligands on the equilibrium distribution of pyridoxal 5′-phosphate-L-serine intermediates. Biochemistry 1996, 35, 1872–1880. [Google Scholar] [CrossRef] [PubMed]
- Hyde, C.C.; Ahmed, S.A.; Padlan, E.A.; Miles, E.W.; Davies, D.R. Three-dimensional structure of the tryptophan synthase alpha 2 beta 2 multienzyme complex from Salmonella typhimurium. J. Biol. Chem. 1988, 263, 17857–17871. [Google Scholar] [PubMed]
- Peracchi, A.; Mozzarelli, A.; Rossi, G.L. Monovalent cations affect dynamic and functional properties of the tryptophan synthase alpha 2 beta 2 complex. Biochemistry 1995, 34, 9459–9465. [Google Scholar] [CrossRef]
- Woehl, E.; Dunn, M.F. Mechanisms of monovalent cation action in enzyme catalysis: The first stage of the tryptophan synthase beta-reaction. Biochemistry 1999, 38, 7118–7130. [Google Scholar] [CrossRef]
- Sarsero, J.P.; Merino, E.; Yanofsky, C. A Bacillus subtilis operon containing genes of unknown function senses tRNATrp charging and regulates expression of the genes of tryptophan biosynthesis. Proc. Natl. Acad. Sci. USA 2000, 97, 2656–2661. [Google Scholar] [CrossRef]
- Gutierrez-Preciado, A.; Yanofsky, C.; Merino, E. Comparison of tryptophan biosynthetic operon regulation in different Gram-positive bacterial species. Trends Genet. 2007, 23, 422–426. [Google Scholar] [CrossRef]
- Wellington, S.; Nag, P.P.; Michalska, K.; Johnston, S.E.; Jedrzejczak, R.P.; Kaushik, V.K.; Clatworthy, A.E.; Siddiqi, N.; McCarren, P.; Bajrami, B.; et al. A small-molecule allosteric inhibitor of Mycobacterium tuberculosis tryptophan synthase. Nat. Chem. Biol. 2017, 13, 943–950. [Google Scholar] [CrossRef] [PubMed]
- Michalska, K.; Chang, C.; Maltseva, N.I.; Jedrzejczak, R.; Robertson, G.T.; Gusovsky, F.; McCarren, P.; Schreiber, S.L.; Nag, P.P.; Joachimiak, A. Allosteric inhibitors of Mycobacterium tuberculosis tryptophan synthase. Protein. Sci. 2020, 29, 779–788. [Google Scholar] [CrossRef] [PubMed]
- Dodd, D.; Spitzer, M.H.; Van Treuren, W.; Merrill, B.D.; Hryckowian, A.J.; Higginbottom, S.K.; Le, A.; Cowan, T.M.; Nolan, G.P.; Fischbach, M.A.; et al. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 2017, 551, 648–652. [Google Scholar] [CrossRef] [PubMed]
- Negatu, D.A.; Liu, J.J.J.; Zimmerman, M.; Kaya, F.; Dartois, V.; Aldrich, C.C.; Gengenbacher, M.; Dick, T. Whole-cell screen of fragment library identifies gut microbiota metabolite indole propionic acid as antitubercular. Antimicrob. Agents. Chemother. 2018, 62, e01571-17. [Google Scholar] [CrossRef] [PubMed]
- Negatu, D.A.; Yamada, Y.; Xi, Y.; Go, M.L.; Zimmerman, M.; Ganapathy, U.; Dartois, V.; Gengenbacher, M.; Dick, T. Gut microbiota metabolite indole propionic acid targets tryptophan biosynthesis in mycobacterium tuberculosis. mBio 2019, 10, e02781-18. [Google Scholar] [CrossRef]
- Nishio, K.; Ogasahara, K.; Morimoto, Y.; Tsukihara, T.; Lee, S.J.; Yutani, K. Large conformational changes in the Escherichia coli tryptophan synthase beta(2) subunit upon pyridoxal 5′-phosphate binding. FEBS J. 2010, 277, 2157–2170. [Google Scholar] [CrossRef]
