Abstract
Cartilage lesions have a multifactorial nature, and genetic factors are their strongest determinants. As biochemical and genetic studies have dramatically progressed over the past decade, the molecular basis of cartilage pathologies has become clearer. Several homeostasis abnormalities within cartilaginous tissue have been found, including various structural changes, differential gene expression patterns, as well as altered epigenetic regulation. However, the efficient treatment of cartilage pathologies represents a substantial challenge. Understanding the complex genetic background pertaining to cartilage pathologies is useful primarily in the context of seeking new pathways leading to disease progression as well as in developing new targeted therapies. A technology utilizing gene transfer to deliver therapeutic genes to the site of injury is quickly becoming an emerging approach in cartilage renewal. The goal of this work is to provide an overview of the genetic basis of chondral lesions and the different approaches of the most recent systems exploiting therapeutic gene transfer in cartilage repair. The integration of tissue engineering with viral gene vectors is a novel and active area of research. However, despite promising preclinical data, this therapeutic concept needs to be supported by the growing body of clinical trials.
1. Introduction
Articular cartilage is a highly specialized connective tissue. Its most important components include the extracellular matrix (ECM), comprising of chondrocytes, collagen fibers (mainly type II and IX), small non-collagen proteins (e.g., aggrecan, high molecular weight proteoglycan and cartilage oligomeric matrix protein (COMP)), a small volume of non-collagenous proteins, glycoproteins such as fibronectin and water [1]. This composition is regulated by chondrocytes in response to the changes in their chemical and mechanical environment.
In addition to structural elements, chondrocytes produce substances that perform physiological functions: enzymes and cytokines. Such enzymes include metalloproteinases, adamalysins (A Disintegrin and Metalloproteinase Domain; ADAMs), serine and cysteine proteases, and their inhibitors [2,3]. Proinflammatory cytokines (e.g., IL-1β, TNFα, IL-6, IL-15, IL-17, and IL-18) participate in the turnover of the cartilaginous matrix, stimulating chondrocytes to increase the production of enzymes to enhance matrix degradation, while IL-4, IL-10, and IL-13 inhibit this process [4,5]. Human articular chondrocytes express a constitutive complex of major histocompatibility system (MHC) class I, which are molecules that regulate complement activation. After their activation, such as under the influence of IFN-α, IL-1, TNF-α, or as a result of inflammatory joint diseases, chondrocytes also express MHC class II and ICAM-1 intercellular adhesion molecules [6]. Numerous studies have shown that chondrocytes also have tissue-specific antigens that induce the production of antibodies in patients with cartilage transplants, as well as in patients with rheumatoid arthritis (RA) and osteoarthritis (OA). The role of the genes coding structural components of cartilage and regulatory enzymes in the pathogenesis of osteochondral pathologies concerning inter- and intracellular signaling pathways is still under investigation.
Understanding and better characterizing the genetic basis of cartilage damage is important for several reasons. First, it can be useful to identify early and specific diagnostic biomarkers signaling the beginning of degradation; identifying such markers can play a crucial role in predicting and monitoring disease progression. As well, the identification of predisposing genes also aids in the stratification of risk groups and the introduction of preventative strategies. However, the most important consideration from a clinical perspective is the therapeutic advantages; the identification of genetic determinants is increasingly an introduction to the development of new individualized therapeutic protocols. Gene therapies are promising for diseases causing cartilage damage, and nanoconstructions containing low molecular RNA or vectors with gene expression modulators are being developed.
Biomechanical, genetic, inflammatory, and hormonal components are important for the development and progression of cartilage tissue diseases. The purpose of this review is to present the diversity of genetic changes in cartilage and the prospective use of gene therapy in joint pathologies.
2. Genetic Basis of Cartilage Pathology
Disorders of cartilage homeostasis are a significant feature of the pathological state of the tissue. In a state of imbalance between catabolic and anabolic processes, degeneration prevails over regeneration, resulting in the loss of cartilage matrix.
An important cause of these cartilaginous pathologies is the successive changes at the genome and transcriptome levels, as well as their effects in the form of proteomic changes. The molecular markers associated with the development of cartilage pathologies can be identified using molecular biology techniques (single-gene analysis by polymerase chain reaction or genome-wide testing, such as with microarrays) or bioinformatics and computational biology methods (e.g., meta-analyzes, association studies) [7,8,9]. The advent of sequencing methodologies (next-generation sequencing or massively parallel high-throughput DNA sequencing) has provided new possibilities for searching, as well as quantitative and functional analysis of genome and transcriptome. The main purposes of large-scale genomic screens are searching for novel biomarkers and their genetic signatures, and these are also associated with function in physiological and pathological conditions [7,10,11,12]. Several scientific studies show a correlation between abnormalities in the behavior of cartilage homeostasis and structural changes within DNA (mainly single nucleotide polymorphisms type), differentiated gene expression, as well as epigenetic regulations (such as microRNA and promotor region methylation). Genome and transcriptome changes have been described for most cartilage diseases; however, they were best characterized for RA and OA. Genetic biomarkers of cartilage structure damage and metabolism disorders, with their therapeutic potential, are presented in Table 1.

Table 1.
Summary of biomarkers of cartilage damage showing the potential to support regenerative therapy. ECM: extracellular matrix.
The genetic changes in cartilage are regulated directly and indirectly via genes associated with tissue metabolism. Quantitative and qualitative changes in key genes trigger a cascade of changes that lead to disorders in several signaling pathways (such as signal transduction, NF-ĸB, JAK/STAT, Wnt, and mTOR) [51]. The initiation and progression of osteoarthritis may be associated with, among others, changes in IL-1β expression levels that affect signal transduction through the NF-ĸB pathway [52]. Cartilage degradation by the proteasome–ubiquitin system and intra-cartilage ossification have been correlated with abnormalities in the Wnt pathway mediated by Sox9 and FRZB [53,54]. In turn, genetic changes in IL-1, IL-6, and TNFα affect, via the NF-ĸB and JAK/STAT pathways, the induction of rheumatoid arthritis [55,56]. Therefore, the introduction of inhibitors of overexpressed transcription factors and proinflammatory cytokines may have clinical benefits in the regulation of chondrocyte proliferation and differentiation [36]. The activity, concentration, or expression of the above-mentioned molecules is relatively easy to determine (at the gene or protein level) in biological fluids such as blood, urine, and joint fluid. Markers of cartilage degeneration have a moderate or good correlation with clinical and radiological changes in the course of degenerative diseases, especially OA and RA [25].
Cartilage diseases are often accompanied by synovitis [57] (Figure 1). Symptoms of the inflammatory state are the proliferation of synoviocytes and tissue hypertrophy. Synoviocytes release inflammatory mediators and matrix-degenerating enzymes into the joint. Their activation occurs due to the action of inflammatory mediators and cartilage matrix molecules, initiating a feedback cycle within the synovium, which results in progressive degeneration of the joint.
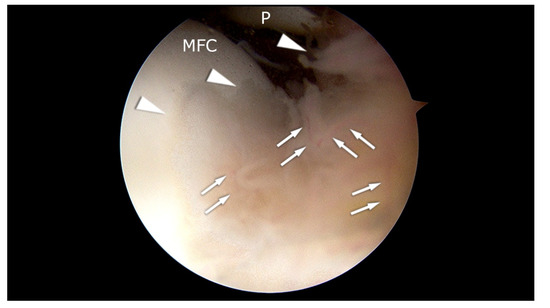
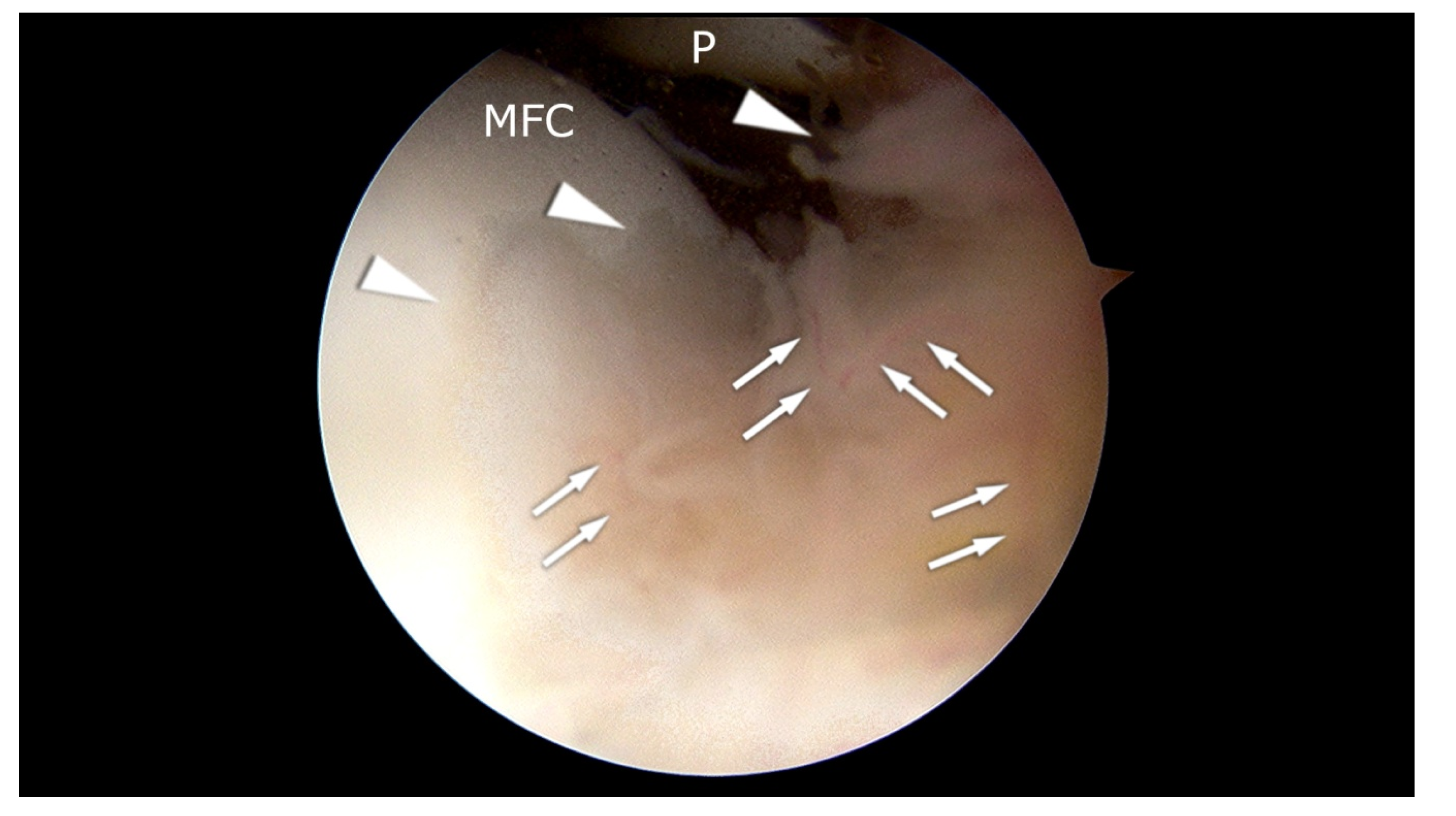
Figure 1.
