Wnt/CTNNB1 Signal Transduction Pathway Inhibits the Expression of ZFP36 in Squamous Cell Carcinoma, by Inducing Transcriptional Repressors SNAI1, SLUG and TWIST
Abstract
1. Introduction
2. Results
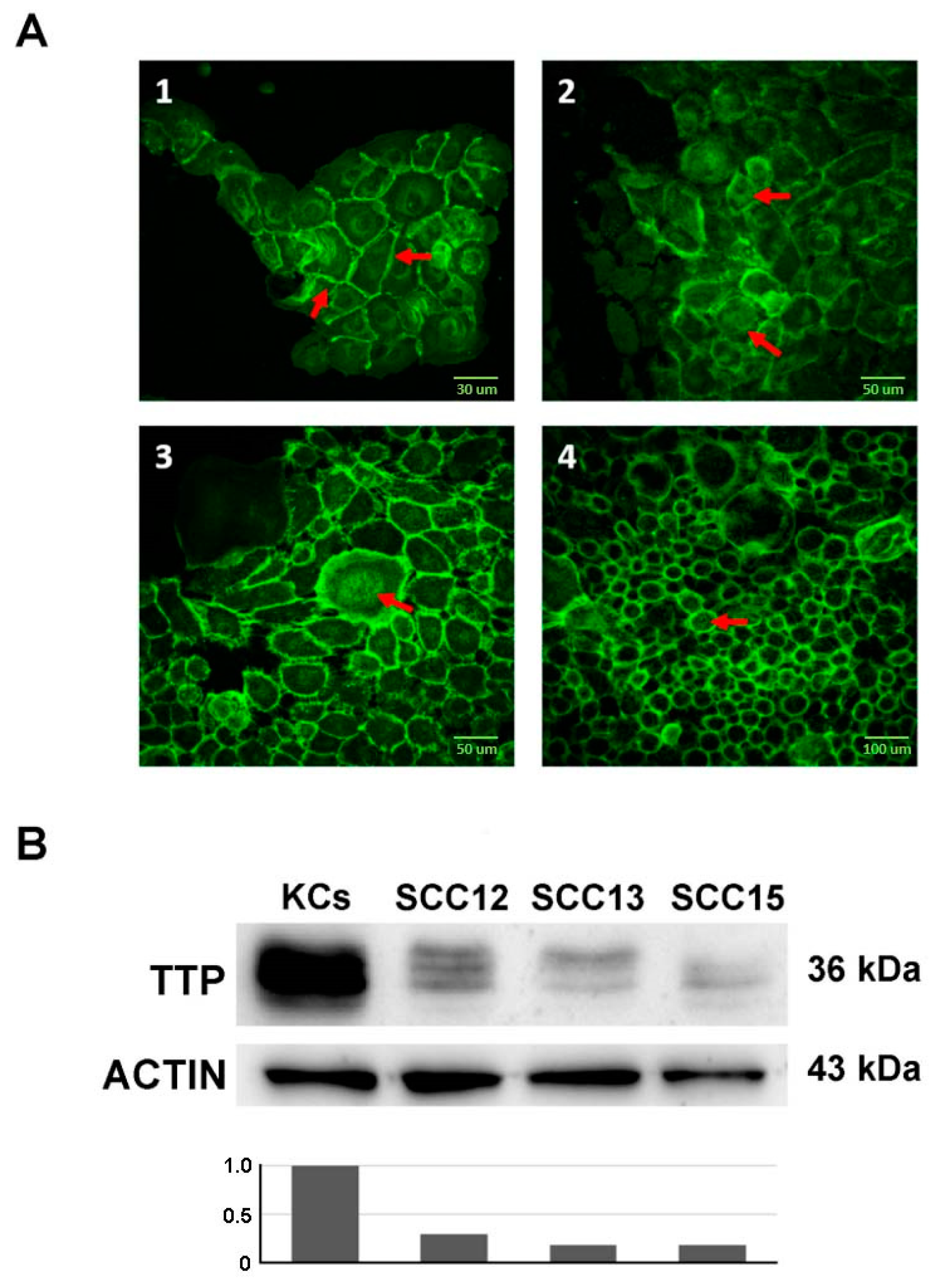
2.1. Wnt/CTNNB1 Pathway Constitutive Activation and ZFP36 Down-Regulation in SCC Cell Lines Compared to Normal Keratinocytes
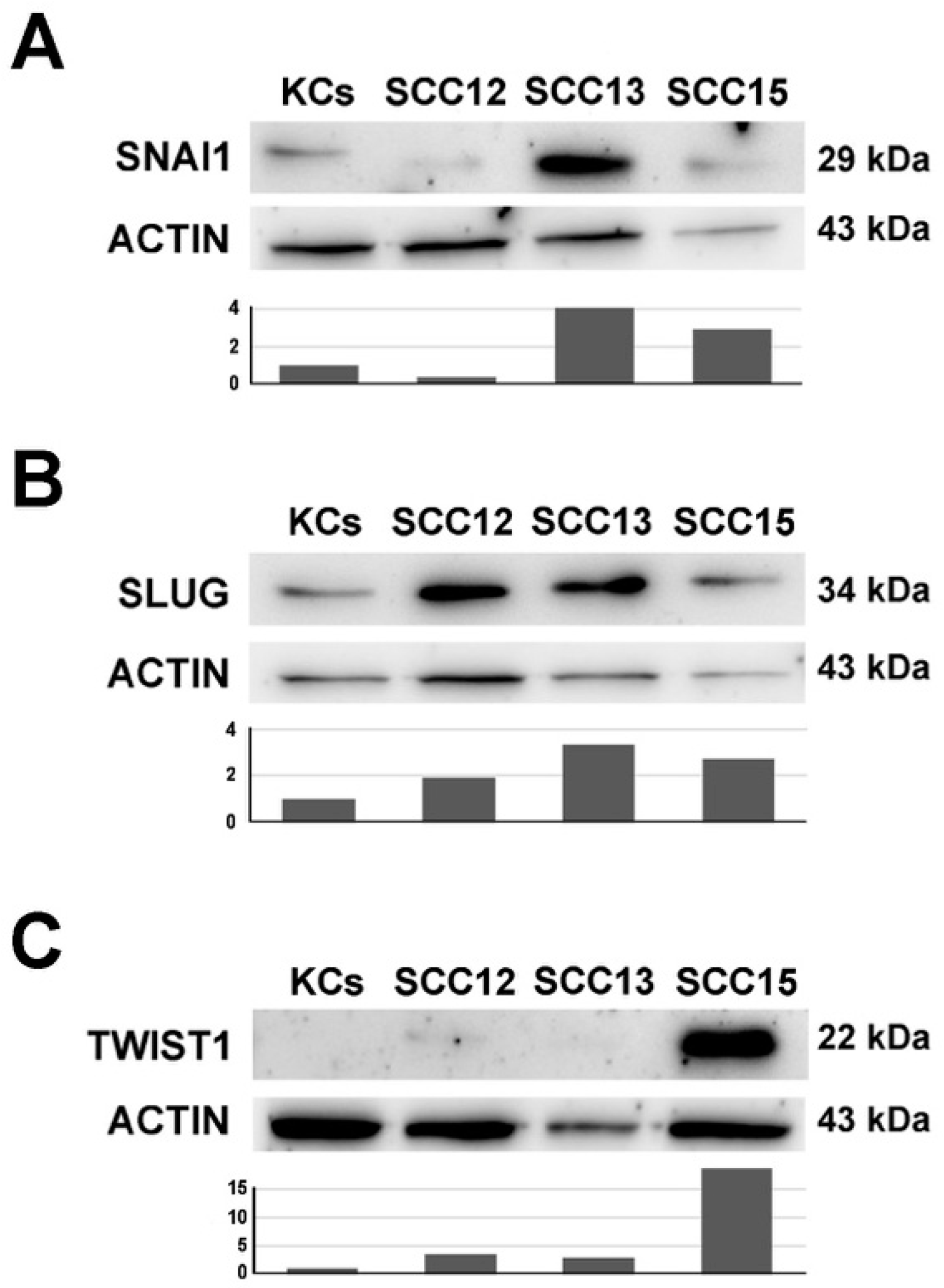
2.2. Expression of SLUG, SNAI1 and TWIST in SCC12, 13 and 15 Cell Lines Compared to Normal Keratinocytes
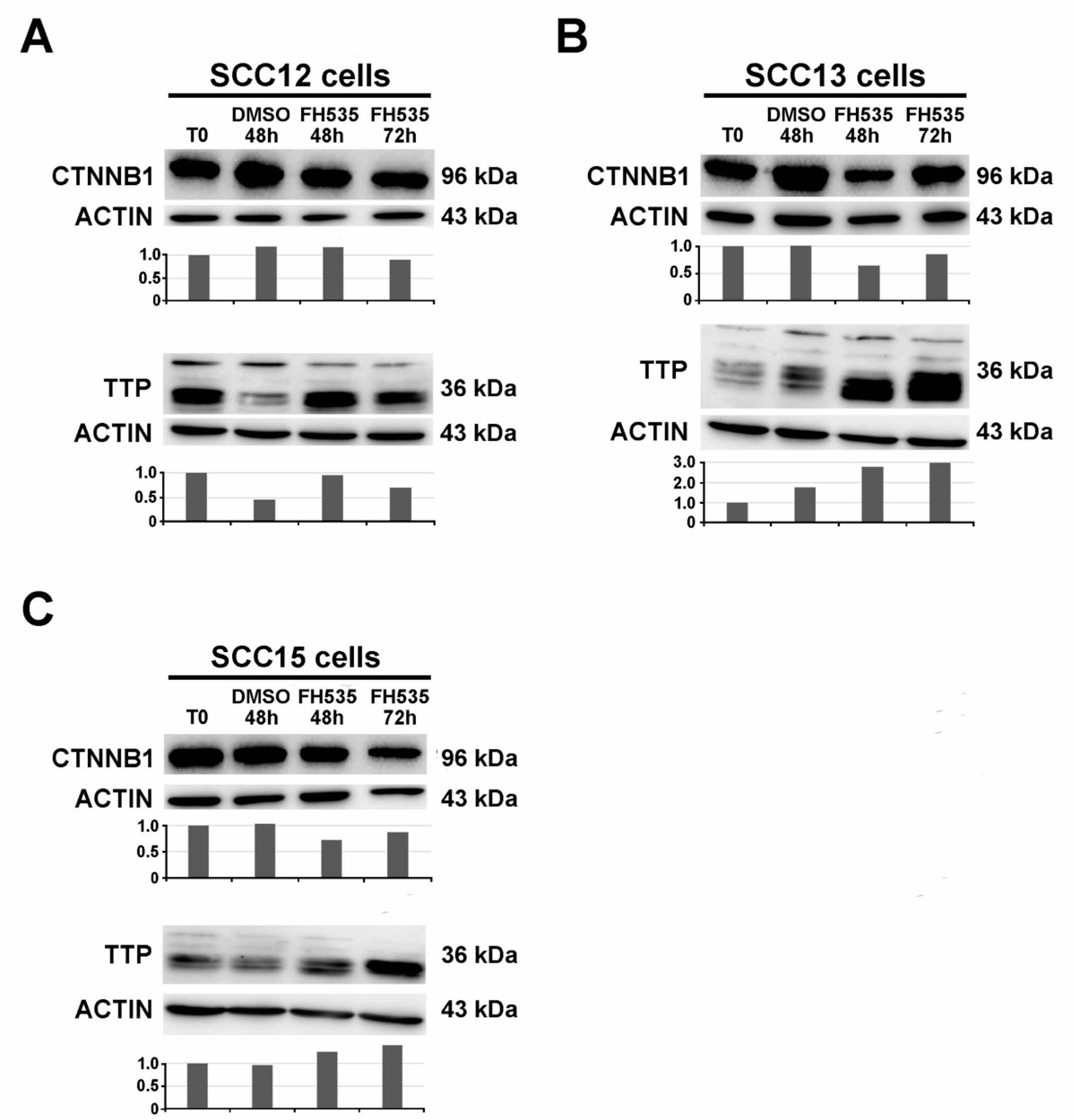
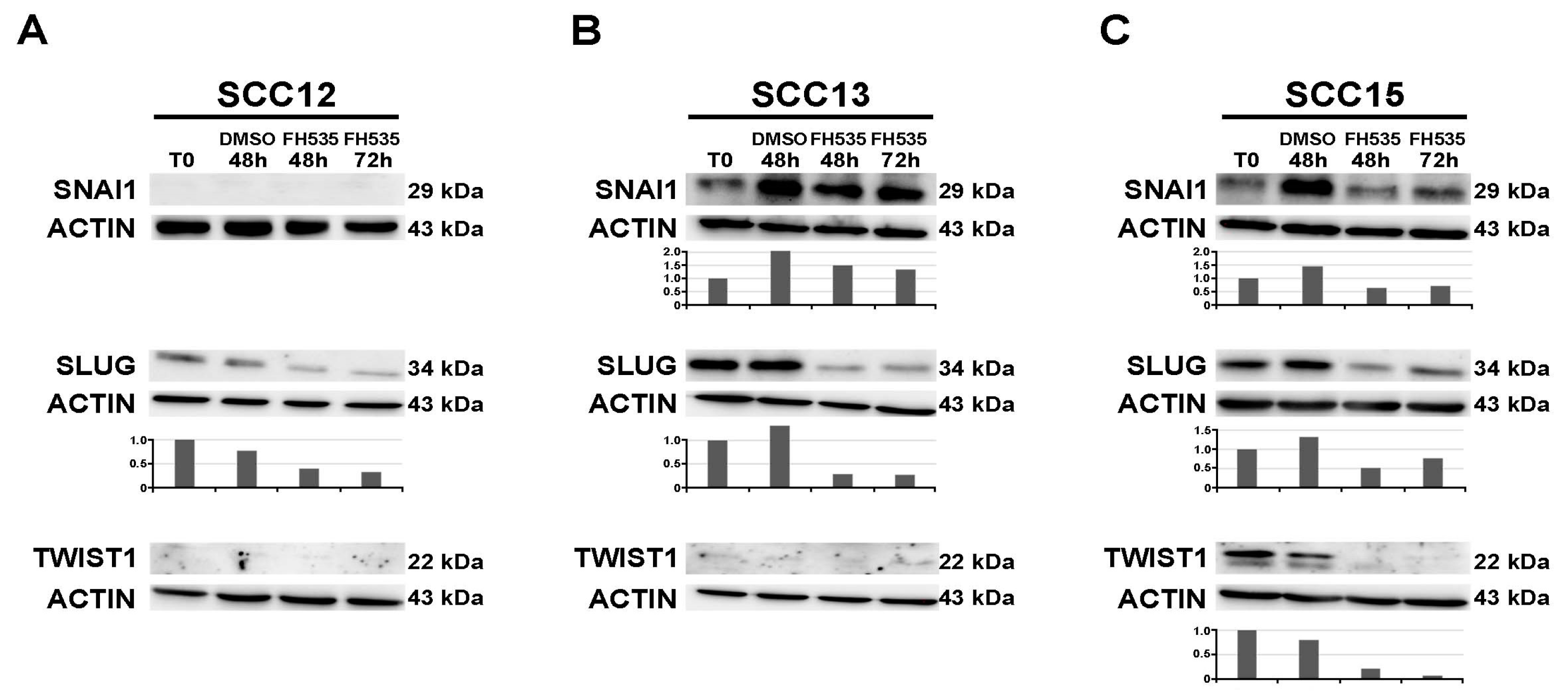
2.3. Treatment with the Wnt/CTNNB1 Inhibitor FH535 Induces ZFP36 Up-Regulation and the Downregulation of Transcriptional Repressors SNAI1, SLUG and TWIST
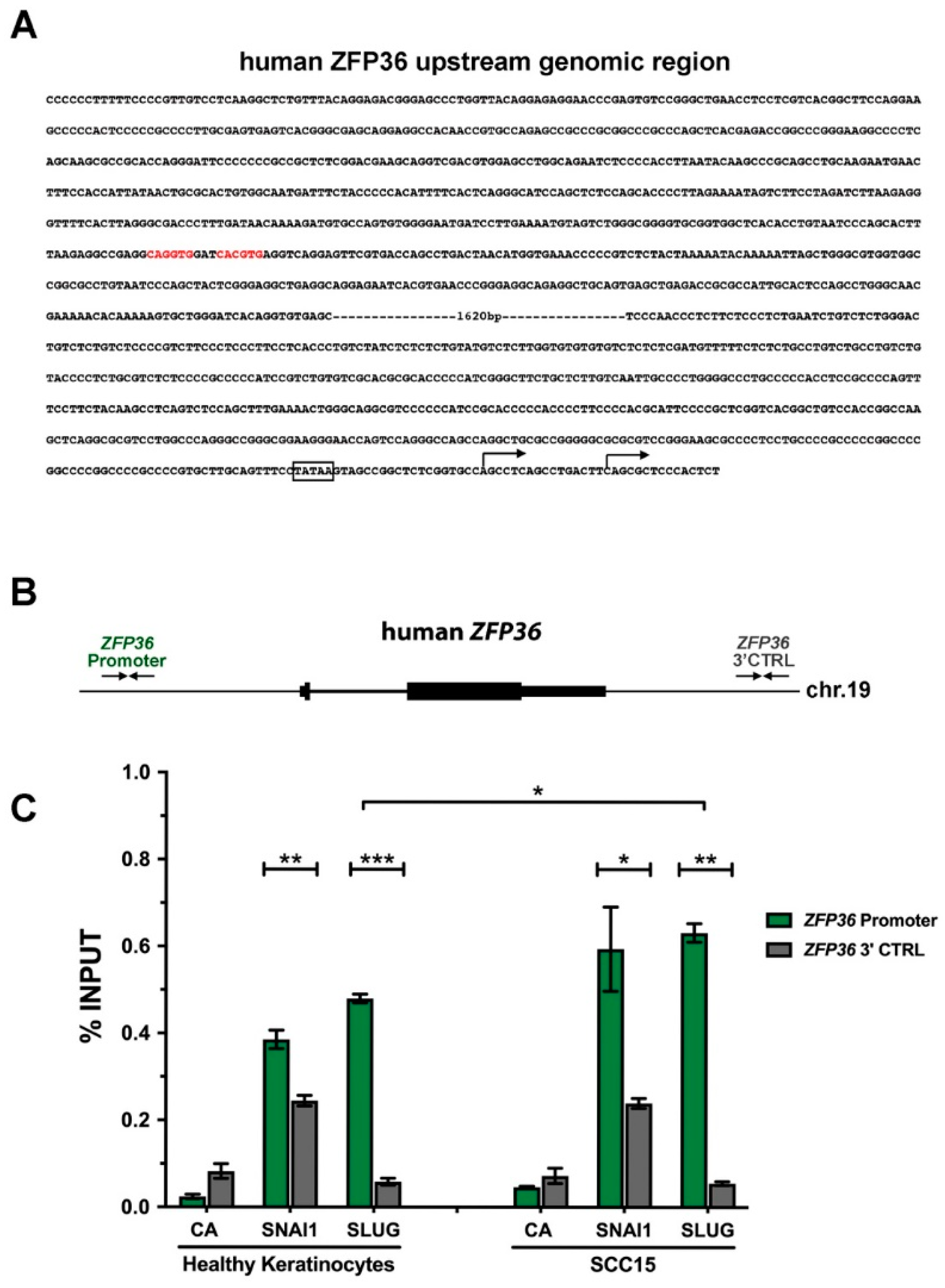
2.4. SNAI1 and SLUG Proteins Bind the ZFP36 Promoter Region Both in Healthy Keratinocytes and in SCC15 Cell Line
3. Discussion
4. Materials and Methods
4.1. Cell Cultures, Treatments and Transfections
4.2. Western Blot Analysis
4.3. Immunofluorescence
4.4. Chromatin Immunoprecipitation
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ARE | AU-rich element |
| ATCC | American Type Culture Collection |
| bHLH | basic Helix-Loop-Helix |
| bp | base pairs |
| cSCC | cutaneus Squamous Cell Carcinoma |
| CTNNB1 | β-catenin |
| DMEM | Dulbecco’s Modified Eagle Medium |
| DMSO | Dimethyl sulfoxide |
| EDTA | Ethylenediaminetetraacetic acid |