- Orozco-Gomez, D.I.; Sosa-Hernandez, J.E.; Gallardo-Navarro, O.A.; Santana-Solano, J.; Santillan, M. Bistable behaviour and medium-dependent post-translational regulation of the tryptophanase operon regulatory pathway in Echerichia coli. Sci. Rep. 2019, 9, 5451. [Google Scholar] [CrossRef]
- Phillips, R.S.; Demidkina, T.V.; Faleev, N.G. Structure and mechanism of tryptophan indole-lyase and tyrosine phenol-lyase. Biochim. Biophys. Acta 2003, 1647, 167–172. [Google Scholar] [CrossRef]
- Demidkina, T.V.; Antson, A.A.; Faleev, N.G.; Phillips, R.S.; Zakomirdina, L.N. Spatial structure and the mechanism of tyrosine phenol-lyase and tryptophan indole-lyase. Molecular. Biol. 2009, 43, 269–283. [Google Scholar] [CrossRef]
- Isupov, M.N.; Antson, A.A.; Dodson, E.J.; Dodson, G.G.; Dementieva, I.S.; Zakomirdina, L.N.; Wilson, K.S.; Dauter, Z.; Lebedev, A.A.; Harutyunyan, E.H. Crystal structure of tryptophanase. J. Mol. Biol. 1998, 276, 603–623. [Google Scholar] [CrossRef]
- Jansonius, J.N. Structure, evolution and action of vitamin B6-dependent enzymes. Curr. Opin. Struct. Biol. 1998, 8, 759–769. [Google Scholar] [CrossRef]
- Honda, T.; Tokushige, M. Effects of temperature and monovalent cations on activity and quaternary structure of tryptophanase. J. Biochem. 1986, 100, 679–685. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Young, K.D. Indole production by the tryptophanase TnaA in Escherichia coli is determined by the amount of exogenous tryptophan. Microbiology 2013, 159, 402–410. [Google Scholar] [CrossRef] [PubMed]
- Does coenzyme binding determine enzyme stability? Nutr. Rev. 1978, 36, 251–254. Available online: https://academic.oup.com/nutritionreviews/article-abstract/36/8/251/1859914?redirectedFrom=fulltext (accessed on 13 August 2020).
- Di Salvo, M.L.; Contestabile, R.; Safo, M.K. Vitamin B(6) salvage enzymes: Mechanism, structure and regulation. Biochim. Biophys. Acta 2011, 1814, 1597–1608. [Google Scholar] [CrossRef] [PubMed]
- Phillips, R.S. Chemistry and diversity of pyridoxal-5′-phosphate dependent enzymes. Biochim. Biophys. Acta 2015, 1854, 1167–1174. [Google Scholar] [CrossRef]
- Hayashi, H.; Inoue, K.; Nagata, T.; Kuramitsu, S.; Kagamiyama, H. Escherichia coli aromatic amino acid aminotransferase: Characterization and comparison with aspartate aminotransferase. Biochemistry 1993, 32, 12229–12239. [Google Scholar] [CrossRef]
- Kulkarni, G.B.; Nayak, A.S.; Sajjan, S.S.; Oblesha, A.; Karegoudar, T.B. Indole-3-acetic acid biosynthetic pathway and aromatic amino acid aminotransferase activities in Pantoea dispersa strain GPK. Lett. Appl. Microbiol. 2013, 56, 340–347. [Google Scholar] [CrossRef]
- Paris, C.G.; Magasanik, B. Purification and properties of aromatic amino acid aminotransferase from Klebsiella aerogenes. J. Bacteriol. 1981, 145, 266–271. [Google Scholar] [CrossRef]
- Fazel, A.M.; Jensen, R.A. Aromatic aminotransferases in coryneform bacteria. J. Bacteriol. 1979, 140, 580–587. [Google Scholar] [CrossRef]
- Mavrides, C.; Orr, W. Multispecific aspartate and aromatic amino acid aminotransferases in Escherichia coli. J. Biol. Chem. 1975, 250, 4128–4133. [Google Scholar]