Arthroscopic appearance of the patient with synovitis and initial pathologic changes in the cartilage of the medial femoral condyle (MFC). Arrowheads: hypertrophic synovium. MFC: cartilage of the medial femoral condyle. P: patella. Arrows: blood vessels. The picture comes from our own material.
3. Diagnostic and Therapeutic Biomarkers
Metalloproteinases, inflammatory factors, signaling molecules, and transcription factors belong to the best-described groups of enzymes and their genes involved in the pathogenesis of cartilage tissue disease [36,58]. Genetic changes within these gene superfamilies are useful diagnostically and also have therapeutic potential.
3.1. Metalloproteinases
Metalloproteinases (MMPs) are responsible for the irreversible proteolytic destruction of cartilage, especially via the breakdown of type II collagen. Seven matrix metalloproteinases are expressed under varying circumstances in articular cartilage [59,60,61]. Among them, only MMP-1, MMP-2, MMP-13, and MMP-14 are constitutively expressed in adult cartilage. Their physiological function is tissue turnover and the level of their expression increases significantly in pathologic states. The presence of MMP-3, MMP-8, and MMP-9 in cartilage appears to be characteristic of pathological circumstances only [59]. Additionally, the soluble collagenases MMP-1, MMP-8, and MMP-13 play a key role in cartilage destruction. The collagenolytic activity of other MMPs (such as MMP-2 and MMP-14) is likely minor. It was experimentally demonstrated that MMP-3, MMP-9, and MMP-10 degrade other ECM components, but in vivo, they are unable to cleave native type II collagen [59,62,63]. The proper regulation of expression of the metalloproteinase family depends on many factors and triggers several intracellular signaling pathways. The expression patterns of MMPs in cartilage depend on proinflammatory and pleiotropic cytokines and growth factors [64,65]. The overexpression of MMPs is an important marker of the progression of osteochondral diseases, regardless of etiology [59]. There is a relationship between the increase in MMP expression and the rapid rate of joint destruction [66].
MMPs are overexpressed in diseased joints, leading to the disintegration of the ECM, thereby reducing flexibility and resistance to tissue injury [67]. The endogenous inhibitors of MMPs are tissue inhibitors of metalloproteinases (TIMPs); however, in OA activity, they are not effective [61,68,69]. The environment of proinflammatory cytokines such as IL-1, IL-6 and TNF-α promotes the increase in MMP expression. Therefore, new therapeutic protocols aimed at restoring joint function are often directed toward the use of small molecule inhibitors of MMP subclasses or inhibitors of the interaction between IL-1 and its receptor [67,70].
Metalloproteinases are a diverse family of genes for which the correlation of cartilage damage with both the presence of polymorphisms [71] (e.g., –1612 5A/6A polymorphism genotypes of MMP-3 gene promoter [72] or rs639752, rs520540, and rs602128 in MMP-3 [73], rs4747096 in ADAMTS14 [74]), changes in expression patterns (expression changes affecting the development and progression of cartilage degradation are related to mRNA of each MMP and TIMP [61,75]), and expression modulation through miRNA (e.g., miR-27b and miR-127-5p regulates MMP-13 expression [76,77], miR-140 regulates ADAMTS-5 expression [78,79]) has been confirmed.
Van Meurs et al. proved that active MMPs play an important role in cartilage degradation in both the immune-mediated complex arthritis model and in antigen-induced arthritis models [80,81]. Leong et al. [82] experimentally demonstrated that MMP-3 expression is dependent on joint mobility. Immobilization of the limb led to an increase in MMP-3 levels (especially in the superficial zone of articular cartilage), but an hour of daily moderate mechanical stress (passive movement) resulted in a decrease in the growth of MMP-3 and ADAMTS-5. Researchers showed that passive motion loading suppresses immobilization-induced overexpression of the MMP-3 and ADAMTS-5 genes, as well as blocking the increase in total articular cartilage MMP-3 activity. Leong et al. [82] also confirmed that intra-articular injections of the MMP-3 inhibitor silenced the effects of immobilization and MMP-3 overexpression. There is no consensus on the scope of the best therapeutic strategy based on targeted enzymes from the metalloproteinases family. Elliot and Cawston [83] point out that it is important to identify the therapeutic benefits of inhibiting proteoglycan or collagen degradation. The most preferred option seems to be the use of a broad spectrum MMP inhibitor that simultaneously inhibits ADAM proteinases (e.g., TACE, ADAM17) [83,84].
The level of MMP expression modulation is probably the most important; it is primarily regulated by various growth factors and cytokines [85]. In the context of the results of the research, this family of genes (especially MMP-3 and MMP-13) can be considered as the main biomarkers of bone and cartilage damage as well as mediators of joint destruction [23,24,59]. The correlation of MMP-13 expression with cartilage damage makes this gene an interesting candidate for pharmacological intervention [24,86].
3.2. Growth Factors
Changes in the expression levels of this group of genes are particularly important, not only in the search of biomarkers to monitor the progression of cartilage disease, but also for regenerative therapy. Growth factors are biologically active polypeptides that are used as stimulators of cell growth and enhancers of chondrogenesis, but they can also support the treatment of cartilage defects [87,88]. The most important in terms of supporting therapy include the transforming growth factor (TGF) superfamily (e.g., TGF-β1, BMP-2, BMP-7, TGF-β3, CDMP-1, and CDMP-2), fibroblast growth factor (FGF) family (e.g., bFGF, FGF-2, and FGF-18), insulin-like growth factor (IGF), and platelet derived growth factor (PDGF) [88,89,90,91]. The role of these factors in restoring cartilage tissue function and structure is varied. Growth factors by stimulating anabolic and catabolic processes affect the local microenvironment of the joint.
Their therapeutic potential in the treatment of cartilage injuries and early arthritis has been demonstrated in both in vitro and animal model studies [87,88,91,92]. TGF-β is responsible for stimulation of ECM synthesis, chondrogenesis in the synovial lining, and the reduction of IL-1 catabolic activity. BMP-7 inhibits catabolic factors (e.g., MMP-1, MMP-13, IL-1, IL-6, and IL-8) and promotes the synthesis of cartilage matrix [87]. Under physiological conditions, FGF-2 plays a chondroprotective role and is responsible for inhibiting interleukin-1-driven aggrecanase activity [93]. Chia et al. [89] in a mouse model showed that the subcutaneous administration of FGF-2 inhibited osteoarthritis. At the same time, an increase in OA symptoms was observed in knockout mice of this gene. FGF-2 suppresses IL-1- or TNF-stimulated aggrecanase activity in a dose-dependent manner. High doses of FGF-2 may promote the intensification of inflammation by antagonizing IGF-1 and increasing MMPs [87,93]. IGF-1 also helps to maintain cartilage integrity, which suppresses anabolic processes that promote cartilage repair [94,95]. The regenerative effect requires cooperation with TGF-β and BMP-7. As shown in a mouse model, low IGF-1 expression results in joint cartilage damage, while high IGF-1 expression promotes synovial membrane protection [90]. The therapeutic application is described in detail later in this review.
3.3. Inflammatory Factors
The role of synovium and adipose tissue hormones in the intensification of inflammation is emphasized in the development and progression of cartilage diseases. The presence of numerous inflammatory mediators, such as chemokines, interleukins, metalloproteinases, and others, shows a positive correlation with the severity of pain and joint dysfunction. It is now known that the role of inflammation of the tissues forming and surrounding the joint is important for development and progression, and the level of inflammatory response shows a positive correlation with the severity of patients’ complaints [96,97,98]. This creates a potential option for new treatments targeted at modifying transmission pathways dependent on numerous inflammatory mediators.
Based on biochemical and genetic findings [25,26,27], markers of the inflammatory process have been identified. The strongest correlations with inflammation of the joints have been found for (1) inflammatory mediators (cyclooxygenase (COX), prostaglandin E2 (PGE2), prostaglandin D2 (PGD2), prostaglandin F2a2 (PGF2a), thromboxane and prostaglandin I2 (PGI2); (2) circulating or locally occurring cytokines: interleukin-1 (IL-1), IL-6, IL-17, IL-18, TNF-α chemokines, such as C-C motif chemokine ligand 5 (CCL5) and IL-8; (3) nitric oxide (NO); and (4) synovial degradation products: hyaluronan or hyaluronic acid (HA) [99,100]. Synovial inflammation is also correlated with the secretion of proinflammatory cytokines, including vascular endothelial growth factor (VEGF), blood vessel formation (factor VIII), intercellular adhesion molecule-1, and proinflammatory cytokines (TNF-α, IL-6, and IL-1ß) [101]. The consequences of changes within this group of genes and their protein products are primarily increased cartilage degradation and bone resorption, inhibited glycoprotein and collagen synthesis, MMP overexpression, the stimulation of other cells to produce proinflammatory cytokines and growth factors, the stimulation of nitric oxide production, and the induction of chondrocyte apoptosis [102]. Therefore, the treatment of this disease should be aimed at reducing inflammation.
Mediators and effectors of active inflammation, most commonly IL-1ß, IL-6, and TNF-α, are detected in the plasma and synovial fluid of patients with the degenerative disease [96,103,104]. They are released by damaged cells of cartilage, bone tissue, and synovium, which secondarily stimulate the production of other mediators with proinflammatory and catabolic effects on joint structures [105]. Increased chondrocyte apoptosis has been observed in cartilage collected from OA patients, which may be related to their shortened survival [106]. An increase in the concentration of proinflammatory prostaglandins, leukotrienes, interleukins, and metalloproteinases is also observed [107,108]. The presence of inflammation has been shown to inhibit the expression of genes involved in the phenotypic differentiation of chondrocytes, which negatively affects their metabolism and regeneration; therefore, the balance between degenerative and regenerative processes in cartilage tissue is disturbed [109,110].
