The N-Terminal Region of Soybean PM1 Protein Protects Liposomes during Freeze-Thaw
Abstract
1. Introduction
2. Results
2.1. PM1 and PM1-N Proteins Inhibit the Freeze-Thaw-Induced Increase in Turbidity of a Liposome Suspension
2.2. PM1 and PM1-N Proteins Can Inhibit or Prevent Freeze-Thaw-Induced Aggregation of Liposomes and Isolated Thylakoid membranes
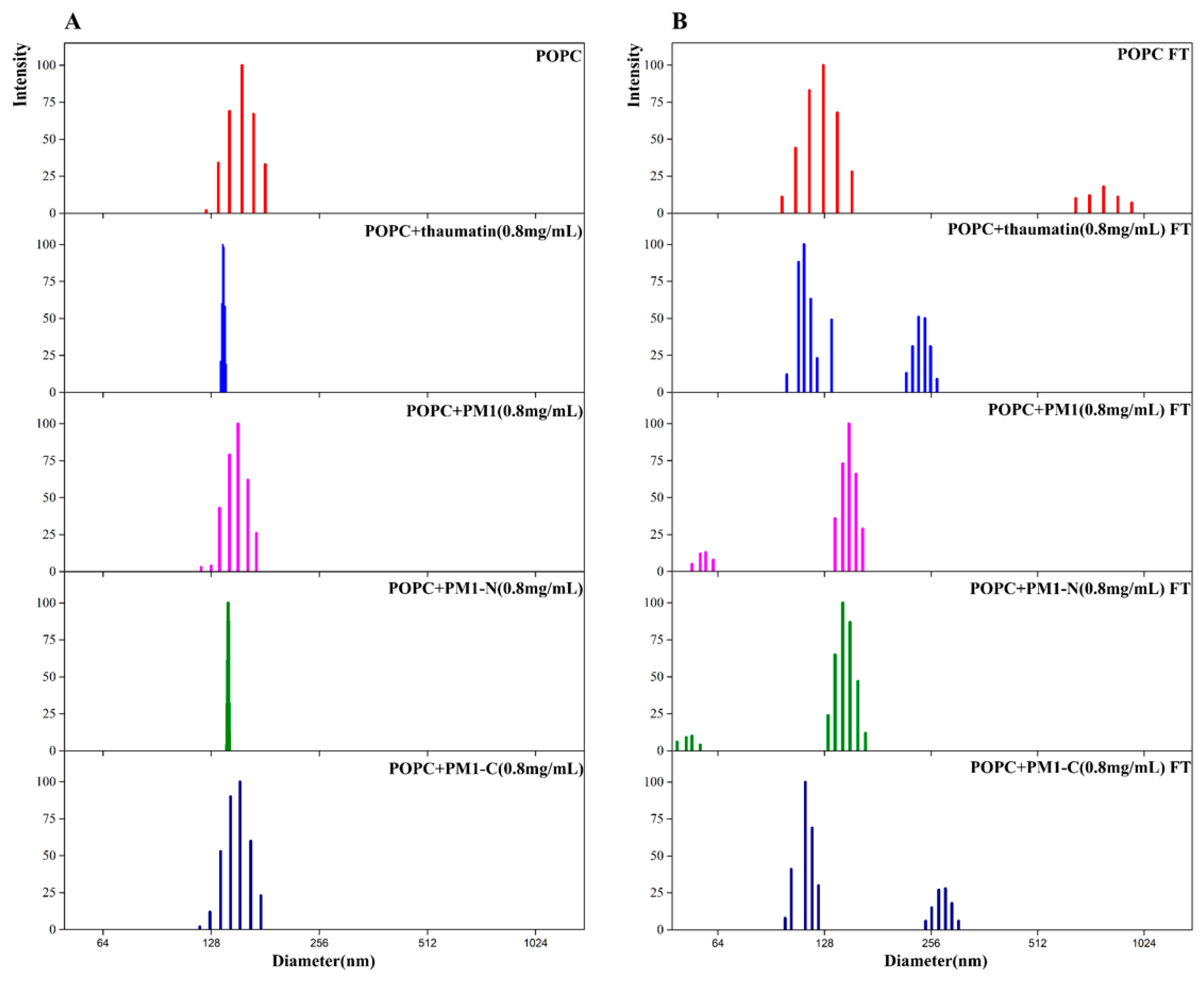
2.3. The Addition of Either PM1 or PM1-N Protein Can Stabilize the Liposome Particle Size
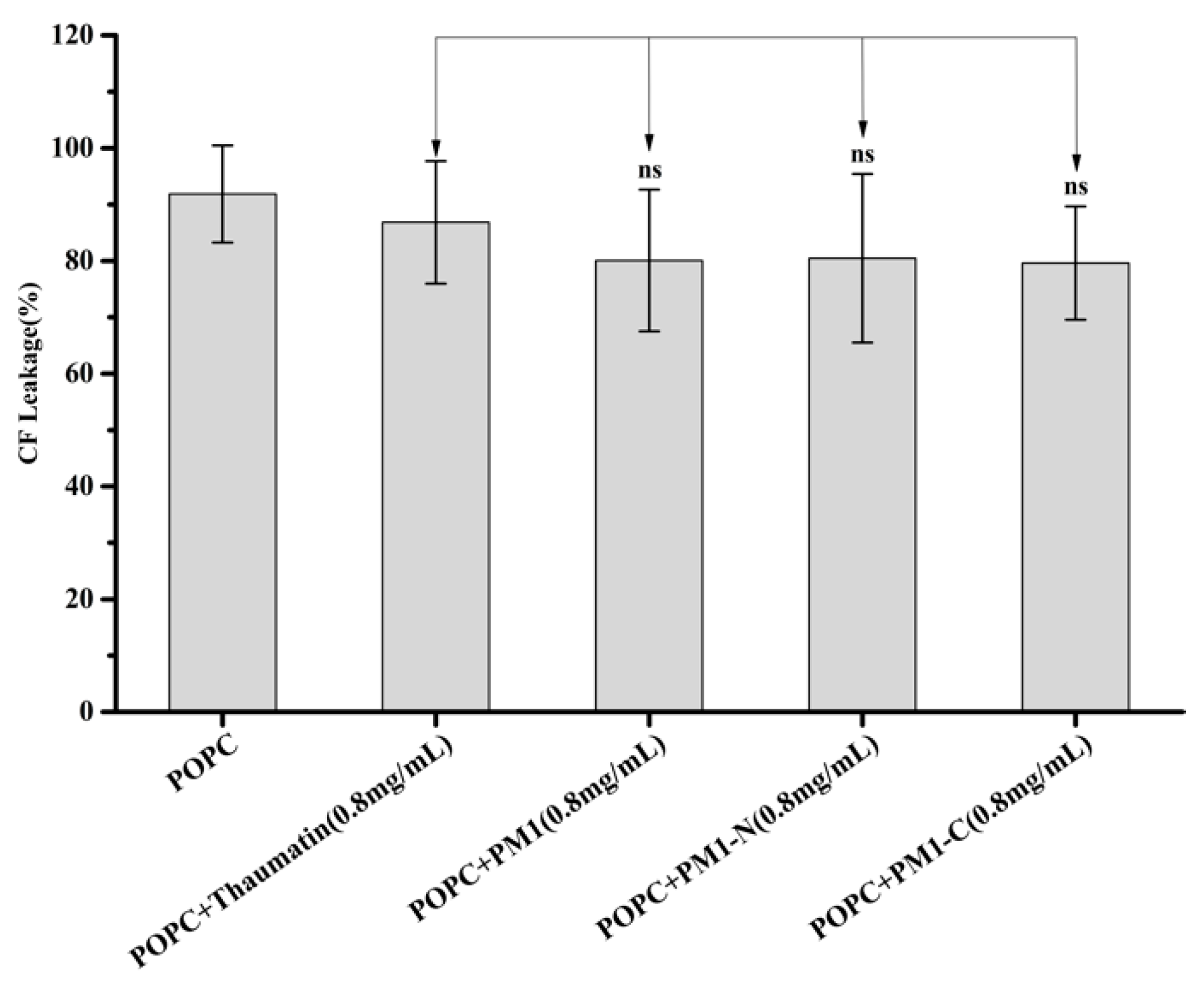
2.4. PM1 and PM1-N Proteins Cannot Prevent Freeze-Thaw-Induced Leakage of Liposomes
2.5. PM1 and PM1-N Proteins Can Effectively Inhibit or Prevent Membrane Fusion
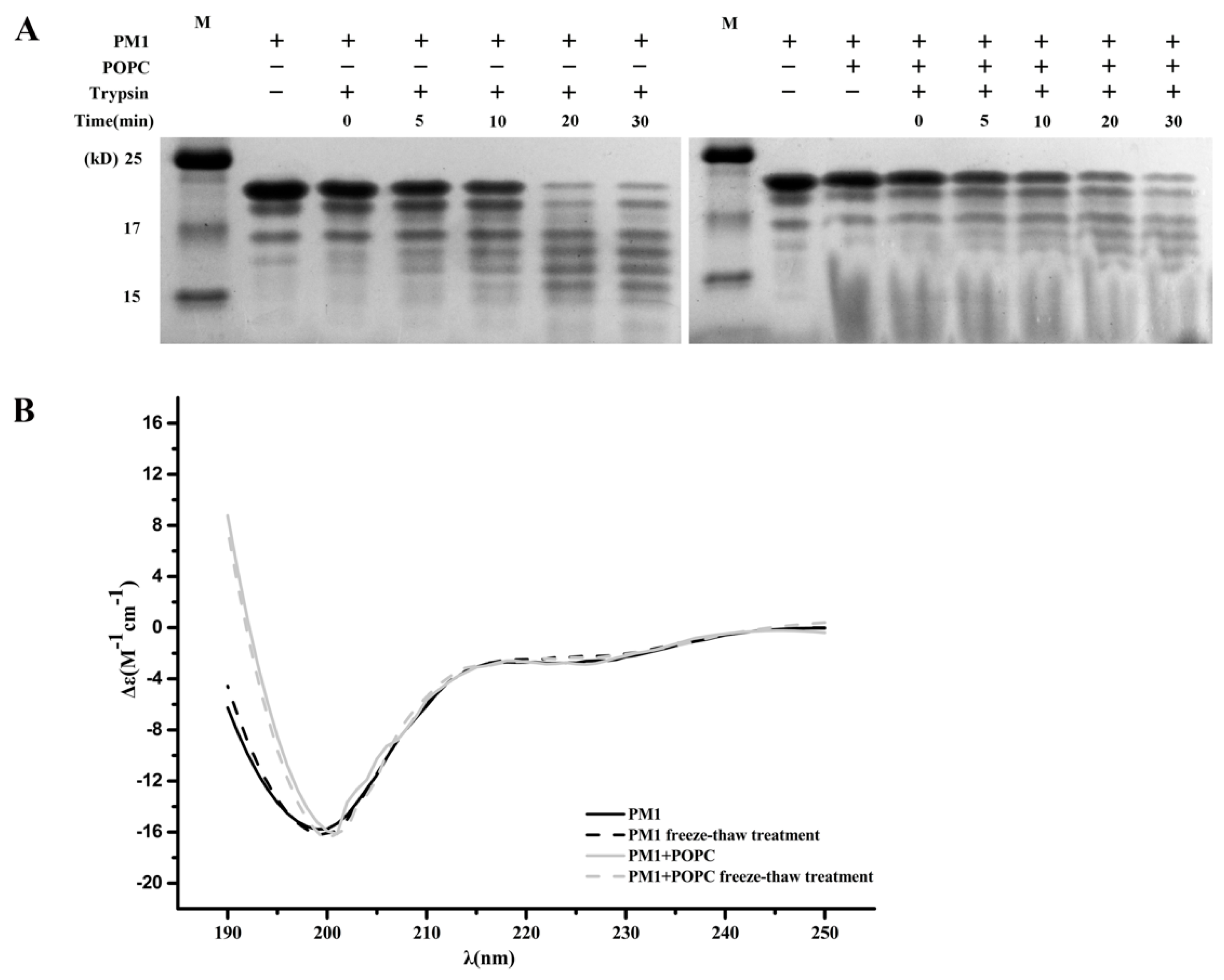
2.6. The Presence of POPC Liposomes Does Not Affect Digestion of PM1 by Trypsin
2.7. The Presence of POPC Liposomes Does Not Change the Secondary Structure of PM1 Protein
3. Discussion
4. Materials and Methods
4.1. Protein Over-Expression and Purification
4.2. Preparation of Liposomes
4.3. Thylakoid Membrane Preparation
4.4. Freeze-Thaw Treatment of Liposomes
4.5. Turbidity Measurement and Dynamic Light Scattering (DLS)
4.6. Light Microscopy
4.7. CF Leakage Experiments
4.8. Membrane Fusion Measurements
4.9. Limited Proteolysis
4.10. Far UV-Circular Dichroism Spectroscopy
4.11. In Vitro Lactate Dehydrogenase Assays
4.12. Statistics
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| LEA | late embryogenesis abundant |
| IDP | intrinsically disordered protein |
| DLS | dynamic light scattering |
| PC | phosphatidylcholine |
| POPC | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine |
| CF | carboxyfluorescein |
| HMW | high molecular weight |
| LUV | large unilaminar vesicle |
| FRET | fluorescence resonance energy transfer |
| CD | circular dichroism |
| LDH | lactate dehydrogenase |