- Whitaker, R.J.; Gaines, C.G.; Jensen, R.A. A multispecific quintet of aromatic aminotransferases that overlap different biochemical pathways in Pseudomonas aeruginosa. J. Biol. Chem. 1982, 257, 13550–13556. [Google Scholar] [PubMed]
- Liu, Y.; Hou, Y.; Wang, G.; Zheng, X.; Hao, H. Gut Microbial Metabolites of Aromatic Amino Acids as Signals in Host-Microbe Interplay. Trends Endocrinol. Metab. 2020. [Google Scholar] [CrossRef] [PubMed]
- Rzad, K.; Milewski, S.; Gabriel, I. Versatility of putative aromatic aminotransferases from Candida albicans. Fungal. Genet. Biol. 2018, 110, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Karsten, W.E.; Reyes, Z.L.; Bobyk, K.D.; Cook, P.F.; Chooback, L. Mechanism of the aromatic aminotransferase encoded by the Aro8 gene from Saccharomyces cerevisiae. Arch. Biochem. Biophys. 2011, 516, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Bulfer, S.L.; Brunzelle, J.S.; Trievel, R.C. Crystal structure of Saccharomyces cerevisiae Aro8, a putative alpha-aminoadipate aminotransferase. Protein. Sci. 2013, 22, 1417–1424. [Google Scholar] [CrossRef] [PubMed]
- Iraqui, I.; Vissers, S.; Cartiaux, M.; Urrestarazu, A. Characterisation of Saccharomyces cerevisiae ARO8 and ARO9 genes encoding aromatic aminotransferases I and II reveals a new aminotransferase subfamily. Mol. Gen. Genet. 1998, 257, 238–248. [Google Scholar] [CrossRef]
- Ohashi, K.; Chaleckis, R.; Takaine, M.; Wheelock, C.E.; Yoshida, S. Kynurenine aminotransferase activity of Aro8/Aro9 engage tryptophan degradation by producing kynurenic acid in Saccharomyces cerevisiae. Sci. Rep. 2017, 7, 12180. [Google Scholar] [CrossRef]
- Stanley, J.C.; Nicholas, A.R.; Dickson, A.J.; Thompson, I.M.; Pogson, C.I. Tryptophan aminotransferase activity in rat liver. Biochem. J. 1984, 220, 341–344. [Google Scholar] [CrossRef]
- Bertoldi, M. Mammalian Dopa decarboxylase: Structure, catalytic activity and inhibition. Arch. Biochem. Biophys. 2014, 546, 1–7. [Google Scholar] [CrossRef]
- Bertoldi, M.; Borri Voltattorni, C. Reaction and substrate specificity of recombinant pig kidney Dopa decarboxylase under aerobic and anaerobic conditions. Biochim. Biophys. Acta 2003, 1647, 42–47. [Google Scholar] [CrossRef]
- Daidone, F.; Montioli, R.; Paiardini, A.; Cellini, B.; Macchiarulo, A.; Giardina, G.; Bossa, F.; Borri Voltattorni, C. Identification by virtual screening and in vitro testing of human DOPA decarboxylase inhibitors. PLoS ONE 2012, 7, e31610. [Google Scholar] [CrossRef] [PubMed]
- Burkhard, P.; Dominici, P.; Borri-Voltattorni, C.; Jansonius, J.N.; Malashkevich, V.N. Structural insight into Parkinson’s disease treatment from drug-inhibited DOPA decarboxylase. Nat. Struct. Biol. 2001, 8, 963–967. [Google Scholar] [CrossRef] [PubMed]
- Bertoldi, M.; Voltattorni, C.B. Multiple roles of the active site lysine of Dopa decarboxylase. Arch. Biochem. Biophys. 2009, 488, 130–139. [Google Scholar] [CrossRef]
- Bertoldi, M.; Gonsalvi, M.; Contestabile, R.; Voltattorni, C.B. Mutation of tyrosine 332 to phenylalanine converts dopa decarboxylase into a decarboxylation-dependent oxidative deaminase. J. Biol. Chem. 2002, 277, 36357–36362. [Google Scholar] [CrossRef]