Inflammation is one of the important factors in the progression of cartilage erosion and the intensification of disease symptoms in patients with confirmed OA [111,112], and it is manifested as synovial membrane mononuclear cell infiltration, which can be seen both at the early and late stages [113]. Nevertheless, there is no definitive evidence as to whether inflammation occurs earlier or is a consequence of OA development [101]. At the molecular level, a synovitis marker in a genome-wide study was analyzed by Scanzello et al. The study included material obtained from patients with no symptoms or OA features in the radiological image, who underwent arthroscopic meniscectomy due to knee injuries. The control group consisted of patients with early or advanced OA diagnosis. Scanzello et al. established chemokine expression patterns correlated with the presence or absence of inflammation. The molecular signature of the compared samples differed in the expression of 28 mediators (including IL-8, CCR7, CCL19, CCL21, and CCL5), most of which were overexpressed in inflammation [101].
4. Types of Genetic Changes
Characterization of the complex conditions of cartilage damage and the resulting identification of markers useful diagnostically and therapeutically requires the integration of data from genomic, transcriptomic, epigenetic, and proteomic experiments. Single nucleotide polymorphisms (SNPs) that are easy to identify are most useful as diagnostic and prognostic markers. Multiple SNPs in genes associated with cartilage diseases have been identified as risk factors (e.g., OA predisposition, Table 2). The second group of markers comprises genes whose changes in expression level are correlated with the induction and severity of cartilage degradation (described above). These biomarkers and their regulators are primarily useful in developing new therapies and as supplementing existing treatments. Among the regulators of gene expression, microRNAs are of particular interest due to their great therapeutic potential.

Table 2.
Polymorphism from selected genes correlated with the risk of osteoarthritis.
4.1. Single Nucleotide Polymorphisms
The effective study of genetic conditions for the initiation and progression of diseases requires progress in understanding their genomic organization; therefore, one of the main directions of research is also the identification of single nucleotide polymorphisms in the structure of selected candidate genes. An interesting research direction is searching for SNPs within the candidate. SNPs, as the most common genetic varieties in the population, have been successfully correlated with the pathogenesis of many diseases of articular cartilage, mainly osteoarthritis (Table 2).
Multiple SNPs in various genes play different roles in the pathogenesis of OA and its subtypes [123,141,151,152,153,154]. For example, Wang et al. [154] in their overview and meta-analysis described 56 SNPs from different genes that are associated with either hip OA (e.g., in COL11A1, TGF-1β, VEGF), knee OA (e.g., IL-6, ASPN, GDF5), or both joints (e.g., IL-8, CALM1, SMAD3) [154]. Guo et al. [73] have experimentally demonstrated that SNPs in the MMP-3 gene contribute to an increase in OA risk and therefore can serve as molecular markers of osteoarthritis susceptibility. Similar observations were made by Tong et al. [114], who confirmed the relationship of two common allelic variants of MMP-3 (rs650108) and TIMP-3 (rs715572) with the risk of developing cartilage disease. Miyamoto et al. [155], Chapman et al. [156], Williams et al. [157], Wang et al. [158], and Southam et al. [159] independently identified a missense mutation (rs143383) in the GDF5 promoter. GDF5 is regarded as a prominent genetic risk factor associated with OA, and rs143383 SNP significantly reduced the transcriptional activity of GDF5 in chondrocytes. This change results in the disruption of cartilage synthesis and maintenance by resident cells [159]. The regulation of GDF5 activity is of therapeutic significance because in scientific and preclinical studies, the effect of this gene on the healing of connective tissue (fibrous and cartilage) has been demonstrated [160,161,162,163]. Parrish et al. [164] showed that intra-articular supplementation of recombinant human GDF5 (rhGDF5) in the osteoarthritic joint leads to a significant slowdown in disease progression.
4.2. Epigenetic Changes
The phenotype of mature chondrocytes is stabilized by numerous epigenetic modifications, including DNA methylation (hypo- and hypermethylation mainly in the promoter sequences of target genes at CpG sites), histone modification (methylation, ubiquitination, acetylation, and phosphorylation), and non-coding RNAs binding to the 3′-untranslated region of the target gene [165,166,167,168,169,170,171]. Epigenetic changes have been described for many groups (transcription factors, proteinases, cytokines, chemokines, growth factors, and ECM proteins) of genes relevant for cartilage destruction, hypertrophic chondrocyte formation, and synovitis. Modifications to the epigenetic pattern can lead to genetic disruptions that result in the overexpression of cartilage-degrading proteases and inflammatory process factors [169].
In recent years, especially microRNAs have enjoyed increasing interest in the scientific world. These low molecular weight non-coding RNAs are used as diagnostic markers and predictors. However, as post-transcriptional regulators of gene expression, they are an important candidate for personalized therapy [172,173]. Identifying the expression signatures of microRNA sets associated with specific degradation phenotypes opens up new opportunities, both in developing new strategies for preventing tissue structure destruction as well as new options for anti-degenerative therapy [174,175]. MicroRNAs in biofluids are primarily minimally invasive biomarkers that are useful diagnostically and prognostically. Therapeutic strategies based on modulating miRNA activity are increasingly emerging because of the ability of these ncRNAs to affect cell phenotype [173,174,176]. The modulation of expression using microRNA has become a new trend in determining the mechanisms regulating various diseases [177], especially those involving inflammation. Various delivery mechanisms and nano constructions are tested in vivo [79,178,179,180,181], but their routine use in clinical practice should still be seen as a promising future perspective.
Epigenetic regulations of cytokine expression are well described in the literature. For example, IL-1β expression levels are modulated by methylation and demethylation of the CpG site of the promoter region [182], as well as the activities of miR-146a and miR-149 [171,183]. In addition, it was found that the use of histone deacetylase inhibitors in chondrocytes results in the reduced secretion of inflammatory cytokines, such as IL-1β, suppressing synovitis and preventing the redifferentiation of dedifferentiated chondrocytes [169,184]. An example of research into the therapeutic role of miRNA is the study by Si et al. [79], which explored the possibility of slowing the progression of induced surgically OA in model rats using intra-articular injections of a miRNA-140 mimic or inhibitor. MiRNA-140 is specifically expressed in cartilage, and its role includes the regulation of ECM-degrading enzymes. The overexpression of miRNA-140 in primary human chondrocytes has been shown to promote collagen II expression and inhibit the expression of target genes: MMP-13 and ADAMTS-5. Si et al. [79] observed that in the group treated with miRNA-140 agomir, significantly higher results were obtained in cartilage thickness, chondrocyte count, and collagen II expression level, while simultaneously reducing expression levels of MMP-13 and ADAMTS-5. Thus, the potential of miRNA-140 in the attenuation of OA progression by modulating ECM homeostasis was demonstrated. Similar observations were made by Wang et al. [179] for miR-483-5p (MMP-2 expression regulator) in a transgenic (TG483) mice model. Researchers, using the appropriate constructs introduced intra-articularly, could both enhance degenerative disease (by introducing LV3-miR-483-5p lentivirus) and delay the progression of experimental OA (using antago-miR-483-5p) [179]. Interesting results have been described by Huang et al. [178], who conducted an intra-articular injection of recombinant AAV for the in vivo overexpression of miR-204 in a surgically induced OA mice model. They showed that two homologous microRNAs, miR-204 and miR-211, are able to maintain joint homeostasis and thus protect the pathogenesis of osteoarthritis. Loss of miR-204 and miR-211 function leads to the accumulation of Runx2 and induction of proteases that degrade the matrix in articular chondrocytes and synoviocytes. This mechanism activates the destruction of articular cartilage. They showed that intra-articular injection of the miR-204-expressing adeno-associated virus significantly slows the progression of OA.
5. Gene Therapy Approaches
Bioactive proteins are difficult to administer effectively; however, gene transfer approaches are being developed to provide their sustained synthesis at sites of repair. The treatment of cartilage lesions is applied by transferring genes, encoding for specific growth factors, into chondrocytes or progenitor cells [185]. Gene delivery into the osteochondral unit is categorized as either in vivo (direct gene delivery into host tissue within the lesion site) or ex vivo (indirect gene delivery, e.g., via stem cells or fibroblasts, following in vitro transfection or transduction). Viral gene vectors (e.g., adenovirus, retrovirus, adeno-associated virus (AAV), and herpes simplex virus) present an efficient method for gene transfer [186]. The direct method is less technically demanding, but indirect gene delivery is safer because the gene manipulation takes place under controlled conditions outside the body. Moreover, safety tests can be performed in the genetically engineered cells before the implantation in the defects. Although the development of effective treatments for osteochondral defects remains complicated, gene transfer might improve repair and regeneration at sites of injury by enabling the local, sustained and, potentially regulated expression of morphogens, growth factors, and anti-inflammatory proteins [187]. Understanding the molecular basis of cartilage and joint diseases is important and useful for the establishment of effective therapies.
5.1. Marrow Stimulation
There are two gene-based techniques of marrow stimulation. Pascher et al. presented an approach to enhance natural repair mechanisms within cartilage lesions by targeting bone marrow-derived cells for genetic modification [188]. As an alternative medium for gene delivery, they investigated the feasibility of using coagulated bone marrow aspirates. Mixing an adenoviral suspension with the fluid phase of freshly aspirated bone marrow resulted in uniform vector dispersion throughout, and rates of transgenic expression in direct proportion to the density of nucleated cells in the corresponding clot. Sieker et al. presented good results with this method using cDNA that encoded bone morphogenetic protein BMP-2 and Indian hedgehog protein and was effective in improving cartilage repair in osteochondral defects in the trochlea of rabbit knees. However, BMP-2 treatment carried the risk of intralesional bone formation [189]. Ivkovic et al. used ovine autologous bone marrow transduced with adenoviral vectors containing cDNA for green fluorescent protein or transforming growth factor (TGF-β1). The marrow was allowed to clot, forming a gene plug that was implanted into partial-thickness defects created on the medial condyle in the sheep model [190]. This method improved the outcome, and TGF-treated defects showed significantly higher amounts of collagen II. In the second approach, the recombinant adeno-associated virus is used directly on the exudate that enters the osteochondral lesion. In a rabbit osteochondral defect model, fibroblast growth factor 2 (FGF-2), insulin-like growth factor 1 (IGF-1), and the transcription factor Sox9 have been delivered by the transgene, with promising results [187,191,192,193].
