AcoMYB4, an Ananas comosus L. MYB Transcription Factor, Functions in Osmotic Stress through Negative Regulation of ABA Signaling
Abstract
1. Introduction
2. Results
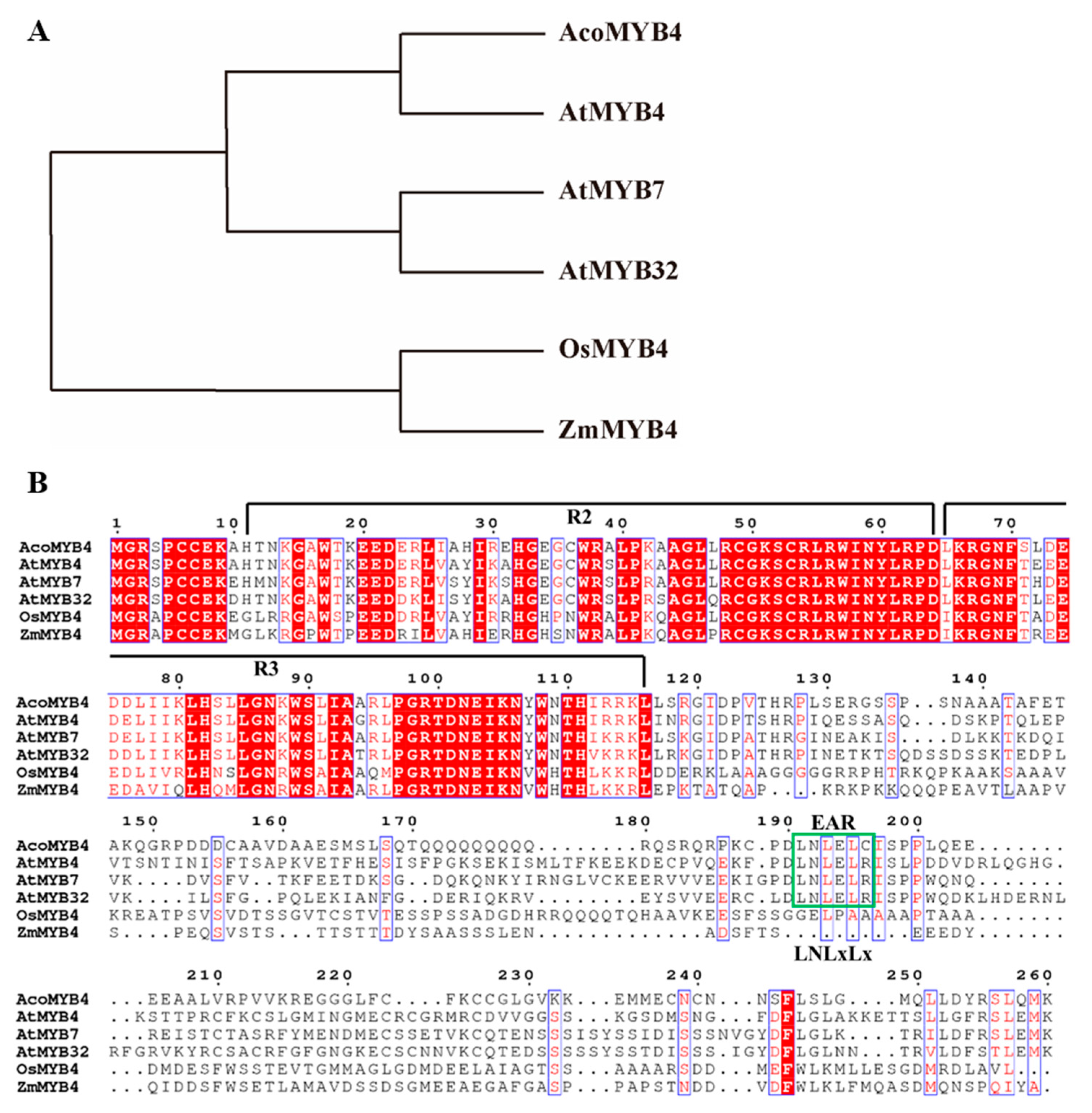
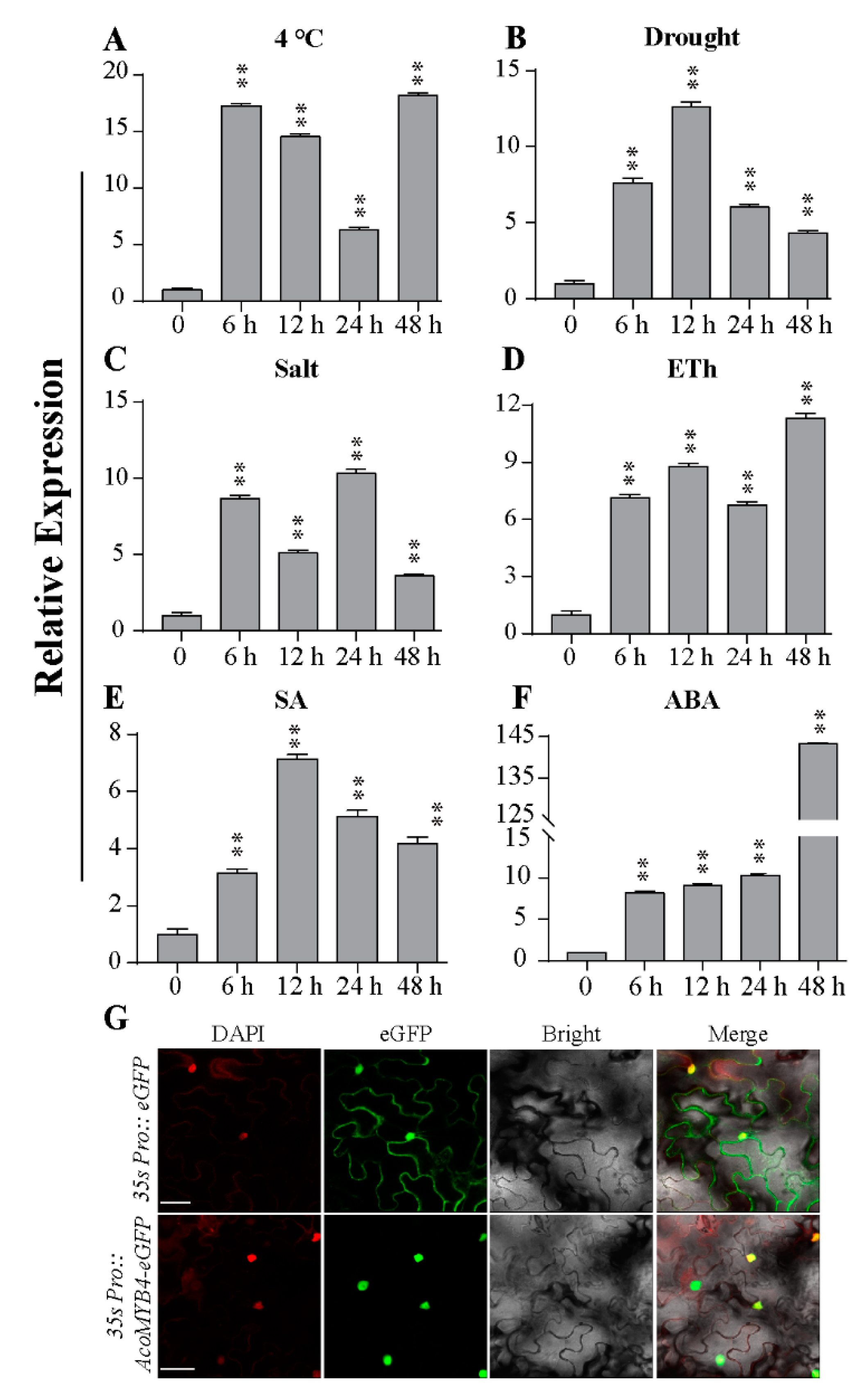
2.1. Expression Profiles of AcoMYB4 Response to Various Abiotic Stresses
2.2. Subcellular Localization of AcoMYB4
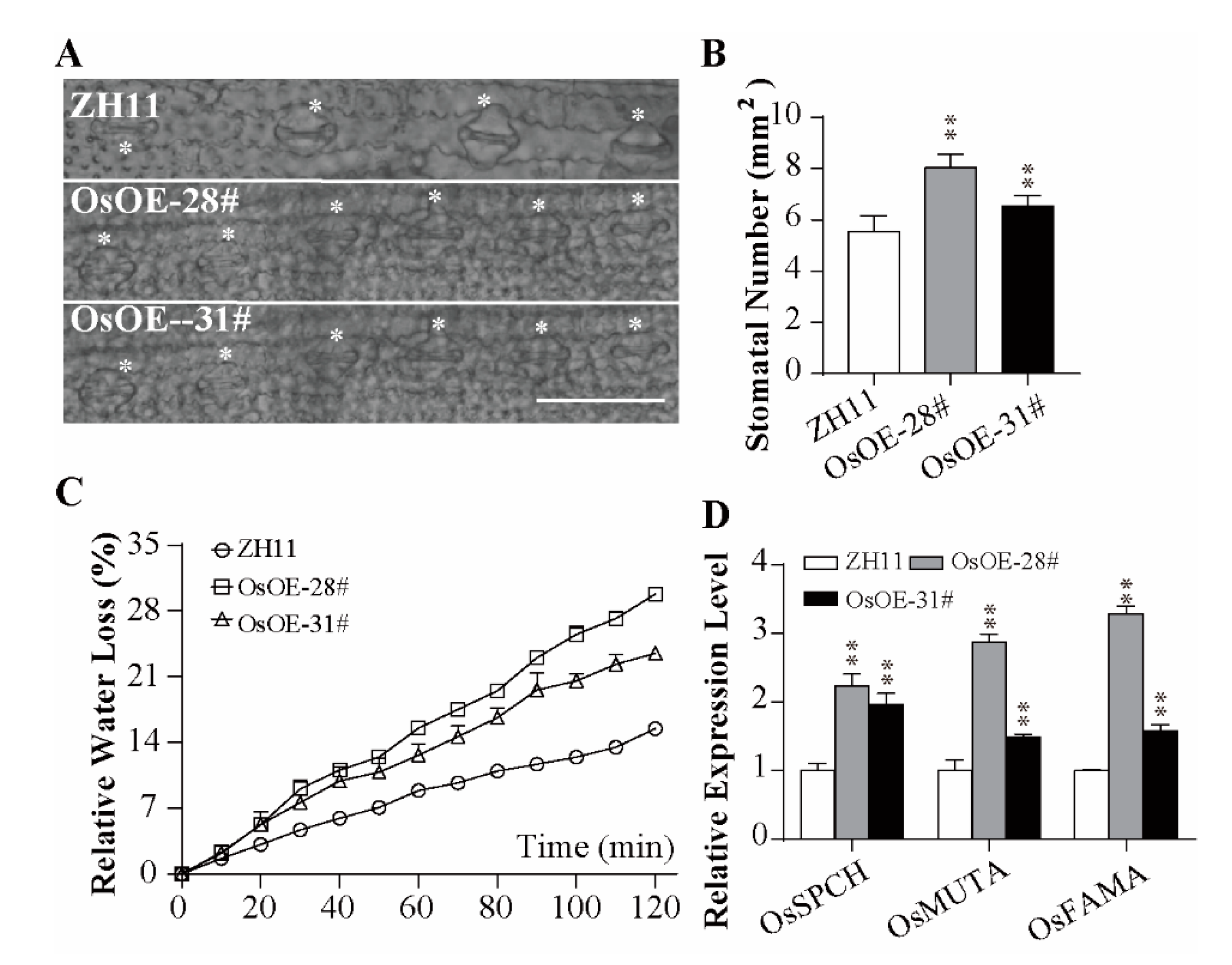
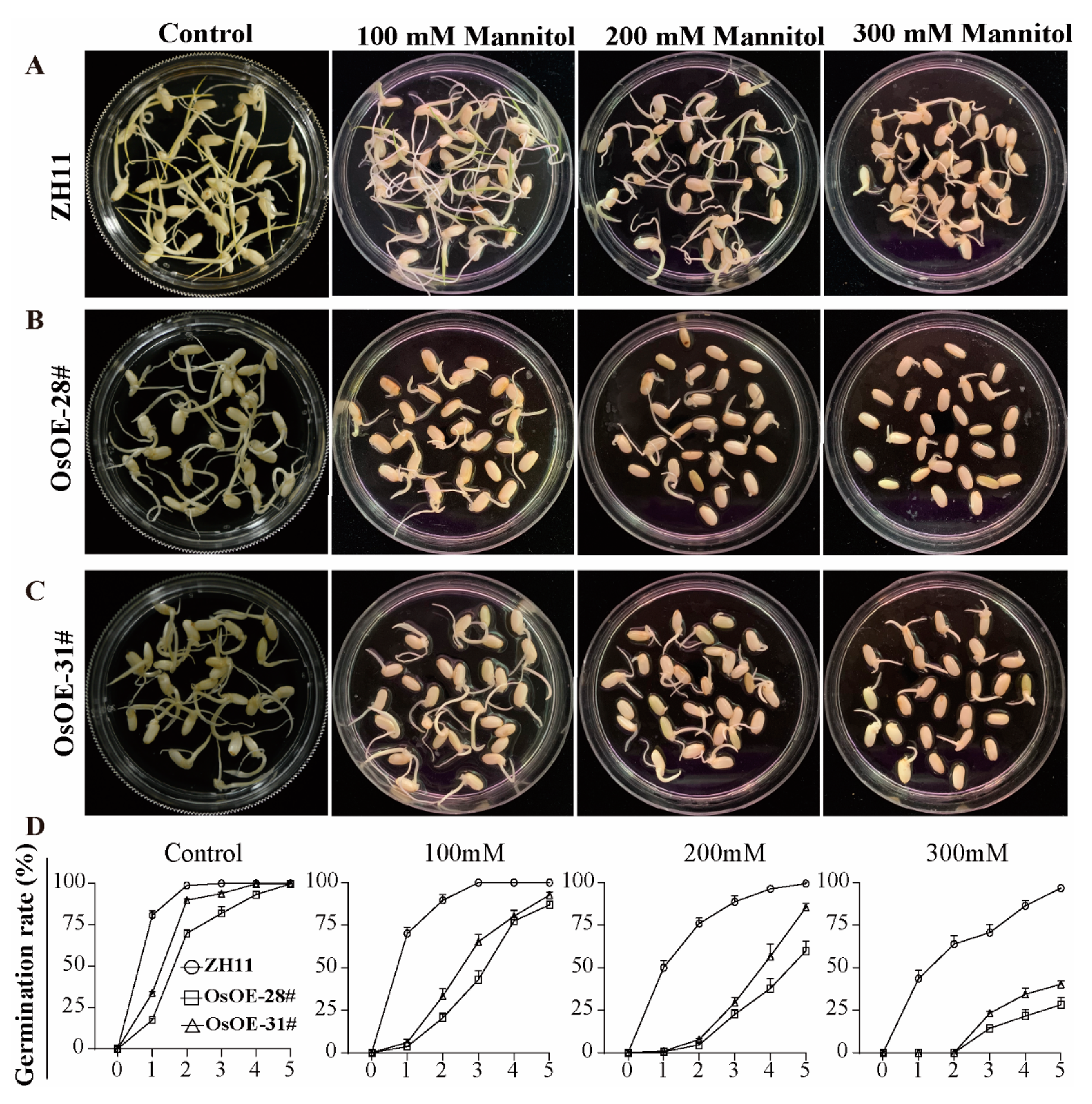
2.3. Overexpression of AcoMYB4 Plants Was Sensitive to Drought and Salt Stress
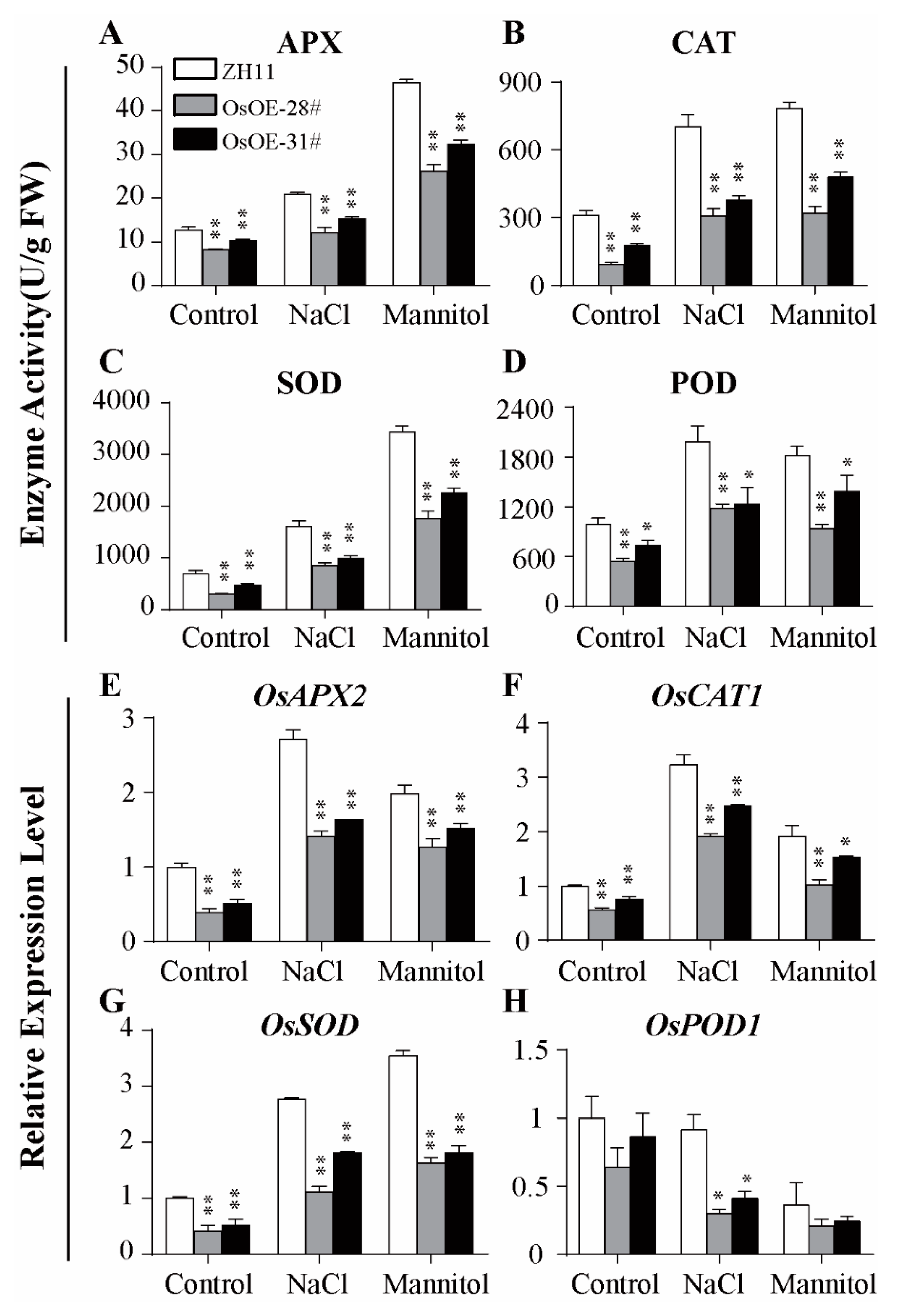
2.4. Overexpression of AcoMYB4 Decreased Antioxidant Enzyme Activities and Enhanced Oxidative Damage
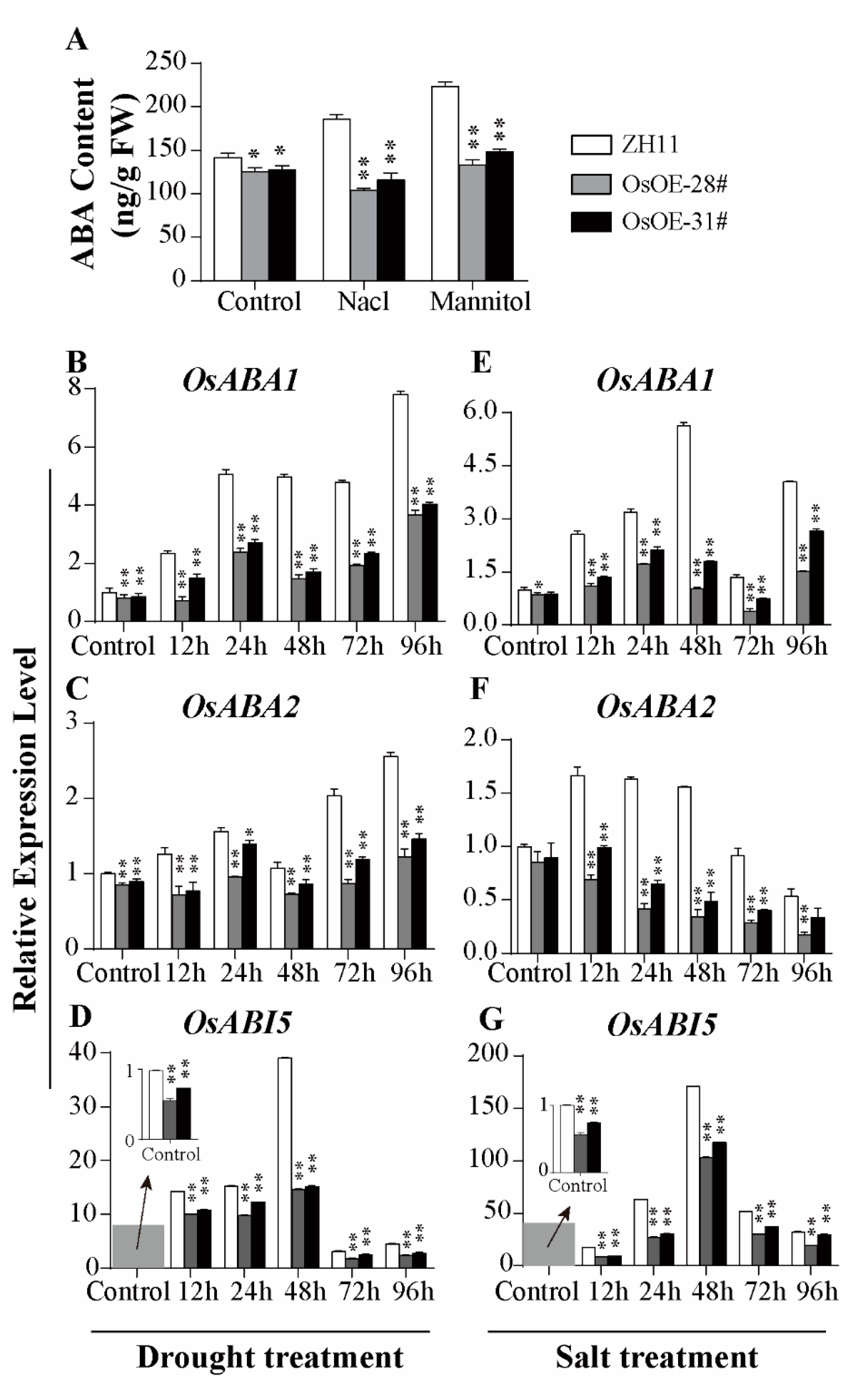
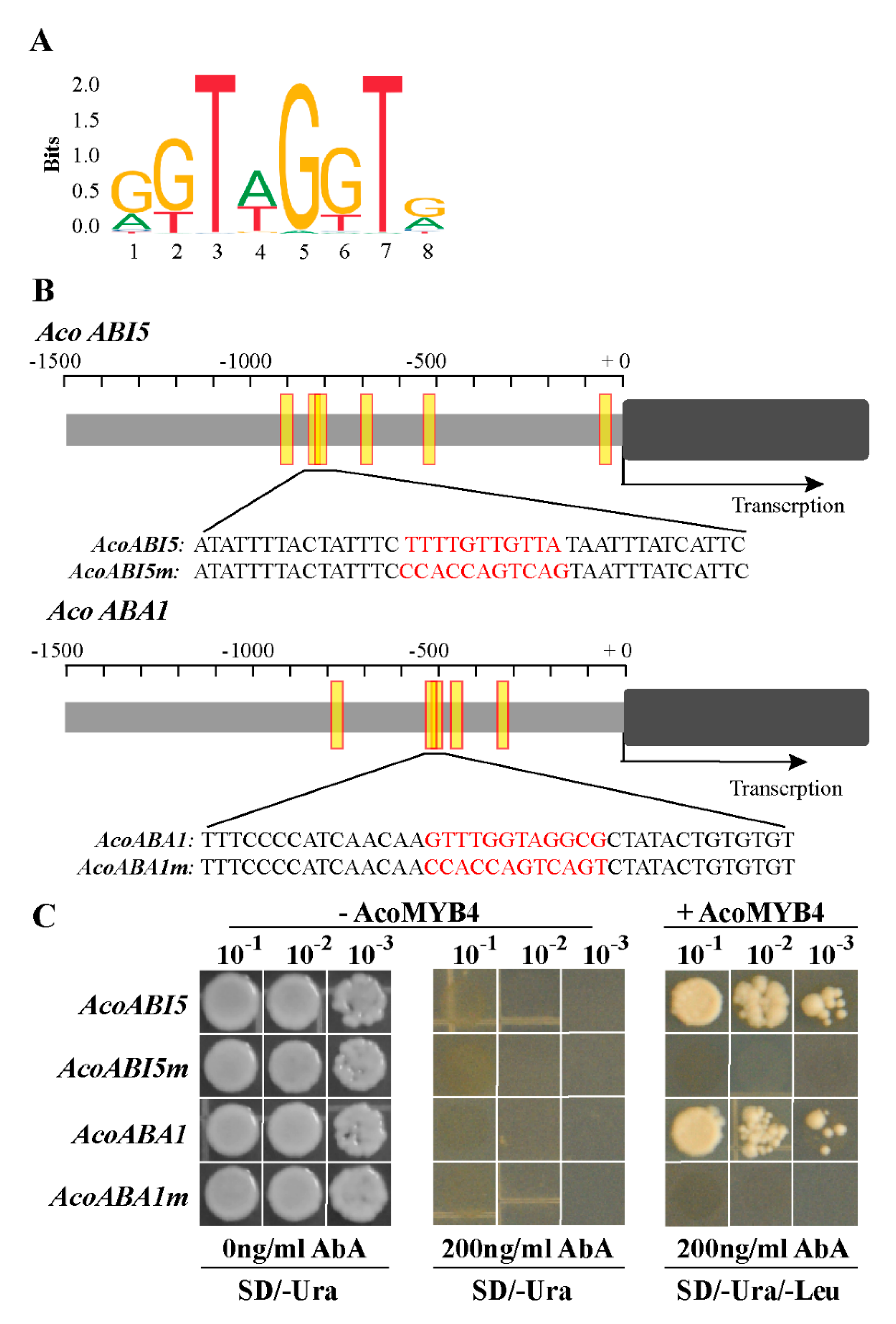
2.5. AcoMYB4 Regulates ABA Biosynthesis by Directly Binding to the Promoters of AcoABI5 and AcoABA1
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Stress Treatment of Pineapple
4.2. Bioinformatics Analysis
4.3. RNA Extraction and qRT-PCR
4.4. Cloning and Subcellular Localization
4.5. Germination, Cotyledon Greening, and Survival Rate of Transgenic Plants
4.6. Assessment of Drought and Salt Tolerance in Transgenic Plants
4.7. Analysis of Proline, Soluble Sugars, ABA Content, and Enzyme Activity
4.8. Yeast One-Hybrid System
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pastori, G.M.; Foyer, C.H. Common components, networks, and pathways of cross-tolerance to stress. The central role of “redox” and abscisic acid-mediated controls. Plant Physiol. 2002, 129, 460–468. [Google Scholar] [CrossRef] [PubMed]