- Williams, B.B.; Van Benschoten, A.H.; Cimermancic, P.; Donia, M.S.; Zimmermann, M.; Taketani, M.; Ishihara, A.; Kashyap, P.C.; Fraser, J.S.; Fischbach, M.A. Discovery and characterization of gut microbiota decarboxylases that can produce the neurotransmitter tryptamine. Cell Host Microbe 2014, 16, 495–503. [Google Scholar] [CrossRef]
- Montioli, R.; Oppici, E.; Cellini, B.; Roncador, A.; Dindo, M.; Voltattorni, C.B. S250F variant associated with aromatic amino acid decarboxylase deficiency: Molecular defects and intracellular rescue by pyridoxine. Hum. Mol. Genet 2013, 22, 1615–1624. [Google Scholar] [CrossRef]
- Giardina, G.; Montioli, R.; Gianni, S.; Cellini, B.; Paiardini, A.; Voltattorni, C.B.; Cutruzzola, F. Open conformation of human DOPA decarboxylase reveals the mechanism of PLP addition to Group II decarboxylases. Proc. Natl. Acad. Sci. USA 2011, 108, 20514–20519. [Google Scholar] [CrossRef]
- Matsuda, N.; Hayashi, H.; Miyatake, S.; Kuroiwa, T.; Kagamiyama, H. Instability of the apo form of aromatic L-amino acid decarboxylase in vivo and in vitro: Implications for the involvement of the flexible loop that covers the active site. J. Biochem. 2004, 135, 33–42. [Google Scholar] [CrossRef]
- Mawe, G.M.; Hoffman, J.M. Serotonin signalling in the gut--functions, dysfunctions and therapeutic targets. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 473–486. [Google Scholar] [CrossRef]
- Bader, M. Inhibition of serotonin synthesis: A novel therapeutic paradigm. Pharmacol. Ther. 2020, 205, 107423. [Google Scholar] [CrossRef]
- Allen, G.F.; Neergheen, V.; Oppenheim, M.; Fitzgerald, J.C.; Footitt, E.; Hyland, K.; Clayton, P.T.; Land, J.M.; Heales, S.J. Pyridoxal 5′-phosphate deficiency causes a loss of aromatic L-amino acid decarboxylase in patients and human neuroblastoma cells, implications for aromatic L-amino acid decarboxylase and vitamin B(6) deficiency states. J. Neurochem. 2010, 114, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Bender, D.A. Effects of oestradiol and vitamin B6 on tryptophan metabolism in the rat: Implications for the interpretation of the tryptophan load test for vitamin B6 nutritional status. Br. J. Nutr. 1983, 50, 33–42. [Google Scholar] [CrossRef]
- Robins, E.; Robins, J.M.; Croninger, A.B.; Moses, S.G.; Spencer, S.J.; Hudgens, R.W. The low level of 5-hydroxytryptophan decarboxylase in human brain. Biochem. Med. 1967, 1, 240–251. [Google Scholar] [CrossRef]
- Bhattarai, Y.; Williams, B.B.; Battaglioli, E.J.; Whitaker, W.R.; Till, L.; Grover, M.; Linden, D.R.; Akiba, Y.; Kandimalla, K.K.; Zachos, N.C.; et al. Gut microbiota-produced tryptamine activates an epithelial g-protein-coupled receptor to increase colonic secretion. Cell Host Microbe 2018, 23, 775–785.e5. [Google Scholar] [CrossRef]
- Young, S.N.; Anderson, G.M.; Gauthier, S.; Purdy, W.C. The origin of indoleacetic acid and indolepropionic acid in rat and human cerebrospinal fluid. J. Neurochem. 1980, 34, 1087–1092. [Google Scholar] [CrossRef]