5.2. Autologous Chondrocyte Implantation
Over the years, Autologous Chondrocyte Implantation (ACI) has been established as a good treatment option to deal with large full-thickness chondral lesions [194,195,196]. This approach requires two surgeries. Firstly, articular cartilage is harvested from a lesser-weight-bearing part of the joint. Then, autologous chondrocytes need to be expanded in culture and implanted into the defect. As the application of ACI has been limited by the high cost of autologous therapy and by the need for two surgeries, using genetically modified allografted chondrocytes could reduce complexity and improve cost-effectiveness. Kang et al. showed for the first time that genetically modified allografted chondrocytes could persist and express transgenes in rabbits’ osteochondral defects [197]. There is also a large body of evidence that confirms that genetically modified allogeneic or autogenous chondrocytes are effective for cartilage repair in animals [90,198]. Ortved et al. presented that transferring IGF-1 by AAV to autologous chondrocytes improved the repair of full-thickness chondral defects in equines [199]. Genetically enhanced allograft chondrocytes were used in human clinical trials [200]. The transduced cells were surgically introduced into cartilage lesions using a fibrin scaffold. A line of human chondrocytes was obtained from a newborn with polydactyly. One cohort of cells was transduced with a retrovirus carrying TGF-β1 cDNA. There were no treatment-related serious adverse events. Although knee evaluation scores seemed to indicate a trend toward efficacy, patient numbers were not sufficient to determine statistical significance.
5.3. Muscle and Fat Grafts
Evans et al. demonstrated another therapy based on the remarkable potential of genetically modified, autologous skeletal muscle and fat grafts to heal large osseous and chondral defects. These tissues can be harvested, genetically modified, and then press-fit into osteochondral lesions within the time frame of a single surgery. The theory behind muscle-based tissue engineering is related to the unique biology of skeletal muscle-derived cells. Skeletal muscle contains satellite cells that are capable of fusing to form post-mitotic, multinucleated myotubes and myofibers. The post-mitotic myofibers are stable cells and theoretically capable of long-lasting gene expression. Their potency is likely to reflect the presence of endogenous progenitor cells, the secretion of morphogenetic signals by the genetically modified cells, and the scaffolding properties of the tissues themselves. When compared to ACI, the complexity of the procedure is reduced, which should lower costs. Moreover, skeletal muscle and fat are easily accessible and available for eventual biopsies. Results from pilot experiments with rabbits showed that using adenovirus vectors carrying BMP2 cDNA is encouraging. The implanted genetically modified fat and muscle grafts formed bone in the subchondral region and cartilage above, indicating the impact of the progenitor cell location on the process of differentiation [201].
5.4. Scaffolds
The main problem to the treatment of focal cartilage defects with the non-scaffolds approach is that the genetically modified cells or gene vectors are diluted by the joint fluid and fail to maintain at the target lesion area [202]. To avoid this obstacle, a promising approach is to deliver modified cells or gene vectors using different types of biomaterials. Scaffolds for cartilage repair present new options to structurally support cartilage repair [203]. Gene therapy, combined with scaffolds, increases the efficiency and duration of transfected genes, forming an efficient system to promote osteochondral regeneration. When the scaffold is degraded, the contents are slowly released to the target area. These biomaterials can be implanted into articular cartilage defects to provide gene transfer that enables the controlled release of the vector over time. The regulated transmission of genetic material via biomaterials could enhance the properties of the gene products and protect these active agents against degradation [204]. Additionally, biomaterial-mediated gene delivery provides biomechanical properties in magnitudes similar to that of native articular cartilage, which is especially important for the repair of large defects [205]. Using scaffolds in gene therapy for osteochondral tissue repair can be applied by two methods: through the incorporation of the vector during scaffold preparation and putting into the chondral lesion to transfect chondrocytes (Figure 2) or by the connection of the vector with cells to a formed scaffold.
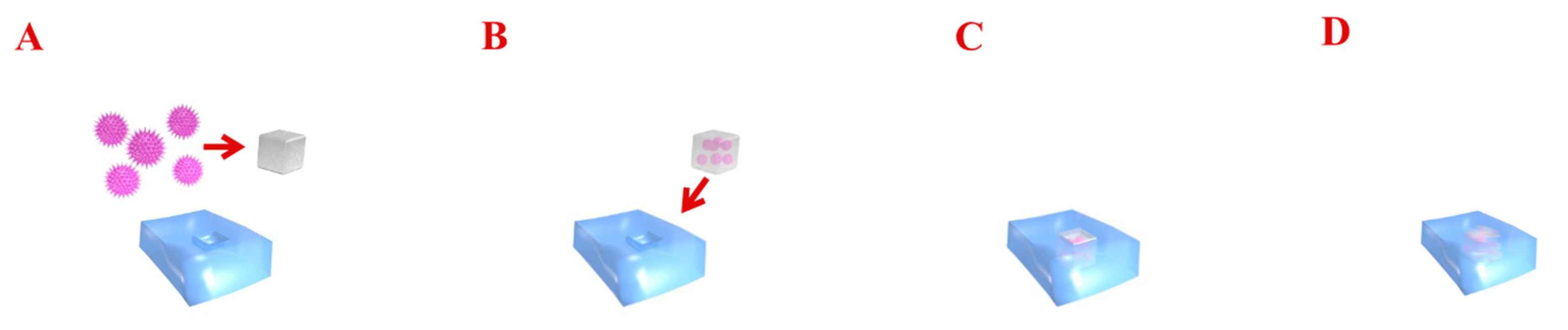
Figure 2.
The diagram presents gene delivery therapy into cartilage. Transferred genes encoding the specific growth factors are combined with viral gene vectors (A,B). Such a construct is implanted into the chondral defect (C). Gene vectors are able to transfect chondrocytes and improve regeneration at sites of injury by enabling the local expression of morphogens, growth factors, and anti-inflammatory proteins (D).
Among the numerous synthetic materials, biocompatible and biodegradable compounds matrices are thought to possess the most promising potential for repair of osteochondral defects [206,207,208]. Different gene delivery approaches include the use of solid scaffolds, hydrogels and micelles (alginate, poloxamer PF68, and poloxamine T908 polymeric micelles based on poly(ethylene oxide)—PEO—and poly(propylene oxide)—PPO—triblock copolymers, self-assembling RAD16-I peptide hydrogels, polypseudorotaxane gels) to create vector-loaded biomaterials [186,209,210]. These biomaterials provide the formation of cartilage tissue with adapted mechanical properties and affording protection against tissue degradation in conditions that enable joint resurfacing. Different scaffold-based approaches are shown in Table 3, taking into account the vector, biomaterial, and transferred genes.

Table 3.
Scaffold-based gene therapy approaches in cartilage repair.
5.5. Mesenchymal Stem Cells
The use of controlled gene delivery approaches to facilitate clinical cartilage repair is still a developing area. Emerging approaches include the use of progenitor cells, rather than chondrocytes, as agents of gene transfer for cartilage repair. Mesenchymal stem cells (MSCs) are the most widely studied due to their high availability and proliferative/differentiation ability. Bone marrow-derived MSCs (BMSCs) and adipose-derived MSCs (ASCs) are commonly employed for osteochondral therapy [210]. Although genetically modified MSCs are not being used in human clinical practice, some experimental studies have been completed. Leng et al. used transfected BMSCs with hIGF-1 cDNA and mixed with calcium alginate gels for transplantation into osteochondral defects on the femoral condyle of goats and showed the improvement in the repair [220]. Yang and al. transfected BMCs with adenoviruses expressing C-type natriuretic peptides and seeded the cells onto silk/chitosan scaffolds to promote chondrogenesis in rats [221].
In another study, Venkatesan et al. designed 3D fibrin–polyurethane scaffolds in a hydrodynamic environment that provided a favorable growth environment for recombinant adeno-associated virus vector (rAAV)-infected Sox9-modified human BMSCs and promoted their differentiation into chondrocytes [222].
Controlled tissue growth and biomimetic cartilage properties were maintained upon seeding the human ASCs into large PCL scaffolds immobilized with Dox-inducible lentiviruses expressing IL-1Ra [205]. Although these techniques have not been confirmed in clinical studies yet, they hold great scientific promise for treating cartilage injuries in the near future. In addition, interest in improving the efficiency and targeting of non-viral vectors continues. For example, Pi et al. identified a chondrocyte-affinity peptide that enhances cartilage-targeting transfection when attached to polyethyleneimine in rabbit knees [223]. These data demonstrate that the potential for gene therapy of cartilage lesions is encouraging; however, the area is still under development. The assessments of possible associated toxicities are also lacking, but they are essential for clinical translation.
6. Conclusions and Future Directions
Genes and their polymorphisms play a key role in the development of the osteochondral pathologies in the general population. The complex background of the genetic heterogeneity of the cartilage diseases consists of a plethora of genes and their epigenetic modifications. It results in changes in their activity, and thus explains the backgrounds of pathological processes. Analyses of allelic expression imbalances, genetic expression signatures, epigenetic regulatory mechanisms, and key cellular pathway disturbances provide comprehensive knowledge that brings us closer to understanding normal tissue metabolism as well as pathological processes. Therefore, the challenge is to find sensitive and specific biomarkers that will help in the early diagnosis and targeted therapy of cartilage ailments. Genetic engineering, a combination of gene transfer techniques and tissue engineering, is one of the potential new strategies for the treatment of osteochondral injuries. The potential advantage of gene transfer into the chondrogenic cells relies on sustained levels of growth factors, which can be reached through transgene expression in situ. Although the field of genetic engineering is young, current research offers the gene transfer approaches developed to provide a sustained synthesis of bioactive reagents at the cartilage repair sites. To augment the regeneration of articular cartilage, therapeutic genes can be delivered directly to the cartilage lesion. Since cartilage injuries are not life-threatening, the safety of gene transfer approaches for repair is of particular importance. Therefore, the harnessing of this technology for clinical use is strongly dependent on the use of safe and efficient vectors, transgenes, and delivery systems. The major considerations for the clinical translation of such methods are their biology, safety, ease of manufacture, and cost-effectiveness [224]. In this regard, combining gene therapy with tissue engineering concepts might overcome the various physiological barriers that impede the safe, effective, and long-term treatment of damaged articular cartilage. Cartilage repair could become a significant domain of gene therapy due to the pervasiveness of osteochondral pathologies, and applications in gene therapy may provide local treatment with a relatively small amount of vector.
Author Contributions
J.S. prepared the first part of the manuscript entitled: “Genetic basis of the cartilage lesions’’, edited the article after reviews, finally approved the article, and corresponded with the editor; D.S. conceived of the presented idea, prepared the second part of the manuscript entitled: ”Genetic therapies approaches”, edited the article after reviews; Ł.P. prepared Figure 2 and corrected the manuscript; J.Z. prepared Figure 1; P.P. provided language correction; J.K. approved the submitted version. All authors discussed the manuscript and contributed to the final manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare that there are no conflict of interest. Paczesny serves as a consultant to Biovico Ltd., but not related to the manuscript.
