Dephosphorylation of LjMPK6 by Phosphatase LjPP2C is Involved in Regulating Nodule Organogenesis in Lotus japonicus
Abstract
1. Introduction
2. Results
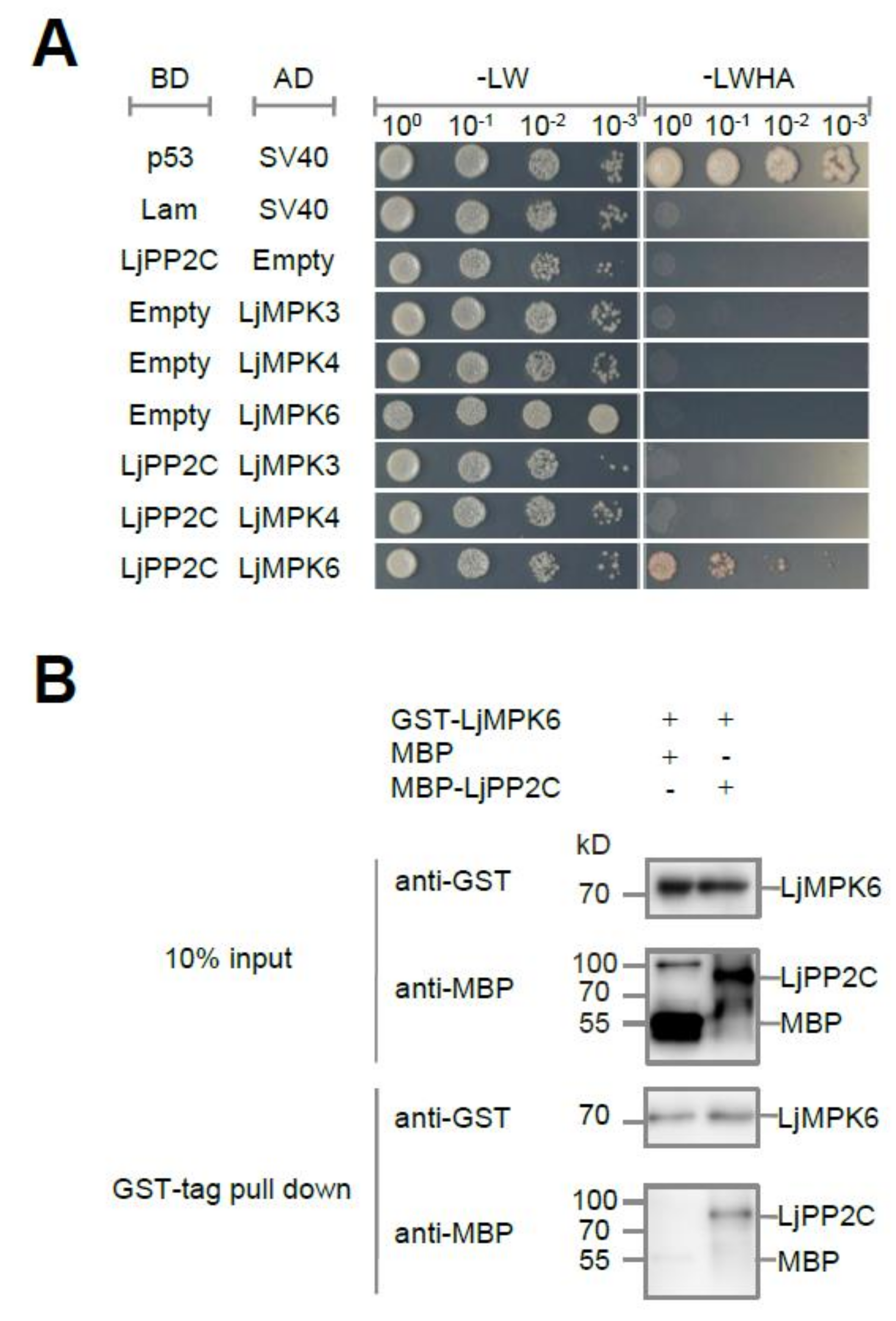
2.1. LjPP2C Interacts with and Dephosphorylates LjMPK6 In Vitro
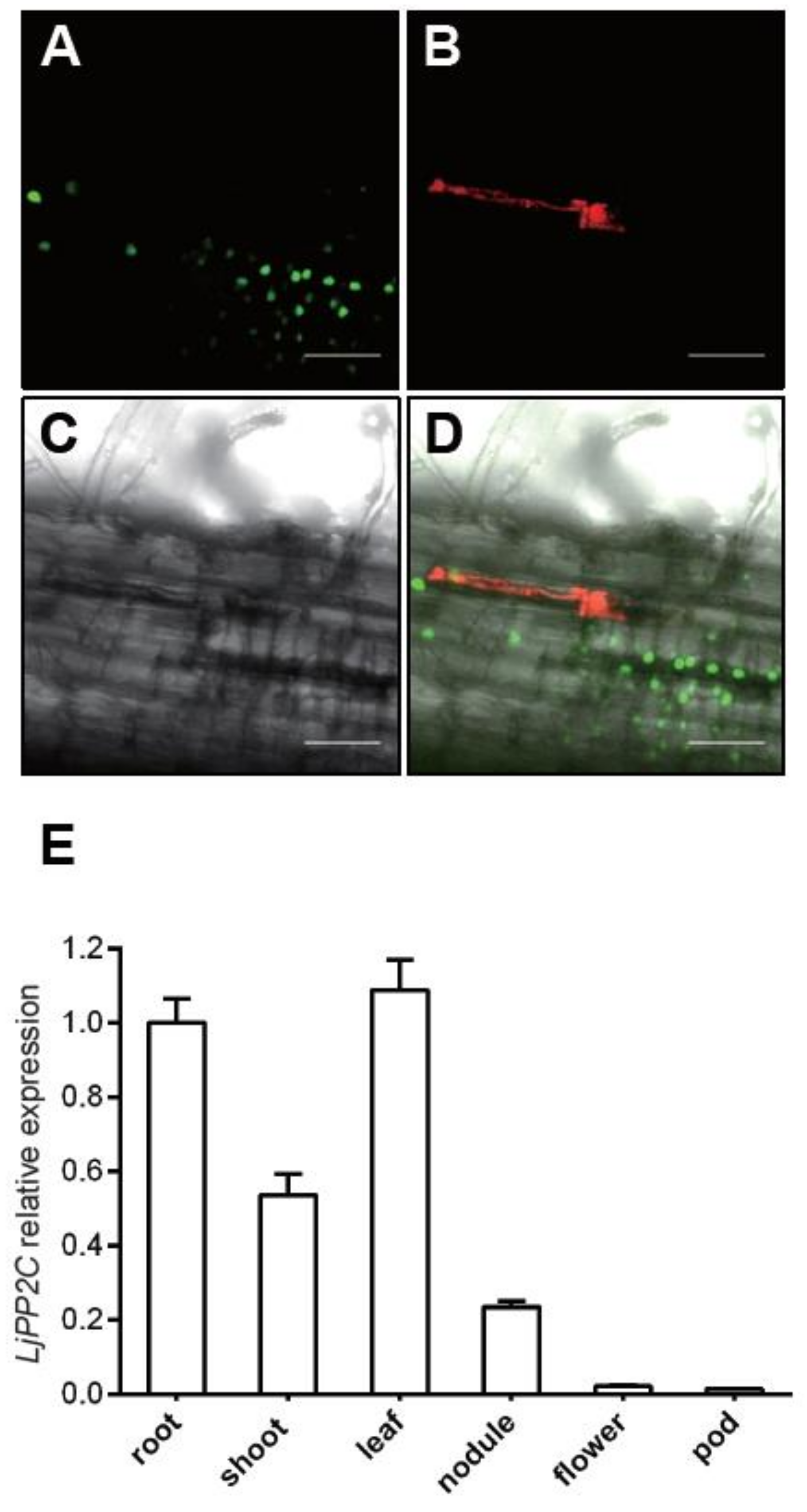
2.2. Expression Pattern of LjPP2C in Both Non-Symbiotic and Symbiotic Tissues
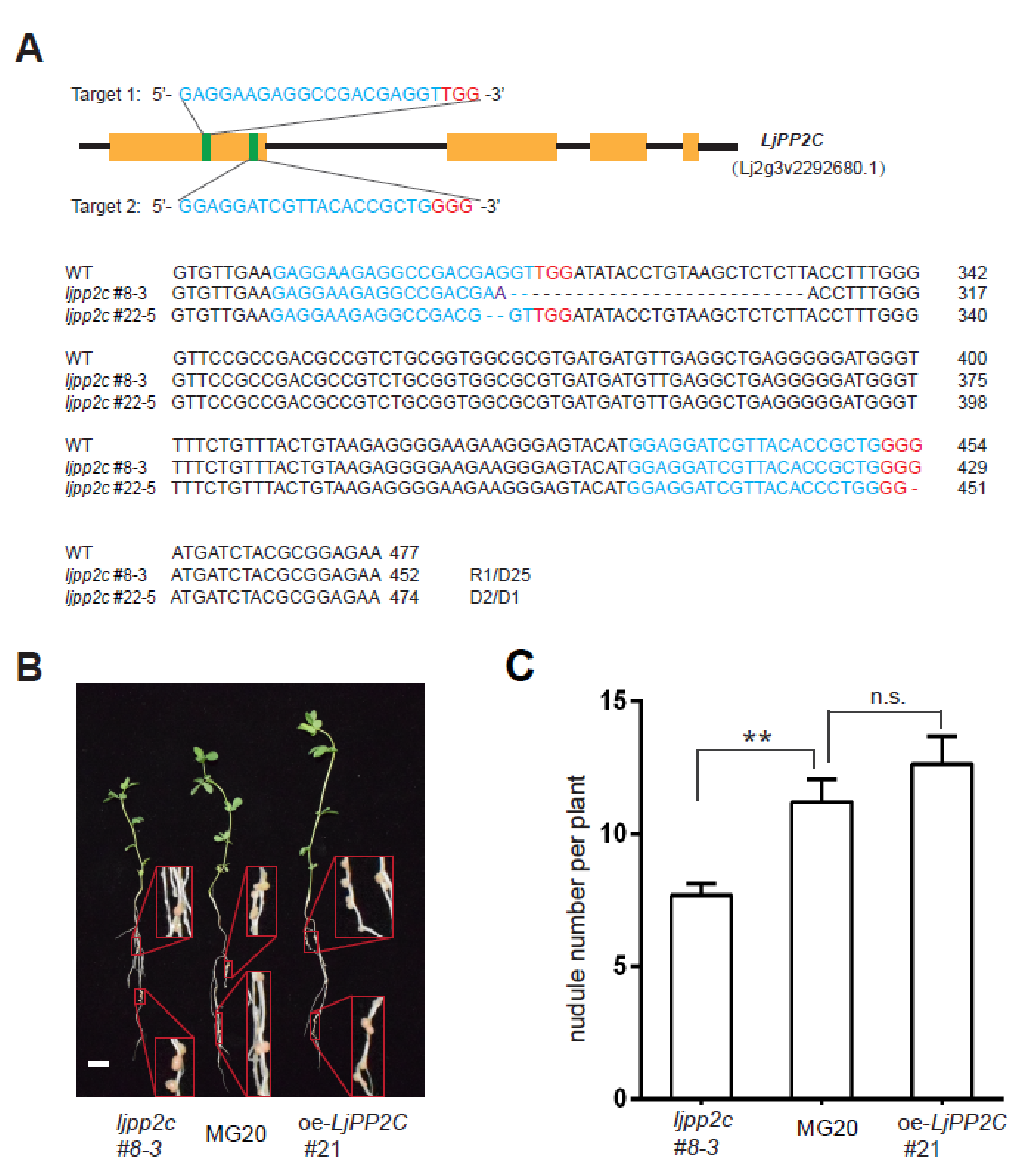
2.3. LjPP2C is Required for Nodule Development
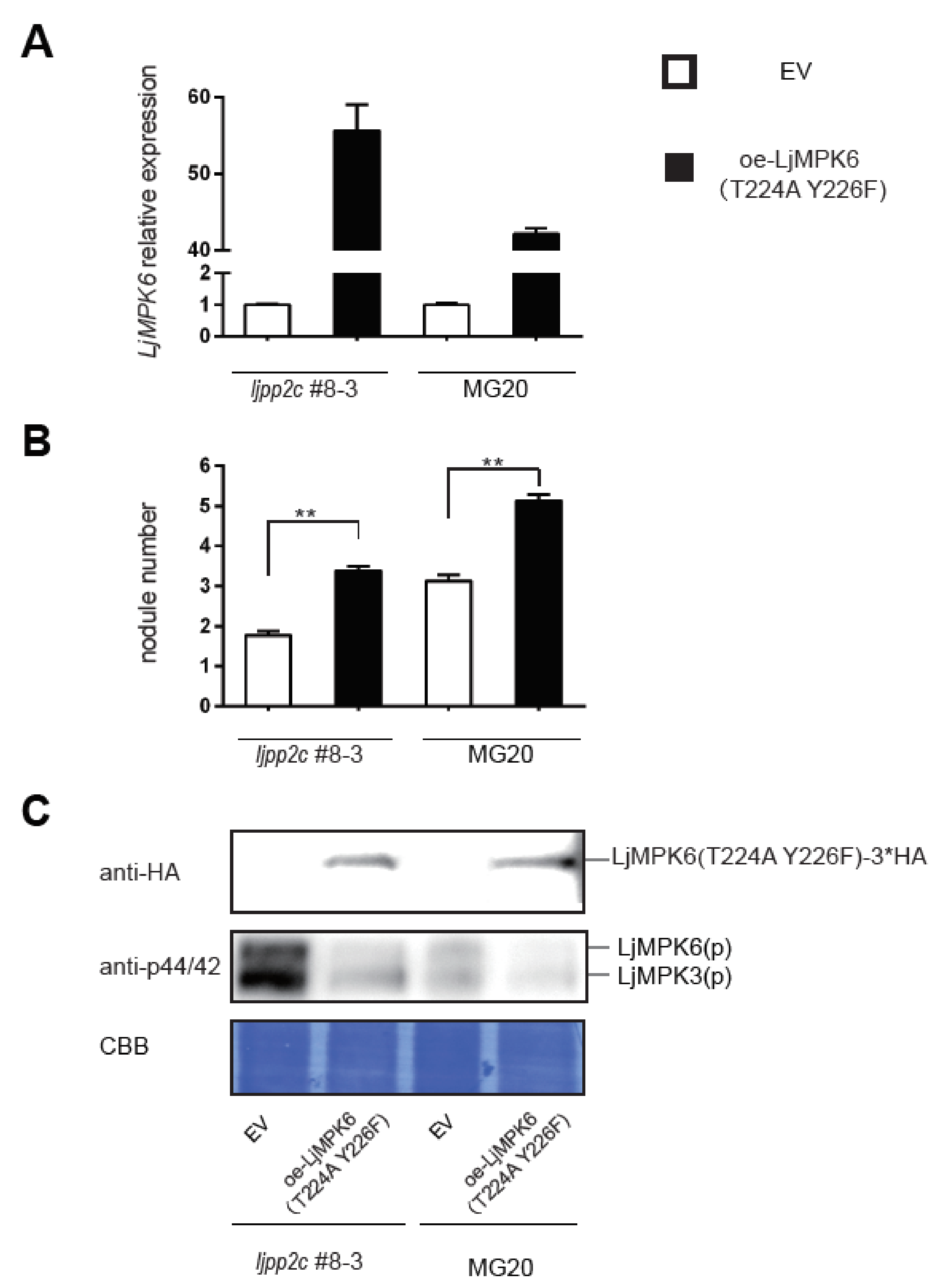
2.4. LjPP2C is Required for MAPK Dephosphorylation In Vivo
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Plasmid Construction
4.3. Yeast Two-Hybrid Assay
4.4. In Vitro Pull-Down Assay
4.5. Phosphatase Assay
4.6. RNA Extraction and Reverse-Transcription Quantitative PCR (qRT-PCR)
4.7. Hairy Root Transformation and Nodulation Assays
4.8. Accession Numbers
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CRISPR | clustered regularly interspaced short palindromic repeats |
| GFP | Green Fluorescent Protein |
| GST | Glutathione S-transferase |
| MAPK | mitogen-activated protein kinase |
| MBP | maltose-binding protein |
| NF | nod factor |
| NIN | nodule inception gene |
| PP2C | type 2C protein phosphatase |
| RLK | receptor-like kinase |
| Y2H | yeast two-hybrid |
References
- Denarie, J.; Debelle, F.; Prome, J.C. Rhizobium lipo-chitooligosaccharide nodulation factors: Signaling molecules mediating recognition and morphogenesis. Annu. Rev. Biochem. 1996, 65, 503–535. [Google Scholar] [CrossRef] [PubMed]
- Broughton, W.J.; Jabbouri, S.; Perret, X. Keys to symbiotic harmony. J. Bacteriol. 2000, 182, 5641–5652. [Google Scholar] [CrossRef] [PubMed]
