The Role of Pre-Clinical 3-Dimensional Models of Osteosarcoma
Abstract
1. Introduction
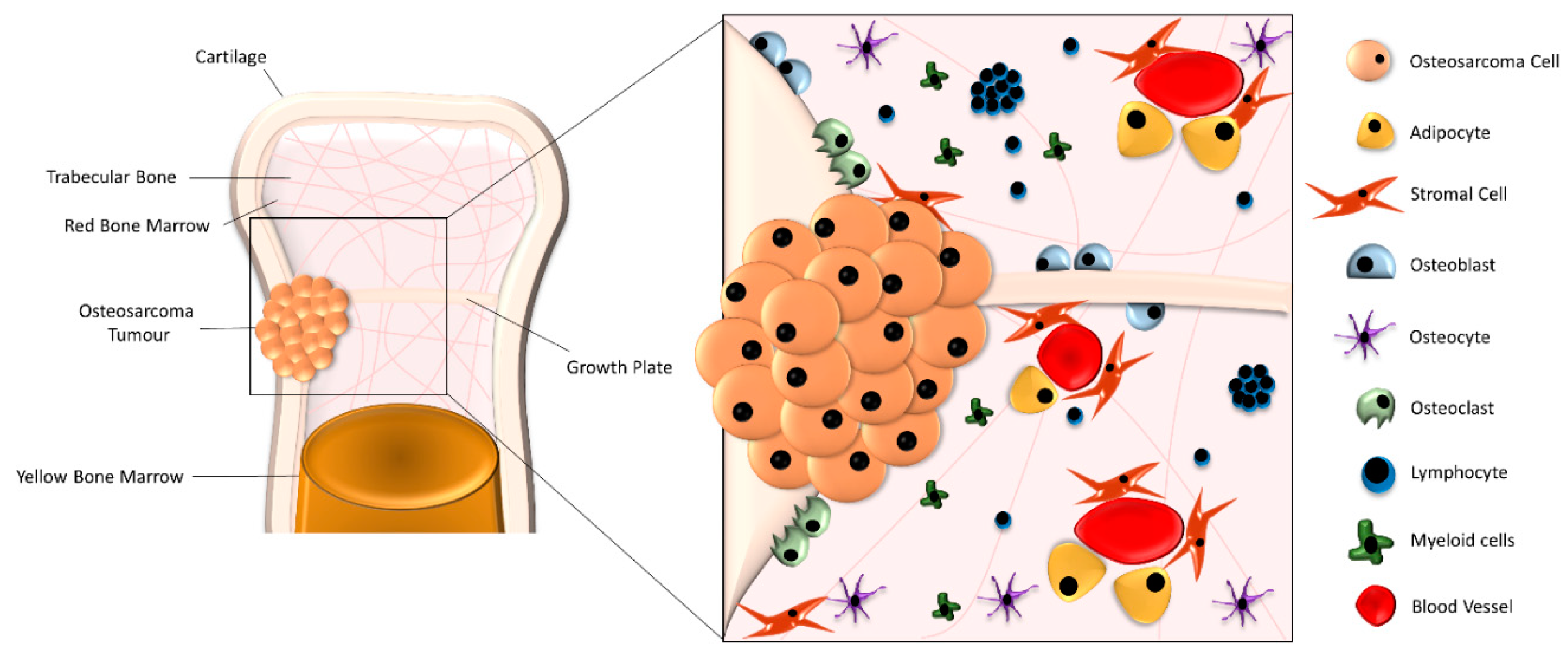
2. Biological Understanding of OS
3. Pre-Clinical Modelling of OS-3D Models
4. In Vivo Animal Models
5. In Vitro 3D Models
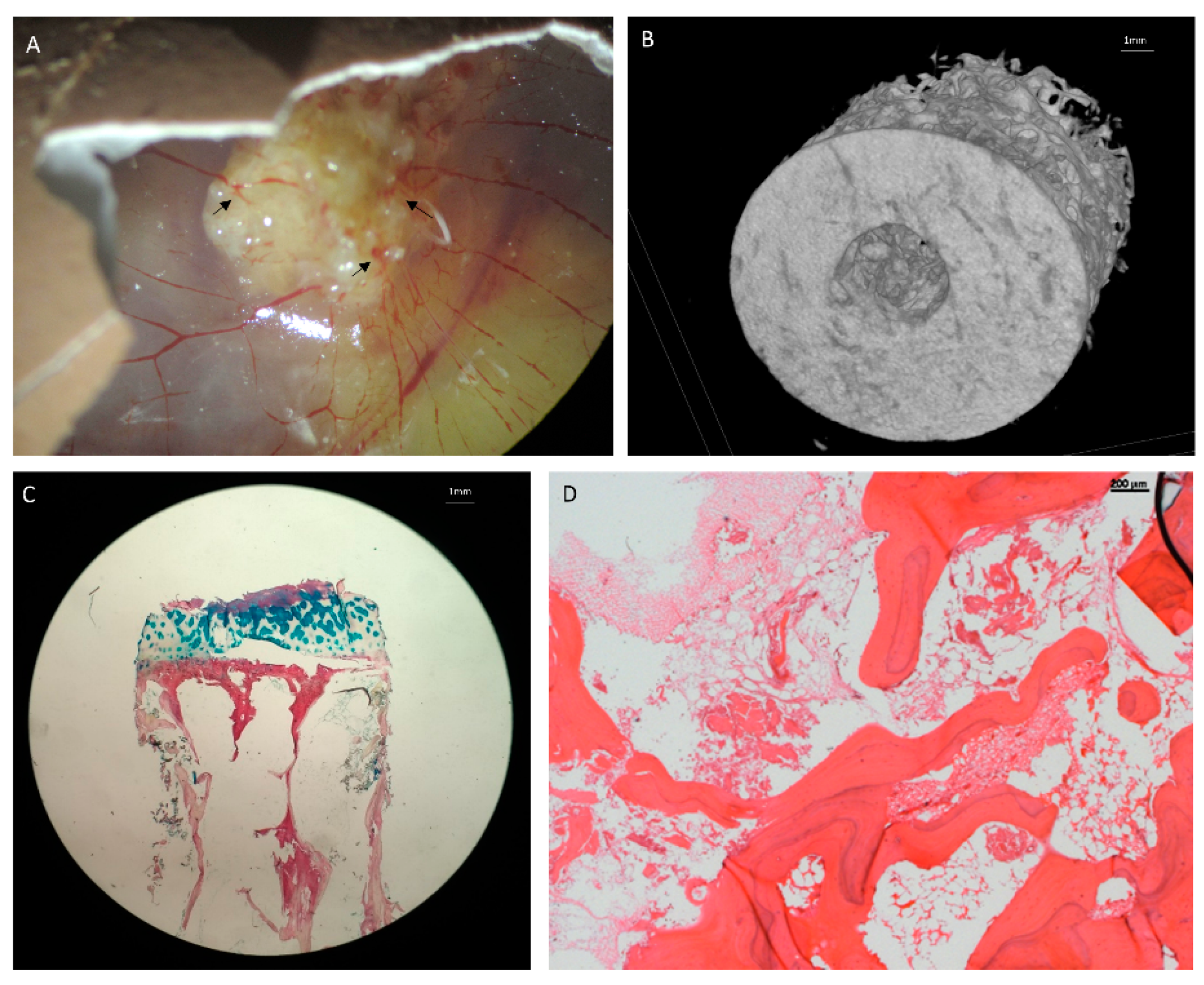
6. In Ovo Chorioallantoic Membrane Models
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| OS | Osteosarcoma |
| 3D | Three dimensional |
| CAM | Chorioallantoic membrane |
| RB | Retinoblastoma |
| FDA | Food and Drug Administration |
| GM-CSF | Granulocyte macrophage colony stimulating factor |
| MSC | Mesenchymal stem cell |
| BMSC | Bone marrow stromal cells |
| RANKL | Receptor activator of nuclear factor κB ligand |
| IL-6 | Interleukin 6 |
| IL-11 | Interleukin 11 |
| TGFβ | Transforming Growth Factor Beta |
| BMP | Bone morphogenetic protein |
| ECM | Extracellular matrix |
| BM | Bone marrow |
| Treg | Regulatory T cell |
| TAM | Tumour associated macrophages |
| 2D | Two-dimensional |
| HEMA | Poly-hydroxyethyl methacrylate |
| CSC | Cancer stem cell |
References
- Bielack, S.; Carrle, D.; Jost, L.; On behalf of the ESMO Guidelines Working Group. Osteosarcoma: ESMO Clinical Recommendations for diagnosis, treatment and follow-up. Ann. Oncol. 2008, 19, ii94–ii96. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Zhao, J.; Bai, J.; Shen, H.; Zhang, B.; Deng, L.; Sun, C.; Liu, Y.; Zhang, J.; Zheng, J. Risk and clinicopathological features of osteosarcoma metastasis to the lung: A population-based study. J. Bone Oncol. 2019, 16, 100230. [Google Scholar] [CrossRef] [PubMed]
- Overholtzer, M.; Rao, P.H.; Favis, R.; Lu, X.-Y.; Elowitz, M.B.; Barany, F.; Ladanyi, M.; Gorlick, R.; Levine, A.J. The presence of p53 mutations in human osteosarcomas correlates with high levels of genomic instability. Proc. Natl. Acad. Sci. USA 2003, 100, 11547–11552. [Google Scholar] [CrossRef] [PubMed]
- Selvarajah, S.; Yoshimoto, M.; Maire, G.; Paderova, J.; Bayani, J.; Squire, J.A.; Zielenska, M. Identification of cryptic microaberrations in osteosarcoma by high-definition oligonucleotide array comparative genomic hybridization. Cancer Genet. Cytogenet 2007, 179, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.W.; Aslo, A.; Tsay, C.; Slamon, D.; Ishizaki, K.; Toguchida, J.; Yamamuro, T.; Lampkin, B.; Koeffler, H.P. Frequency and structure of p53 rearrangements in human osteosarcoma. Cancer Res. 1990, 50, 7950–7954. [Google Scholar] [PubMed]