References
- Fox, A.J.S.; Bedi, A.; Rodeo, S.A. The basic science of articular cartilage: Structure, composition, and function. Sports Heal. A Multidiscip. Approach 2009, 1, 461–468. [Google Scholar] [CrossRef]
- Cudic, M.; Fields, G.B. Extracellular proteases as targets for drug development. Curr. Protein Pept. Sci. 2009, 10, 297–307. [Google Scholar] [CrossRef]
- Gill, S.E.; Parks, W.C. Metalloproteinases and their inhibitors: Regulators of wound healing. Int. J. Biochem. Cell Biol. 2008, 40, 1334–1347. [Google Scholar] [CrossRef]
- Kany, S.; Vollrath, J.T.; Relja, B. Cytokines in Inflammatory Disease. Int. J. Mol. Sci. 2019, 20. [Google Scholar] [CrossRef]
- Wojdasiewicz, P.; Poniatowski, L.A.; Szukiewicz, D. The role of inflammatory and anti-inflammatory cytokines in the pathogenesis of osteoarthritis. Mediat. Inflamm. 2014, 2014, 1–19. [Google Scholar] [CrossRef]
- Osiecka-Iwan, A.; Hyc, A.; Radomska-Lesniewska, D.M.; Rymarczyk, A.; Skopinski, P. Antigenic and immunogenic properties of chondrocytes. Implications for chondrocyte therapeutic transplantation and pathogenesis of inflammatory and degenerative joint diseases. Cent Eur. J. Immunol. 2018, 43, 209–219. [Google Scholar] [CrossRef]
- Musumeci, G.; Aiello, F.; Szychlinska, M.; Di Rosa, M.; Castrogiovanni, P.; Mobasheri, A. Osteoarthritis in the XXIst Century: Risk Factors and Behaviours that Influence Disease Onset and Progression. Int. J. Mol. Sci. 2015, 16, 6093–6112. [Google Scholar] [CrossRef]
- Dwivedi, S.; Purohit, P.; Misra, R.; Pareek, P.; Goel, A.; Khattri, S.; Pant, K.K.; Misra, S.; Sharma, P. Diseases and Molecular Diagnostics: A Step Closer to Precision Medicine. Indian J. Clin. Biochem. 2017, 32, 374–398. [Google Scholar] [CrossRef]
- Gasperskaja, E.; Kučinskas, V. The most common technologies and tools for functional genome analysis. Acta Med. Litu. 2017, 24, 1–11. [Google Scholar] [CrossRef]
- Peffers, M.; Liu, X.; Clegg, P. Transcriptomic signatures in cartilage ageing. Arthritis Res. Ther. 2013, 15. [Google Scholar] [CrossRef]
- Reuter, J.A.; Spacek, D.V.; Snyder, M.P. High-Throughput Sequencing Technologies. Mol. Cell 2015, 58, 586–597. [Google Scholar] [CrossRef]
- Coutinho de Almeida, R.; Ramos, Y.F.M.; Mahfouz, A.; den Hollander, W.; Lakenberg, N.; Houtman, E.; van Hoolwerff, M.; Suchiman, H.E.D.; Rodríguez Ruiz, A.; Slagboom, P.E.; et al. RNA sequencing data integration reveals an miRNA interactome of osteoarthritis cartilage. Ann. Rheum. Dis. 2019, 78, 270–277. [Google Scholar] [CrossRef]
- Lotz, M.; Martel-Pelletier, J.; Christiansen, C.; Brandi, M.L.; Bruyere, O.; Chapurlat, R.; Collette, J.; Cooper, C.; Giacovelli, G.; Kanis, J.A.; et al. Value of biomarkers in osteoarthritis: Current status and perspectives. Ann. Rheum. Dis. 2013, 72, 1756–1763. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Sharma, A.R.; Chakraborty, C.; Saibaba, B.; Ahn, M.E.; Lee, S.S. Review of Prospects of Biological Fluid Biomarkers in Osteoarthritis. Int. J. Mol. Sci. 2017, 18. [Google Scholar] [CrossRef]
- Furumatsu, T.; Matsumoto-Ogawa, E.; Tanaka, T.; Lu, Z.; Ozaki, T. ROCK inhibition enhances aggrecan deposition and suppresses matrix metalloproteinase-3 production in human articular chondrocytes. Connect Tissue Res. 2014, 55, 89–95. [Google Scholar] [CrossRef]
- Long, D.; Blake, S.; Song, X.Y.; Lark, M.; Loeser, R.F. Human articular chondrocytes produce IL-7 and respond to IL-7 with increased production of matrix metalloproteinase-13. Arthritis Res. 2008, 10, R23. [Google Scholar] [CrossRef]
- Park, J.S.; Kim, D.K.; Shin, H.D.; Lee, H.J.; Jo, H.S.; Jeong, J.H.; Choi, Y.L.; Lee, C.J.; Hwang, S.C. Apigenin Regulates Interleukin-1beta-Induced Production of Matrix Metalloproteinase Both in the Knee Joint of Rat and in Primary Cultured Articular Chondrocytes. Biomol. Ther. (Seoul) 2016, 24, 163–170. [Google Scholar] [CrossRef]
- Prasadam, I.; Crawford, R.; Xiao, Y. Aggravation of ADAMTS and matrix metalloproteinase production and role of ERK1/2 pathway in the interaction of osteoarthritic subchondral bone osteoblasts and articular cartilage chondrocytes -- possible pathogenic role in osteoarthritis. J. Rheumatol. 2012, 39, 621–634. [Google Scholar] [CrossRef] [PubMed]
- Tetlow, L.C.; Woolley, D.E. Histamine stimulates matrix metalloproteinase-3 and -13 production by human articular chondrocytes in vitro. Ann. Rheum. Dis. 2002, 61, 737–740. [Google Scholar] [CrossRef] [PubMed]
- Yammani, R.R.; Carlson, C.S.; Bresnick, A.R.; Loeser, R.F. Increase in production of matrix metalloproteinase 13 by human articular chondrocytes due to stimulation with S100A4: Role of the receptor for advanced glycation end products. Arthritis Rheum. 2006, 54, 2901–2911. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ma, S.; Su, H.; Cheng, J. Isoliquiritigenin Inhibits IL-1beta-Induced Production of Matrix Metalloproteinase in Articular Chondrocytes. Mol. Ther. Methods Clin. Dev. 2018, 9, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Forsyth, C.B.; Cole, A.; Murphy, G.; Bienias, J.L.; Im, H.J.; Loeser, R.F., Jr. Increased matrix metalloproteinase-13 production with aging by human articular chondrocytes in response to catabolic stimuli. J. Gerontol. A Biol. Sci. Med. Sci. 2005, 60, 1118–1124. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, D.; Yuan, Y.; Min, J. New insights on the MMP-13 regulatory network in the pathogenesis of early osteoarthritis. Arthritis Res. Ther. 2017, 19. [Google Scholar] [CrossRef] [PubMed]
- Li, N.G.; Shi, Z.H.; Tang, Y.P.; Wang, Z.J.; Song, S.L.; Qian, L.H.; Qian, D.W.; Duan, J.A. New Hope for the Treatment of Osteoarthritis Through Selective Inhibition of MMP-13. Curr. Med. Chem. 2011, 18, 977–1001. [Google Scholar] [CrossRef]
- Lee, A.S.; Ellman, M.B.; Yan, D.; Kroin, J.S.; Cole, B.J.; van Wijnen, A.J.; Im, H.J. A current review of molecular mechanisms regarding osteoarthritis and pain. Gene 2013, 527, 440–447. [Google Scholar] [CrossRef]
- Goldring, M.B.; Berenbaum, F. The regulation of chondrocyte function by proinflammatory mediators: Prostaglandins and nitric oxide. Clin. Orthop. Relat. Res. 2004, S37–S46. [Google Scholar] [CrossRef]
- Wang, X.; Hunter, D.; Xu, J.; Ding, C. Metabolic triggered inflammation in osteoarthritis. Osteoarthr. Cartil. 2015, 23, 22–30. [Google Scholar] [CrossRef]
- Arai, Y.; Kubo, T.; Kobayashi, K.; Takeshita, K.; Takahashi, K.; Ikeda, T.; Imanishi, J.; Takigawa, M.; Hirasawa, Y. Adenovirus vector-mediated gene transduction to chondrocytes: In vitro evaluation of therapeutic efficacy of transforming growth factor-beta 1 and heat shock protein 70 gene transduction. J. Rheumatol. 1997, 24, 1787–1795. [Google Scholar]
- Alaaeddine, N.; Di Battista, J.A.; Pelletier, J.-P.; Kiansa, K.; Cloutier, J.-M.; Martel-Pelletier, J. Differential Effects of Il-8, Lif (Pro-Inflammatory) and Il-11 (Anti-Inflammatory) on Tnf-α-Induced Pge2release and on Signalling Pathways in Human Oa Synovial Fibroblasts. Cytokine 1999, 11, 1020–1030. [Google Scholar] [CrossRef]
- Fleischmann, R.M.; Schechtman, J.; Bennett, R.; Handel, M.L.; Burmester, G.-R.; Tesser, J.; Modafferi, D.; Poulakos, J.; Sun, G. Anakinra, a recombinant human interleukin-1 receptor antagonist (r-metHuIL-1ra), in patients with rheumatoid arthritis: A large, international, multicenter, placebo-controlled trial. Arthritis Rheum. 2003, 48, 927–934. [Google Scholar] [CrossRef]
- Kafienah, W.; Al-Fayez, F.; Hollander, A.P.; Barker, M.D. Inhibition of cartilage degradation: A combined tissue engineering and gene therapy approach. Arthritis Rheum. 2003, 48, 709–718. [Google Scholar] [CrossRef]
- Lotz, M. Cytokines in Cartilage Injury and Repair. Clin. Orthop. Relat. Res. 2001, 391, S108–S115. [Google Scholar] [CrossRef]
- Robbins, P.D.; Evans, C.H.; Chernajovsky, Y. Gene therapy for arthritis. Gene Ther. 2003, 10, 902–911. [Google Scholar] [CrossRef]
- Rudolphi, K.; Gerwin, N.; Verzijl, N.; van der Kraan, P.; van den Berg, W. Pralnacasan, an inhibitor of interleukin-1β converting enzyme, reduces joint damage in two murine models of osteoarthritis. Osteoarthr. Cartil. 2003, 11, 738–746. [Google Scholar] [CrossRef]