- Furukawa, S.; Usuda, K.; Abe, M.; Ogawa, I. Effect of indole-3-acetic acid derivatives on neuroepithelium in rat embryos. J. Toxicol. Sci. 2005, 30, 165–174. [Google Scholar] [CrossRef]
- Badawy, A.A. Kynurenine pathway of tryptophan metabolism: Regulatory and functional aspects. Int. J. Tryptophan. Res. 2017, 10, 1178646917691938. [Google Scholar] [CrossRef]
- DiNatale, B.C.; Murray, I.A.; Schroeder, J.C.; Flaveny, C.A.; Lahoti, T.S.; Laurenzana, E.M.; Omiecinski, C.J.; Perdew, G.H. Kynurenic acid is a potent endogenous aryl hydrocarbon receptor ligand that synergistically induces interleukin-6 in the presence of inflammatory signaling. Toxicol. Sci. 2010, 115, 89–97. [Google Scholar] [CrossRef]
- Han, Q.; Cai, T.; Tagle, D.A.; Li, J. Structure, expression, and function of kynurenine aminotransferases in human and rodent brains. Cell Mol. Life Sci. 2010, 67, 353–368. [Google Scholar] [CrossRef]
- Han, Q.; Li, J.; Li, J. pH dependence, substrate specificity and inhibition of human kynurenine aminotransferase I. Eur. J. Biochem. 2004, 271, 4804–4814. [Google Scholar] [CrossRef]
- Han, Q.; Robinson, H.; Cai, T.; Tagle, D.A.; Li, J. Biochemical and structural properties of mouse kynurenine aminotransferase III. Mol. Cell. Biol. 2009, 29, 784–793. [Google Scholar] [CrossRef]
- Han, Q.; Cai, T.; Tagle, D.A.; Robinson, H.; Li, J. Substrate specificity and structure of human aminoadipate aminotransferase/kynurenine aminotransferase II. Biosci. Rep. 2008, 28, 205–215. [Google Scholar] [CrossRef]
- Xu, H.; Andi, B.; Qian, J.; West, A.H.; Cook, P.F. The alpha-aminoadipate pathway for lysine biosynthesis in fungi. Cell. Biochem. Biophys. 2006, 46, 43–64. [Google Scholar] [CrossRef]
- Miyazaki, T.; Miyazaki, J.; Yamane, H.; Nishiyama, M. Alpha-Aminoadipate aminotransferase from an extremely thermophilic bacterium, Thermus thermophilus. Microbiology 2004, 150, 2327–2334. [Google Scholar] [CrossRef]
- Kiss, C.; Vécsei, L. Kynurenines in the Brain: Preclinical and clinical studies, therapeutic considerations. In Handbook of Neurochemistry and Molecular Neurobiology: Brain and Spinal Cord Trauma; Lajtha, A., Banik, N., Ray, S.K., Eds.; Springer Publishing: Boston, MA, USA, 2009; pp. 91–105. [Google Scholar]
- Yu, P.; Li, Z.; Zhang, L.; Tagle, D.A.; Cai, T. Characterization of kynurenine aminotransferase III, a novel member of a phylogenetically conserved KAT family. Gene 2006, 365, 111–118. [Google Scholar] [CrossRef]
- Yang, C.; Zhang, L.; Han, Q.; Liao, C.; Lan, J.; Ding, H.; Zhou, H.; Diao, X.; Li, J. Kynurenine aminotransferase 3/glutamine transaminase L/cysteine conjugate beta-lyase 2 is a major glutamine transaminase in the mouse kidney. Biochem. Biophys. Rep. 2016, 8, 234–241. [Google Scholar] [CrossRef][Green Version]
- Han, Q.; Robinson, H.; Cai, T.; Tagle, D.A.; Li, J. Biochemical and structural characterization of mouse mitochondrial aspartate aminotransferase, a newly identified kynurenine aminotransferase-IV. Biosci. Rep. 2011, 31, 323–332. [Google Scholar] [CrossRef]
