Flexible NAD+ Binding in Deoxyhypusine Synthase Reflects the Dynamic Hypusine Modification of Translation Factor IF5A
Abstract
1. Introduction
2. Results
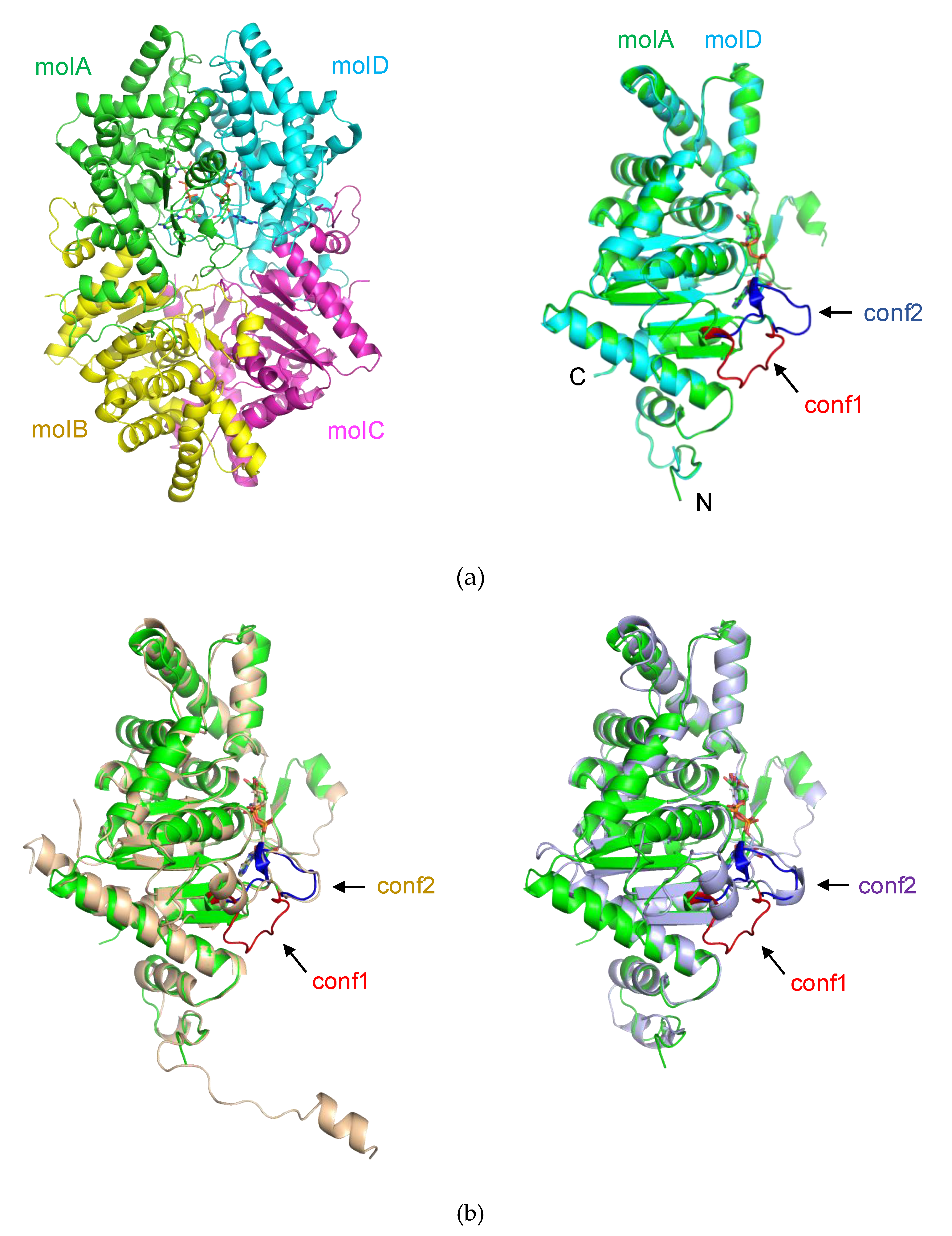
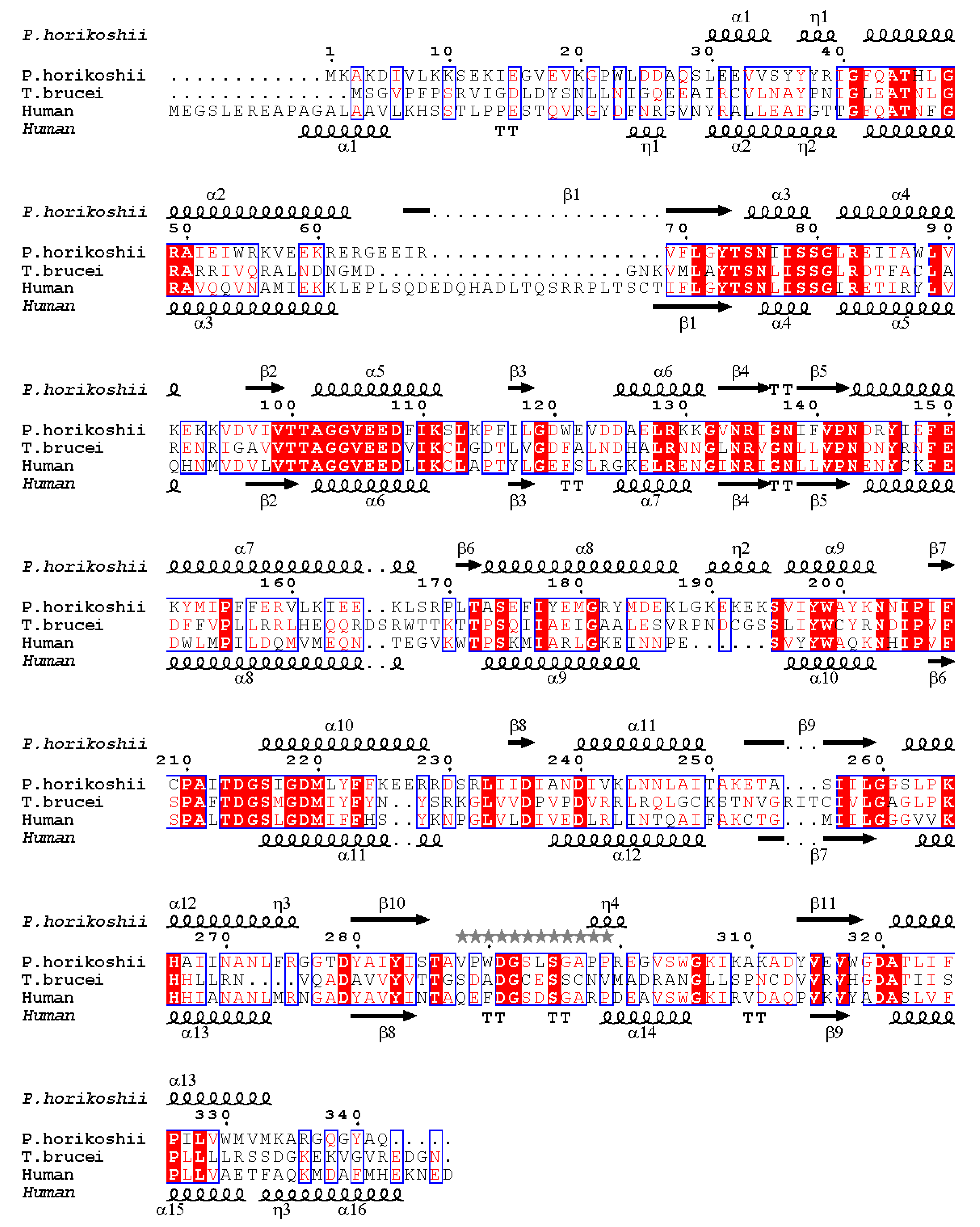
2.1. The Overall Structure of PhoDHS
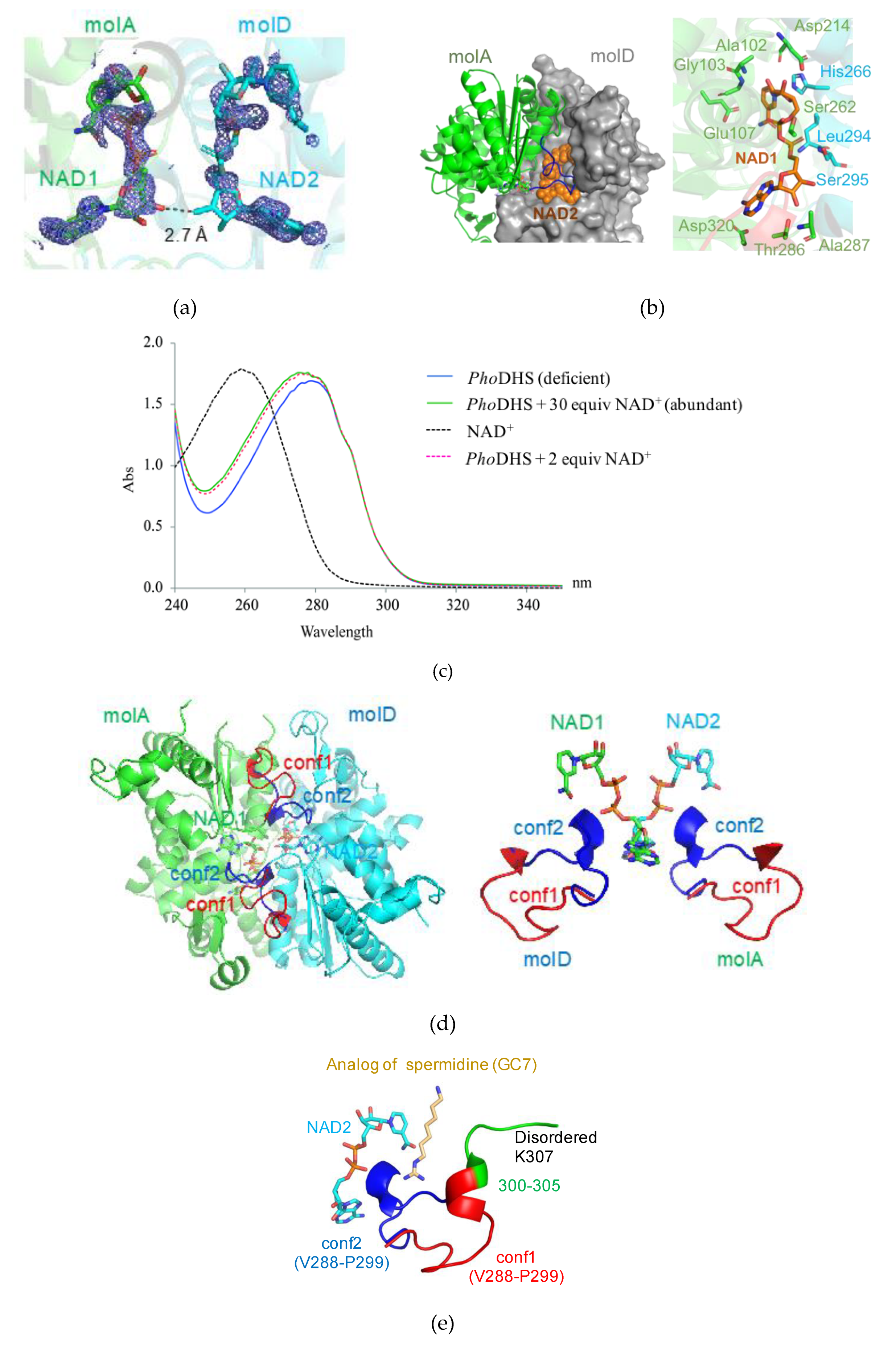
2.2. NAD+ Binding Manner of PhoDHS
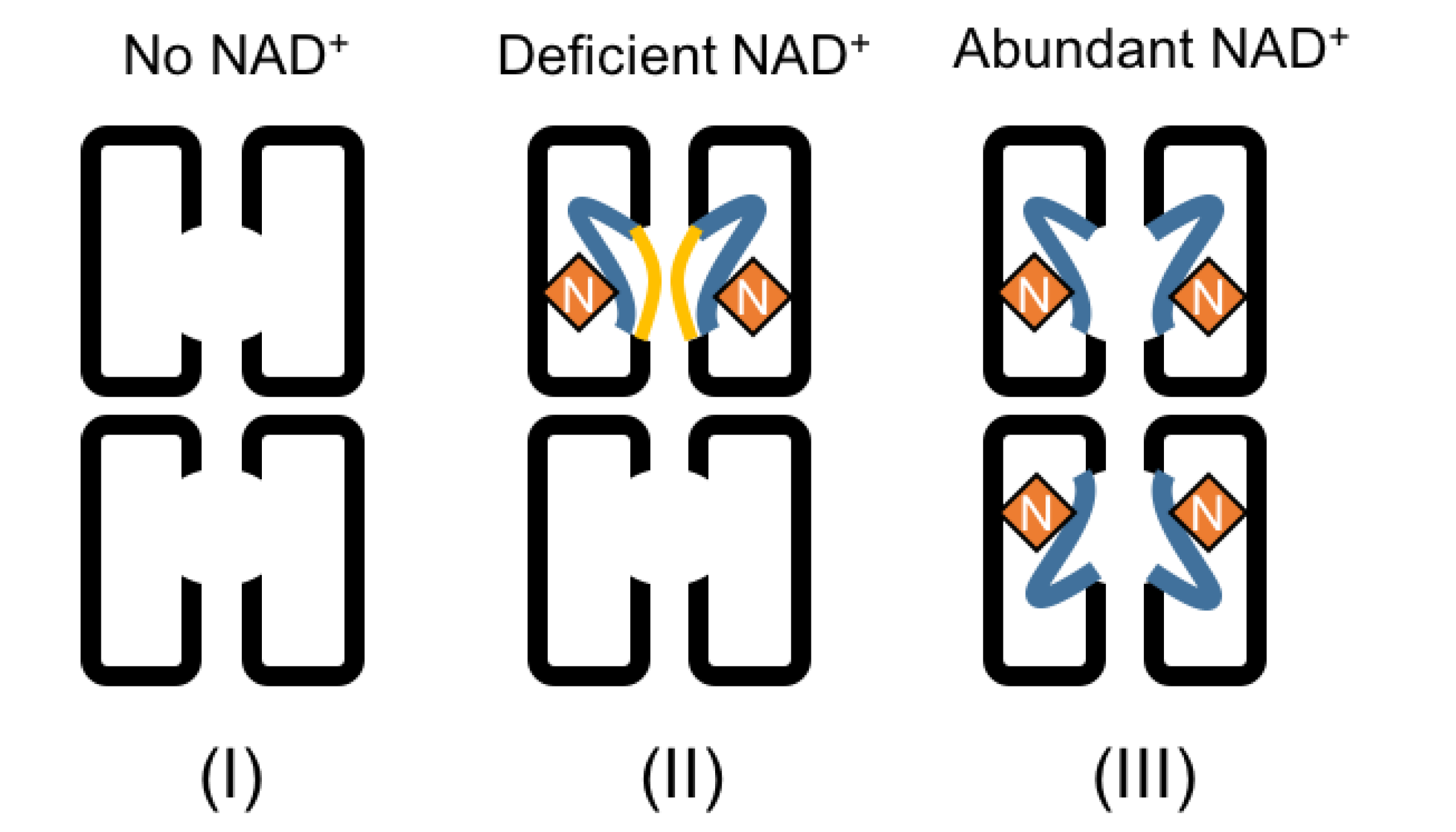
2.3. Dynamic Conformation of the Region for Both NAD+ and Spermidine Binding
3. Discussion
4. Materials and Methods
4.1. Expression and Purification of PhoDHS
4.2. Crystallization and Data Collection
4.3. Structural Determination and Refinement
4.4. Analysis of NAD+ Binding by UV Absorbance Spectroscopy
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Greganova, E.; Altmann, M.; Buetikofer, P. Unique modifications of translation elongation factors. FEBS J. 2011, 278, 2613–2624. [Google Scholar] [CrossRef] [PubMed]
- Saini, P.; Eyler, D.E.; Green, R.; Dever, T.E. Hypusine-containing protein eIF5A promotes translation elongation. Nature 2009, 459, 118–121. [Google Scholar] [CrossRef] [PubMed]
- Landau, G.; Bercovich, Z.; Park, M.H.; Kahana, C. The Role of Polyamines in Supporting Growth of Mammalian Cells Is Mediated through Their Requirement for Translation Initiation and Elongation. J. Biol. Chem. 2010, 285, 12474–12481. [Google Scholar] [CrossRef] [PubMed]
- Jao, D.L.E.; Chen, K.Y. Tandem affinity purification revealed the hypusine-dependent binding of eukaryotic initiation factor 5A to the translating 80S ribosomal complex. J. Cell. Biochem. 2006, 97, 583–598. [Google Scholar] [CrossRef] [PubMed]