- Neumann, E.; Judex, M.; Kullmann, F.; Grifka, J.; Robbins, P.D.; Pap, T.; Gay, R.E.; Evans, C.H.; Gay, S.; Schölmerich, J.; et al. Inhibition of cartilage destruction by double gene transfer of IL-1Ra and IL-10 involves the activin pathway. Gene Ther. 2002, 9, 1508–1519. [Google Scholar] [CrossRef]
- Nishimura, R.; Hata, K.; Takahata, Y.; Murakami, T.; Nakamura, E.; Ohkawa, M.; Ruengsinpinya, L. Role of Signal Transduction Pathways and Transcription Factors in Cartilage and Joint Diseases. Int. J. Mol. Sci. 2020, 21. [Google Scholar] [CrossRef]
- Tsuchiya, H.; Kitoh, H.; Sugiura, F.; Ishiguro, N. Chondrogenesis enhanced by overexpression of sox9 gene in mouse bone marrow-derived mesenchymal stem cells. Biochem. Biophys. Res. Commun. 2003, 301, 338–343. [Google Scholar] [CrossRef]
- Tew, S.R.; Li, Y.; Pothacharoen, P.; Tweats, L.M.; Hawkins, R.E.; Hardingham, T.E. Retroviral transduction with SOX9 enhances re-expression of the chondrocyte phenotype in passaged osteoarthritic human articular chondrocytes. Osteoarthr. Cartil. 2005, 13, 80–89. [Google Scholar] [CrossRef]
- Li, Y.; Tew, S.R.; Russell, A.M.; Gonzalez, K.R.; Hardingham, T.E.; Hawkins, R.E. Transduction of Passaged Human Articular Chondrocytes with Adenoviral, Retroviral, and Lentiviral Vectors and the Effects of Enhanced Expression of SOX9. Tissue Eng. 2004, 10, 575–584. [Google Scholar] [CrossRef]
- Cucchiarini, M.; Thurn, T.; Weimer, A.; Kohn, D.; Terwilliger, E.F.; Madry, H. Restoration of the extracellular matrix in human osteoarthritic articular cartilage by overexpression of the transcription factorSOX9. Arthritis Rheum. 2007, 56, 158–167. [Google Scholar] [CrossRef]
- Minina, E.; Wenzel, H.M.; Kreschel, C.; Karp, S.; Gaffield, W.; McMahon, A.P.; Vortkamp, A. BMP and Ihh/PTHrP signaling interact to coordinate chondrocyte proliferation and differentiation. Development 2001, 128, 4523–4534. [Google Scholar] [PubMed]
- Grande, D.A.; Mason, J.; Light, E.; Dines, D. Stem cells as platforms for delivery of genes to enhance cartilage repair. J. Bone Jt Surg. Am. 2003, 85-A (Suppl. S2), 111–116. [Google Scholar] [CrossRef] [PubMed]
- Tavella, S.; Biticchi, R.; Schito, A.; Minina, E.; Di Martino, D.; Pagano, A.; Vortkamp, A.; Horton, W.A.; Cancedda, R.; Garofalo, S. Targeted expression of SHH affects chondrocyte differentiation, growth plate organization, and Sox9 expression. J. Bone Min. Res. 2004, 19, 1678–1688. [Google Scholar] [CrossRef] [PubMed]
- Vortkamp, A. Interaction of growth factors regulating chondrocyte differentiation in the developing embryo. Osteoarthr. Cartil. 2001, 9 (Suppl. SA), S109–S117. [Google Scholar] [PubMed]
- Scharstuhl, A.; Diepens, R.; Lensen, J.; Vitters, E.; van Beuningen, H.; van der Kraan, P.; van den Berg, W. Adenoviral overexpression of Smad-7 and Smad-6 differentially regulates TGF-beta-mediated chondrocyte proliferation and proteoglycan synthesis. Osteoarthr. Cartil. 2003, 11, 773–782. [Google Scholar] [CrossRef]
- Scharstuhl, A.; Vitters, E.L.; van der Kraan, P.M.; van den Berg, W.B. Reduction of osteophyte formation and synovial thickening by adenoviral overexpression of transforming growth factor beta/bone morphogenetic protein inhibitors during experimental osteoarthritis. Arthritis Rheum. 2003, 48, 3442–3451. [Google Scholar] [CrossRef]
- D’Lima, D.D.; Hashimoto, S.; Chen, P.C.; Lotz, M.K.; Colwell, C.W. Cartilage Injury Induces Chondrocyte Apoptosis. J. Bone Jt. Surg. Am. 2001, 83, 19–21. [Google Scholar] [CrossRef]
- Yao, Q.; Wang, S.; Gambotto, A.; Glorioso, J.C.; Evans, C.H.; Robbins, P.D.; Ghivizzani, S.C.; Oligino, T.J. Intra-articular adenoviral-mediated gene transfer of trail induces apoptosis of arthritic rabbit synovium. Gene Ther. 2003, 10, 1055–1060. [Google Scholar] [CrossRef][Green Version]
- Lotz, M.; Hashimoto, S.; Kühn, K. Mechanisms of chondrocyte apoptosis. Osteoarthr. Cartil. 1999, 7, 389–391. [Google Scholar] [CrossRef]
- Yao, Q.; Glorioso, J.C.; Evans, C.H.; Robbins, P.D.; Kovesdi, I.; Oligino, T.J.; Ghivizzani, S.C. Adenoviral mediated delivery of FAS ligand to arthritic joints causes extensive apoptosis in the synovial lining. J. Gene Med. 2000, 2, 210–219. [Google Scholar] [CrossRef]
- Kobayashi, H.; Hirata, M.; Saito, T.; Itoh, S.; Chung, U.I.; Kawaguchi, H. Transcriptional induction of ADAMTS5 protein by nuclear factor-kappaB (NF-kappaB) family member RelA/p65 in chondrocytes during osteoarthritis development. J. Biol. Chem. 2013, 288, 28620–28629. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, M.; Martel-Pelletier, J.; Lajeunesse, D.; Pelletier, J.P.; Fahmi, H. Role of proinflammatory cytokines in the pathophysiology of osteoarthritis. Nat. Rev. Rheumatol. 2011, 7, 33–42. [Google Scholar] [CrossRef]
- Akiyama, H. Interactions between Sox9 and -catenin control chondrocyte differentiation. Genes Dev. 2004, 18, 1072–1087. [Google Scholar] [CrossRef] [PubMed]
- Loughlin, J.; Dowling, B.; Chapman, K.; Marcelline, L.; Mustafa, Z.; Southam, L.; Ferreira, A.; Ciesielski, C.; Carson, D.A.; Corr, M. Functional variants within the secreted frizzled-related protein 3 gene are associated with hip osteoarthritis in females. Proc. Natl. Acad. Sci. USA 2004, 101, 9757–9762. [Google Scholar] [CrossRef] [PubMed]
- Terashima, A.; Takayanagi, H. Overview of Osteoimmunology. Calcif. Tissue Int. 2018, 102, 503–511. [Google Scholar] [CrossRef]
- Romas, E.; Gillespie, M.T.; Martin, T.J. Involvement of receptor activator of NFkappaB ligand and tumor necrosis factor-alpha in bone destruction in rheumatoid arthritis. Bone 2002, 30, 340–346. [Google Scholar] [CrossRef]
- Babaei, M.; Javadian, Y.; Narimani, H.; Ranaei, M.; Heidari, B.; Basereh, H.; Gholinia, H.; Firouzjahi, A. Correlation between systemic markers of inflammation and local synovitis in knee osteoarthritis. Casp. J. Intern Med. 2019, 10, 383–387. [Google Scholar] [CrossRef]
- Nishimura, R.; Hata, K.; Nakamura, E.; Murakami, T.; Takahata, Y. Transcriptional network systems in cartilage development and disease. Histochem. Cell Biol. 2018, 149, 353–363. [Google Scholar] [CrossRef]
- Rose, B.J.; Kooyman, D.L. A Tale of Two Joints: The Role of Matrix Metalloproteases in Cartilage Biology. Dis. Markers 2016, 2016, 4895050. [Google Scholar] [CrossRef]
- Yang, C.Y.; Chanalaris, A.; Troeberg, L. ADAMTS and ADAM metalloproteinases in osteoarthritis–looking beyond the ‘usual suspects’. Osteoarth. Cart 2017, 25, 1000–1009. [Google Scholar] [CrossRef]
- Kevorkian, L.; Young, D.A.; Darrah, C.; Donell, S.T.; Shepstone, L.; Porter, S.; Brockbank, S.M.; Edwards, D.R.; Parker, A.E.; Clark, I.M. Expression profiling of metalloproteinases and their inhibitors in cartilage. Arthritis Rheum. 2004, 50, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Murphy, G.; Lee, M.H. What are the roles of metalloproteinases in cartilage and bone damage? Ann. Rheum. Dis. 2005, 64 (Suppl. S4), iv44–iv47. [Google Scholar] [CrossRef]
- Young, D.A.; Barter, M.J.; Wilkinson, D.J. Recent advances in understanding the regulation of metalloproteinases. F1000Research 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Liacini, A.; Sylvester, J.; Li, W.Q.; Huang, W.; Dehnade, F.; Ahmad, M.; Zafarullah, M. Induction of matrix metalloproteinase-13 gene expression by TNF-alpha is mediated by MAP kinases, AP-1, and NF-kappaB transcription factors in articular chondrocytes. Exp. Cell Res. 2003, 288, 208–217. [Google Scholar] [CrossRef]
- Vincenti, M.P.; Brinckerhoff, C.E. Transcriptional regulation of collagenase (MMP-1, MMP-13) genes in arthritis: Integration of complex signaling pathways for the recruitment of gene-specific transcription factors. Arthritis Res. 2002, 4, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Burrage, P.S.; Mix, K.S.; Brinckerhoff, C.E. Matrix metalloproteinases: Role in arthritis. Front Biosci. 2006, 11, 529–543. [Google Scholar] [CrossRef] [PubMed]
- Meszaros, E.; Malemud, C.J. Prospects for treating osteoarthritis: Enzyme–protein interactions regulating matrix metalloproteinase activity. Ther. Adv. Chronic Dis. 2012, 3, 219–229. [Google Scholar] [CrossRef]
- Moore, C.S.; Crocker, S.J. An alternate perspective on the roles of TIMPs and MMPs in pathology. Am. J. Pathol. 2012, 180, 12–16. [Google Scholar] [CrossRef]
- Clutterbuck, A.L.; Asplin, K.E.; Harris, P.; Allaway, D.; Mobasheri, A. Targeting matrix metalloproteinases in inflammatory conditions. Curr. Drug Targets 2009, 10, 1245–1254. [Google Scholar] [CrossRef]