- Guillemin, G.J.; Kerr, S.J.; Smythe, G.A.; Smith, D.G.; Kapoor, V.; Armati, P.J.; Croitoru, J.; Brew, B.J. Kynurenine pathway metabolism in human astrocytes: A paradox for neuronal protection. J. Neurochem. 2001, 78, 842–853. [Google Scholar] [CrossRef]
- Rossi, F.; Miggiano, R.; Ferraris, D.M.; Rizzi, M. The synthesis of kynurenic acid in mammals: An updated kynurenine aminotransferase structural KATalogue. Front. Mol. Biosci. 2019, 6, 7. [Google Scholar] [CrossRef]
- Han, Q.; Robinson, H.; Cai, T.; Tagle, D.A.; Li, J. Structural insight into the inhibition of human kynurenine aminotransferase I/glutamine transaminase K. J. Med. Chem. 2009, 52, 2786–2793. [Google Scholar]
- Nematollahi, A.; Sun, G.; Harrop, S.J.; Hanrahan, J.R.; Church, W.B. Structure of the PLP-form of the human kynurenine aminotransferase II in a novel spacegroup at 1.83 a resolution. Int. J. Mol. Sci. 2016, 17, 446. [Google Scholar] [CrossRef]
- Nematollahi, A.; Sun, G.; Jayawickrama, G.S.; Church, W.B. Kynurenine aminotransferase isozyme inhibitors: A review. Int. J. Mol. Sci. 2016, 17, 946. [Google Scholar] [CrossRef]
- Soda, K.; Tanizawa, K. Kynureninases: Enzymological properties and regulation mechanism. Adv. Enzymol. Relat. Areas. Mol. Biol. 1979, 49, 1–40. [Google Scholar]
- Phillips, R.S. Structure and mechanism of kynureninase. Arch. Biochem. Biophys. 2014, 544, 69–74. [Google Scholar] [CrossRef]
- Phillips, R.S. Structure, mechanism, and substrate specificity of kynureninase. Biochim. Biophys. Acta 2011, 1814, 1481–1488. [Google Scholar] [CrossRef]
- Lima, S.; Kumar, S.; Gawandi, V.; Momany, C.; Phillips, R.S. Crystal structure of the Homo sapiens kynureninase-3-hydroxyhippuric acid inhibitor complex: Insights into the molecular basis of kynureninase substrate specificity. J. Med. Chem. 2009, 52, 389–396. [Google Scholar]
- Lima, S.; Khristoforov, R.; Momany, C.; Phillips, R.S. Crystal structure of Homo sapiens kynureninase. Biochemistry 2007, 46, 2735–2744. [Google Scholar]
- Masisi, K.; Suidasari, S.; Zhang, P.; Okazaki, Y.; Yanaka, N.; Kato, N. Comparative study on the responses of concentrations of B(6)-vitamers in several tissues of mice to the dietary level of pyridoxine. J. Nutr. Sci. Vitaminol. 2012, 58, 446–451. [Google Scholar] [CrossRef][Green Version]
- Magnusdottir, S.; Ravcheev, D.; de Crecy-Lagard, V.; Thiele, I. Systematic genome assessment of B-vitamin biosynthesis suggests co-operation among gut microbes. Front. Genet. 2015, 6, 148. [Google Scholar] [CrossRef]
- Fitzpatrick, T.B.; Amrhein, N.; Kappes, B.; Macheroux, P.; Tews, I.; Raschle, T. Two independent routes of de novo vitamin B6 biosynthesis: Not that different after all. Biochem. J. 2007, 407, 1–13. [Google Scholar] [CrossRef]
- Gurwara, S.; Ajami, N.J.; Jang, A.; Hessel, F.C.; Chen, L.; Plew, S.; Wang, Z.; Graham, D.Y.; Hair, C.; White, D.L.; et al. Dietary nutrients involved in one-carbon metabolism and colonic mucosa-associated gut microbiome in individuals with an endoscopically normal colon. Nutrients 2019, 11, 613. [Google Scholar] [CrossRef]
