Comprehensive Analysis of Expression, Clinicopathological Association and Potential Prognostic Significance of RABs in Pancreatic Cancer
Abstract
1. Introduction
2. Results
2.1. Differential Expression of RAB Transcripts in Pancreatic Tumor Tissues and Association with Clinical Progression
2.2. Association of RABs Expression with Race, Gender, and Age of Pancreatic Cancer Patients
2.3. Association of RABs Expression in Pancreatic Tumors with Drinking Habits and Diagnoses of Diabetes and Pancreatitis in Patients
2.4. Prognostic Potential of RABs Expression in Pancreatic Ductal Adenocarcinoma
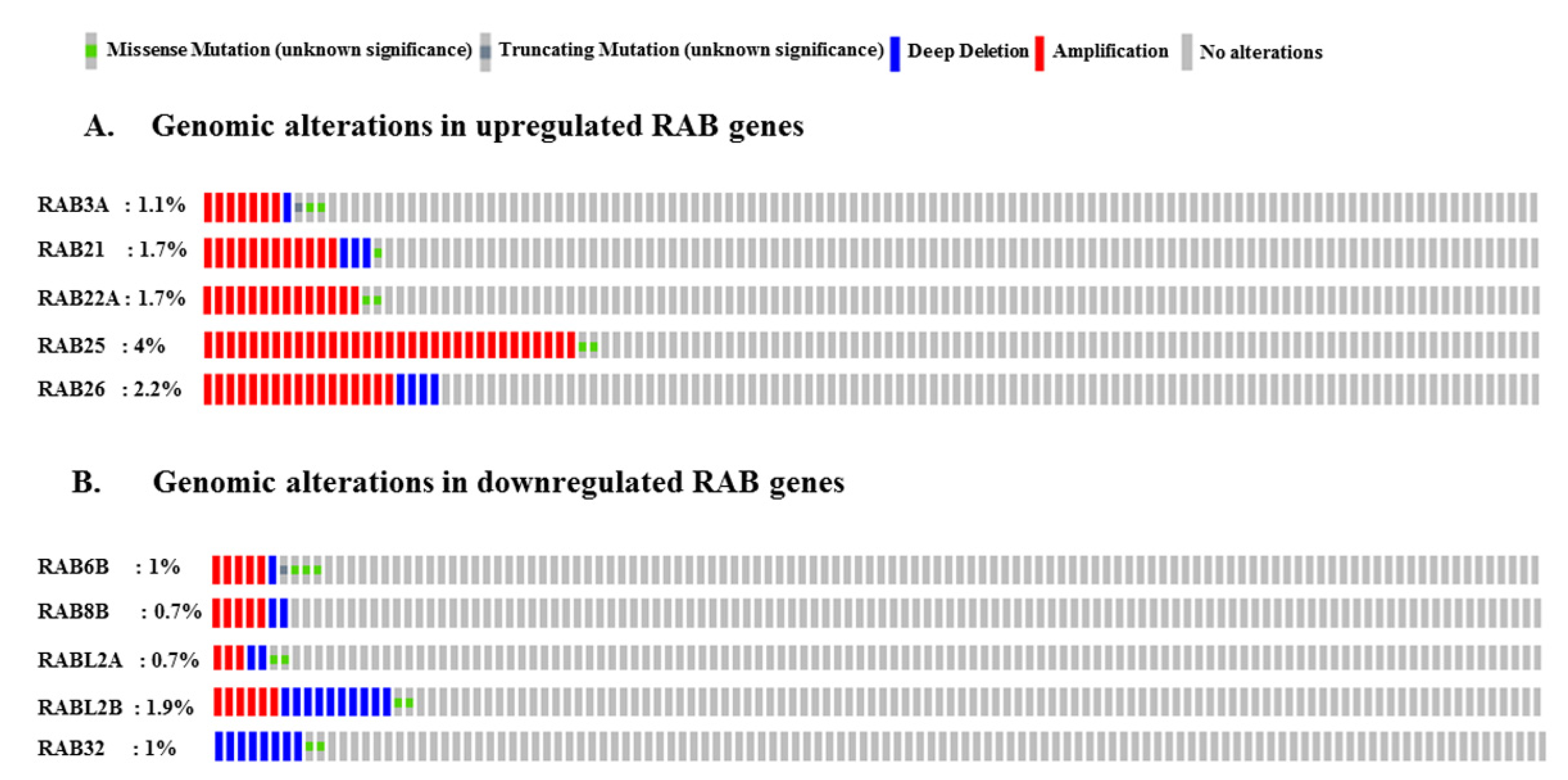
2.5. Genomic Alterations in RAB Genes Associated with Pancreatic Cancer
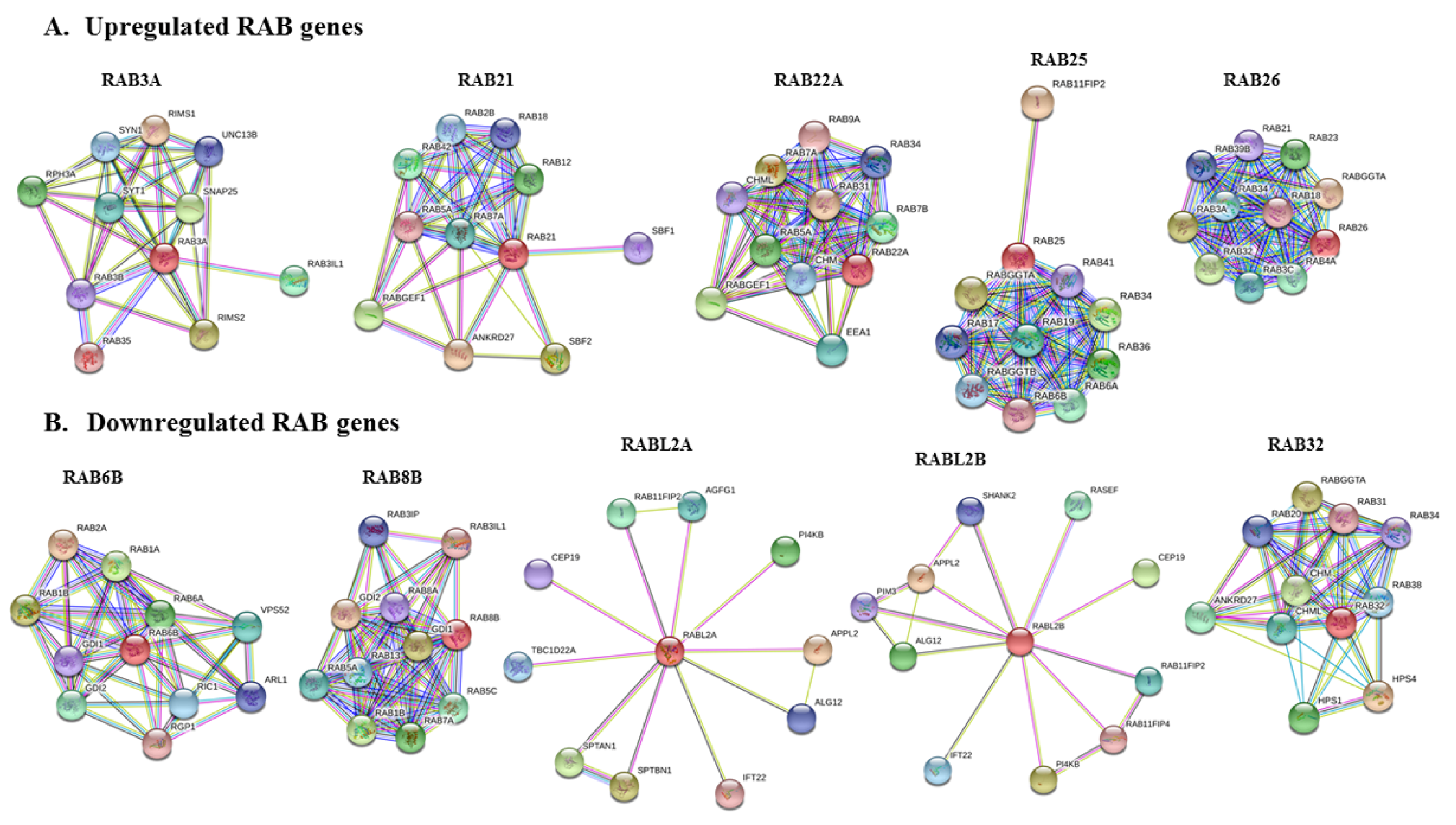
2.6. Protein–Protein Interaction Network of Selected RABs
3. Discussion
4. Materials and Methods
4.1. Gene Expression Analysis Using UALCAN
4.2. Prognostic Significance and In Situ Expression of Differentially Expressed RAB Genes
4.3. Genomic Alterations Analysis and Protein–Protein Interaction Network of Selected RAB Genes
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AA | African American |
| CA | Caucasian American |
| PDAC | Pancreatic ductal adenocarcinoma |
| TCGA | The Cancer Genome Atlas |
References
- Rawla, P.; Sunkara, T.; Gaduputi, V. Epidemiology of Pancreatic Cancer: Global Trends, Etiology and Risk Factors. World J. Oncol. 2019, 10, 10–27. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef] [PubMed]
- Wolfgang, C.L.; Herman, J.M.; Laheru, D.A.; Klein, A.P.; Erdek, M.A.; Fishman, E.K.; Hruban, R.H. Recent progress in pancreatic cancer. CA Cancer J. Clin. 2013, 63, 318–348. [Google Scholar] [CrossRef]
- Chand, S.; O’Hayer, K.; Blanco, F.F.; Winter, J.M.; Brody, J.R. The Landscape of Pancreatic Cancer Therapeutic Resistance Mechanisms. Int. J. Biol. Sci. 2016, 12, 273–282. [Google Scholar] [CrossRef]
- Capurso, G.; Sette, C. Drug resistance in pancreatic cancer: New player caught in act. EBiomed 2019, 40, 39–40. [Google Scholar] [CrossRef]
- Khan, M.A.; Srivastava, S.K.; Zubair, H.; Patel, G.K.; Arora, S.; Khushman, M.; Carter, J.E.; Gorman, G.S.; Singh, S.; Singh, A.P. Co-targeting of CXCR4 and hedgehog pathways disrupts tumor-stromal crosstalk and improves chemotherapeutic efficacy in pancreatic cancer. J. Biol. Chem. 2020, 295, 8413–8424. [Google Scholar] [CrossRef]
- Chakravarthi, B.V.; Nepal, S.; Varambally, S. Genomic and Epigenomic Alterations in Cancer. Am. J. Pathol. 2016, 186, 1724–1735. [Google Scholar] [CrossRef]
- Khan, M.A.; Azim, S.; Zubair, H.; Bhardwaj, A.; Patel, G.K.; Khushman, M.; Singh, S.; Singh, A.P. Molecular Drivers of Pancreatic Cancer Pathogenesis: Looking Inward to Move Forward. Int. J. Mol. Sci. 2017, 18, 779. [Google Scholar] [CrossRef]
- Rojas, A.M.; Fuentes, G.; Rausell, A.; Valencia, A. The Ras protein superfamily: Evolutionary tree and role of conserved amino acids. J. Cell Biol. 2012, 196, 189–201. [Google Scholar] [CrossRef]