- Radutoiu, S.; Madsen, L.H.; Madsen, E.B.; Felle, H.H.; Umehara, Y.; Gronlund, M.; Sato, S.; Nakamura, Y.; Tabata, S.; Sandal, N.; et al. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 2003, 425, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Madsen, E.B.; Madsen, L.H.; Radutoiu, S.; Olbryt, M.; Rakwalska, M.; Szczyglowski, K.; Sato, S.; Kaneko, T.; Tabata, S.; Sandal, N.; et al. A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 2003, 425, 637–640. [Google Scholar] [CrossRef]
- Tsikou, D.; Ramirez, E.E.; Psarrakou, I.S.; Wong, J.E.; Jensen, D.B.; Isono, E.; Radutoiu, S.; Papadopoulou, K.K. A Lotus japonicus E3 ligase interacts with the Nod Factor Receptor 5 and positively regulates nodulation. BMC Plant Biol. 2018, 18, 217. [Google Scholar] [CrossRef]
- Stracke, S.; Kistner, C.; Yoshida, S.; Mulder, L.; Sato, S.; Kaneko, T.; Tabata, S.; Sandal, N.; Stougaard, J.; Szczyglowski, K.; et al. A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 2002, 417, 959–962. [Google Scholar] [CrossRef]
- Riely, B.K.; Ane, J.M.; Penmetsa, R.V.; Cook, D.R. Genetic and genomic analysis in model legumes bring Nod-factor signaling to center stage. Curr. Opin. Plant Biol. 2004, 7, 408–413. [Google Scholar] [CrossRef]
- Ried, M.K.; Antolin-Llovera, M.; Parniske, M. Spontaneous symbiotic reprogramming of plant roots triggered by receptor-like kinases. Elife 2014, e03891. [Google Scholar] [CrossRef]
- Holsters, M. SYMRK, an enigmatic receptor guarding and guiding microbial endosymbioses with plant roots. Proc. Natl. Acad. Sci. USA 2008, 105, 4537–4538. [Google Scholar] [CrossRef]