- Osakabe, Y.; Osakabe, K.; Shinozaki, K.; Tran, L.S. Response of plants to water stress. Front. Plant Sci. 2014, 5, 86. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Zheng, Z.; Chinnusamy, V.; Zhu, J.; Cui, X.; Iida, K.; Zhu, J.K. RAS1, a quantitative trait locus for salt tolerance and ABA sensitivity in Arabidopsis. Proc. Natl. Acad. Sci. USA 2010, 107, 5669–5674. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Finkelstein, R.R.; Rock, C.D. Abscisic Acid biosynthesis and response. Arab. Book 2002, 1, e0058. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, R.R.; Gampala, S.S.; Rock, C.D. Abscisic acid signaling in seeds and seedlings. Plant Cell 2002, 14, S15–S45. [Google Scholar] [CrossRef]
- Choi, H.; Hong, J.; Ha, J.; Kang, J.; Kim, S.Y. ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 2000, 275, 1723–1730. [Google Scholar] [CrossRef]
- Fujita, Y.; Fujita, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant. Res. 2011, 124, 509–525. [Google Scholar] [CrossRef]
- Ding, Z.J.; Yan, J.Y.; Xu, X.Y.; Yu, D.Q.; Li, G.X.; Zhang, S.Q.; Zheng, S.J. Transcription factor WRKY46 regulates osmotic stress responses and stomatal movement independently in Arabidopsis. Plant J. 2014, 79, 13–27. [Google Scholar] [CrossRef]
- Schwartz, S.H.; Leon-Kloosterziel, K.M.; Koornneef, M.; Zeevaart, J.A. Biochemical characterization of the aba2 and aba3 mutants in Arabidopsis thaliana. Plant Physiol. 1997, 114, 161–166. [Google Scholar] [CrossRef]
- Barrero, J.M.; Rodriguez, P.L.; Quesada, V.; Piqueras, P.; Ponce, M.R.; Micol, J.L. Both abscisic acid (ABA)-dependent and ABA-independent pathways govern the induction of NCED3, AAO3 and ABA1 in response to salt stress. Plant Cell Environ. 2006, 29, 2000–2008. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C Phosphatase Activity Function as Abscisic Acid Sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.F.; et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Song, Z.; Li, C.; Jiang, J.; Zhou, Y.; Wang, R.; Wang, Q.; Ni, C.; Liang, Q.; Chen, H.; et al. RSM1, an Arabidopsis MYB protein, interacts with HY5/HYH to modulate seed germination and seedling development in response to abscisic acid and salinity. PLoS Genet. 