| EMT | Epithelial to Mesenchymal Transition |
| FAK | Focal Adhesion Kinase |
| FH535 | C13H10Cl2N2O4S |
| Ham’s | F12 Ham’s Nutrient Mixture F12 |
| HEPES | 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid |
| HRP | Horseradish peroxidase |
| KSC | Keratinocyte Stem Cells |
| PBS | Phosphate-Buffered Saline Phosphate-Buffered Saline |
| PCR | Polymerase Chain Reaction |
| RIPA | Radioimmunoprecipitation Assay |
| SDS | Sodium Dodecyl Sulfate |
| SEM | Standard Error of the Mean |
| SCC | Squamous Cell Carcinoma |
| TBST | Tris-buffered saline, 0.1% Tween 20 |
| TCF/LEF | T Cell Factor/Lymphoid Enhancer-binding Factor |
| TGF-β | Transforming Growth Factor β |
| TTP | Tristetraprolin |
| UTR | Un-Traslated Region |
| WREs | Wnt-Response-Element |
| ZFP36 | Zinc Finger Protein of 36 kDa |
References
- Taylor, G.A.; Lai, W.S.; Oakey, R.J.; Seldin, M.F.; Shows, T.B.; Eddy, R.L.; Blackshear, P.J. The human TTP protein: Sequence, alignment with related proteins, and chromosomal localization of the mouse and human genes. Nucleic Acids Res. 1991, 19, 3454. [Google Scholar] [CrossRef]
- Tiedje, C.; Diaz-Munoz, M.D.; Trulley, P.; Ahlfors, H.; Laaß, K.; Blackshear, P.J.; Turner, M.; Gaestel, M. The RNA-binding protein TTP is a global post-transcriptional regulator of feedback control in inflammation. Nucleic Acids Res. 2016, 44, 7418–7440. [Google Scholar] [CrossRef]