- Toguchida, J.; Yamaguchi, T.; Ritchie, B.; Beauchamp, R.L.; Dayton, S.H.; Herrera, G.E.; Yamamuro, T.; Kotoura, Y.; Sasaki, M.S.; Little, J.B. Mutation spectrum of the p53 gene in bone and soft tissue sarcomas. Cancer Res. 1992, 52, 6194–6199. [Google Scholar] [PubMed]
- Wadayama, B.; Toguchida, J.; Shimizu, T.; Ishizaki, K.; Sasaki, M.S.; Kotoura, Y.; Yamamuro, T. Mutation spectrum of the retinoblastoma gene in osteosarcomas. Cancer Res. 1994, 54, 3042–3048. [Google Scholar]
- Hansen, M.F.; Koufos, A.; Gallie, B.; Phillips, R.A.; Fodstad, O.; Brögger, A.; Gedde-Dahl, T.; Cavenee, W.K. Osteosarcoma and retinoblastoma: A shared chromosomal mechanism revealing recessive predisposition. Proc. Natl. Acad. Sci. USA 1985, 82, 6216–6220. [Google Scholar] [CrossRef]
- Mirabello, L.; Troisi, R.J.; Savage, S.A. Osteosarcoma incidence and survival rates from 1973 to 2004: Data from the Surveillance, Epidemiology, and End Results Program. Cancer 2009, 115, 1531–1543. [Google Scholar] [CrossRef]
- Guijarro, M.V.; Ghivizzani, C.S.; Gibbs, P.C. Corrigendum: Animal models in osteosarcoma. Front. Genet. 2015, 5, 475. [Google Scholar] [CrossRef]
- Smeland, S.; Whelan, J.; Bielack, S.S.; Marina, N.; Jovic, G.; Butterfass-Bahloul, T.; Calaminus, G.; Eriksson, M.; Gelderblom, H.; Gorlick, R.G.; et al. Event-free survival and overall survival in 2,253 patients with osteosarcoma registered to EURAMOS-1. J. Clin. Oncol. 2015, 33, 10512. [Google Scholar] [CrossRef]
- Jimmy, R.; Stern, C.; Lisy, K.; White, S. Effectiveness of mifamurtide in addition to standard chemotherapy for high-grade osteosarcoma. JBI Database Syst. Rev. Implement. Rep. 2017, 15, 2113–2152. [Google Scholar] [CrossRef] [PubMed]
- Meyers, P.A.; Healey, J.H.; Chou, A.J.; Wexler, L.H.; Merola, P.R.; Morris, C.D.; Laquaglia, M.P.; Kellick, M.G.; Abramson, S.J.; Gorlick, R. Addition of pamidronate to chemotherapy for the treatment of osteosarcoma. Cancer 2010, 117, 1736–1744. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yang, R.; Roth, M.; Piperdi, S.; Zhang, W.; Dorfman, H.; Rao, P.; Park, A.; Tripathi, S.; Freeman, C.; et al. Genetically transforming human osteoblasts to sarcoma: Development of an osteosarcoma model. Genes Cancer 2017, 8, 484–494. [Google Scholar] [CrossRef]
