Spectroscopic Investigation of the Kinetic Mechanism Involved in the Association of Potyviral VPg with the Host Plant Translation Initiation Factor eIF4E
Abstract
1. Introduction
2. Results and Discussions
2.1. Investigation of the eIF4E–VPg Interaction Domains
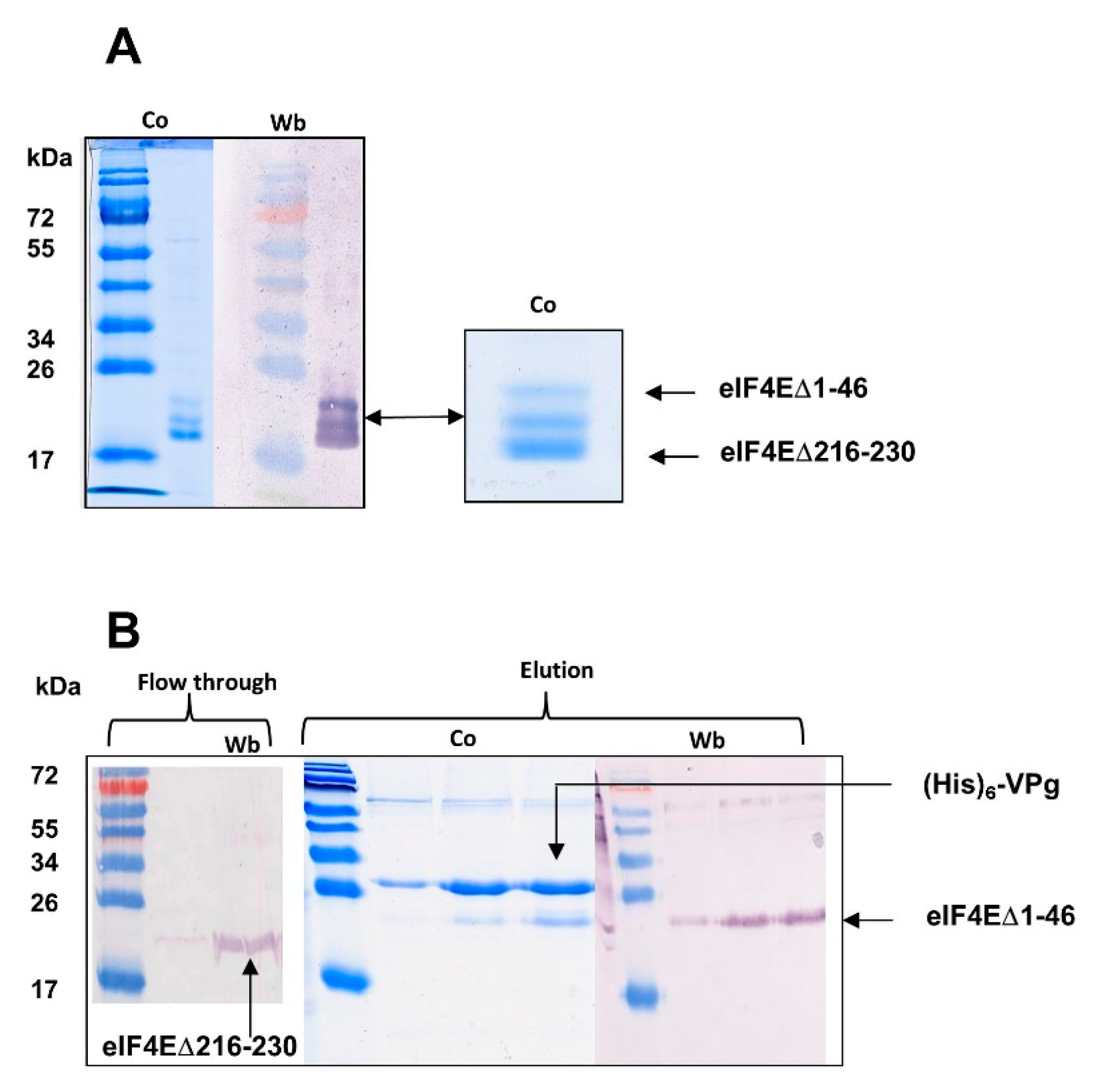
2.1.1. The eIF4E C-ter Domain Is Involved in the Interaction with VPg
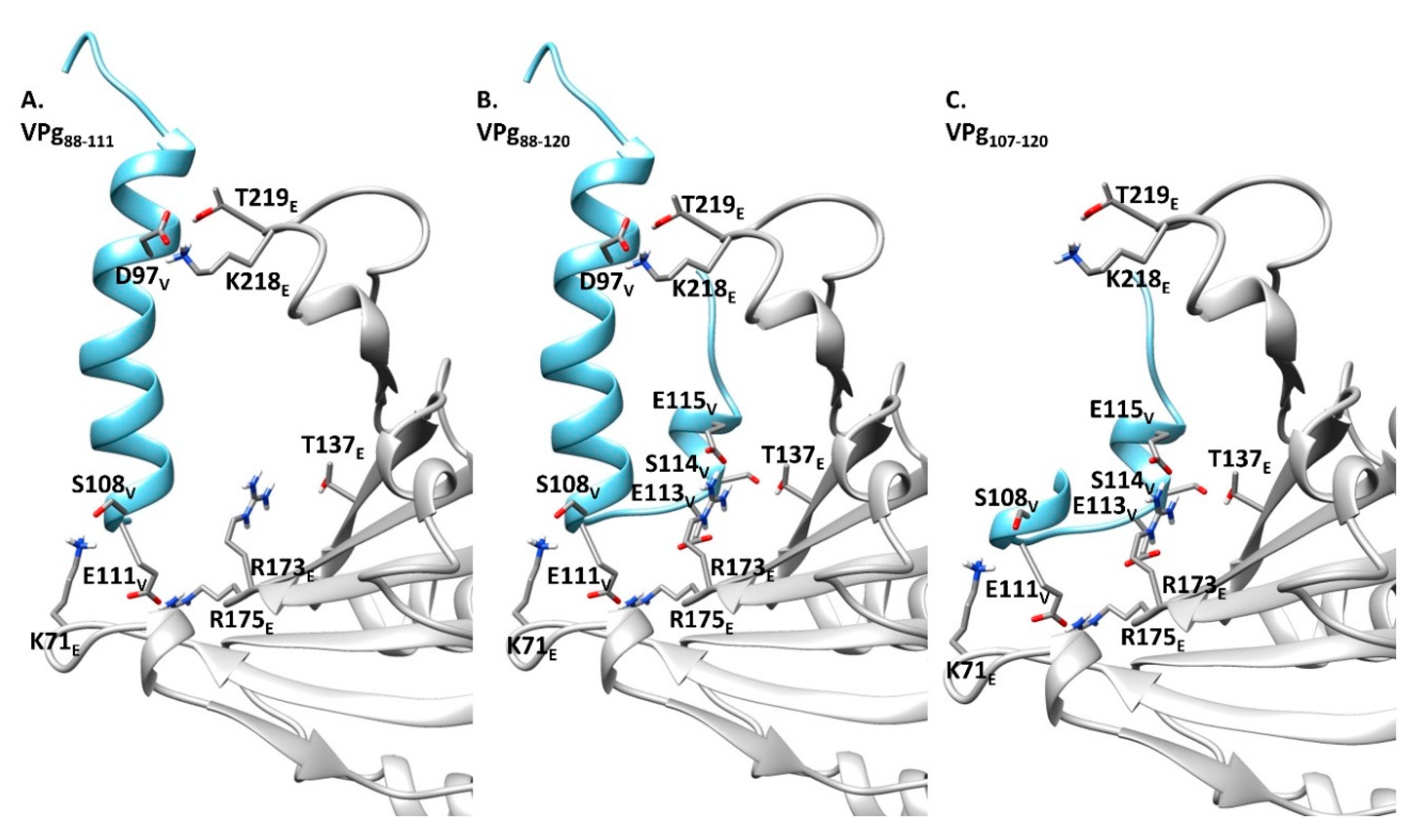
2.1.2. Sites, Which in the VPg Central Region Determine Host Resistance Breakdown, Contribute to the Mechanism of VPg-eIF4E Interaction
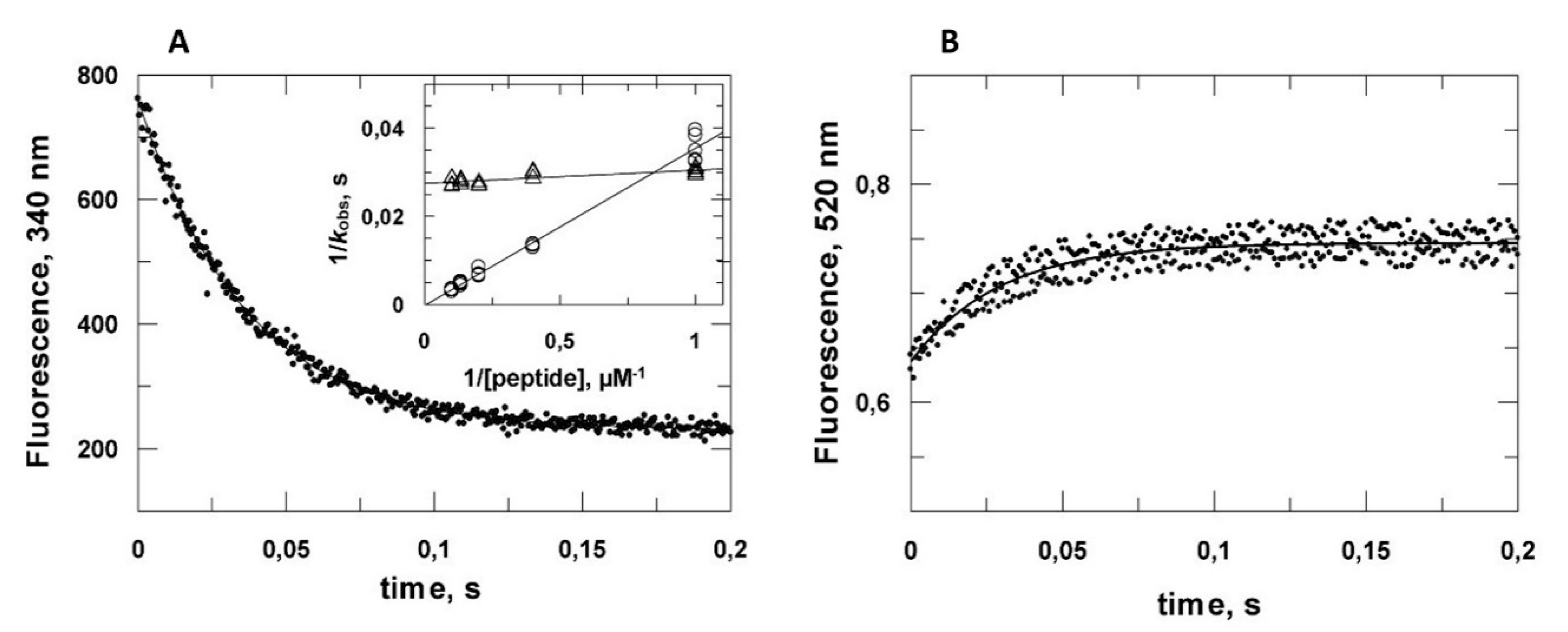
2.2. Kinetic Analysis of the VPg Central Domain Association with eIF4E
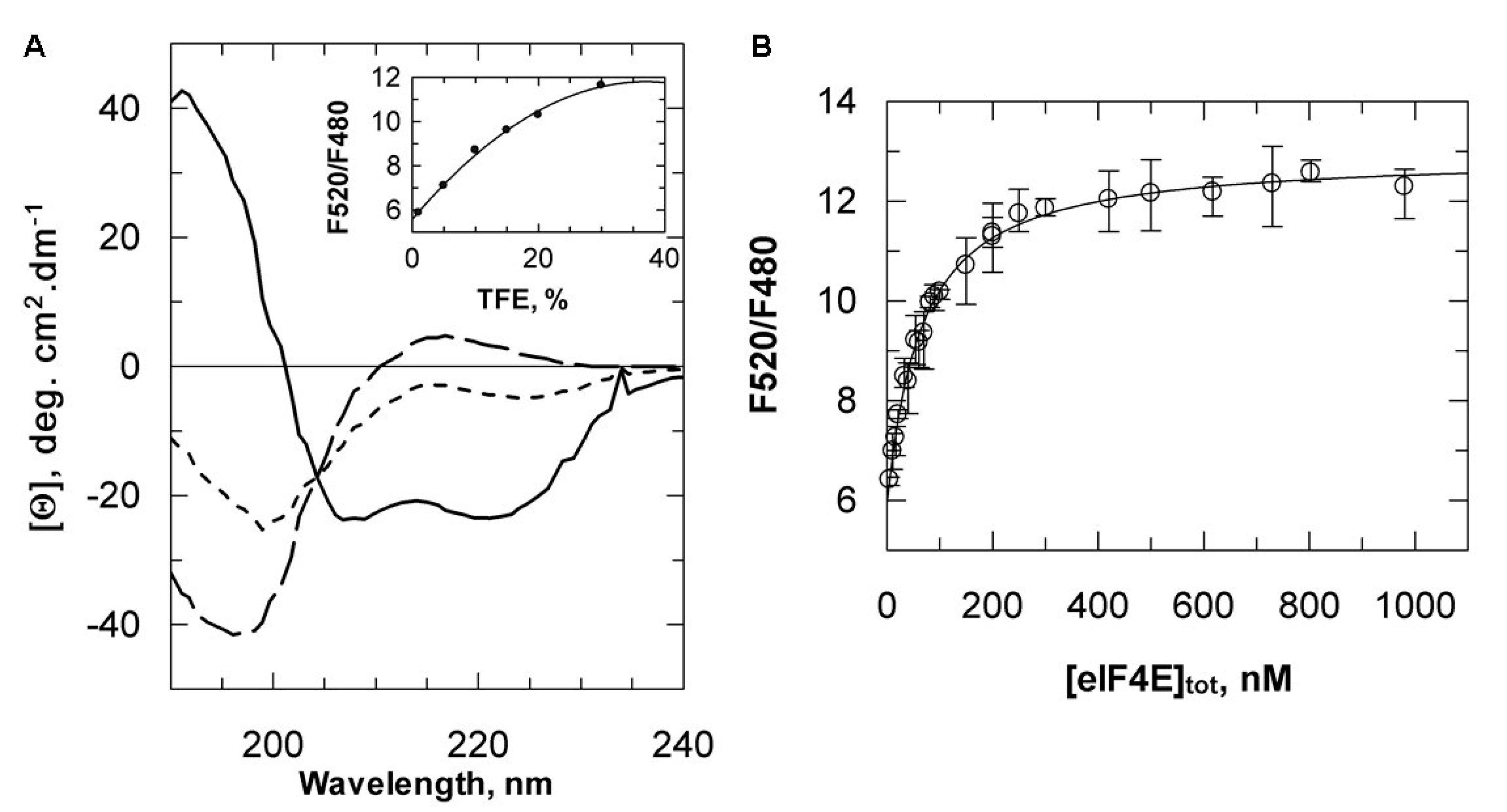
2.3. Reassessment of the Binding Properties of the VPg Central Region with eIF4E
3. Experimental Procedures
3.1. Protein Preparation
3.2. VPg(88–111) Dual Labeling
3.3. Circular Dichroism
3.4. Fluorescence Measurements
3.5. Fret Measurements
3.6. Data Analysis
3.7. Protein Modeling
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| LMV | lettuce mosaic virus |
| PVY | potato virus Y |
| TEV | tobacco etch virus |
| TuMV | turnip mosaic virus |
| IAEDANS | N′-(iodoacetyl)-N′-(5-sulfo-1-naphtyl)ethylenediamine |
| His6 eIF4E | N-ter hexahistidine-tagged lettuce eIF4E |
| His6 VPg | N-ter hexahistidine-tagged LMV VPg |
| eIF4EΔ1−46 | untagged lettuce eIF4E deleted from its first 46 N-ter amino acids |
| His6 VPg* | IAEDANS-labeled N-ter hexahistidine-tagged LMV VPg |
| His6 eIF4E* | IAEDANS-labeled N-ter hexahistidine-tagged lettuce eIF4E |
| eIF4E* | IAEDANS-labeled untagged lettuce eIF4E |
| eIF4EΔ1−46* | IAEDANS-labeled untagged lettuce eIF4E deleted from its first 46 N-ter amino acids |
| [His6 VPg*–His6 eIF4E] | IAEDANS-labeled binary complex |
References