- Zelante, T.; Iannitti, R.G.; Cunha, C.; De Luca, A.; Giovannini, G.; Pieraccini, G.; Zecchi, R.; D’Angelo, C.; Massi-Benedetti, C.; Fallarino, F.; et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 2013, 39, 372–385. [Google Scholar] [CrossRef] [PubMed]
- Pagano, L.; Busca, A.; Candoni, A.; Cattaneo, C.; Cesaro, S.; Fanci, R.; Nadali, G.; Potenza, L.; Russo, D.; Tumbarello, M.; et al. Risk stratification for invasive fungal infections in patients with hematological malignancies: SEIFEM recommendations. Blood. Rev. 2017, 31, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Yano, J.M.; Yu, K.; Donaldson, G.P.; Shastri, G.G.; Ann, P.; Ma, L.; Nagler, C.R.; Ismagilov, R.F.; Mazmanian, S.K.; Hsiao, E.Y. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 2015, 161, 264–276. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, F.; Tsubouchi, R.; Shibata, Y. Effect of tryptophan metabolites on the activities of rat liver pyridoxal kinase and pyridoxamine 5-phosphate oxidase in vitro. Biochem. J. 1985, 227, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Saibeni, S.; Cattaneo, M.; Vecchi, M.; Zighetti, M.L.; Lecchi, A.; Lombardi, R.; Meucci, G.; Spina, L.; de Franchis, R. Low vitamin B(6) plasma levels, a risk factor for thrombosis, in inflammatory bowel disease: Role of inflammation and correlation with acute phase reactants. Am. J. Gastroenterol. 2003, 98, 112–117. [Google Scholar] [CrossRef]
- Ligaarden, S.C.; Farup, P.G. Low intake of Vitamin B6 is associated with irritable bowel syndrome symptoms. Nutr. Res. 2011, 31, 356–361. [Google Scholar] [CrossRef]
- Kowlessar, O.D.; Haeffner, L.J.; Benson, G.D. Abnormal tryptophan metabolism in patients with adult celiac disease, with evidence for deficiency of Vitamin B6. J. Clin. Investig. 1964, 43, 894–903. [Google Scholar] [CrossRef]
- Reinken, L.; Zieglauer, H.; Berger, H. Vitamin B6 nutriture of children with acute celiac disease, celiac disease in remission, and of children with normal duodenal mucosa. Am. J. Clin. Nutr. 1976, 29, 750–753. [Google Scholar] [CrossRef]
- Ghishan, F.K.; Kiela, P.R. Vitamins and minerals in inflammatory bowel disease. Gastroenterol. Clin. N. Am. 2017, 46, 797–808. [Google Scholar] [CrossRef]
- Benight, N.M.; Stoll, B.; Chacko, S.; da Silva, V.R.; Marini, J.C.; Gregory, J.F., 3rd; Stabler, S.P.; Burrin, D.G. B-vitamin deficiency is protective against DSS-induced colitis in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2011, 301, G249–G259. [Google Scholar] [CrossRef] [PubMed]
- Selhub, J.; Byun, A.; Liu, Z.; Mason, J.B.; Bronson, R.T.; Crott, J.W. Dietary vitamin B6 intake modulates colonic inflammation in the IL10-/- model of inflammatory bowel disease. J. Nutr. Biochem. 2013, 24, 2138–2143. [Google Scholar] [CrossRef] [PubMed]
- Kau, A.L.; Ahern, P.P.; Griffin, N.W.; Goodman, A.L.; Gordon, J.I. Human nutrition, the gut microbiome and the immune system. Nature 2011, 474, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Qian, B.; Shen, S.; Zhang, J.; Jing, P. Effects of Vitamin B6 Deficiency on the Composition and Functional Potential of T Cell Populations. J. Immunol. Res. 2017. [Google Scholar] [CrossRef]