Appendix A

References
- Tunnacliffe, A.; Wise, M.J. The continuing conundrum of the LEA proteins. Naturwissenschaften 2007, 94, 791–812. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, M.; Covarrubias, A.A. Late Embryogenesis Abundant (LEA) proteins in legumes. Front. Plant Sci. 2013, 4, 190. [Google Scholar] [CrossRef] [PubMed]
- Artur, M.A.S.; Zhao, T.; Ligterink, W.; Schranz, E.; Hilhorst, H.W.M. Dissecting the Genomic Diversification of Late Embryogenesis Abundant (LEA) Protein Gene Families in Plants. Genome Biol. Evol. 2019, 11, 459–471. [Google Scholar] [CrossRef] [PubMed]
- Roberts, J.K.; DeSimone, N.A.; Lingle, W.L.; Dure, L., 3rd. Cellular Concentrations and Uniformity of Cell-Type Accumulation of Two Lea Proteins in Cotton Embryos. Plant Cell 1993, 5, 769–780. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, Z.; Wang, L.; Wu, R.; Phillips, J.; Deng, X. LEA 4 group genes from the resurrection plant Boeahygrometrica confer dehydration tolerance in transgenic tobacco. Plant Sci. 2009, 176, 90–98. [Google Scholar] [CrossRef]
- Olvera-Carrillo, Y.; Campos, F.; Reyes, J.L.; Garciarrubio, A.; Covarrubias, A.A. Functional Analysis of the Group 4 Late Embryogenesis Abundant proteins reveals their relevance in the adaptive response during water deficit in Arabidopsis thaliana. Plant Physiol. 2010, 154, 373–390. [Google Scholar] [CrossRef]
- Artur, M.A.S.; Rienstra, J.; Dennis, T.J.; Farrant, J.M.; Ligterink, W.; Hilhorst, H. Structural Plasticity of Intrinsically Disordered LEA Proteins from Xerophytaschlechteri Provides Protection In Vitro and In Vivo. Front. Plant Sci. 2019, 10, 1272. [Google Scholar] [CrossRef]
- Zeng, X.; Ling, H.; Yang, J.; Li, Y.; Guo, S. LEA proteins from Gastrodiaelata enhance tolerance to low temperature stress in Escherichia coli. Gene 2018, 646, 136–142. [Google Scholar] [CrossRef]