- Zanelli, C.F.; Maragno, A.L.; Gregio, A.P.; Komili, S.; Pandolfi, J.R.; Mestriner, C.A.; Lustri, W.R.; Valentini, S.R. eIF5A binds to translational machinery components and affects translation in yeast. Biochem. Biophys. Res. Commun. 2006, 348, 1358–1366. [Google Scholar] [CrossRef]
- Park, M.H.; Joe, Y.A.; Kang, K.R. Deoxyhypusine synthase activity is essential for cell viability in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1998, 273, 1677–1683. [Google Scholar] [CrossRef]
- Nishimura, K.; Lee, S.B.; Park, J.H.; Park, M.H. Essential role of eIF5A-1 and deoxyhypusine synthase in mouse embryonic development. Amino Acids 2012, 42, 703–710. [Google Scholar] [CrossRef]
- Patel, P.H.; Costa-Mattioli, M.; Schulze, K.L.; Bellen, H.J. The Drosophila deoxyhypusine hydroxylase homologue nero and its target elF5A are required for cell growth and the regulation of autophagy. J. Cell Biol. 2009, 185, 1181–1194. [Google Scholar] [CrossRef]
- Lee, S.B.; Park, J.H.; Kaevel, J.; Sramkova, M.; Weigert, R.; Park, M.H. The effect of hypusine modification on the intracellular localization of eIF5A. Biochem. Biophys. Res. Commun. 2009, 383, 497–502. [Google Scholar] [CrossRef]
- Gentz, P.M.; Blatch, G.L.; Dorrington, R.A. Dimerization of the yeast eukaryotic translation initiation factor 5A requires hypusine and is RNA dependent. FEBS J. 2009, 276, 695–706. [Google Scholar] [CrossRef]