- Adams, M.; Zerbini, F.; French, R.; Rabenstein, F.; Stenger, D.; Valkonen, J. Potyviridae. In Virus Taxonomy, 9th Report of the International Committee for Taxonomy of Viruses; King, A.M.Q., Adams, M.J., Carstens, E.B., Lefkowitz, E.J., Eds.; Elsevier Academic Press: San Diego, CA, USA, 2011. [Google Scholar]
- Nicaise, V. Crop immunity against viruses: Outcomes and future challenges. Front Plant Sci. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Revers, F.; García, J.A. Molecular Biology of Potyviruses. In Advances in Virus Research; Maramorosch, K., Murphy, F.A., Shatkin, A.J., Eds.; Academic Press: Cambridge, MA, USA, 2015; Volume 92, pp. 101–199. [Google Scholar]
- Murphy, J.F.; Klein, P.G.; Hunt, A.G.; Shaw, J.G. Replacement of the tyrosine residue that links a potyviral VPg to the viral RNA is lethal. Virology 1996, 220, 535–538. [Google Scholar] [CrossRef] [PubMed]
- Murphy, J.F.; Rychlik, W.; Rhoads, R.E.; Hunt, A.G.; Shaw, J.G. A tyrosine residue in the small nuclear inclusion protein of tobacco vein mottling virus links the VPg to the viral RNA. J. Virol 1991, 65, 511–513. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Laliberte, J.F. The genome-linked protein VPg of plant viruses-a protein with many partners. Curr. Opin. Virol. 2011, 1, 347–354. [Google Scholar] [CrossRef]
- Martínez, F.; Rodrigo, G.; Aragonés, V.; Ruiz, M.; Lodewijk, I.; Fernández, U.; Santiago, F.E.; Daros, J.-A. Interaction network of tobacco etch potyvirus NIa protein with the host proteome during infection. BMC Genom. 2016, 17, 87. [Google Scholar] [CrossRef]
- Charron, C.; Nicolaï, M.; Gallois, J.L.; Robaglia, C.; Moury, B.; Palloix, A.; Caranta, C. Natural variation and functional analyses provide evidence for co-evolution between plant eIF4E and potyviral VPg. Plant J. 2008, 54, 56–68. [Google Scholar] [CrossRef]