2018, 14, e1007839. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Yamaguchi-Shinozaki, K. ABA signaling in stress-response and seed development. Plant Cell Rep. 2013, 32, 959–970. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef]
- Waszczak, C.; Carmody, M.; Kangasjarvi, J. Reactive Oxygen Species in Plant Signaling. Annu. Rev. Plant Biol. 2018, 69, 209–236. [Google Scholar] [CrossRef]
- Zhang, H.; Han, B.; Wang, T.; Chen, S.; Li, H.; Zhang, Y.; Dai, S. Mechanisms of plant salt response: Insights from proteomics. J. Proteome Res. 2012, 11, 49–67. [Google Scholar] [CrossRef]
- Guan, L.M.; Scandalios, J.G. Catalase transcript accumulation in response to dehydration and osmotic stress in leaves of maize viviparous mutants. Redox Rep. 2000, 5, 377–383. [Google Scholar] [CrossRef][Green Version]
- Hu, X.; Jiang, M.; Zhang, A.; Lu, J. Abscisic acid-induced apoplastic H2O2 accumulation up-regulates the activities of chloroplastic and cytosolic antioxidant enzymes in maize leaves. Planta 2005, 223, 57–68. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, L.; Dong, F.; Gao, J.; Galbraith, D.W.; Song, C.P. Hydrogen Peroxide Is Involved in Abscisic Acid-Induced Stomatal Closure in Vicia faba. Plant Physiol. 2001, 126, 1438–1448. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yang, T.; Lin, Z.; Gu, B.; Xing, C.; Zhao, L.; Dong, H.; Gao, J.; Xie, Z.; Zhang, S.; et al. A WRKY transcription factor PbrWRKY53 from Pyrus betulaefolia is involved in drought tolerance and AsA accumulation. Plant Biotechnol. J. 2019, 17, 1770–1787. [Google Scholar] [CrossRef] [PubMed]