- Reyes, J.L.; Campos, F.; Wei, H.; Arora, R.; Yang, Y.; Karlson, D.T.; Covarrubias, A.A. Functional dissection of hydrophilins during in vitro freeze protection. Plant Cell Environ. 2008, 31, 1781–1790. [Google Scholar] [CrossRef]
- Cuevas-Velazquez, C.L.; Saab-Rincon, G.; Reyes, J.L.; Covarrubias, A.A. The Unstructured N-terminal Region of Arabidopsis Group 4 Late Embryogenesis Abundant (LEA) Proteins Is Required for Folding and for Chaperone-like Activity under Water Deficit. J. Biol. Chem. 2016, 291, 10893–10903. [Google Scholar] [CrossRef]
- Hundertmark, M.; Dimova, R.; Lengefeld, J.; Seckler, R.; Hincha, D.K. The intrinsically disordered late embryogenesis abundant protein LEA18 from Arabidopsis thaliana modulates membrane stability through binding and folding. Biochim. Biophys. Acta 2011, 1808, 446–453. [Google Scholar] [CrossRef] [PubMed]
- Shih, M.D.; Hsieh, T.Y.; Lin, T.P.; Hsing, Y.I.; Hoekstra, F.A. Characterization of two soybean (Glycine max L.) LEA IV proteins by circular dichroism and Fourier transform infrared spectrometry. Plant Cell Physiol. 2010, 51, 395–407. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Xu, H.; Zhang, L.; Zheng, Y. Fe binding properties of two soybean (Glycine max L.) LEA4 proteins associated with antioxidant activity. Plant Cell Physiol. 2011, 52, 994–1002. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Liu, K.; Gao, Y.; Zheng, Y. Involvement of C-Terminal Histidines in Soybean PM1 Protein Oligomerization and Cu2+ Binding. Plant Cell Physiol. 2017, 58, 1018–1029. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, S.K.; Kutzer, M.; Procek, J.; Grobner, G.; Harryson, P. Tunable membrane binding of the intrinsically disordered dehydrin lti30, a cold-induced plant stress protein. Plant Cell 2011, 23, 2391–2404. [Google Scholar] [CrossRef]