- Sanduja, S. The role of tristetraprolin in cancer and inflammation. Front. Biosci. 2012, 17, 174. [Google Scholar] [CrossRef]
- Tu, Y.; Wu, X.; Yu, F.; Dang, J.; Wang, J.; Wei, Y.; Cai, Z.; Zhou, Z.; Liao, W.; Li, L.; et al. Tristetraprolin specifically regulates the expression and alternative splicing of immune response genes in HeLa cells. BMC Immunol. 2019, 20, 13. [Google Scholar] [CrossRef]
- Brooks, S.A.; Blackshear, P.J. Tristetraprolin (TTP): Interactions with mRNA and proteins, and current thoughts on mechanisms of action. Biochim. Biophys. Acta (BBA)-Bioenerg. 2013, 1829, 666–679. [Google Scholar] [CrossRef]
- Montorsi, L.; Guizzetti, F.; Alecci, C.; Caporali, A.; Martello, A.; Atene, C.G.; Parenti, S.; Pizzini, S.; Zanovello, P.; Bortoluzzi, S.; et al. Loss of zfp36 expression in colorectal cancer correlates to wnt/ β-catenin activity and enhances epithelial-to-mesenchymal transition through upregulation of zeb1, sox9 and macc1. Oncotarget 2016, 7, 59144–59157. [Google Scholar] [CrossRef] [PubMed]
- Abe, A.; Emi, N.; Kanie, T.; Imagama, S.; Kuno, Y.; Takahashi, M.; Saito, H.; Naoe, T. Expression cloning of oligomerization-activated genes with cell-proliferating potency by pseudotype retrovirus vector. Biochem. Biophys. Res. Commun. 2004, 320, 920–926. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, L.; Moghrabi, W.; Khabar, K.S.; Hitti, E.G. Bi-phased regulation of the post-transcriptional inflammatory response by Tristetraprolin levels. RNA Boil. 2019, 16, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Ning, H.; Peng, H.; Wei, L.; Hou, R.; Hoft, D.F.; Liu, J. Tristetraprolin inhibits macrophage IL-27-induced activation of antitumour cytotoxic T cell responses. Nat. Commun. 2017, 8, 867. [Google Scholar] [CrossRef] [PubMed]
- Ross, C.R.; Brennan-Laun, S.E.; Wilson, G.M. Tristetraprolin: Roles in cancer and senescence. Ageing Res. Rev. 2012, 11, 473–484. [Google Scholar] [CrossRef]
- Park, J.-M.; Lee, T.-H.; Kang, T.H. Roles of Tristetraprolin in Tumorigenesis. Int. J. Mol. Sci. 2018, 19, 3384. [Google Scholar] [CrossRef] [PubMed]
- Palazzo, E.; Marconi, A.; Pincelli, C.; Morasso, M.I. Do DLX3 and CD271 Protect Human Keratinocytes from Squamous Tumor Development? Int. J. Mol. Sci. 2019, 20, 3541. [Google Scholar] [CrossRef] [PubMed]
- Dallaglio, K.; Marconi, A.; Pincelli, C. Survivin: A Dual Player in Healthy and Diseased Skin. J. Investig. Dermatol. 2012, 132, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, E.; Raghavan, S. Getting under the skin of epidermal morphogenesis. Nat. Rev. Genet. 2002, 3, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Dallaglio, K.; Petrachi, T.; Marconi, A.; Truzzi, F.; Lotti, R.; Saltari, A.; Morandi, P.; Puviani, M.; Maiorana, A.; Pincelli, C. Expression of nuclear survivin in normal skin and squamous cell carcinoma: A possible role in tumour invasion. Br. J. Cancer 2013, 110, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Dallaglio, K.; Petrachi, T.; Marconi, A.; Truzzi, F.; Lotti, R.; Saltari, A.; Morandi, P.; Puviani, M.; Maiorana, A.; Roop, D.R.; et al. Isolation and Characterization of Squamous Cell Carcinoma-Derived Stem-like Cells: Role in Tumor Formation. Int. J. Mol. Sci. 2013, 14, 19540–19555. [Google Scholar] [CrossRef] [PubMed]
- Morris, R.J. Keratinocyte stem cells: Targets for cutaneous carcinogens. J. Clin. Investig. 2000, 106, 3–8. [Google Scholar] [CrossRef]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Boil. 2014, 15, 178–196. [Google Scholar] [CrossRef]
- Thomson, T.M.; Balcells, C.; Cascante, M. Metabolic Plasticity and Epithelial-Mesenchymal Transition. J. Clin. Med. 2019, 8, 967. [Google Scholar] [CrossRef]
- Kalluri, R.; Robert, A.W. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef]
- Sánchez-Tilló, E.; Liu, Y.; De Barrios, O.; Siles, L.; Fanlo, L.; Cuatrecasas, M.; Darling, D.S.; Dean, D.C.; Castells, A.; Postigo, A. EMT-activating transcription factors in cancer: Beyond EMT and tumor invasiveness. Cell. Mol. Life Sci. 2012, 69, 3429–3456. [Google Scholar] [CrossRef] [PubMed]
- Schramm, H.M. The Epithelial-Myeloid-Transition (EMyeT) of cancer cells as a wrongly perceived primary inflammatory process eventually progressing to a bone remodeling malignancy: The alternative pathway for Epithelial- Mesenchymal-Transition hypothesis (EMT)? J. Cancer 2019, 10, 3798–3809. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Xiao, L.; Sun, L.; Liu, F. Wnt/β-Catenin Signaling: A Promising New Target for Fibrosis Diseases. Physiol. Res. 2012, 61, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Voronkov, A.; Krauss, S. Wnt/beta-Catenin Signaling and Small Molecule Inhibitors. Curr. Pharm. Des. 2012, 19, 634–664. [Google Scholar] [CrossRef]
- Daugherty, R.L.; Gottardi, C.J. Phospho-regulation of Beta-catenin adhesion and signaling functions. Physiology 2007, 22, 303–309. [Google Scholar] [CrossRef]
- Huber, O.; Korn, R.; McLaughlin, J.; Ohsugi, M.; Herrmann, B.G.; Kemler, R. Nuclear localization of beta-catenin by interaction with transcription factor LEF-1. Mech. Dev. 1996, 59, 3–10. [Google Scholar] [CrossRef]
- Gradl, D.; Kühl, M.; Wedlich, D. The Wnt/Wg Signal Transducer β-Catenin Controls Fibronectin Expression. Mol. Cell. Boil. 1999, 19, 5576–5587. [Google Scholar] [CrossRef]
- Shimizu, H.; Julius, M.A.; Giarré, M.; Zheng, Z.; Brown, A.M.; Kitajewski, J. Transformation by Wnt family proteins correlates with regulation of beta-catenin. Cell Growth Differ. Mol. Boil. J. Am. Assoc. Cancer Res. 1997, 8, 1349–1358. [Google Scholar]
- Hinck, L.; Nelson, W.; Papkoff, J. Wnt-1 modulates cell-cell adhesion in mammalian cells by stabilizing beta-catenin binding to the cell adhesion protein cadherin. J. Cell Boil. 1994, 124, 729–741. [Google Scholar] [CrossRef]