- Pittenger, M.F. Multilineage Potential of Adult Human Mesenchymal Stem Cells. Science 1999, 284, 143–147. [Google Scholar] [CrossRef]
- Poggi, A.; Giuliani, M. Mesenchymal Stromal Cells Can Regulate the Immune Response in the Tumor Microenvironment. Vaccines 2016, 4, 41. [Google Scholar] [CrossRef]
- Karnoub, A.E.; Dash, A.B.; Vo, A.P.; Sullivan, A.; Brooks, M.W.; Bell, G.W.; Richardson, A.L.; Polyak, K.; Tubo, R.; Weinberg, R.A. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature 2007, 449, 557–563. [Google Scholar] [CrossRef]
- Suzuki, K.; Sun, R.; Origuchi, M.; Kanehira, M.; Takahata, T.; Itoh, J.; Umezawa, A.; Kijima, H.; Fukuda, S.; Saijo, Y. Mesenchymal Stromal Cells Promote Tumor Growth through the Enhancement of Neovascularization. Mol. Med. 2011, 17, 579–587. [Google Scholar] [CrossRef]
- Ragland, B.D.; Bell, W.C.; Lopez, R.R.; Siegal, G.P. Cytogenetics and molecular biology of osteosarcoma. Lab. Investig. 2002, 82, 365–373. [Google Scholar] [CrossRef][Green Version]
- Krzeszinski, J.Y.; Wan, Y. New therapeutic targets for cancer bone metastasis. Trends Pharm. Sci. 2015, 36, 360–373. [Google Scholar] [CrossRef]
- Kinpara, K.; Mogi, M.; Kuzushima, M.; Togari, A. Osteoclast Differentiation Factor in Human Osteosarcoma Cell Line. J. Immunoass. 2000, 21, 327–340. [Google Scholar] [CrossRef] [PubMed]
- Broadhead, M.L.; Clark, J.C.M.; Myers, D.E.; Dass, C.R.; Choong, P. The Molecular Pathogenesis of Osteosarcoma: A Review. Sarcoma 2011, 2011, 12. [Google Scholar] [CrossRef]
- Weilbaecher, K.N.; Guise, T.A.; McCauley, L.K. Cancer to bone: A fatal attraction. Nat. Rev. Cancer 2011, 11, 411–425. [Google Scholar] [CrossRef] [PubMed]
- Zhao, E.; Xu, H.; Wang, L.; Kryczek, I.; Wu, K.; Hu, Y.; Wang, G.; Zou, W. Bone marrow and the control of immunity. Cell. Mol. Immunol. 2011, 9, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Cao, X. Immunosuppressive cells in tumor immune escape and metastasis. J. Mol. Med. 2015, 94, 509–522. [Google Scholar] [CrossRef]
- Kapałczyńska, M.; Kolenda, T.; Przybyła, W.; Zajączkowska, M.; Teresiak, A.; Filas, V.; Ibbs, M.; Bliźniak, R.; Łuczewski, Ł.; Lamperska, K. 2D and 3D cell cultures—A comparison of different types of cancer cell cultures. Arch. Med. Sci. 2016, 14, 910–919. [Google Scholar] [CrossRef]
- Uluckan, O.; Segaliny, A.; Botter, S.; Santiago, J.M.; Mutsaers, A. Preclinical mouse models of osteosarcoma. Bonekey Rep. 2015, 4, 670. [Google Scholar] [CrossRef]