- Lotto, V.; Choi, S.W.; Friso, S. Vitamin B6: A challenging link between nutrition and inflammation in CVD. Br. J. Nutr. 2011, 106, 183–195. [Google Scholar] [CrossRef]
- Chiang, E.P.; Bagley, P.J.; Selhub, J.; Nadeau, M.; Roubenoff, R. Abnormal vitamin B(6) status is associated with severity of symptoms in patients with rheumatoid arthritis. Am. J. Med. 2003, 114, 283–287. [Google Scholar] [CrossRef]
- Zhang, P.; Tsuchiya, K.; Kinoshita, T.; Kushiyama, H.; Suidasari, S.; Hatakeyama, M.; Imura, H.; Kato, N.; Suda, T. Vitamin B6 Prevents IL-1beta Protein Production by Inhibiting NLRP3 Inflammasome Activation. J. Biol. Chem. 2016, 291, 24517–24527. [Google Scholar] [CrossRef]
- Jones, L.A.; Sun, E.W.; Martin, A.M.; Keating, D.J. The ever-changing roles of serotonin. Int. J. Biochem. Cell. Biol. 2020, 125, 105776. [Google Scholar] [CrossRef]
- Tullius, S.G.; Biefer, H.R.; Li, S.; Trachtenberg, A.J.; Edtinger, K.; Quante, M.; Krenzien, F.; Uehara, H.; Yang, X.; Kissick, H.T.; et al. NAD+ protects against EAE by regulating CD4+ T-cell differentiation. Nat. Commun. 2014, 5, 5101. [Google Scholar] [CrossRef]
- Nowak, E.C.; de Vries, V.C.; Wasiuk, A.; Ahonen, C.; Bennett, K.A.; Le Mercier, I.; Ha, D.G.; Noelle, R.J. Tryptophan hydroxylase-1 regulates immune tolerance and inflammation. J. Exp. Med. 2012, 209, 2127–2135. [Google Scholar] [CrossRef]
- Gershon, M.D.; Tack, J. The serotonin signaling system: From basic understanding to drug development for functional GI disorders. Gastroenterology 2007, 132, 397–414. [Google Scholar] [CrossRef] [PubMed]
- Nemazannikova, N.; Mikkelsen, K.; Stojanovska, L.; Blatch, G.L.; Apostolopoulos, V. Is there a Link between Vitamin B and Multiple Sclerosis? Med. Chem. 2018, 14, 170–180. [Google Scholar] [PubMed]
- Romani, L.; Puccetti, P. Protective tolerance to fungi: The role of IL-10 and tryptophan catabolism. Trends Microbiol. 2006, 14, 183–189. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cellini, B.; Zelante, T.; Dindo, M.; Bellet, M.M.; Renga, G.; Romani, L.; Costantini, C. Pyridoxal 5′-Phosphate-Dependent Enzymes at the Crossroads of Host–Microbe Tryptophan Metabolism. Int. J. Mol. Sci. 2020, 21, 5823. https://doi.org/10.3390/ijms21165823
Cellini B, Zelante T, Dindo M, Bellet MM, Renga G, Romani L, Costantini C. Pyridoxal 5′-Phosphate-Dependent Enzymes at the Crossroads of Host–Microbe Tryptophan Metabolism. International Journal of Molecular Sciences. 2020; 21(16):5823. https://doi.org/10.3390/ijms21165823
Chicago/Turabian StyleCellini, Barbara, Teresa Zelante, Mirco Dindo, Marina M. Bellet, Giorgia Renga, Luigina Romani, and Claudio Costantini. 2020. "Pyridoxal 5′-Phosphate-Dependent Enzymes at the Crossroads of Host–Microbe Tryptophan Metabolism" International Journal of Molecular Sciences 21, no. 16: 5823. https://doi.org/10.3390/ijms21165823
APA StyleCellini, B., Zelante, T., Dindo, M., Bellet, M. M., Renga, G., Romani, L., & Costantini, C. (2020). Pyridoxal 5′-Phosphate-Dependent Enzymes at the Crossroads of Host–Microbe Tryptophan Metabolism. International Journal of Molecular Sciences, 21(16), 5823. https://doi.org/10.3390/ijms21165823