- Wolff, E.C.; Kang, K.R.; Kim, Y.S.; Park, M.H. Posttranslational synthesis of hypusine: Evolutionary progression and specificity of the hypusine modification. Amino Acids 2007, 33, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Park, M.H.; Lee, Y.B.; Joe, Y.A. Hypusine is essential for eukaryotic cell proliferation. Biol. Signals 1997, 6, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Park, M.H.; Cooper, H.L.; Folk, J.E. The biosynthesis of protein-bound hypusine (N epsilon—(4-amino-2-hydroxybutyl)lysine). Lysine as the amino acid precursor and the intermediate role of deoxyhypusine (N epsilon—(4-aminobutyl) lysine). J. Biol. Chem. 1982, 257, 7217–7222. [Google Scholar] [PubMed]
- Park, M.H.; Wolff, E.C. Cell-free synthesis of deoxyhypusine-separation of protein substrate and enzyme and identification of 1,3-diaminopropane as aproduct of spermidine cleavage. J. Biol. Chem. 1988, 263, 15264–15269. [Google Scholar]
- Wolff, E.C.; Lee, Y.B.; Chung, S.I.; Folk, J.E.; Park, M.H. Deoxyhypusine synthase from rat testis: Purification and characterization. J. Biol. Chem. 1995, 270, 8660–8666. [Google Scholar] [CrossRef]
- Kaiser, A. Translational control of eIF5A in various diseases. Amino Acids 2012, 42, 679–684. [Google Scholar] [CrossRef]
- Von Koschitzky, I.; Gerhardt, H.; Lämmerhofer, M.; Kohout, M.; Gehringer, M.; Laufer, S.; Pink, M.; Schmitz-Spanke, S.; Strube, C.; Kaiser, A. New insights into novel inhibitors against deoxyhypusine hydroxylase from plasmodium falciparum: Compounds with an iron chelating potential. Amino Acids 2015, 47, 1155–1166. [Google Scholar] [CrossRef]