- Khanna, C.; Khan, J.; Nguyen, P.; Prehn, J.; Caylor, J.; Yeung, C.; Trepel, J.; Meltzer, P.; Helman, L. Metastasis-associated differences in gene expression in a murine model of osteosarcoma. Cancer Res. 2001, 61, 3750–3759. [Google Scholar]
- Kim, W.Y.; Sharpless, N.E. Drug Efficacy Testing in Mice. Curr. Top. Microbiol. Immunol. 2010, 355, 19–38. [Google Scholar] [CrossRef]
- Teicher, B.A.; Bagley, R.G.; Rouleau, C.; Kruger, A.; Ren, Y.; Kurtzberg, L. Characteristics of human Ewing/PNET sarcoma models. Ann. Saudi Med. 2011, 31, 174–182. [Google Scholar] [CrossRef]
- Scott, M.C.; Tomiyasu, H.; Garbe, J.R.; Cornax, I.; Amaya, C.; O’Sullivan, M.G.; Subramanian, S.; Bryan, B.A.; Modiano, J.F. Heterotypic mouse models of canine osteosarcoma recapitulate tumor heterogeneity and biological behavior. Dis. Model. Mech. 2016, 9, 1435–1444. [Google Scholar] [CrossRef] [PubMed]
- Shimozaki, S.; Yamamoto, N.; Domoto, T.; Nishida, H.; Hayashi, K.; Kimura, H.; Takeuchi, A.; Miwa, S.; Igarashi, K.; Kato, T.; et al. Efficacy of glycogen synthase kinase-3β targeting against osteosarcoma via activation of β-catenin. Oncotarget 2016, 7, 77038–77051. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Becher, O.J.; Holland, E.C.; Sausville, E.A.; Burger, A.M. Genetically engineered models have advantages over xenografts for preclinical studies. Cancer Res. 2006, 66, 3355–3359. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.Y.-H.P.; Chang, C.-Y.; Hu, N.; Wang, Y.-C.J.; Lai, C.-C.; Herrup, K.; Lee, W.-H.; Bradley, A. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature 1992, 359, 288–294. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.P.; Pandey, M.K.; Jin, F.; Raymond, A.; Akiyama, H.; Lozano, G. Targeted mutation of p53 and Rb in mesenchymal cells of the limb bud produces sarcomas in mice. Carcinogen 2009, 30, 1789–1795. [Google Scholar] [CrossRef]
- Taneja, P.; Zhu, S.; Maglic, D.; Fry, E.A.; Kendig, R.D.; Inoue, K. Transgenic and Knockout Mice Models to Reveal the Functions of Tumor Suppressor Genes. Clin. Med. Insights Oncol. 2011, 5, 235–257. [Google Scholar] [CrossRef]
- Walkley, C.R.; Qudsi, R.; Sankaran, V.G.; Perry, J.A.; Gostissa, M.; Roth, S.I.; Rodda, S.J.; Snay, E.; Dunning, P.; Fahey, F.H.; et al. Conditional mouse osteosarcoma, dependent on p53 loss and potentiated by loss of Rb, mimics the human disease. Genes Dev. 2008, 22, 1662–1676. [Google Scholar] [CrossRef]