- Oldroyd, G.E. Speak, friend, and enter: Signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 2013, 11, 252–263. [Google Scholar] [CrossRef]
- Miller, J.B.; Pratap, A.; Miyahara, A.; Zhou, L.; Bornemann, S.; Morris, R.J.; Oldroyd, G.E. Calcium/Calmodulin-dependent protein kinase is negatively and positively regulated by calcium, providing a mechanism for decoding calcium responses during symbiosis signaling. Plant Cell 2013, 25, 5053–5066. [Google Scholar] [CrossRef] [PubMed]
- Mitra, R.M.; Gleason, C.A.; Edwards, A.; Hadfield, J.; Downie, J.A.; Oldroyd, G.E.; Long, S.R. A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: Gene identification by transcript-based cloning. Proc. Natl. Acad. Sci. USA 2004, 101, 4701–4705. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Katzer, K.; Lambert, J.; Cerri, M.; Parniske, M. CYCLOPS, a DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe. 2014, 15, 139–152. [Google Scholar] [CrossRef] [PubMed]
- Yano, K.; Yoshida, S.; Muller, J.; Singh, S.; Banba, M.; Vickers, K.; Markmann, K.; White, C.; Schuller, B.; Sato, S.; et al. CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc. Natl. Acad. Sci. USA 2008, 105, 20540–20545. [Google Scholar] [CrossRef]
- Schauser, L.; Roussis, A.; Stiller, J.; Stougaard, J. A plant regulator controlling development of symbiotic root nodules. Nature 1999, 402, 191–195. [Google Scholar] [CrossRef]
- Soyano, T.; Kouchi, H.; Hirota, A.; Hayashi, M. Nodule inception directly targets NF-Y subunit genes to regulate essential processes of root nodule development in Lotus japonicus. PLoS Genet. 2013, 9, e1003352. [Google Scholar] [CrossRef]