- Murphy, G.; Knauper, V.; Atkinson, S.; Butler, G.; English, W.; Hutton, M.; Stracke, J.; Clark, I. Matrix metalloproteinases in arthritic disease. Arthritis Res. 2002, 4 (Suppl. S3), S39–S49. [Google Scholar] [CrossRef]
- Ye, S.; Henney, A.M. Detecting polymorphisms in MMP genes. Methods Mol. Biol. 2001, 151, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Honsawek, S.; Malila, S.; Yuktanandana, P.; Tanavalee, A.; Deepaisarnsakul, B.; Parvizi, J. Association of MMP-3 (-1612 5A/6A) polymorphism with knee osteoarthritis in Thai population. Rheumatol. Int. 2013, 33, 435–439. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Xu, P.; Jin, T.; Wang, J.; Fan, D.; Hao, Z.; Ji, Y.; Jing, S.; Han, C.; Du, J.; et al. MMP-3 gene polymorphisms are associated with increased risk of osteoarthritis in Chinese men. Oncotarget 2017, 8, 79491–79497. [Google Scholar] [CrossRef]
- Chen, W.; Wang, Y.; Zhao, J.; Bao, N. The effect of single nucleotide polymorphism on susceptibility of osteoarthritis: Recent progression and implications. Ann. Jt. 2017, 2, 52. [Google Scholar] [CrossRef]
- Salminen, H.J.; Saamanen, A.M.; Vankemmelbeke, M.N.; Auho, P.K.; Perala, M.P.; Vuorio, E.I. Differential expression patterns of matrix metalloproteinases and their inhibitors during development of osteoarthritis in a transgenic mouse model. Ann. Rheum. Dis. 2002, 61, 591–597. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Cheon, E.J.; Lee, M.H.; Kim, H.A. MicroRNA-127-5p Regulates Matrix Metalloproteinase 13 Expression and Interleukin-1β-Induced Catabolic Effects in Human Chondrocytes. Arthritis Rheum. 2013, 65, 3141–3152. [Google Scholar] [CrossRef]
- Akhtar, N.; Rasheed, Z.; Ramamurthy, S.; Anbazhagan, A.N.; Voss, F.R.; Haqqi, T.M. MicroRNA-27b regulates the expression of matrix metalloproteinase 13 in human osteoarthritis chondrocytes. Arthritis Rheum. 2010, 62, 1361–1371. [Google Scholar] [CrossRef]
- Miyaki, S.; Sato, T.; Inoue, A.; Otsuki, S.; Ito, Y.; Yokoyama, S.; Kato, Y.; Takemoto, F.; Nakasa, T.; Yamashita, S.; et al. MicroRNA-140 plays dual roles in both cartilage development and homeostasis. Genes Dev. 2010, 24, 1173–1185. [Google Scholar] [CrossRef]
- Si, H.B.; Zeng, Y.; Liu, S.Y.; Zhou, Z.K.; Chen, Y.N.; Cheng, J.Q.; Lu, Y.R.; Shen, B. Intra-articular injection of microRNA-140 (miRNA-140) alleviates osteoarthritis (OA) progression by modulating extracellular matrix (ECM) homeostasis in rats. Osteoarthr. Cartil. 2017, 25, 1698–1707. [Google Scholar] [CrossRef]
- van Meurs, J.; van Lent, P.; Stoop, R.; Holthuysen, A.; Singer, I.; Bayne, E.; Mudgett, J.; Poole, R.; Billinghurst, C.; van der Kraan, P.; et al. Cleavage of aggrecan at the Asn341-Phe342 site coincides with the initiation of collagen damage in murine antigen-induced arthritis: A pivotal role for stromelysin 1 in matrix metalloproteinase activity. Arthritis Rheum. 1999, 42, 2074–2084. [Google Scholar] [CrossRef]
- van Meurs, J.; van Lent, P.; Holthuysen, A.; Lambrou, D.; Bayne, E.; Singer, I.; van den Berg, W. Active matrix metalloproteinases are present in cartilage during immune complex-mediated arthritis: A pivotal role for stromelysin-1 in cartilage destruction. J. Immunol. 1999, 163, 5633–5639. [Google Scholar]
- Leong, D.J.; Gu, X.I.; Li, Y.; Lee, J.Y.; Laudier, D.M.; Majeska, R.J.; Schaffler, M.B.; Cardoso, L.; Sun, H.B. Matrix metalloproteinase-3 in articular cartilage is upregulated by joint immobilization and suppressed by passive joint motion. Matrix Biol. 2010, 29, 420–426. [Google Scholar] [CrossRef]
- Elliott, S.; Cawston, T. The clinical potential of matrix metalloproteinase inhibitors in the rheumatic disorders. Drugs Aging 2001, 18, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Conway, J.G.; Andrews, R.C.; Beaudet, B.; Bickett, D.M.; Boncek, V.; Brodie, T.A.; Clark, R.L.; Crumrine, R.C.; Leenitzer, M.A.; McDougald, D.L.; et al. Inhibition of tumor necrosis factor-alpha (TNF-alpha) production and arthritis in the rat by GW3333, a dual inhibitor of TNF-alpha-converting enzyme and matrix metalloproteinases. J. Pharm. Exp. Ther. 2001, 298, 900–908. [Google Scholar]
- Borden, P.; Heller, R.A. Transcriptional control of matrix metalloproteinases and the tissue inhibitors of matrix metalloproteinases. Crit. Rev. Eukaryot Gene Expr. 1997, 7, 159–178. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, A.; Guibert, M.; Cailotto, F.; Gasser, A.; Presle, N.; Mainard, D.; Netter, P.; Kempf, H.; Jouzeau, J.Y.; Reboul, P. Fibroblast Growth Factor 23 drives MMP13 expression in human osteoarthritic chondrocytes in a Klotho-independent manner. Osteoarthr. Cartil. 2016, 24, 1961–1969. [Google Scholar] [CrossRef] [PubMed]
- Tuan, R.S.; Chen, A.F.; Klatt, B.A. Cartilage regeneration. J. Am. Acad. Orthop. Surg. 2013, 21, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Fortier, L.A.; Barker, J.U.; Strauss, E.J.; McCarrel, T.M.; Cole, B.J. The role of growth factors in cartilage repair. Clin. Orthop. Relat. Res. 2011, 469, 2706–2715. [Google Scholar] [CrossRef]
- Chia, S.-L.; Sawaji, Y.; Burleigh, A.; McLean, C.; Inglis, J.; Saklatvala, J.; Vincent, T. Fibroblast growth factor 2 is an intrinsic chondroprotective agent that suppresses ADAMTS-5 and delays cartilage degradation in murine osteoarthritis. Arthritis Rheum. 2009, 60, 2019–2027. [Google Scholar] [CrossRef]
- Goodrich, L.R.; Hidaka, C.; Robbins, P.D.; Evans, C.H.; Nixon, A.J. Genetic modification of chondrocytes with insulin-like growth factor-1 enhances cartilage healing in an equine model. J. Bone Jt. Surg. Br. Vol. 2007, 89-B, 672–685. [Google Scholar] [CrossRef]
- Longo, U.G.; Petrillo, S.; Franceschetti, E.; Berton, A.; Maffulli, N.; Denaro, V. Stem Cells and Gene Therapy for Cartilage Repair. Stem Cells Int. 2012, 2012, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.B.; Chen, E.H.; Lynch, S.E. A review of the effects of insulin-like growth factor and platelet derived growth factor on in vivo cartilage healing and repair. Osteoarthr. Cartil. 2006, 14, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Sawaji, Y.; Hynes, J.; Vincent, T.; Saklatvala, J. Fibroblast growth factor 2 inhibits induction of aggrecanase activity in human articular cartilage. Arthritis Rheum. 2008, 58, 3498–3509. [Google Scholar] [CrossRef]
- Fortier, L.A.; Balkman, C.E.; Sandell, L.J.; Ratcliffe, A.; Nixon, A.J. Insulin-like growth factor-I gene expression patterns during spontaneous repair of acute articular cartilage injury. J. Orthop. Res. 2001, 19, 720–728. [Google Scholar] [CrossRef]
- Morisset, S.; Frisbie, D.D.; Robbins, P.D.; Nixon, A.J.; McIlwraith, C.W. IL-1ra/IGF-1 Gene Therapy Modulates Repair of Microfractured Chondral Defects. Clin. Orthop. Relat. Res. 2007, 462, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Sokolove, J.; Lepus, C.M. Role of inflammation in the pathogenesis of osteoarthritis: Latest findings and interpretations. Adv. Musculoskelet. Dis. 2013, 5, 77–94. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Deng, H.; Cui, H.; Fang, J.; Zuo, Z.; Deng, J.; Li, Y.; Wang, X.; Zhao, L. Inflammatory responses and inflammation-associated diseases in organs. Oncotarget 2018, 9, 7204–7218. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Hunter, D.J.; Jin, X.; Ding, C. The importance of synovial inflammation in osteoarthritis: Current evidence from imaging assessments and clinical trials. Osteoarthr. Cartil. 2018, 26, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Ricciotti, E.; FitzGerald, G.A. Prostaglandins and inflammation. Arter. Thromb. Vasc. Biol. 2011, 31, 986–1000. [Google Scholar] [CrossRef]
- Abdulkhaleq, L.A.; Assi, M.A.; Abdullah, R.; Zamri-Saad, M.; Taufiq-Yap, Y.H.; Hezmee, M.N.M. The crucial roles of inflammatory mediators in inflammation: A review. Vet World 2018, 11, 627–635. [Google Scholar] [CrossRef]
- Scanzello, C.R.; McKeon, B.; Swaim, B.H.; DiCarlo, E.; Asomugha, E.U.; Kanda, V.; Nair, A.; Lee, D.M.; Richmond, J.C.; Katz, J.N.; et al. Synovial inflammation in patients undergoing arthroscopic meniscectomy: Molecular characterization and relationship to symptoms. Arthritis Rheum. 2011, 63, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Mobasheri, A.; Henrotin, Y.; Biesalski, H.K.; Shakibaei, M. Scientific evidence and rationale for the development of curcumin and resveratrol as nutraceutricals for joint health. Int. J. Mol. Sci. 2012, 13, 4202–4232. [Google Scholar] [CrossRef] [PubMed]