- Leonard, S.; Plante, D.; Wittmann, S.; Daigneault, N.; Fortin, M.G.; Laliberte, J.F. Complex formation between potyvirus VPg and translation eukaryotic initiation factor 4E correlates with virus infectivity. J. Virol. 2000, 74, 7730–7737. [Google Scholar] [CrossRef]
- Robaglia, C.; Caranta, C. Translation initiation factors: A weak link in plant RNA virus infection. Trends Plant Sci. 2006, 11, 40–45. [Google Scholar] [CrossRef]
- Gallie, D.R. Cap-independent translation conferred by the 5′ leader of tobacco etch virus is eukaryotic initiation factor 4G dependent. J. Virol. 2001, 75, 12141–12152. [Google Scholar] [CrossRef]
- Eskelin, K.; Hafren, A.; Rantalainen, K.I.; Makinen, K. Potyviral VPg enhances viral RNA Translation and inhibits reporter mRNA translation in planta. J. Virol. 2011, 85, 9210–9221. [Google Scholar] [CrossRef]
- Khan, M.A. Phosphorylation of Translation Initiation Factor eIFiso4E Promotes Translation Through Enhanced Binding to Potyvirus VPg. J. Biochem. 2018, 165, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Tavert-Roudet, G.; Anne, A.; Barra, A.; Chovin, A.; Demaille, C.; Michon, T. The potyvirus particle recruits the plant translation initiation factor eIF4E by means of the VPg covalently linked to the viral RNA. Mol. Plant-Microbe Interact. 2017, 30, 754–762. [Google Scholar] [CrossRef] [PubMed]
- Moury, B.; Charron, C.; Janzac, B.; Simon, V.; Gallois, J.L.; Palloix, A.; Caranta, C. Evolution of plant eukaryotic initiation factor 4E (eIF4E) and potyvirus genome-linked protein (VPg): A game of mirrors impacting resistance spectrum and durability. Infect. Genet. Evol. 2014, 27, 472–480. [Google Scholar] [CrossRef] [PubMed]