- Qu, L.; Pan, C.; He, S.M.; Lang, B.; Gao, G.D.; Wang, X.L.; Wang, Y. The Ras Superfamily of Small GTPases in Non-neoplastic Cerebral Diseases. Front. Mol. Neurosci. 2019, 12, 121. [Google Scholar] [CrossRef] [PubMed]
- Pereira-Leal, J.B.; Seabra, M.C. Evolution of the Rab family of small GTP-binding proteins. J. Mol. Biol. 2001, 313, 889–901. [Google Scholar] [CrossRef] [PubMed]
- Pylypenko, O.; Hammich, H.; Yu, I.M.; Houdusse, A. Rab GTPases and their interacting protein partners: Structural insights into Rab functional diversity. Small GTPases 2018, 9, 22–48. [Google Scholar] [CrossRef] [PubMed]
- Segev, N. Ypt/rab gtpases: Regulators of protein trafficking. Sci. STKE 2001. [Google Scholar] [CrossRef]
- Zerial, M.; McBride, H. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2001, 2, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Stenmark, H. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 2009, 10, 513–525. [Google Scholar] [CrossRef]
- Zhen, Y.; Stenmark, H. Cellular functions of Rab GTPases at a glance. J. Cell Sci. 2015, 128, 3171–3176. [Google Scholar] [CrossRef]
- Hutagalung, A.H.; Novick, P.J. Role of Rab GTPases in membrane traffic and cell physiology. Physiol. Rev. 2011, 91, 119–149. [Google Scholar] [CrossRef]
- Tzeng, H.T.; Wang, Y.C. Rab-mediated vesicle trafficking in cancer. J. Biomed. Sci. 2016, 23, 70. [Google Scholar] [CrossRef]
- Yang, J.; Liu, W.; Lu, X.; Fu, Y.; Li, L.; Luo, Y. High expression of small GTPase Rab3D promotes cancer progression and metastasis. Oncotarget 2015, 6, 11125–11138. [Google Scholar] [CrossRef]
- Silva, P.; Mendoza, P.; Rivas, S.; Diaz, J.; Moraga, C.; Quest, A.F.G.; Torres, V.A. Hypoxia promotes Rab5 activation, leading to tumor cell migration, invasion and metastasis. Oncotarget 2016, 7, 29548–29562. [Google Scholar] [CrossRef] [PubMed]
- Villagomez, F.R.; Medina-Contreras, O.; Cerna-Cortes, J.F.; Patino-Lopez, G. The role of the oncogenic Rab35 in cancer invasion, metastasis, and immune evasion, especially in leukemia. Small GTPases 2018, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Cheng, K.W.; Lahad, J.P.; Gray, J.W.; Mills, G.B. Emerging role of RAB GTPases in cancer and human disease. Cancer Res. 2005, 65, 2516–2519. [Google Scholar] [CrossRef] [PubMed]
- Arnold, L.D.; Patel, A.V.; Yan, Y.; Jacobs, E.J.; Thun, M.J.; Calle, E.E.; Colditz, G.A. Are racial disparities in pancreatic cancer explained by smoking and overweight/obesity. Cancer Epidemiol. Biomarkers Prev. 2009, 18, 2397–2405. [Google Scholar] [CrossRef]
- Huang, B.Z.; Stram, D.O.; Le Marchand, L.; Haiman, C.A.; Wilkens, L.R.; Pandol, S.J.; Zhang, Z.F.; Monroe, K.R.; Setiawan, V.W. Interethnic differences in pancreatic cancer incidence and risk factors: The Multiethnic Cohort. Cancer Med. 2019, 8, 3592–3603. [Google Scholar] [CrossRef]
- Michaud, D.S.; Giovannucci, E.; Willett, W.C.; Colditz, G.A.; Fuchs, C.S. Coffee and alcohol consumption and the risk of pancreatic cancer in two prospective United States cohorts. Cancer Epidemiol. Biomarkers Prev. 2001, 10, 429–437. [Google Scholar]