- Lavigueur, A.; Maltby, V.; Mock, D.; Rossant, J.; Pawson, T.; Bernstein, A. High incidence of lung, bone, and lymphoid tumors in transgenic mice overexpressing mutant alleles of the p53 oncogene. Mol. Cell. Boil. 1989, 9, 3982–3991. [Google Scholar] [CrossRef]
- Rowell, J.L.; McCarthy, D.O.; Alvarez, C.E. Dog models of naturally occurring cancer. Trends Mol. Med. 2011, 17, 380–388. [Google Scholar] [CrossRef]
- O’Donoghue, L.E.; Ptitsyn, A.; Kamstock, D.A.; Siebert, J.C.; Thomas, R.S.; Duval, D.L. Expression profiling in canine osteosarcoma: Identification of biomarkers and pathways associated with outcome. BMC Cancer 2010, 10, 506. [Google Scholar] [CrossRef]
- Paoloni, M.C.; Davis, S.; Lana, S.E.; Withrow, S.; Sangiorgi, L.; Picci, P.; Hewitt, S.M.; Triche, T.J.; Meltzer, P.S.; Khanna, C. Canine tumor cross-species genomics uncovers targets linked to osteosarcoma progression. BMC Genom. 2009, 10, 625. [Google Scholar] [CrossRef] [PubMed]
- De Luca, A.; Raimondi, L.; Salamanna, F.; Carina, V.; Costa, V.; Bellavia, D.; Alessandro, R.; Fini, M.; Giavaresi, G. Relevance of 3d culture systems to study osteosarcoma environment. J. Exp. Clin. Cancer Res. 2018, 37, 2. [Google Scholar] [CrossRef] [PubMed]
- Hickman, J.A.; Graeser, R.; De Hoogt, R.; Vidic, S.; Brito, C.; Gutekunst, M.; Van Der Kuip, H.; IMI PREDECT Consortium; Brito, C. Three-dimensional models of cancer for pharmacology and cancer cell biology: Capturing tumor complexity in vitro/ex vivo. Biotechnol. J. 2014, 9, 1115–1128. [Google Scholar] [CrossRef] [PubMed]
- Martins-Neves, S.R.; O Lopes, Á.; Carmo, A.D.; Paiva, A.; Simões, P.C.; Abrunhosa, A.; Gomes, C. Therapeutic implications of an enriched cancer stem-like cell population in a human osteosarcoma cell line. BMC Cancer 2012, 12, 139. [Google Scholar] [CrossRef]
- Indovina, P.; Rainaldi, G.; Santini, M.T. Hypoxia increases adhesion and spreading of MG-63 three-dimensional tumor spheroids. Anticancer. Res. 2008, 28, 1013–1022. [Google Scholar]
- Baek, N.; Seo, O.W.; Kim, M.; Hulme, J.; An, S.S.A. Monitoring the effects of doxorubicin on 3D-spheroid tumor cells in real-time. Oncotargets 2016, 9, 7207–7218. [Google Scholar] [CrossRef]
- Guo, X.; Yu, L.; Zhang, Z.; Dai, G.; Gao, T.; Guo, W. miR-335 negatively regulates osteosarcoma stem cell-like properties by targeting POU5F1. Cancer Cell Int. 2017, 17, 29. [Google Scholar] [CrossRef]