- Wang, C.; Wang, G.; Zhang, C.; Zhu, P.; Dai, H.; Yu, N.; He, Z.; Xu, L.; Wang, E. OsCERK1-Mediated Chitin Perception and Immune Signaling Requires Receptor-like Cytoplasmic Kinase 185 to Activate an MAPK Cascade in Rice. Mol Plant. 2017, 10, 619–633. [Google Scholar] [CrossRef]
- Pitzschke, A. Modes of MAPK substrate recognition and control. Trends Plant Sci. 2015, 20, 49–55. [Google Scholar] [CrossRef]
- Berriri, S.; Garcia, A.V.; Frei dit Frey, N.; Rozhon, W.; Pateyron, S.; Leonhardt, N.; Montillet, J.L.; Leung, J.; Hirt, H.; Colcombet, J. Constitutively active mitogen-activated protein kinase versions reveal functions of Arabidopsis MPK4 in pathogen defense signaling. Plant Cell 2012, 24, 4281–4293. [Google Scholar] [CrossRef]
- Sarma, U.; Ghosh, I. Different designs of kinase-phosphatase interactions and phosphatase sequestration shapes the robustness and signal flow in the MAPK cascade. BMC Syst. Biol. 2012, 6, 82. [Google Scholar] [CrossRef]
- Xu, R.; Duan, P.; Yu, H.; Zhou, Z.; Zhang, B.; Wang, R.; Li, J.; Zhang, G.; Zhuang, S.; Lyu, J.; et al. Control of Grain Size and Weight by the OsMKKK10-OsMKK4-OsMAPK6 Signaling Pathway in Rice. Mol. Plant. 2018, 11, 860–873. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Hua, L.; Dong, S.; Chen, H.; Zhu, X.; Jiang, J.; Zhang, F.; Li, Y.; Fang, X.; Chen, F. OsMAPK6, a mitogen-activated protein kinase, influences rice grain size and biomass production. Plant J. 2015, 84, 672–681. [Google Scholar] [CrossRef] [PubMed]
- Shubchynskyy, V.; Boniecka, J.; Schweighofer, A.; Simulis, J.; Kvederaviciute, K.; Stumpe, M.; Mauch, F.; Balazadeh, S.; Mueller-Roeber, B.; Boutrot, F.; et al. Protein phosphatase AP2C1 negatively regulates basal resistance and defense responses to Pseudomonas syringae. J. Exp. Bot. 2017, 68, 1169–1183. [Google Scholar] [PubMed]