- Andersen, D.K.; Korc, M.; Petersen, G.M.; Eibl, G.; Li, D.; Rickels, M.R.; Chari, S.T.; Abbruzzese, J.L. Diabetes, Pancreatogenic Diabetes, and Pancreatic Cancer. Diabetes 2017, 66, 1103–1110. [Google Scholar] [CrossRef]
- Vieira, O.V. Rab3a and Rab10 are regulators of lysosome exocytosis and plasma membrane repair. Small GTPases 2018, 9, 349–351. [Google Scholar] [CrossRef]
- van Weering, J.R.; Toonen, R.F.; Verhage, M. The role of Rab3a in secretory vesicle docking requires association/dissociation of guanidine phosphates and Munc18-1. PLoS ONE 2007, 2, 616. [Google Scholar] [CrossRef]
- Zhang, X.; He, X.; Fu, X.Y.; Chang, Z. Varp is a Rab21 guanine nucleotide exchange factor and regulates endosome dynamics. J. Cell Sci. 2006, 119, 1053–1062. [Google Scholar] [CrossRef]
- Pellinen, T.; Tuomi, S.; Arjonen, A.; Wolf, M.; Edgren, H.; Meyer, H.; Grosse, R.; Kitzing, T.; Rantala, J.K.; Kallioniemi, O.; et al. Integrin trafficking regulated by Rab21 is necessary for cytokinesis. Dev. Cell 2008, 15, 371–385. [Google Scholar] [CrossRef] [PubMed]
- Magadan, J.G.; Barbieri, M.A.; Mesa, R.; Stahl, P.D.; Mayorga, L.S. Rab22a regulates the sorting of transferrin to recycling endosomes. Mol. Cell Biol. 2006, 26, 2595–2614. [Google Scholar] [CrossRef] [PubMed]
- Shakya, S.; Sharma, P.; Bhatt, A.M.; Jani, R.A.; Delevoye, C.; Setty, S.R. Rab22A recruits BLOC-1 and BLOC-2 to promote the biogenesis of recycling endosomes. EMBO Rep. 2018, 19, 45918. [Google Scholar] [CrossRef] [PubMed]
- Casanova, J.E.; Wang, X.; Kumar, R.; Bhartur, S.G.; Navarre, J.; Woodrum, J.E.; Altschuler, Y.; Ray, G.S.; Goldenring, J.R. Association of Rab25 and Rab11a with the apical recycling system of polarized Madin-Darby canine kidney cells. Mol. Biol. Cell 1999, 10, 47–61. [Google Scholar] [CrossRef]
- Caswell, P.T.; Spence, H.J.; Parsons, M.; White, D.P.; Clark, K.; Cheng, K.W.; Mills, G.B.; Humphries, M.J.; Messent, A.J.; Anderson, K.I.; et al. Rab25 associates with alpha5beta1 integrin to promote invasive migration in 3D microenvironments. Dev. Cell 2007, 13, 496–510. [Google Scholar] [CrossRef]
- Binotti, B.; Pavlos, N.J.; Riedel, D.; Wenzel, D.; Vorbruggen, G.; Schalk, A.M.; Kühnel, K.; Boyken, J.; Erck, C.; Martens, H.; et al. The GTPase Rab26 links synaptic vesicles to the autophagy pathway. Elife 2015, 4, 05597. [Google Scholar] [CrossRef]
- Jin, R.U.; Mills, J.C. RAB26 coordinates lysosome traffic and mitochondrial localization. J. Cell Sci. 2014, 127, 1018–1032. [Google Scholar] [CrossRef]
- Opdam, F.J.; Echard, A.; Croes, H.J.; van den Hurk, J.A.; van de Vorstenbosch, R.A.; Ginsel, L.A.; Goud, B.; Fransen, J.A. The small GTPase Rab6B, a novel Rab6 subfamily member, is cell-type specifically expressed and localised to the Golgi apparatus. J. Cell Sci. 2000, 113, 2725–2735. [Google Scholar]