- Ayme, V.; Souche, S.; Caranta, C.; Jacquemond, M.; Chadœuf, J.; Palloix, A.; Moury, B. Different Mutations in the Genome-Linked Protein VPg of Potato virus Y Confer Virulence on the pvr2 3 Resistance in Pepper. Mol. Plant-Microbe Interact. 2006, 19, 557–563. [Google Scholar] [CrossRef]
- Ayme, V.; Petit-Pierre, J.; Souche, S.; Palloix, A.; Moury, B. Molecular dissection of the potato virus Y VPg virulence factor reveals complex adaptations to the pvr2 resistance allelic series in pepper. J. Gen. Virol. 2007, 88, 1594–1601. [Google Scholar] [CrossRef]
- Roudet-Tavert, G.; Michon, T.; Walter, J.; Delaunay, T.; Redondo, E.; Le Gall, O. Central domain of a potyvirus VPg is involved in the interaction with the host translation initiation factor eIF4E and the viral protein HcPro. J. Gen. Virol. 2007, 88, 1029–1033. [Google Scholar] [CrossRef]
- Walter, J.; Barra, A.; Doublet, B.; Céré, N.; Charon, J.; Michon, T. Hydrodynamic Behavior of the Intrinsically Disordered Potyvirus Protein VPg, of the Translation Initiation Factor eIF4E and of their Binary Complex. Int. J. Mol. Sci. 2019, 20, 1794. [Google Scholar] [CrossRef]
- Sabharwal, P.; Srinivas, S.; Savithri, H.S. Mapping the domain of interaction of PVBV VPg with NIa-Pro: Role of N-terminal disordered region of VPg in the modulation of structure and function. Virology 2018, 524, 18–31. [Google Scholar] [CrossRef]
- Coutinho de Oliveira, L.; Volpon, L.; Rahardjo, A.K.; Osborne, M.J.; Culjkovic-Kraljacic, B.; Trahan, C.; Oeffinger, M.; Kwok, B.H.; Borden, K.L.B. Structural studies of the eIF4E–VPg complex reveal a direct competition for capped RNA: Implications for translation. Proc. Natl. Acad. Sci. USA 2019, 116, 24056–24065. [Google Scholar] [CrossRef]