- Golldack, D.; Li, C.; Mohan, H.; Probst, N. Tolerance to drought and salt stress in plants: Unraveling the signaling networks. Front. Plant Sci. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef] [PubMed]
- Krasensky, J.; Jonak, C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 2012, 63, 1593–1608. [Google Scholar] [CrossRef]
- Miller, G.; Shulaev, V.; Mittler, R. Reactive oxygen signaling and abiotic stress. Physiol. Plant. 2008, 133, 481–489. [Google Scholar] [CrossRef]
- Thompson, M.A.; Ramsay, R.G. Myb: An old oncoprotein with new roles. Bioessays 1995, 17, 341–350. [Google Scholar] [CrossRef]
- Shen-Ong, G.L. The myb oncogene. Biochim. Biophys. Acta 1990, 1032, 39–52. [Google Scholar] [CrossRef]
- Nakagoshi, H.; Nagase, T.; Kanei-Ishii, C.; Ueno, Y.; Ishii, S. Binding of the c-myb proto-oncogene product to the simian virus 40 enhancer stimulates transcription. J. Biol. Chem. 1990, 265, 3479–3483. [Google Scholar]
- Nishina, Y.; Nakagoshi, H.; Imamoto, F.; Gonda, T.J.; Ishii, S. Trans-activation by the c-myb proto-oncogene. Nucleic Acids Res. 1989, 17, 107–117. [Google Scholar] [CrossRef]
- Schubert, R.; Dobritzsch, S.; Gruber, C.; Hause, G.; Athmer, B.; Schreiber, T.; Marillonnet, S.; Okabe, Y.; Ezura, H.; Acosta, I.F.; et al. Tomato MYB21 Acts in Ovules to Mediate Jasmonate-Regulated Fertility. Plant Cell 2019, 31, 1043–1062. [Google Scholar] [CrossRef] [PubMed]
- Volpe, V.; Dell’Aglio, E.; Giovannetti, M.; Ruberti, C.; Costa, A.; Genre, A.; Guether, M.; Bonfante, P. An AM-induced, MYB-family gene of Lotus japonicus (LjMAMI) affects root growth in an AM-independent manner. Plant J. 2013, 73, 442–455. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.K.; Dubos, C.; Dodd, I.C.; Holroyd, G.H.; Hetherington, A.M.; Campbell, M.M. AtMYB61, an R2R3-MYB transcription factor controlling stomatal aperture in Arabidopsis thaliana. Curr. Biol. 2005, 15, 1201–1206. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.G.; Seo, P.J. The MYB96-HHP module integrates cold and abscisic acid signaling to activate the CBF-COR pathway in Arabidopsis. Plant J. 2015, 82, 962–977. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Li, Y.; Zhang, L.; Wang, X.; Zhao, Z.; Tao, Z.; Wang, J.; Wang, J.; Lin, M.; Li, X.; et al. Arabidopsis ABA receptor RCAR1/PYL9 interacts with an R2R3-type MYB transcription factor, AtMYB44. Int. J. Mol. Sci. 2014, 15, 8473–8490. [Google Scholar] [CrossRef]
- Piao, W.; Kim, S.H.; Lee, B.D.; An, G.; Sakuraba, Y.; Paek, N.C. Rice transcription factor OsMYB102 delays leaf senescence by down-regulating abscisic acid accumulation and signaling. J. Exp. Bot. 2019, 70, 2699–2715. [Google Scholar] [CrossRef]
- Yan, J.; Wang, B.; Zhong, Y.; Yao, L.; Cheng, L.; Wu, T. The soybean R2R3 MYB transcription factor GmMYB100 negatively regulates plant flavonoid biosynthesis. Plant Mol. Biol. 2015, 89, 35–48. [Google Scholar] [CrossRef]
- Fang, Q.; Jiang, T.; Xu, L.; Liu, H.; Mao, H.; Wang, X.; Jiao, B.; Duan, Y.; Wang, Q.; Dong, Q.; et al. A salt-stress-regulator from the Poplar R2R3 MYB family integrates the regulation of lateral root emergence and ABA signaling to mediate salt stress tolerance in Arabidopsis. Plant Physiol. Biochem. 2017, 114, 100–110. [Google Scholar] [CrossRef]
- Liu, C.Y.; Xie, T.; Chen, C.J.; Luan, A.P.; Long, J.M.; Li, C.H.; Ding, Y.Q.; He, Y.H. Genome-wide organization and expression profiling of the R2R3-MYB transcription factor family in pineapple (Ananas comosus). BMC Genom. 2017, 18. [Google Scholar] [CrossRef]
- Liu, T.; Ohashi-Ito, K.; Bergmann, D.C. Orthologs of Arabidopsis thaliana stomatal bHLH genes and regulation of stomatal development in grasses. Development 2009, 136, 2265–2276. [Google Scholar] [CrossRef] [PubMed]