- Preukschas, M.; Hagel, C.; Schulte, A.; Weber, K.; Lamszus, K.; Sievert, H.; Pällmann, N.; Bokemeyer, C.; Hauber, J.; Braig, M.; et al. Expression of eukaryotic initiation factor 5A and hypusine forming enzymes in glioblastoma patient samples: Implications for new targeted therapies. PLoS ONE 2012, 7, e43468. [Google Scholar] [CrossRef]
- Tanaka, Y.; Kurasawa, O.; Yokota, A.; Klein, M.G.; Ono, K.; Saito, B.; Matsumoto, S.; Okaniwa, M.; Ambrus-Aikelin, G.; Morishita, D.; et al. Discovery of Novel Allosteric Inhibitors of Deoxyhypusine Synthase. J. Med. Chem. 2020, 63, 3215–3226. [Google Scholar]
- Wolff, E.C.; Folk, J.E.; Park, M.H. Enzyme-substrate intermediate formation at lysine 329 of human deoxyhypusine synthase. J. Biol. Chem. 1997, 272, 15865–15871. [Google Scholar] [CrossRef]
- Liao, D.I.; Wolff, E.C.; Park, M.H.; Davies, D.R. Crystal structure of the NAD complex of human deoxyhypusine synthase: An enzyme with a ball-and-chain mechanism for blocking the active site. Structure 1998, 6, 23–32. [Google Scholar] [CrossRef]
- Umland, T.C.; Wolff, E.C.; Park, M.H.; Davies, D.R. A new crystal structure of deoxyhypusine synthase reveals the configuration of the active enzyme and of an enzyme center dot NAD center dot inhibitor ternary complex. J. Biol. Chem. 2004, 279, 28697–28705. [Google Scholar] [CrossRef] [PubMed]