- Dang, N.X.; Popova, A.V.; Hundertmark, M.; Hincha, D.K. Functional characterization of selected LEA proteins from Arabidopsis thaliana in yeast and in vitro. Planta 2014, 240, 325–336. [Google Scholar] [CrossRef]
- Oliver, A.E.; Hincha, D.K.; Crowe, J.H. Looking beyond sugars: The role of amphiphilic solutes in preventing adventitious reactions in anhydrobiotes at low water contents. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2002, 131, 515–525. [Google Scholar] [CrossRef]
- Hincha, D.K.; Zuther, E.; Hellwege, E.M.; Heyer, A.G. Specific effects of fructo- and gluco-oligosaccharides in the preservation of liposomes during drying. Glycobiology 2002, 12, 103–110. [Google Scholar] [CrossRef]
- Kovacs, D.; Kalmar, E.; Torok, Z.; Tompa, P. Chaperone activity of ERD10 and ERD14, two disordered stress-related plant proteins. Plant Physiol. 2008, 147, 381–390. [Google Scholar] [CrossRef]
- Rantalainen, K.I.; Christensen, P.A.; Hafren, A.; Otzen, D.E.; Kalkkinen, N.; Makinen, K. Interaction of a potyviralVPg with anionic phospholipid vesicles. Virology 2009, 395, 114–120. [Google Scholar] [CrossRef]
- Welker, S.; Rudolph, B.; Frenzel, E.; Hagn, F.; Liebisch, G.; Schmitz, G.; Scheuring, J.; Kerth, A.; Blume, A.; Weinkauf, S.; et al. Hsp12 is an intrinsically unstructured stress protein that folds upon membrane association and modulates membrane function. Mol. Cell 2010, 39, 507–520. [Google Scholar] [CrossRef] [PubMed]
- Koag, M.C.; Fenton, R.D.; Wilkens, S.; Close, T.J. The binding of maize DHN1 to lipid vesicles. Gain of structure and lipid specificity. Plant Physiol. 2003, 131, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Koag, M.C.; Wilkens, S.; Fenton, R.D.; Resnik, J.; Vo, E.; Close, T.J. The K-segment of maize DHN1 mediates binding to anionic phospholipid vesicles and concomitant structural changes. Plant Physiol. 2009, 150, 1503–1514. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, S.; Eremina, N.; Barth, A.; Danielsson, J.; Harryson, P. Membrane-Induced Folding of the Plant Stress Dehydrin Lti30. Plant Physiol. 2016, 171, 932–943. [Google Scholar] [CrossRef]