- Peterson, K.M.; Rychel, A.L.; Torii, K.U. Out of the mouths of plants: The molecular basis of the evolution and diversity of stomatal development. Plant Cell 2010, 22, 296–306. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, R.R.; Lynch, T.J. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 2000, 12, 599–609. [Google Scholar] [CrossRef]
- Lopez-Molina, L.; Mongrand, S.; McLachlin, D.T.; Chait, B.T.; Chua, N.H. ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 2002, 32, 317–328. [Google Scholar] [CrossRef]
- Shen, X.J.; Guo, X.W.; Guo, X.; Zhao, D.; Zhao, W.; Chen, J.S.; Li, T.H. PacMYBA, a sweet cherry R2R3-MYB transcription factor, is a positive regulator of salt stress tolerance and pathogen resistance. Plant Physiol. Biochem. 2017, 112, 302–311. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ding, Y.L.; Li, Z.Y.; Shi, Y.T.; Wang, J.L.; Hua, J.; Gong, Z.Z.; Zhou, J.M.; Yang, S.H. PUB25 and PUB26 Promote Plant Freezing Tolerance by Degrading the Cold Signaling Negative Regulator MYB15. Dev. Cell 2019, 51, 222–246. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Fornale, S.; Lopez, E.; Salazar-Henao, J.E.; Fernandez-Nohales, P.; Rigau, J.; Caparros-Ruiz, D. AtMYB7, a new player in the regulation of UV-sunscreens in Arabidopsis thaliana. Plant Cell Physiol. 2014, 55, 507–516. [Google Scholar] [CrossRef]
- Kim, J.H.; Hyun, W.Y.; Nguyen, H.N.; Jeong, C.Y.; Xiong, L.; Hong, S.W.; Lee, H. AtMyb7, a subgroup 4 R2R3 Myb, negatively regulates ABA-induced inhibition of seed germination by blocking the expression of the bZIP transcription factor ABI5. Plant Cell Environ. 2015, 38, 559–571. [Google Scholar] [CrossRef]