- Yoon, N.A.; Jo, H.G.; Lee, U.H.; Park, J.H.; Yoon, J.E.; Ryu, J.; Kang, S.S.; Min, Y.J.; Ju, S.-A.; Seo, E.H.; et al. Tristetraprolin suppresses the EMT through the down-regulation of Twist1 and Snail1 in cancer cells. Oncotarget 2016, 7, 8931–8943. [Google Scholar] [CrossRef]
- Xia, W.; Liu, Y.; Du, Y.; Cheng, T.; Hu, X.; Li, X. MicroRNA-423 Drug Resistance and Proliferation of Breast Cancer Cells by Targeting ZFP36. OncoTargets Ther. 2020, 13, 769–782. [Google Scholar] [CrossRef] [PubMed]
- O’Neil, J.; Ammit, A.; Clark, A.R. MAPK p38 regulates inflammatory gene expression via tristetraprolin: Doing good by stealth. Int. J. Biochem. Cell Boil. 2017, 94, 6–9. [Google Scholar] [CrossRef] [PubMed]
- Benassi, L.; Ottani, D.; Fantini, F.; Marconi, A.; Chiodino, C.; Giannetti, A.; Pincelli, C. 1,25-Dihydroxyvitamin D3, Transforming Growth Factor β1, Calcium, and Ultraviolet B Radiation Induce Apoptosis in Cultured Human Keratinocytes. J. Investig. Dermatol. 1997, 109, 276–282. [Google Scholar] [CrossRef] [PubMed]
- Fantini, S.; Salsi, V.; Vitobello, A.; Rijli, F.M.; Zappavigna, V. MicroRNA-196b is transcribed from an autonomous promoter and is directly regulated by Cdx2 and by posterior Hox proteins during embryogenesis. Biochim. Biophys. Acta (BBA)-Bioenerg. 2015, 1849, 1066–1080. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zanfi, E.D.; Fantini, S.; Lotti, R.; Bertesi, M.; Marconi, A.; Grande, A.; Manfredini, R.; Pincelli, C.; Zanocco-Marani, T. Wnt/CTNNB1 Signal Transduction Pathway Inhibits the Expression of ZFP36 in Squamous Cell Carcinoma, by Inducing Transcriptional Repressors SNAI1, SLUG and TWIST. Int. J. Mol. Sci. 2020, 21, 5692. https://doi.org/10.3390/ijms21165692