- Tavert-Roudet, G.; Abdul-Razzak, A.; Doublet, B.; Walter, J.; Delaunay, T.; German-Retana, S.; Michon, T.; Le Gall, O.; Candresse, T. The C terminus of lettuce mosaic potyvirus cylindrical inclusion helicase interacts with the viral VPg and with lettuce translation eukaryotic initiation factor 4E. J. Gen. Virol. 2012, 93, 184–193. [Google Scholar] [CrossRef]
- Nicaise, V.; German-Retana, S.; Sanjuán, R.; Dubrana, M.P.; Mazier, M.; Maisonneuve, B.; Candresse, T.; Caranta, C.; Le Gall, O. The eukaryotic translation initiation factor 4E controls lettuce susceptibility to the potyvirus Lettuce mosaic virus. Plant Physiol. 2003, 132, 1272–1282. [Google Scholar] [CrossRef] [PubMed]
- Okade, H.; Fujita, Y.; Miyamoto, S.; Tomoo, K.; Muto, S.; Miyoshi, H.; Natsuaki, T.; Rhoads, D.E.; Ishida, T. Turnip mosaic virus genome-linked protein VPg binds C-terminal region of cap-bound initiation factor 4E orthologue without exhibiting host cellular specificity. J. Biochem. 2009, 145, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Grzela, R.; Strokovska, L.; Andrieu, J.P.; Dublet, B.; Zagorski, W.; Chroboczek, J. Potyvirus terminal protein VPg, effector of host eukaryotic initiation factor eIF4E. Biochimie 2006, 88, 887–896. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Miyoshi, H.; Gallie, D.R.; Goss, D.J. Potyvirus genome-linked protein, VPg, directly affects wheat germ in vitro translation: Interactions with translation initiation factors eIF4F and eIFiso4F. J. Biol. Chem. 2008, 283, 1340–1349. [Google Scholar] [CrossRef]
- Perez, K.; Yeam, I.; Kang, B.C.; Ripoll, D.R.; Kim, J.; Murphy, J.F.; Jahn, M.M. Tobacco etch virus infectivity in Capsicum spp. is determined by a maximum of three amino acids in the viral virulence determinant VPg. Mol. Plant-Microbe Interact. 2012, 25, 1562–1573. [Google Scholar] [CrossRef]