- Wątor, E.; Wilk, P.; Grudnik, P. Half Way to Hypusine—Structural Basis for Substrate Recognition by Human Deoxyhypusine Synthase. Biomolecules 2020, 10, 522. [Google Scholar] [CrossRef] [PubMed]
- Afanador, G.A.; Tomchick, D.R.; Phillips, A.M. Trypanosomatid Deoxyhypusine Synthase Activity Is Dependent on Shared Active-Site Complementation between Pseudoenzyme Paralogs. Structure 2018, 26, 1499–1512.e5. [Google Scholar] [CrossRef] [PubMed]
- Jansson, B.P.M.; Malandrin, L.; Johansson, H.E. Cell cycle arrest in archaea by the hypusination inhibitor N-1-guanyl-1,7-diaminoheptane. J. Bacteriol. 2000, 182, 1158–1161. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.R.; Chung, S.I. Characterization of yeast deoxyhypusine synthase: PKC-dependent phosphorylation in vitro and functional domain identification. Exp. Mol. Med. 1999, 31, 210–216. [Google Scholar] [CrossRef]
- Joe, Y.A.; Wolff, E.C.; Park, M.H. Cloning and expression of human deoxyhypusine synthase cDNA—Structure-function studies with the recombinant enzyme and mutant proteins. J. Biol. Chem. 1995, 270, 22386–22392. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef]
- Lee, C.H.; Park, M.H. Human deoxyhypusine synthase: Interrelationship between binding of NAD and substrates. Biochem. J. 2000, 352, 851–857. [Google Scholar] [CrossRef]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. In Macromolecular Crystallography, PT A. Methods Enzymol. 1997, 276, 307–326. [Google Scholar]
- Vagin, A.; Teplyakov, A. MOLREP: An Automated Program for Molecular Replacement. J. Appl. Crystallogr. 1997, 30, 1022–1025. [Google Scholar] [CrossRef]