- Martinez, O.; Schmidt, A.; Salamero, J.; Hoflack, B.; Roa, M.; Goud, B. The small GTP-binding protein rab6 functions in intra-Golgi transport. J. Cell Biol. 1994, 127, 1575–1588. [Google Scholar] [CrossRef]
- Sato, T.; Iwano, T.; Kunii, M.; Matsuda, S.; Mizuguchi, R.; Jung, Y.; Hagiwara, H.; Yoshihara, Y.; Yuzaki, M.; Harada, R.; et al. Rab8a and Rab8b are essential for several apical transport pathways but insufficient for ciliogenesis. J. Cell Sci. 2014, 127, 422–431. [Google Scholar] [CrossRef]
- Lau, A.S.; Mruk, D.D. Rab8B GTPase and junction dynamics in the testis. Endocrinology 2003, 144, 1549–1563. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, S.; Suzuki, T.; Kawaguchi, A.; Phongphaew, W.; Yoshii, K.; Iwano, T.; Harada, A.; Kariwa, H.; Orba, Y.; Sawa, H. Rab8b Regulates Transport of West Nile Virus Particles from Recycling Endosomes. J. Biol. Chem. 2016, 291, 6559–6568. [Google Scholar] [CrossRef] [PubMed]
- Dateyama, I.; Sugihara, Y.; Chiba, S.; Ota, R.; Nakagawa, R.; Kobayashi, T.; Itoh, H. RABL2 positively controls localization of GPCRs in mammalian primary cilia. J. Cell Sci. 2019, 132, jcs.224428. [Google Scholar] [CrossRef] [PubMed]
- Kanie, T.; Abbott, K.L.; Mooney, N.A.; Plowey, E.D.; Demeter, J.; Jackson, P.K. The CEP19-RABL2 GTPase Complex Binds IFT-B to Initiate Intraflagellar Transport at the Ciliary Base. Dev. Cell 2017, 42, 22–36.e12. [Google Scholar] [CrossRef]
- Alto, N.M.; Soderling, J.; Scott, J.D. Rab32 is an A-kinase anchoring protein and participates in mitochondrial dynamics. J. Cell Biol. 2002, 158, 659–668. [Google Scholar] [CrossRef]
- Wasmeier, C.; Romao, M.; Plowright, L.; Bennett, D.C.; Raposo, G.; Seabra, M.C. Rab38 and Rab32 control post-Golgi trafficking of melanogenic enzymes. J. Cell Biol. 2006, 175, 271–281. [Google Scholar] [CrossRef]
- Albertson, D.G. Gene amplification in cancer. Trends Genet. 2006, 22, 447–455. [Google Scholar] [CrossRef]
- Sasaki, M.S.; Kato, M.; Toguchida, J.; Yamaguchi, T.; Ejima, Y.; Ishizaki, K.; Kaneko, A.; Tanooka, H. Somatic and germinal mutations of tumor-suppressor genes in the development of cancer. J. Radiat. Res. 1991, 32, 266–276. [Google Scholar] [CrossRef]
- Baylin, S.B.; Jones, P.A. Epigenetic Determinants of Cancer. Cold Spring Harb. Perspect. Biol. 2016, 8, 019505. [Google Scholar] [CrossRef]
- Sanchez-Tapia, C.; Wan, F.Y. Fastest time to cancer by loss of tumor suppressor genes. Bull. Math. Biol. 2014, 76, 2737–2784. [Google Scholar] [CrossRef][Green Version]
- Dozynkiewicz, M.A.; Jamieson, N.B.; Macpherson, I.; Grindlay, J.; van den Berghe, P.V.E.; von Thun, A.; Morton, J.P.; Gourley, C.; Timpson, P.; Nixon, C.; et al. Rab25 and CLIC3 collaborate to promote integrin recycling from late endosomes/lysosomes and drive cancer progression. Dev. Cell 2012, 22, 131–145. [Google Scholar] [CrossRef] [PubMed]