- Charon, J.; Barra, A.; Walter, J.; Millot, P.; Hébrard, E.; Moury, B.; Michon, T. First experimental assessment of protein intrinsic disorder involvement in an RNA virus natural adaptive process. Mol. Biol. Evol. 2018, 35, 38–49. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 3rd ed.; Springer: New York, NY, USA, 2006; pp. 443–453. [Google Scholar]
- Wu, P.; Brand, L. Resonance energy transfer: Methods and applications. Anal. Biochem. 1994, 218, 1–13. [Google Scholar] [CrossRef]
- Muñoz, V.; Ramanathan, R. Waltzing α-helices. Proc. Natl. Acad. Sci. USA 2009, 106, 1299–1300. [Google Scholar] [CrossRef]
- Thompson, P.A.; Muñoz, V.; Jas, G.S.; Henry, E.R.; Eaton, W.A.; Hofrichter, J. The Helix-Coil Kinetics of a Heteropeptide. J. Phys. Chem. B 2000, 104, 378–389. [Google Scholar] [CrossRef]
- Habchi, J.; Longhi, S. Structural disorder within paramyxovirus nucleoproteins and phosphoproteins. Mol. Biosyst. 2012, 8, 69–81. [Google Scholar] [CrossRef]
- Dosnon, M.; Bonetti, D.; Morrone, A.; Erales, J.; di Silvio, E.; Longhi, S.; Gianny, S. Demonstration of a folding after binding mechanism in the recognition between the measles virus NTAIL and X domains. ACS Chem. Biol. 2015, 10, 795–802. [Google Scholar] [CrossRef] [PubMed]
- German-Retana, S.; Walter, J.; Doublet, B.; Roudet-Tavert, G.; Nicaise, V.; Lecampion, C.; Houvenaghel, M.-C.; Robaglia, C.; Michon, T.; Le Gall, O. Mutational analysis of plant cap-binding protein eIF4E reveals key amino acids involved in biochemical functions and potyvirus infection. J. Virol. 2008, 82, 7601–7612. [Google Scholar] [CrossRef] [PubMed]
- Michon, T.; Estevez, Y.; Walter, J.; German-Retana, S.; Le Gall, O. The potyviral virus genome-linked protein VPg forms a ternary complex with the eukaryotic initiation factors eIF4E and eIF4G and reduces eIF4E affinity for a mRNA cap analogue. FEBS J. 2006, 273, 1312–1322. [Google Scholar] [CrossRef] [PubMed]