- Agarwal, P.; Mitra, M.; Banerjee, S.; Roy, S. MYB4 transcription factor, a member of R2R3-subfamily of MYB domain protein, regulates cadmium tolerance via enhanced protection against oxidative damage and increases expression of PCS1 and MT1C in Arabidopsis. Plant Sci. 2020, 297, 110501. [Google Scholar] [CrossRef]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef]
- Xiong, L.; Ishitani, M.; Lee, H.; Zhu, J.K. The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell 2001, 13, 2063–2083. [Google Scholar] [PubMed]
- Araus, J.L.; Slafer, G.A.; Reynolds, M.P.; Royo, C. Plant breeding and drought in C3 cereals: What should we breed for? Ann. Bot. 2002, 89, 925–940. [Google Scholar] [CrossRef] [PubMed]
- Dittrich, M.; Mueller, H.M.; Bauer, H.; Peirats-Llobet, M.; Rodriguez, P.L.; Geilfus, C.M.; Carpentier, S.C.; Al Rasheid, K.A.S.; Kollist, H.; Merilo, E.; et al. The role of Arabidopsis ABA receptors from the PYR/PYL/RCAR family in stomatal acclimation and closure signal integration. Nat. Plants 2019, 5, 1002–1011. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Hiei, Y.; Komari, T. Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nat. Protoc. 2008, 3, 824–834. [Google Scholar] [CrossRef]
- Bartholomew, D.P.; Paull, R.E.; Rohrbach, K.G. The pineapple: Botany, production and uses. CAB Int. 2003. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlic, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef]
- Chen, H.; Hu, B.; Zhao, L.; Shi, D.; She, Z.; Huang, X.; Priyadarshani, S.V.G.N.; Niu, X.; Qin, Y. Differential Expression Analysis of Reference Genes in Pineapple (Ananas comosus L.) during Reproductive Development and Response to Abiotic Stress, Hormonal Stimuli. Trop. Plant Biol. 2019, 12, 67–77. [Google Scholar] [CrossRef]
- Yoshida, S.; Forno, D.A.; Cock, J.H.; Gomez, K.A. Laboratory Manual for Physiological Studies of Rice. Int. Rice Res. Inst. 1971, 18, 62–65. [Google Scholar]