- Caswell, P.T.; Chan, M.; Lindsay, A.J.; McCaffrey, M.W.; Boettiger, D.; Norman, J.C. Rab-coupling protein coordinates recycling of alpha5beta1 integrin and EGFR1 to promote cell migration in 3D microenvironments. J. Cell Biol. 2008, 183, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Bravo-Cordero, J.J.; Marrero-Diaz, R.; Megias, D.; Genis, L.; Garcia-Grande, A.; Garcia, M.A.; Arroyo, A.G.; Montoya, M.C. MT1-MMP proinvasive activity is regulated by a novel Rab8-dependent exocytic pathway. EMBO J. 2007, 26, 1499–1510. [Google Scholar] [CrossRef] [PubMed]
- Hendrix, A.; Maynard, D.; Pauwels, P.; Braems, G.; Denys, H.; Van den Broecke, R.; Lambert, J.; Van Belle, S.; Cocquyt, V.; Gespach, C.; et al. Effect of the secretory small GTPase Rab27B on breast cancer growth, invasion, and metastasis. J. Natl. Cancer Inst. 2010, 102, 866–880. [Google Scholar] [CrossRef] [PubMed]
- Recchi, C.; Seabra, M.C. Novel functions for Rab GTPases in multiple aspects of tumour progression. Biochem. Soc. Trans. 2012, 40, 1398–1403. [Google Scholar] [CrossRef]
- Hooper, S.; Gaggioli, C.; Sahai, E. A chemical biology screen reveals a role for Rab21-mediated control of actomyosin contractility in fibroblast-driven cancer invasion. Br. J. Cancer 2010, 102, 392–402. [Google Scholar] [CrossRef]
- Peinado, H.; Aleckovic, M.; Lavotshkin, S.; Matei, I.; Costa-Silva, B.; Moreno-Bueno, G.; Hergueta-Redondo, M.; Williams, C.; García-Santos, G.; Ghajar, C.; et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012, 18, 883–891. [Google Scholar] [CrossRef]
- Pfeffer, S.R. Two Rabs for exosome release. Nat. Cell Biol. 2010, 12, 3–4. [Google Scholar] [CrossRef]
- Cheng, K.W.; Lahad, J.P.; Kuo, W.L.; Lapuk, A.; Yamada, K.; Auersperg, N.; Liu, J.; Smith-McCune, K.; Lu, K.H.; Fishman, D.; et al. The RAB25 small GTPase determines aggressiveness of ovarian and breast cancers. Nat. Med. 2004, 10, 1251–1256. [Google Scholar] [CrossRef]
- Zhang, J.; Wei, J.; Lu, J.; Tong, Z.; Liao, B.; Yu, B.; Zheng, F.; Huang, X.; Chen, Z.; Fang, Y.; et al. Overexpression of Rab25 contributes to metastasis of bladder cancer through induction of epithelial-mesenchymal transition and activation of Akt/GSK-3beta/Snail signaling. Carcinogenesis 2013, 34, 2401–2408. [Google Scholar] [CrossRef]
- Ren, P.; Yang, X.Q.; Zhai, X.L.; Zhang, Y.Q.; Huang, J.F. Overexpression of Rab27B is correlated with distant metastasis and poor prognosis in ovarian cancer. Oncol. Lett. 2016, 12, 1539–1545. [Google Scholar] [CrossRef]
- Seven, D.; Dogan, S.; Kilic, E.; Karaman, E.; Koseoglu, H.; Buyru, N. Downregulation of Rab25 activates Akt1 in head and neck squamous cell carcinoma. Oncol. Lett. 2015, 10, 1927–1931. [Google Scholar] [CrossRef]
- Wang, M.; Wang, W.; Ding, J.; Wang, J.; Zhang, J. Downregulation of Rab17 promotes cell proliferation and invasion in non-small cell lung cancer through STAT3/HIF-1alpha/VEGF signaling. Thorac. Cancer 2020, 11, 379–388. [Google Scholar] [CrossRef]
- Kim, J.K.; Lee, S.Y.; Park, C.W.; Park, S.H.; Yin, J.; Kim, J.; Park, J.B.; Lee, J.Y.; Kim, H.; Kim, S.C. Rab3a promotes brain tumor initiation and progression. Mol. Biol. Rep. 2014, 41, 5903–5911. [Google Scholar] [CrossRef] [PubMed]