- Chaddad, H.; Kuchler, S.; Fuhrmann, G.; Gégout, H.; Ubeaud-Sequier, G.; Schwinté, P.; Bornert, F.; Benkirane-Jessel, N.; Idoux-Gillet, Y. Combining 2D angiogenesis and 3D osteosarcoma microtissues to improve vascularization. Exp. Cell Res. 2017, 360, 138–145. [Google Scholar] [CrossRef]
- Sitarski, A.M.; Fairfield, H.; Falank, C.; Reagan, M.R. 3D Tissue Engineered in Vitro Models of Cancer in Bone. ACS Biomater. Sci. Eng. 2017, 4, 324–336. [Google Scholar] [CrossRef]
- Tan, P.H.S.; Chia, S.S.; Toh, S.L.; Goh, J.C.H.; Nathan, S.S. Three-dimensional spatial configuration of tumour cells confers resistance to chemotherapy independent of drug delivery. J. Tissue Eng. Regen. Med. 2013, 10, 637–646. [Google Scholar] [CrossRef]
- O’Brien, F.J. Biomaterials & scaffolds for tissue engineering. Mater. Today 2011, 14, 88–95. [Google Scholar] [CrossRef]
- Kim, J.A.; Yun, H.-S.; Choi, Y.A.; Kim, J.-E.; Choi, S.-Y.; Kwon, T.-G.; Kim, Y.K.; Bae, M.A.; Kim, N.J.; Bae, Y.C.; et al. Magnesium phosphate ceramics incorporating a novel indene compound promote osteoblast differentiation in vitro and bone regeneration in vivo. Biomaterials 2018, 157, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Wang, M. Developing bioactive composite materials for tissue replacement. Biomaterials 2003, 24, 2133–2151. [Google Scholar] [CrossRef]
- Oh, S.H.; Kang, S.G.; Kim, E.S.; Cho, S.H.; Lee, J.H. Fabrication and characterization of hydrophilic poly(lactic-co-glycolic acid)/poly(vinyl alcohol) blend cell scaffolds by melt-molding particulate-leaching method. Biomaterials 2003, 24, 4011–4021. [Google Scholar] [CrossRef]
- Lisi, A.; Briganti, E.; Ledda, M.; Losi, P.; Grimaldi, S.; Marchese, R.; Soldani, G. A Combined Synthetic-Fibrin Scaffold Supports Growth and Cardiomyogenic Commitment of Human Placental Derived Stem Cells. PloS ONE 2012, 7, e34284. [Google Scholar] [CrossRef] [PubMed]
- Pavlou, M.; Shah, M.; Gikas, P.; Briggs, T.; Roberts, S.; Cheema, U. Osteomimetic matrix components alter cell migration and drug response in a 3D tumour-engineered osteosarcoma model. Acta Biomater. 2019, 96, 247–257. [Google Scholar] [CrossRef]
- Ivanov, V.; Ivanova, S.; Roomi, M.W.; Kalinovsky, T.; Niedzwiecki, A.; Rath, M. Naturally produced extracellular matrix inhibits growth rate and invasiveness of human osteosarcoma cancer cells. Med. Oncol. 2007, 24, 209–217. [Google Scholar] [CrossRef]