- Artus, N.N.; Uemura, M.; Steponkus, P.L.; Gilmour, S.J.; Lin, C.; Thomashow, M.F. Constitutive expression of the cold-regulated Arabidopsis thaliana COR15a gene affects both chloroplast and protoplast freezing tolerance. Proc. Natl. Acad. Sci. USA 1996, 93, 13404–13409. [Google Scholar] [CrossRef]
- Thalhammer, A.; Bryant, G.; Sulpice, R.; Hincha, D.K. Disordered cold regulated15 proteins protect chloroplast membranes during freezing through binding and folding, but do not stabilize chloroplast enzymes in vivo. Plant Physiol. 2014, 166, 190–201. [Google Scholar] [CrossRef]
- Hara, M.; Terashima, S.; Fukaya, T.; Kuboi, T. Enhancement of cold tolerance and inhibition of lipid peroxidation by citrus dehydrin in transgenic tobacco. Planta 2003, 217, 290–298. [Google Scholar] [CrossRef]
- Tolleter, D.; Hincha, D.K.; Macherel, D. A mitochondrial late embryogenesis abundant protein stabilizes model membranes in the dry state. Biochim. Biophys. Acta 2010, 1798, 1926–1933. [Google Scholar] [CrossRef]
- Moore, D.S.; Hand, S.C. Cryopreservation of lipid bilayers by LEA proteins from Artemiafranciscana and trehalose. Cryobiology 2016, 73, 240–247. [Google Scholar] [CrossRef]
- Wise, M.J.; Tunnacliffe, A. POPP the question: What do LEA proteins do? Trends Plant Sci. 2004, 9, 13–17. [Google Scholar] [CrossRef]
- Rahman, L.N.; Chen, L.; Nazim, S.; Bamm, V.V.; Yaish, M.W.; Moffatt, B.A.; Dutcher, J.R.; Harauz, G. Interactions of intrinsically disordered Thellungiellasalsugineadehydrins TsDHN-1 and TsDHN-2 with membranes—Synergistic effects of lipid composition and temperature on secondary structure. Biochem. Cell Biol. 2010, 88, 791–807. [Google Scholar] [CrossRef] [PubMed]