- Iezzi, M.; Regazzi, R.; Wollheim, C.B. The Rab3-interacting molecule RIM is expressed in pancreatic beta-cells and is implicated in insulin exocytosis. FEBS Lett. 2000, 474, 66–70. [Google Scholar] [CrossRef]
- Guo, W.; Chen, Z.; Chen, Z.; Yu, J.; Liu, H.; Li, T.; Lin, T.; Chen, H.; Zhao, M.; Li, G.; et al. Promotion of Cell Proliferation through Inhibition of Cell Autophagy Signalling Pathway by Rab3IP is Restrained by MicroRNA-532-3p in Gastric Cancer. J. Cancer 2018, 9, 4363–4373. [Google Scholar] [CrossRef] [PubMed]
- Pellinen, T.; Arjonen, A.; Vuoriluoto, K.; Kallio, K.; Fransen, J.A.; Ivaska, J. Small GTPase Rab21 regulates cell adhesion and controls endosomal traffic of beta1-integrins. J. Cell Biol. 2006, 173, 767–780. [Google Scholar] [CrossRef] [PubMed]
- Ge, J.; Chen, Q.; Liu, B.; Wang, L.; Zhang, S.; Ji, B. Knockdown of Rab21 inhibits proliferation and induces apoptosis in human glioma cells. Cell Mol. Biol. Lett. 2017, 22, 30. [Google Scholar] [CrossRef]
- Ye, F.; Tang, H.; Liu, Q.; Xie, X.; Wu, M.; Liu, X.; Chen, B.; Xie, X. miR-200b as a prognostic factor in breast cancer targets multiple members of RAB family. J. Transl. Med. 2014, 12, 17. [Google Scholar] [CrossRef]
- Wang, N.; Meng, W.; Jia, R.; Xiang, S. Rab GTPase 21 mediates caerulin-induced TRAF3-MKK3-p38 activation and acute pancreatitis response. Biochem. Biophys. Res. Commun. 2019, 518, 50–58. [Google Scholar] [CrossRef]
- Weigert, R.; Yeung, A.C.; Li, J.; Donaldson, J.G. Rab22a regulates the recycling of membrane proteins internalized independently of clathrin. Mol. Biol. Cell 2004, 15, 3758–3770. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Shen, L.; Jiang, C.; Gao, G.; Wang, K.; Jiao, Y.; Sun, L.; Cui, Y.; Ke, Z.; Yang, Z. Rab22a is a novel prognostic marker for cell progression in breast cancer. Int. J. Mol. Med. 2020, 45, 1037–1046. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Gilkes, D.M.; Takano, N.; Xiang, L.; Luo, W.; Bishop, C.J.; Chaturvedi, P.; Green, J.J.; Semeza, G.L. Hypoxia-inducible factors and RAB22A mediate formation of microvesicles that stimulate breast cancer invasion and metastasis. Proc. Natl. Acad. Sci. USA 2014, 111, 3234–3242. [Google Scholar] [CrossRef]
- Fang, Z.; Li, C.; Li, S. MicroRNA-193b acts as a tumor suppressor in colon cancer progression via targeting RAB22A. Exp. Ther. Med. 2019, 17, 3921–3928. [Google Scholar] [CrossRef] [PubMed]
- Xiong, F.; Liu, K.; Zhang, F.; Sha, K.; Wang, X.; Guo, X.; Huang, N. MiR-204 inhibits the proliferation and invasion of renal cell carcinoma by inhibiting RAB22A expression. Oncol. Rep. 2016, 35, 3000–3008. [Google Scholar] [CrossRef]
- Yang, D.; Liu, G.; Wang, K. miR-203 Acts as a Tumor Suppressor Gene in Osteosarcoma by Regulating RAB22A. PLoS ONE 2015, 10, e0132225. [Google Scholar] [CrossRef]