- Rubio, R.; Abarrategi, A.; García-Castro, J.; Martinez-Cruzado, L.; Suárez, C.; Tornin, J.; Santos, L.; Astudillo, A.; Colmenero, I.; Mulero, F.; et al. Bone Environment is Essential for Osteosarcoma Development from Transformed Mesenchymal Stem Cells. Stem Cells 2014, 32, 1136–1148. [Google Scholar] [CrossRef]
- Balke, M.; Neumann, A.; Kersting, C.; Agelopoulos, K.; Gebert, C.; Gosheger, G.; Buerger, H.; Hagedorn, M. Morphologic characterization of osteosarcoma growth on the chick chorioallantoic membrane. BMC Res. Notes 2010, 3, 58. [Google Scholar] [CrossRef]
- Murphy, J.B. Transplantability of tissues to the embryo of foreign species. J. Exp. Med. 1913, 17, 482–493. [Google Scholar] [CrossRef]
- Ribatti, D. The chick embryo chorioallantoic membrane (CAM). A multifaceted experimental model. Mech. Dev. 2016, 141, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.-Y.; Tsai, H.-C.; Chou, P.-Y.; Wang, S.-W.; Chen, H.-T.; Lin, Y.-M.; Chiang, I.-P.; Chang, T.-M.; Hsu, S.-K.; Chou, M.-C.; et al. CCL3 promotes angiogenesis by dysregulation of miR-374b/ VEGF-A axis in human osteosarcoma cells. Oncotarget 2015, 7, 4310–4325. [Google Scholar] [CrossRef] [PubMed]
- Deryugina, E.I.; Quigley, J.P. Chapter 2 Chick Embryo Chorioallantoic Membrane Models to Quantify Angiogenesis Induced by Inflammatory and Tumor Cells or Purified Effector Molecules. Methods Enzymol. 2008, 444, 21–41. [Google Scholar] [CrossRef] [PubMed]
- Lokman, N.A.; Elder, A.S.F.; Ricciardelli, C.; Oehler, M.K. Chick Chorioallantoic Membrane (CAM) Assay as an In Vivo Model to Study the Effect of Newly Identified Molecules on Ovarian Cancer Invasion and Metastasis. Int. J. Mol. Sci. 2012, 13, 9959–9970. [Google Scholar] [CrossRef]
- Zuo, Z.; Syrovets, T.; Wu, Y.; Hafner, S.; Vernikouskaya, I.; Liu, W.; Ma, G.; Weil, T.; Simmet, T.; Rasche, V. The CAM cancer xenograft as a model for initial evaluation of MR labelled compounds. Sci. Rep. 2017, 7, 46690. [Google Scholar] [CrossRef]
- Manjunathan, R.; Ragunathan, M. Chicken chorioallantoic membrane as a reliable model to evaluate osteosarcoma-an experimental approach using SaOS2 cell line. Boil. Proced. Online 2015, 17, 10. [Google Scholar] [CrossRef][Green Version]
- Dohle, D.S.; Pasa, S.D.; Gustmann, S.; Laub, M.; Wissler, J.H.; Jennissen, H.P.; Dünker, N. Chick ex ovo Culture and ex ovo CAM Assay: How it Really Works. J. Vis. Exp. 2009, 33, 1620. [Google Scholar] [CrossRef]