- Hincha, D.K.; Oliver, A.E.; Crowe, J.H. Lipid composition determines the effects of arbutin on the stability of membranes. Biophys. J. 1999, 77, 2024–2034. [Google Scholar] [CrossRef]
- Furuki, T.; Sakurai, M. Group 3 LEA protein model peptides protect liposomes during desiccation. Biochim. Biophys. Acta 2014, 1838, 2757–2766. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wu, J.; Sun, N.; Tu, C.; Shi, X.; Cheng, H.; Liu, S.; Li, S.; Wang, Y.; Zheng, Y.; et al. Intrinsically Disordered Proteins as Important Players during Desiccation Stress of Soybean Radicles. J. Proteome Res. 2017, 16, 2393–2409. [Google Scholar] [CrossRef] [PubMed]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, L.; Sun, Y.; Liu, Y.; Zou, Y.; Huang, J.; Zheng, Y.; Liu, G. The N-Terminal Region of Soybean PM1 Protein Protects Liposomes during Freeze-Thaw. Int. J. Mol. Sci. 2020, 21, 5552. https://doi.org/10.3390/ijms21155552
Chen L, Sun Y, Liu Y, Zou Y, Huang J, Zheng Y, Liu G. The N-Terminal Region of Soybean PM1 Protein Protects Liposomes during Freeze-Thaw. International Journal of Molecular Sciences. 2020; 21(15):5552. https://doi.org/10.3390/ijms21155552
Chicago/Turabian StyleChen, Liyi, Yajun Sun, Yun Liu, Yongdong Zou, Jianzi Huang, Yizhi Zheng, and Guobao Liu. 2020. "The N-Terminal Region of Soybean PM1 Protein Protects Liposomes during Freeze-Thaw" International Journal of Molecular Sciences 21, no. 15: 5552. https://doi.org/10.3390/ijms21155552
APA StyleChen, L., Sun, Y., Liu, Y., Zou, Y., Huang, J., Zheng, Y., & Liu, G. (2020). The N-Terminal Region of Soybean PM1 Protein Protects Liposomes during Freeze-Thaw. International Journal of Molecular Sciences, 21(15), 5552. https://doi.org/10.3390/ijms21155552