- Li, J.; Besseau, S.; Toronen, P.; Sipari, N.; Kollist, H.; Holm, L.; Palva, E.T. Defense-related transcription factors WRKY70 and WRKY54 modulate osmotic stress tolerance by regulating stomatal aperture in Arabidopsis. New Phytol. 2013, 200, 457–472. [Google Scholar] [CrossRef] [PubMed]
- Perin, E.C.; Crizel, R.L.; Galli, V.; Messias, R.D.; Rombaldi, C.V.; Chaves, F.C. Extraction and Quantification of Abscisic Acid and Derivatives in Strawberry by LC-MS. Food Anal. Method 2018, 11, 2547–2552. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, H.; Lai, L.; Li, L.; Liu, L.; Jakada, B.H.; Huang, Y.; He, Q.; Chai, M.; Niu, X.; Qin, Y. AcoMYB4, an Ananas comosus L. MYB Transcription Factor, Functions in Osmotic Stress through Negative Regulation of ABA Signaling. Int. J. Mol. Sci. 2020, 21, 5727. https://doi.org/10.3390/ijms21165727
Chen H, Lai L, Li L, Liu L, Jakada BH, Huang Y, He Q, Chai M, Niu X, Qin Y. AcoMYB4, an Ananas comosus L. MYB Transcription Factor, Functions in Osmotic Stress through Negative Regulation of ABA Signaling. International Journal of Molecular Sciences. 2020; 21(16):5727. https://doi.org/10.3390/ijms21165727
Chicago/Turabian StyleChen, Huihuang, Linyi Lai, Lanxin Li, Liping Liu, Bello Hassan Jakada, Youmei Huang, Qing He, Mengnan Chai, Xiaoping Niu, and Yuan Qin. 2020. "AcoMYB4, an Ananas comosus L. MYB Transcription Factor, Functions in Osmotic Stress through Negative Regulation of ABA Signaling" International Journal of Molecular Sciences 21, no. 16: 5727. https://doi.org/10.3390/ijms21165727
APA StyleChen, H., Lai, L., Li, L., Liu, L., Jakada, B. H., Huang, Y., He, Q., Chai, M., Niu, X., & Qin, Y. (2020). AcoMYB4, an Ananas comosus L. MYB Transcription Factor, Functions in Osmotic Stress through Negative Regulation of ABA Signaling. International Journal of Molecular Sciences, 21(16), 5727. https://doi.org/10.3390/ijms21165727