- Whitmore, L.; Wallace, B.A. Protein secondary structure analyses from circular dichroism spectroscopy: Methods and reference databases. Biopolymers 2008, 89, 392–400. [Google Scholar] [CrossRef]
- Stryer, L. Fluorescence Energy Transfer as a Spectroscopic Ruler. Ann. Rev. Biochem. 1978, 47, 819–846. [Google Scholar] [CrossRef]
- Wang, C.-K.; Cheung, H.C. Proximity relationship in the binary complex formed between troponin I and troponin C. J. Mol. Biol. 1986, 191, 509–521. [Google Scholar] [CrossRef]
- Marsh, D.J.; Lowey, S. Fluorescence energey transfer in myosin subfragment-1. Biochemistry 1980, 19, 774–784. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
- Schymkowitz, J.; Borg, J.; Stricher, F.; Nys, R.; Rousseau, F.; Serrano, L. The FoldX web server: An online force field. Nucleic Acids Res. 2005, 33, W382–W388. [Google Scholar] [CrossRef]
- Van Zundert, G.C.P.; Rodrigues, J.P.G.L.M.; Trellet, M.; Schmitz, C.; Kastritis, P.L.; Karaca, E.; Melquiond, A.S.J.; van Dijk, M.; de Vries, S.J.; Bonvin, A.M.J.J. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J. Mol. Biol. 2016, 428, 720–725. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]


| Dissociation Constants (KD × 109, mol/L) at 25 °C | ||
|---|---|---|
| VPg | m7GTP | |
| eIF4E0 | 242 ± 70 | 307 ± 27 |
| eIF4E1 | 340 ± 80 | 261 ± 43 |
| eIF4E2 | 288 ± 30 | 351 ± 18 |
| eIF4E0 [∆1–46] | 276 ± 17 | 242 ± 33 |
| eIF4E0-m7GTP ♦ | 746 ± 98 | nd |
| eIF4E0–VPg ♦ | nd | 3000 ± 239 |
| eIF4E1-m7GTP ♦ | 632 ± 29 | nd |
| eIF4E2-m7GTP ♦ | 703 ± 55 | nd |
| Sequence | KD (nmol/L) | |
|---|---|---|
| VPg88–111 | VFSDIGLVQDAFGKERLKLLSGGE | 72 ± 11 |
| VPg88–120 | VFSDIGLVQDAFGKERLKLLSGGEIESEHMRSG | 67 ± 7 |