- Scapin, S.M.; Carneiro, F.R.; Alves, A.C.; Medrano, F.J.; Guimaraes, B.G.; Zanchin, N.I. The crystal structure of the small GTPase Rab11b reveals critical differences relative to the Rab11a isoform. J. Struct. Biol. 2006, 154, 260–268. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, P.; Wang, X.; Yu, Y.; Zhu, G.; Zheng, L.; Xu, Z.; Li, F.; You, Q.; Yang, Q.; et al. Rab25 promotes erlotinib resistance by activating the β1 integrin/AKT/β-catenin pathway in NSCLC. Cell Prolif. 2019, 52, 12592. [Google Scholar] [CrossRef]
- Xiong, J.; Li, S.; Zeng, X. High Rab25 expression associates with Ki67/TP53/CD133/VEGFR expression predicts poor prognosis in gastric cancer. Int. J. Clin. Exp. Pathol. 2017, 10, 7792–7800. [Google Scholar]
- Gomez-Roman, N.; Sahasrabudhe, N.M.; McGregor, F.; Chalmers, A.J.; Cassidy, J.; Plumb, J. Hypoxia-inducible factor 1 alpha is required for the tumourigenic and aggressive phenotype associated with Rab25 expression in ovarian cancer. Oncotarget 2016, 7, 22650–22664. [Google Scholar] [CrossRef]
- Cheng, J.M.; Volk, L.; Janaki, D.K.; Vyakaranam, S.; Ran, S.; Rao, K.A. Tumor suppressor function of Rab25 in triple-negative breast cancer. Int. J. Cancer. 2010, 126, 2799–2812. [Google Scholar] [CrossRef] [PubMed]
- Goldenring, J.R.; Nam, K.T. Rab25 as a tumour suppressor in colon carcinogenesis. Br. J. Cancer 2011, 104, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, K.J.; Wang, G.; Schmued, L.C. Temporal progression of kainic acid induced neuronal and myelin degeneration in the rat forebrain. Brain Res. 2000, 864, 69–80. [Google Scholar] [CrossRef]
- Chen, D.; Guo, J.; Gahl, W.A. RAB GTPases expressed in human melanoma cells. Biochim. Biophys. Acta 1997, 1355, 1–6. [Google Scholar] [CrossRef]
- Wang, C.; Liu, Z.; Huang, X. Rab32 is important for autophagy and lipid storage in Drosophila. PLoS ONE 2012, 7, e32086. [Google Scholar] [CrossRef]
- Drizyte-Miller, K.; Chen, J.; Cao, H.; Schott, M.B.; McNiven, M.A. The small GTPase Rab32 resides on lysosomes to regulate mTORC1 signaling. J. Cell Sci. 2020, 133. [Google Scholar] [CrossRef]
- Esnaola, N.F.; Ford, M.E. Racial differences and disparities in cancer care and outcomes: Where’s the rub. Surg. Oncol. Clin. N. Am. 2012, 21, 417–437. [Google Scholar] [CrossRef]
- Deshmukh, S.K.; Azim, S.; Ahmad, A.; Zubair, H.; Tyagi, N.; Srivastava, S.K.; Bhardwaj, A.; Singh, S.; Rocconi, R.P.; Singh, A.P. Biological basis of cancer health disparities: Resources and challenges for research. Am. J. Cancer Res. 2017, 7, 1–12. [Google Scholar]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef]
- Wang, Y.T.; Gou, Y.W.; Jin, W.W.; Xiao, M.; Fang, H.Y. Association between alcohol intake and the risk of pancreatic cancer: A dose-response meta-analysis of cohort studies. BMC Cancer 2016, 16, 212. [Google Scholar] [CrossRef]
- Dragomir, M.; Calin, G.A. Circular RNAs in Cancer—Lessons Learned From microRNAs. Front. Oncol. 2018, 8, 179. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Kjems, J.; Damgaard, C.K. Circular RNA and miR-7 in cancer. Cancer Res. 2013, 73, 5609–5612. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Hureta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