Zanfi ED, Fantini S, Lotti R, Bertesi M, Marconi A, Grande A, Manfredini R, Pincelli C, Zanocco-Marani T. Wnt/CTNNB1 Signal Transduction Pathway Inhibits the Expression of ZFP36 in Squamous Cell Carcinoma, by Inducing Transcriptional Repressors SNAI1, SLUG and TWIST. International Journal of Molecular Sciences. 2020; 21(16):5692. https://doi.org/10.3390/ijms21165692
Chicago/Turabian StyleZanfi, Emma D., Sebastian Fantini, Roberta Lotti, Matteo Bertesi, Alessandra Marconi, Alexis Grande, Rossella Manfredini, Carlo Pincelli, and Tommaso Zanocco-Marani. 2020. "Wnt/CTNNB1 Signal Transduction Pathway Inhibits the Expression of ZFP36 in Squamous Cell Carcinoma, by Inducing Transcriptional Repressors SNAI1, SLUG and TWIST" International Journal of Molecular Sciences 21, no. 16: 5692. https://doi.org/10.3390/ijms21165692
APA StyleZanfi, E. D., Fantini, S., Lotti, R., Bertesi, M., Marconi, A., Grande, A., Manfredini, R., Pincelli, C., & Zanocco-Marani, T. (2020). Wnt/CTNNB1 Signal Transduction Pathway Inhibits the Expression of ZFP36 in Squamous Cell Carcinoma, by Inducing Transcriptional Repressors SNAI1, SLUG and TWIST. International Journal of Molecular Sciences, 21(16), 5692. https://doi.org/10.3390/ijms21165692