- Chen, T.; Zhu, H.; Ke, D.; Cai, K.; Wang, C.; Gou, H.; Hong, Z.; Zhang, Z. A MAP kinase kinase interacts with SymRK and regulates nodule organogenesis in Lotus japonicus. Plant Cell 2012, 24, 823–838. [Google Scholar] [CrossRef]
- Yin, J.; Guan, X.; Zhang, H.; Wang, L.; Li, H.; Zhang, Q.; Chen, T.; Xu, Z.; Hong, Z.; Cao, Y.; et al. An MAP kinase interacts with LHK1 and regulates nodule organogenesis in Lotus japonicus. Sci. China Life Sci. 2019, 62, 1203–1217. [Google Scholar] [CrossRef]
- Wang, L.; Tan, Q.; Fan, Q.; Zhu, H.; Hong, Z.; Zhang, Z.; Duanmu, D. Efficient Inactivation of Symbiotic Nitrogen Fixation Related Genes in Lotus japonicus Using CRISPR-Cas9. Front. Plant Sci. 2016, 7, 1333. [Google Scholar] [CrossRef]
- Lu, X.; Xiong, Q.; Cheng, T.; Li, Q.T.; Liu, X.L.; Bi, Y.D.; Li, W.; Zhang, W.K.; Ma, B.; Lai, Y.C.; et al. A PP2C-1 Allele Underlying a Quantitative Trait Locus Enhances Soybean 100-Seed Weight. Mol. Plant. 2017, 10, 670–684. [Google Scholar] [CrossRef]
- Couto, D.; Niebergall, R.; Liang, X.; Bucherl, C.A.; Sklenar, J.; Macho, A.P.; Ntoukakis, V.; Derbyshire, P.; Altenbach, D.; Maclean, D.; et al. The Arabidopsis Protein Phosphatase PP2C38 Negatively Regulates the Central Immune Kinase BIK1. PLoS Pathog. 2016, 12, e1005811. [Google Scholar] [CrossRef]
- Park, C.J.; Peng, Y.; Chen, X.; Dardick, C.; Ruan, D.; Bart, R.; Canlas, P.E.; Ronald, P.C. Rice XB15, a protein phosphatase 2C, negatively regulates cell death and XA21-mediated innate immunity. PLOS Biol. 2008, 6, e231. [Google Scholar]