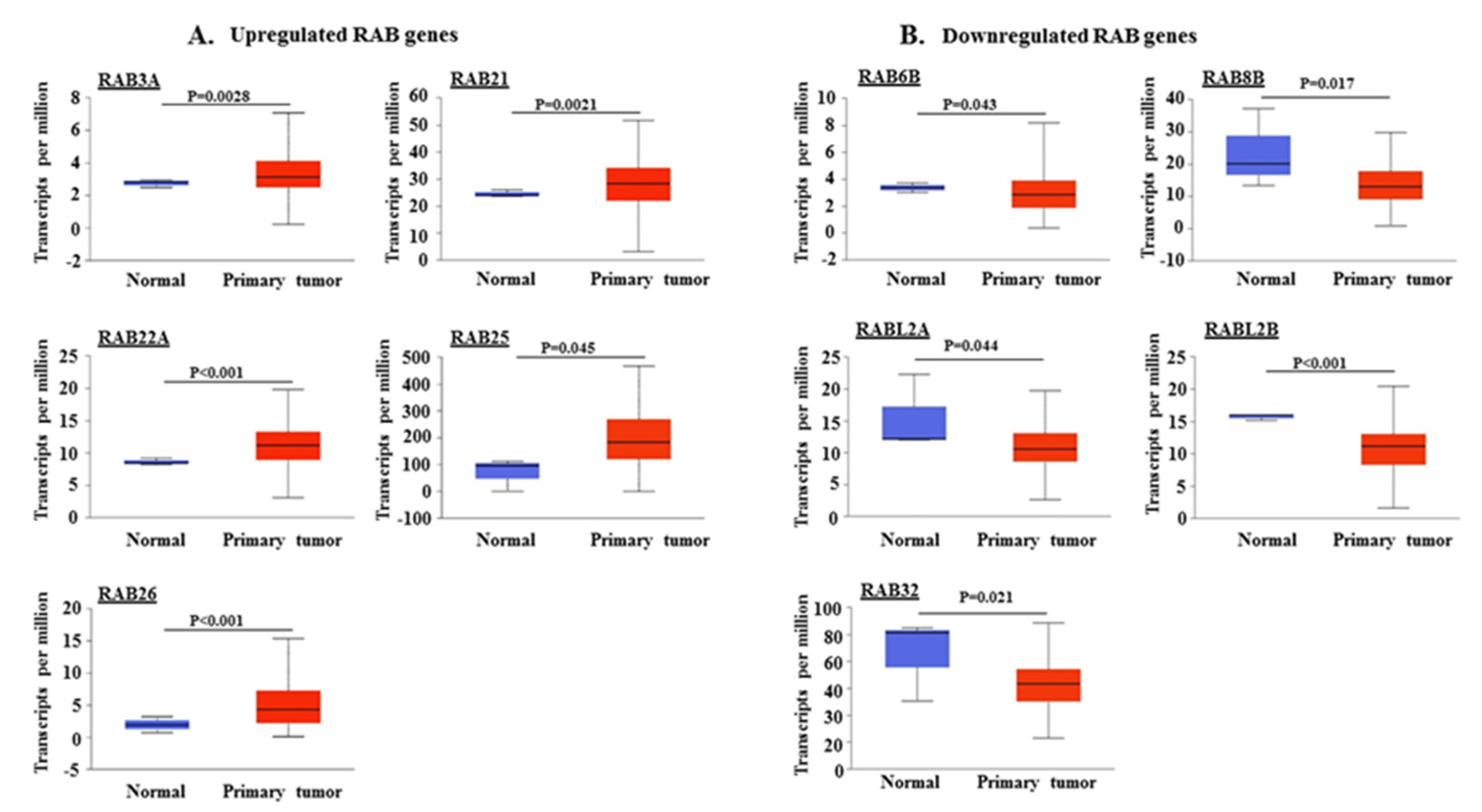

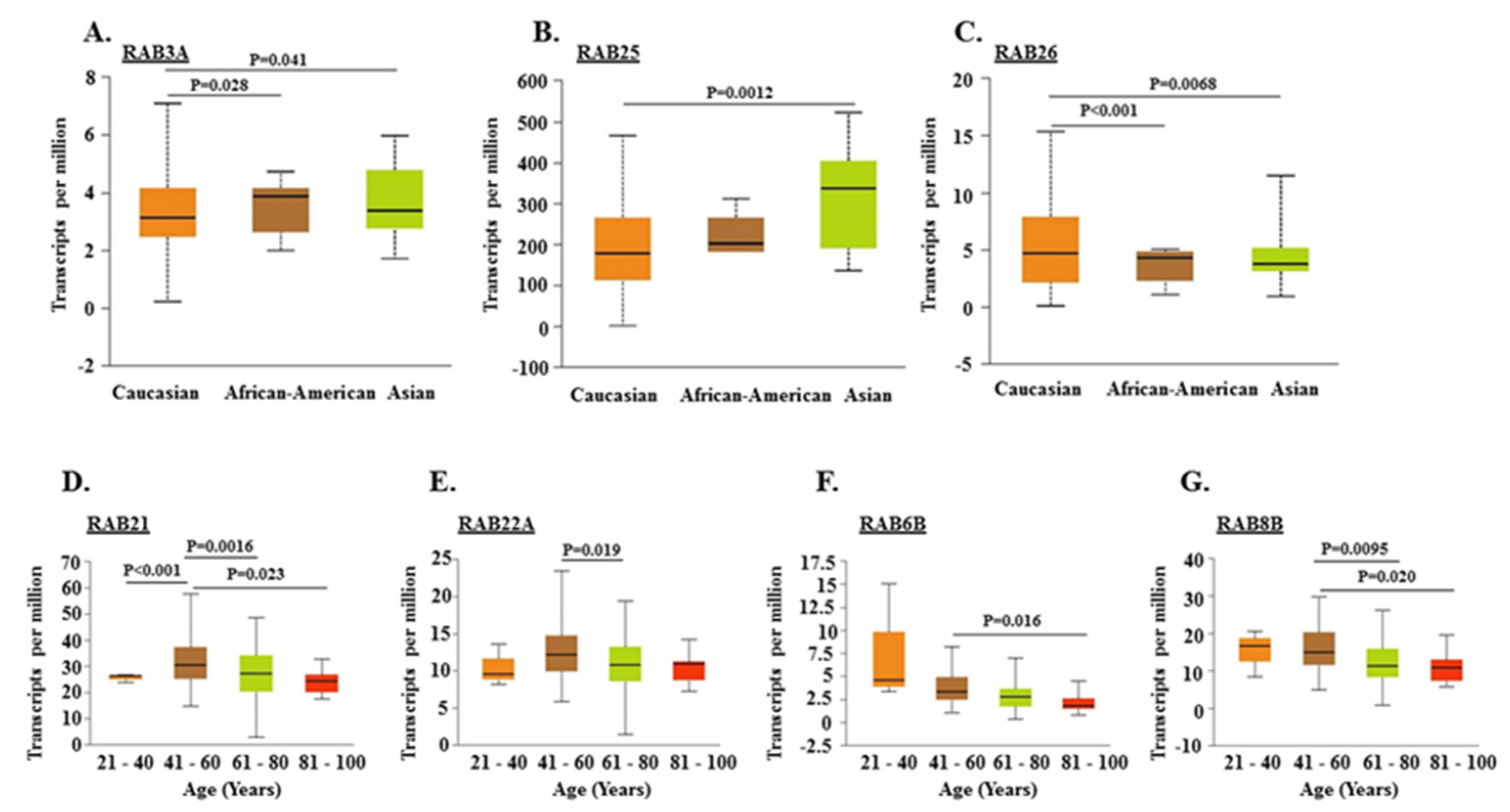
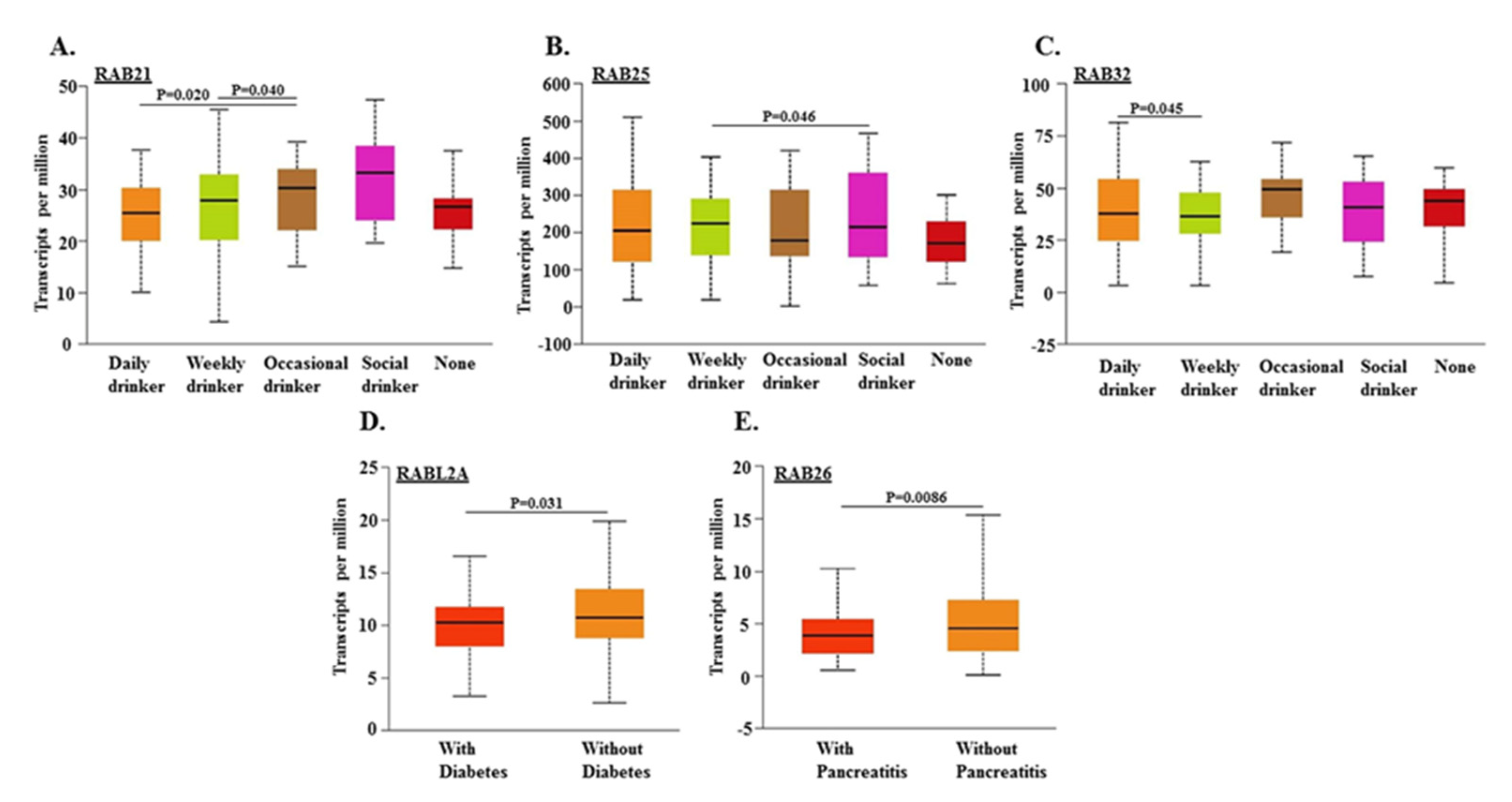
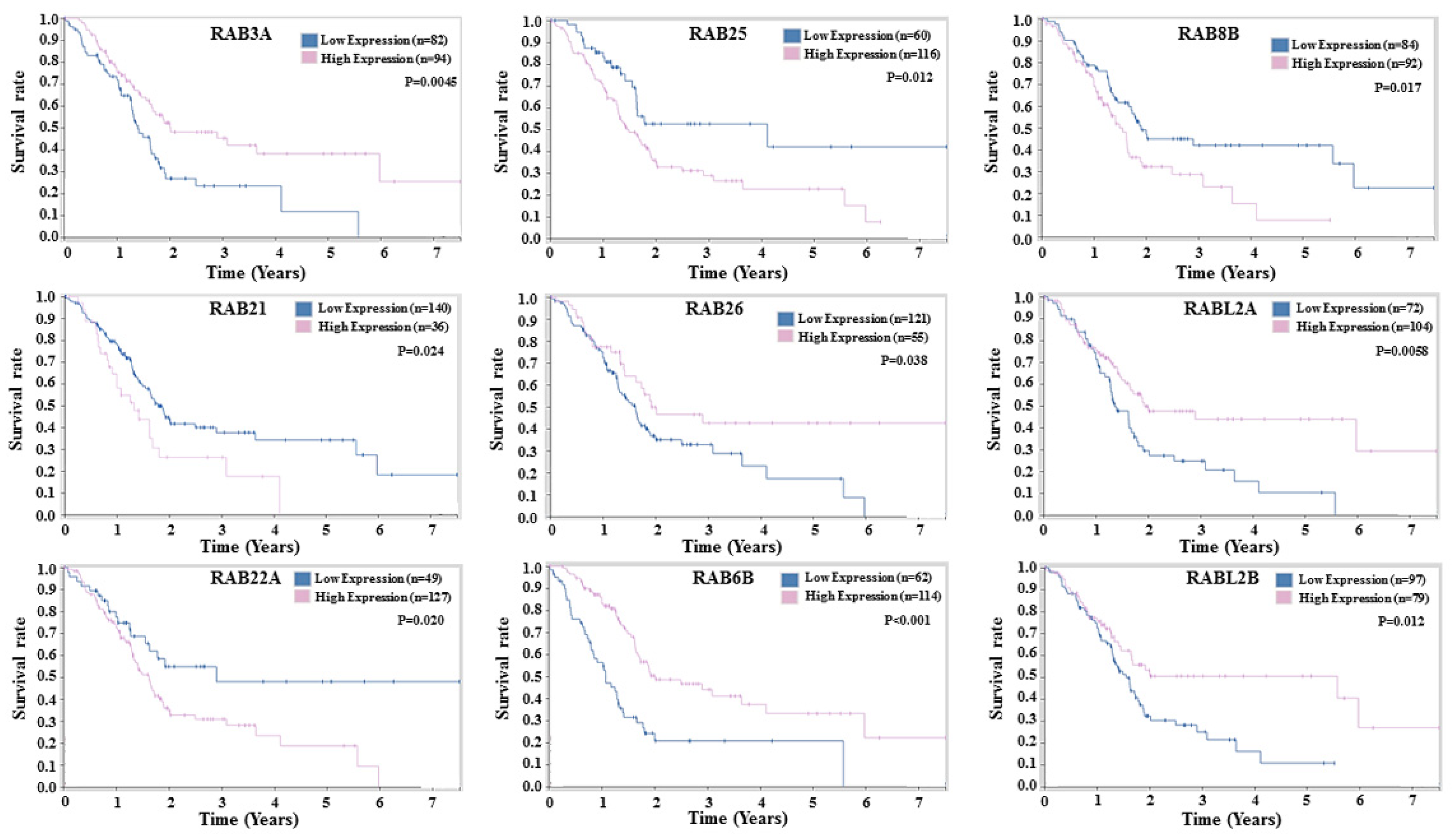
| RAB Genes | Location | Function | References |
|---|---|---|---|
| Upregulated RAB Genes | |||
| RAB3A | Synaptotagmins and secretory vesicles | Regulation of exocytosis (Ca+2 dependent), secretory vesicle docking, fusion and transport of synaptic vesicles | [28,29] |
| RAB21 | Early endosomes, trans Golgi network, cytoplasmic vesicles | Endosomal transport of integrins, cell adhesion and migration, endolysosomal transport | [30,31] |
| RAB22A | Early endosomes, phagosomal membrane, and trans Golgi network | Protein trafficking from early endosomes to late endosomes, phagosome maturation, recycling endosomal biogenesis | [32,33] |
| RAB25 | Recycling endosomes | Protein trafficking through recycling endosomes to the plasma membrane, integrin transport | [34,35] |
| RAB26 | Synaptic and secretory vesicles | Regulated exocytosis of secretory granules and synaptic vesicles | [36,37] |
| Downregulated RAB Genes | |||
| RAB6B | Golgi | Retrograde transport from late endosomes via Golgi to ER | [38,39] |
| RAB8B | Golgi, plasma membrane, phagosome membrane | Apical transport pathways, adherens junction dynamics, from trans Golgi network and recycling endosomes to plasma membrane | [40,41,42] |
| RABL2A/RABL2B | Primary cilia, centriole | Intraflagellar transport and ciliary assembly | [43,44] |
| RAB32 | Mitochondria, melanosomes, autophagosomes | Mitochondrial dynamics regulation, transport from trans Golgi network to melanosomes, autophagy | [45,46] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anand, S.; Khan, M.A.; Khushman, M.; Dasgupta, S.; Singh, S.; Singh, A.P. Comprehensive Analysis of Expression, Clinicopathological Association and Potential Prognostic Significance of RABs in Pancreatic Cancer. Int. J. Mol. Sci. 2020, 21, 5580. https://doi.org/10.3390/ijms21155580
Anand S, Khan MA, Khushman M, Dasgupta S, Singh S, Singh AP. Comprehensive Analysis of Expression, Clinicopathological Association and Potential Prognostic Significance of RABs in Pancreatic Cancer. International Journal of Molecular Sciences. 2020; 21(15):5580. https://doi.org/10.3390/ijms21155580
Chicago/Turabian StyleAnand, Shashi, Mohammad Aslam Khan, Moh’d Khushman, Santanu Dasgupta, Seema Singh, and Ajay Pratap Singh. 2020. "Comprehensive Analysis of Expression, Clinicopathological Association and Potential Prognostic Significance of RABs in Pancreatic Cancer" International Journal of Molecular Sciences 21, no. 15: 5580. https://doi.org/10.3390/ijms21155580
APA StyleAnand, S., Khan, M. A., Khushman, M., Dasgupta, S., Singh, S., & Singh, A. P. (2020). Comprehensive Analysis of Expression, Clinicopathological Association and Potential Prognostic Significance of RABs in Pancreatic Cancer. International Journal of Molecular Sciences, 21(15), 5580. https://doi.org/10.3390/ijms21155580