| VPg107–120 | LSGGEIESEHMRSG | 852 ± 57 |
| PVY VPg–Human eIF4E | LMV VPg–Lettuce eIF4E | ||
|---|---|---|---|
| eIF4E | VPg | VPg | eIF4E |
| K206 | E98 | D97 | K218 |
| S207 | E98 | D97 | T219 |
| K52 | E109 | S108 | K71 |
| K159 | D111 | E111 | R175 |
| R157 | E114 | E113 | R173 |
| K159 | E114 | E113 | R175 |
| R157 | L118 | E115 | R173 |
| R157 T116 | M115 M115 | S114 S114 | R173 T137 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Walter, J.; Barra, A.; Charon, J.; Tavert-Roudet, G.; Michon, T. Spectroscopic Investigation of the Kinetic Mechanism Involved in the Association of Potyviral VPg with the Host Plant Translation Initiation Factor eIF4E. Int. J. Mol. Sci. 2020, 21, 5618. https://doi.org/10.3390/ijms21165618
Walter J, Barra A, Charon J, Tavert-Roudet G, Michon T. Spectroscopic Investigation of the Kinetic Mechanism Involved in the Association of Potyviral VPg with the Host Plant Translation Initiation Factor eIF4E. International Journal of Molecular Sciences. 2020; 21(16):5618. https://doi.org/10.3390/ijms21165618
Chicago/Turabian StyleWalter, Jocelyne, Amandine Barra, Justine Charon, Geneviève Tavert-Roudet, and Thierry Michon. 2020. "Spectroscopic Investigation of the Kinetic Mechanism Involved in the Association of Potyviral VPg with the Host Plant Translation Initiation Factor eIF4E" International Journal of Molecular Sciences 21, no. 16: 5618. https://doi.org/10.3390/ijms21165618
APA StyleWalter, J., Barra, A., Charon, J., Tavert-Roudet, G., & Michon, T. (2020). Spectroscopic Investigation of the Kinetic Mechanism Involved in the Association of Potyviral VPg with the Host Plant Translation Initiation Factor eIF4E. International Journal of Molecular Sciences, 21(16), 5618. https://doi.org/10.3390/ijms21165618