- Murshudov, G.N.; Skubák, P.; Lebedev, A.A.; Pannu, N.S.; Steiner, R.A.; Nicholls, R.A.; Winn, M.D.; Long, F.; Vagin, A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. Sect. D Struct. Biol. 2011, 67, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.M.; Krissinel, E.B.; Leslie, A.G.; McCoy, A.; et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Yao, M.; Tanaka, I. New algorithm for protein model building: Extending a partial model in a map segment. J. Appl. Crystallogr. 2006, 39, 57–63. [Google Scholar] [CrossRef]
- Brünger, A.T.; Adams, P.D.; Clore, G.M.; DeLano, W.L.; Gros, P.; Grosse-Kunstleve, R.W.; Jiang, J.S.; Kuszewski, J.; Nilges, M.; Pannu, N.S.; et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 1998, 54, 905–921. [Google Scholar]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef]
- Emsley, P.; Coot, K.C. Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef]
- Krissinel, E.; Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef]
| Data Collection | |
| Crystal | Native DHS |
| Space group | P212121 |
| Beamline | Spring 8 BL41XU |
| Wavelength (Å) | 1.0000 |
| Unit-cell parameter | a = 87.3 Å, b = 89.9 Å, c = 164.8 Å, α = β= γ = 90° |
| Resolution range (Å) | 50–2.20 (2.28–2.20) |
| Total number of reflections | 666,150 (6550) |
| Completeness (%) | 100.0 (100.0) |
| Redundancy | 7.4 (7.4) |
| I/σ(I) | 29.1 (5.2) |
| R-merge | 0.110 (0.479) |
| Refinement Statistics | |
| Resolution range (Å) | 34.42–2.20 |
| Rwork/Rfree (%) | 17.52/22.95 |