- Schuh, J.C.L. Trials, Tribulations, and Trends in Tumor Modeling in Mice. Toxicol. Pathol. 2004, 32, 53–66. [Google Scholar] [CrossRef]
- Liu, Z.; Vunjak-Novakovic, G. Modeling tumor microenvironments using custom-designed biomaterial scaffolds. Curr. Opin. Chem. Eng. 2016, 11, 94–105. [Google Scholar] [CrossRef]
- Nowak-Sliwinska, P.; Segura, T.; Iruela-Arispe, M.L. The chicken chorioallantoic membrane model in biology, medicine and bioengineering. Angiogenesis 2014, 17, 779–804. [Google Scholar] [CrossRef]
- Debord, L.C.; Pathak, R.R.; Villaneuva, M.; Liu, H.-C.; Harrington, D.A.; Yu, W.; Lewis, M.T.; Sikora, A.G. The chick chorioallantoic membrane (CAM) as a versatile patient-derived xenograft (PDX) platform for precision medicine and preclinical research. Am. J. Cancer Res. 2018, 8, 1642–1660. [Google Scholar] [PubMed]
- Li, M.; Pathak, R.R.; López-Rivera, E.; Friedman, S.L.; Aguirre-Ghiso, J.A.; Sikora, A.G. The In Ovo Chick Chorioallantoic Membrane (CAM) Assay as an Efficient Xenograft Model of Hepatocellular Carcinoma. J. Vis. Exp. 2015, 10, 52411. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Jimenez, I.; Lanham, S.A.; Kanczler, J.M.; Hulsart-Billstrom, G.; Evans, N.D.; Oreffo, R.O.C. Remodelling of human bone on the chorioallantoic membrane of the chicken egg:De novobone formation and resorption. J. Tissue Eng. Regen. Med. 2018, 12, 1877–1890. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smith, H.L.; Beers, S.A.; Gray, J.C.; Kanczler, J.M. The Role of Pre-Clinical 3-Dimensional Models of Osteosarcoma. Int. J. Mol. Sci. 2020, 21, 5499. https://doi.org/10.3390/ijms21155499
Smith HL, Beers SA, Gray JC, Kanczler JM. The Role of Pre-Clinical 3-Dimensional Models of Osteosarcoma. International Journal of Molecular Sciences. 2020; 21(15):5499. https://doi.org/10.3390/ijms21155499
Chicago/Turabian StyleSmith, Hannah L., Stephen A. Beers, Juliet C. Gray, and Janos M. Kanczler. 2020. "The Role of Pre-Clinical 3-Dimensional Models of Osteosarcoma" International Journal of Molecular Sciences 21, no. 15: 5499. https://doi.org/10.3390/ijms21155499
APA StyleSmith, H. L., Beers, S. A., Gray, J. C., & Kanczler, J. M. (2020). The Role of Pre-Clinical 3-Dimensional Models of Osteosarcoma. International Journal of Molecular Sciences, 21(15), 5499. https://doi.org/10.3390/ijms21155499