- Kapranov, P.; Jensen, T.J.; Poulsen, C.; de Bruijn, F.J.; Szczyglowski, K. A protein phosphatase 2C gene, LjNPP2C1, from Lotus japonicus induced during root nodule development. Proc. Natl. Acad. Sci. USA 1999, 96, 1738–1743. [Google Scholar] [CrossRef]
- Schweighofer, A.; Kazanaviciute, V.; Scheikl, E.; Teige, M.; Doczi, R.; Hirt, H.; Schwanninger, M.; Kant, M.; Schuurink, R.; Mauch, F.; et al. The PP2C-type phosphatase AP2C1, which negatively regulates MPK4 and MPK6, modulates innate immunity, jasmonic acid, and ethylene levels in Arabidopsis. Plant Cell 2007, 19, 2213–2224. [Google Scholar] [CrossRef] [PubMed]
- Maekawa, T.; Kusakabe, M.; Shimoda, Y.; Sato, S.; Tabata, S.; Murooka, Y.; Hayashi, M. Polyubiquitin promoter-based binary vectors for overexpression and gene silencing in Lotus japonicus. Mol. Plant Microbe Interact. 2008, 21, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Tirichine, L.; Herrera-Cervera, J.A.; Stougaard, J. Transformation-regeneration procedure for Lotus japonicus. In Lotus Japonicus Handbook; Springer: Dordrecht, The Netherlands, 2005; pp. 279–284. [Google Scholar]
- Reid, D.E.; Heckmann, A.B.; Novak, O.; Kelly, S.; Stougaard, J. CYTOKININ OXIDASE/DEHYDROGENASE3 Maintains Cytokinin Homeostasis during Root and Nodule Development in Lotus japonicus. Plant Physiol. 2016, 170, 1060–1074. [Google Scholar] [CrossRef] [PubMed]
- Diaz, C.L.; Gronlund, M.; Schlaman, H.R.M.; Spaink, H.P. Induction of hairy roots for symbiotic gene expression studies. In Lotus Japonicus Handbook; Springer: Dordrecht, The Netherlands, 2005; pp. 261–277. [Google Scholar]