| R.m.s.d bond lengths (Å) | 0.07 |
| R.m.s.d angles (°) | 0.93 |
| No. non-H atoms | |
| Proteins | 9817 |
| Water molecules | 408 |
| NAD+ molecules | 88 |
| Average B value | |
| Protein | 35.17 |
| Water molecules | 41.81 |
| NAD+ molecules | 71.98 |
| Ramachandran Plot (%) | |
| Favored | 98.91 |
| Allowed | 1.09 |
| Outline | 0.00 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, M.; Gai, Z.; Okada, C.; Ye, Y.; Yu, J.; Yao, M. Flexible NAD+ Binding in Deoxyhypusine Synthase Reflects the Dynamic Hypusine Modification of Translation Factor IF5A. Int. J. Mol. Sci. 2020, 21, 5509. https://doi.org/10.3390/ijms21155509
Chen M, Gai Z, Okada C, Ye Y, Yu J, Yao M. Flexible NAD+ Binding in Deoxyhypusine Synthase Reflects the Dynamic Hypusine Modification of Translation Factor IF5A. International Journal of Molecular Sciences. 2020; 21(15):5509. https://doi.org/10.3390/ijms21155509
Chicago/Turabian StyleChen, Meirong, Zuoqi Gai, Chiaki Okada, Yuxin Ye, Jian Yu, and Min Yao. 2020. "Flexible NAD+ Binding in Deoxyhypusine Synthase Reflects the Dynamic Hypusine Modification of Translation Factor IF5A" International Journal of Molecular Sciences 21, no. 15: 5509. https://doi.org/10.3390/ijms21155509
APA StyleChen, M., Gai, Z., Okada, C., Ye, Y., Yu, J., & Yao, M. (2020). Flexible NAD+ Binding in Deoxyhypusine Synthase Reflects the Dynamic Hypusine Modification of Translation Factor IF5A. International Journal of Molecular Sciences, 21(15), 5509. https://doi.org/10.3390/ijms21155509