- Gage, D.J. Analysis of infection thread development using Gfp- and DsRed-expressing Sinorhizobium meliloti. J. Bacteriol. 2002, 184, 7042–7046. [Google Scholar] [CrossRef]
- Haney, C.H.; Riely, B.K.; Tricoli, D.M.; Cook, D.R.; Ehrhardt, D.W.; Long, S.R. Symbiotic rhizobia bacteria trigger a change in localization and dynamics of the Medicago truncatula receptor kinase LYK3. Plant Cell 2011, 23, 2774–2787. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, Z.; Cao, J.; Fan, Q.; Chao, H.; Guan, X.; Zhang, Z.; Duanmu, D. Dephosphorylation of LjMPK6 by Phosphatase LjPP2C is Involved in Regulating Nodule Organogenesis in Lotus japonicus. Int. J. Mol. Sci. 2020, 21, 5565. https://doi.org/10.3390/ijms21155565
Yan Z, Cao J, Fan Q, Chao H, Guan X, Zhang Z, Duanmu D. Dephosphorylation of LjMPK6 by Phosphatase LjPP2C is Involved in Regulating Nodule Organogenesis in Lotus japonicus. International Journal of Molecular Sciences. 2020; 21(15):5565. https://doi.org/10.3390/ijms21155565
Chicago/Turabian StyleYan, Zhongyuan, Jingjing Cao, Qiuling Fan, Hongmin Chao, Xiaomin Guan, Zhongming Zhang, and Deqiang Duanmu. 2020. "Dephosphorylation of LjMPK6 by Phosphatase LjPP2C is Involved in Regulating Nodule Organogenesis in Lotus japonicus" International Journal of Molecular Sciences 21, no. 15: 5565. https://doi.org/10.3390/ijms21155565
APA StyleYan, Z., Cao, J., Fan, Q., Chao, H., Guan, X., Zhang, Z., & Duanmu, D. (2020). Dephosphorylation of LjMPK6 by Phosphatase LjPP2C is Involved in Regulating Nodule Organogenesis in Lotus japonicus. International Journal of Molecular Sciences, 21(15), 5565. https://doi.org/10.3390/ijms21155565