- Madson, K.L.; Moore, T.L.; Lawrence, J.M., 3rd; Osborn, T.G. Cytokine levels in serum and synovial fluid of patients with juvenile rheumatoid arthritis. J. Rheumatol. 1994, 21, 2359–2363. [Google Scholar] [PubMed]
- Perry, M.G.; Jessop, D.S.; Hunt, L.P.; Sharif, M.; Kirwan, J.R. Overnight variations in IL-6 in synovial fluid and plasma in patients with active rheumatoid arthritis. Int. J. Clin. Rheumatol. 2010, 5, 593–600. [Google Scholar] [CrossRef]
- Goldring, M.B.; Otero, M. Inflammation in osteoarthritis. Curr. Opin. Rheumatol. 2011, 23, 471–478. [Google Scholar] [CrossRef]
- Hwang, H.S.; Kim, H.A. Chondrocyte Apoptosis in the Pathogenesis of Osteoarthritis. Int. J. Mol. Sci. 2015, 16, 26035–26054. [Google Scholar] [CrossRef]
- Fingleton, B. Matrix metalloproteinases as regulators of inflammatory processes. Biochim. Biophys. Acta Mol. Cell. Res. 2017, 1864, 2036–2042. [Google Scholar] [CrossRef]
- Robinson, W.H.; Lepus, C.M.; Wang, Q.; Raghu, H.; Mao, R.; Lindstrom, T.M.; Sokolove, J. Low-grade inflammation as a key mediator of the pathogenesis of osteoarthritis. Nat. Rev. Rheumatol. 2016, 12, 580–592. [Google Scholar] [CrossRef]
- Maldonado, M.; Nam, J. The role of changes in extracellular matrix of cartilage in the presence of inflammation on the pathology of osteoarthritis. Biomed. Res. Int. 2013, 2013, 1–10. [Google Scholar] [CrossRef]
- Goldring, M.B.; Otero, M.; Tsuchimochi, K.; Ijiri, K.; Li, Y. Defining the roles of inflammatory and anabolic cytokines in cartilage metabolism. Ann. Rheum. Dis. 2008, 67 (Suppl. S3), iii75–iii82. [Google Scholar] [CrossRef]
- Ayral, X.; Pickering, E.H.; Woodworth, T.G.; Mackillop, N.; Dougados, M. Synovitis: A potential predictive factor of structural progression of medial tibiofemoral knee osteoarthritis-results of a 1 year longitudinal arthroscopic study in 422 patients. Osteoarthr. Cartil. 2005, 13, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Torres, L.; Dunlop, D.D.; Peterfy, C.; Guermazi, A.; Prasad, P.; Hayes, K.W.; Song, J.; Cahue, S.; Chang, A.; Marshall, M.; et al. The relationship between specific tissue lesions and pain severity in persons with knee osteoarthritis. Osteoarthr. Cartil. 2006, 14, 1033–1040. [Google Scholar] [CrossRef] [PubMed]
- Benito, M.J.; Veale, D.J.; FitzGerald, O.; van den Berg, W.B.; Bresnihan, B. Synovial tissue inflammation in early and late osteoarthritis. Ann. Rheum. Dis. 2005, 64, 1263–1267. [Google Scholar] [CrossRef]
- Tong, Z.; Liu, Y.; Chen, B.; Yan, L.; Hao, D. Association between MMP3 and TIMP3 polymorphisms and risk of osteoarthritis. Oncotarget 2017, 8, 83563–83569. [Google Scholar] [CrossRef] [PubMed]
- Nakki, A.; Rodriguez-Fontenla, C.; Gonzalez, A.; Harilainen, A.; Leino-Arjas, P.; Heliovaara, M.; Eriksson, J.G.; Tallroth, K.; Videman, T.; Kaprio, J.; et al. Association study of MMP8 gene in osteoarthritis. Connect Tissue Res. 2016, 57, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Su, S.L.; Yang, H.Y.; Lee, H.S.; Huang, G.S.; Lee, C.H.; Liu, W.S.; Wang, C.C.; Peng, Y.J.; Lai, C.H.; Chen, C.Y.; et al. Gene-gene interactions between TGF-beta/Smad3 signalling pathway polymorphisms affect susceptibility to knee osteoarthritis. BMJ Open 2015, 5, e007931. [Google Scholar] [CrossRef]
- Rodriguez-Fontenla, C.; Calaza, M.; Evangelou, E.; Valdes, A.M.; Arden, N.; Blanco, F.J.; Carr, A.; Chapman, K.; Deloukas, P.; Doherty, M.; et al. Assessment of Osteoarthritis Candidate Genes in a Meta-Analysis of Nine Genome-Wide Association Studies. Arthritis Rheumatol. 2014, 66, 940–949. [Google Scholar] [CrossRef]
- Nakki, A.; Videman, T.; Kujala, U.M.; Suhonen, M.; Mannikko, M.; Peltonen, L.; Battie, M.C.; Kaprio, J.; Saarela, J. Candidate gene association study of magnetic resonance imaging-based hip osteoarthritis (OA): Evidence for COL9A2 gene as a common predisposing factor for hip OA and lumbar disc degeneration. J. Rheumatol. 2011, 38, 747–752. [Google Scholar] [CrossRef]
- Zhu, L.; Shi, D.; Dai, J.; Qin, J.; Fan, J.; Wang, Z.; Qiu, X.; Xu, Z.; Chen, D.; Jiang, Q. Lack of evidence for association between DVWA gene polymorphisms and developmental dysplasia of the hip in Chinese Han population. Rheumatol. Int. 2011, 31, 883–887. [Google Scholar] [CrossRef]
- Valdes, A.M.; Spector, T.D.; Doherty, S.; Wheeler, M.; Hart, D.J.; Doherty, M. Association of the DVWA and GDF5 polymorphisms with osteoarthritis in UK populations. Ann. Rheum. Dis. 2009, 68, 1916–1920. [Google Scholar] [CrossRef]
- Meulenbelt, I.; Chapman, K.; Dieguez-Gonzalez, R.; Shi, D.; Tsezou, A.; Dai, J.; Malizos, K.N.; Kloppenburg, M.; Carr, A.; Nakajima, M.; et al. Large replication study and meta-analyses of DVWA as an osteoarthritis susceptibility locus in European and Asian populations. Hum. Mol. Genet. 2009, 18, 1518–1523. [Google Scholar] [CrossRef] [PubMed]
- Baker-Lepain, J.C.; Lynch, J.A.; Parimi, N.; McCulloch, C.E.; Nevitt, M.C.; Corr, M.; Lane, N.E. Variant alleles of the Wnt antagonist FRZB are determinants of hip shape and modify the relationship between hip shape and osteoarthritis. Arthritis Rheum. 2012, 64, 1457–1465. [Google Scholar] [CrossRef] [PubMed]
- Evangelou, E.; Chapman, K.; Meulenbelt, I.; Karassa, F.B.; Loughlin, J.; Carr, A.; Doherty, M.; Doherty, S.; Gomez-Reino, J.J.; Gonzalez, A.; et al. Large-scale analysis of association between GDF5 and FRZB variants and osteoarthritis of the hip, knee, and hand. Arthritis Rheum. 2009, 60, 1710–1721. [Google Scholar] [CrossRef]
- Zhang, R.; Yao, J.; Xu, P.; Ji, B.; Luck, J.V.; Chin, B.; Lu, S.; Kelsoe, J.R.; Ma, J. A comprehensive meta-analysis of association between genetic variants of GDF5 and osteoarthritis of the knee, hip and hand. Inflamm. Res. 2015, 64, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Mototani, H.; Mabuchi, A.; Saito, S.; Fujioka, M.; Iida, A.; Takatori, Y.; Kotani, A.; Kubo, T.; Nakamura, K.; Sekine, A.; et al. A functional single nucleotide polymorphism in the core promoter region of CALM1 is associated with hip osteoarthritis in Japanese. Hum. Mol. Genet. 2005, 14, 1009–1017. [Google Scholar] [CrossRef] [PubMed]
- Poulou, M.; Kaliakatsos, M.; Tsezou, A.; Kanavakis, E.; Malizos, K.N.; Tzetis, M. Association of the CALM1 core promoter polymorphism with knee osteoarthritis in patients of Greek origin. Genet Test 2008, 12, 263–265. [Google Scholar] [CrossRef]
- Loughlin, J.; Sinsheimer, J.S.; Carr, A.; Chapman, K. The CALM1 core promoter polymorphism is not associated with hip osteoarthritis in a United Kingdom Caucasian population. Osteoarthr. Cartil. 2006, 14, 295–298. [Google Scholar] [CrossRef]
- Shi, D.; Ni, H.; Dai, J.; Qin, J.; Xu, Y.; Zhu, L.; Yao, C.; Shao, Z.; Chen, D.; Xu, Z.; et al. Lack of association between the CALM1 core promoter polymorphism (-16C/T) and susceptibility to knee osteoarthritis in a Chinese Han population. BMC Med. Genet 2008, 9, 91. [Google Scholar] [CrossRef]
- Liu, C.; Sun, J.; Zhang, H.; Li, L. TGF beta1 gene polymorphisms correlate with the susceptibility of osteoarthritis. Int. J. Clin. Exp. Pathol. 2017, 10, 8780–8785. [Google Scholar]
- Urano, T.; Narusawa, K.; Shiraki, M.; Usui, T.; Sasaki, N.; Hosoi, T.; Ouchi, Y.; Nakamura, T.; Inoue, S. Association of a single nucleotide polymorphism in the insulin-like growth factor-1 receptor gene with spinal disc degeneration in postmenopausal Japanese women. Spine (Phila Pa 1976) 2008, 33, 1256–1261. [Google Scholar] [CrossRef]
- Claessen, K.M.; Ramautar, S.R.; Pereira, A.M.; Smit, J.W.; Biermasz, N.R.; Kloppenburg, M. Relationship between insulin-like growth factor-1 and radiographic disease in patients with primary osteoarthritis: A systematic review. Osteoarthr. Cartil. 2012, 20, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Limer, K.L.; Tosh, K.; Bujac, S.R.; McConnell, R.; Doherty, S.; Nyberg, F.; Zhang, W.; Doherty, M.; Muir, K.R.; Maciewicz, R.A. Attempt to replicate published genetic associations in a large, well-defined osteoarthritis case-control population (the GOAL study). Osteoarthr. Cartil. 2009, 17, 782–789. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Meulenbelt, I.; Bijkerk, C.; Miedema, H.S.; Breedveld, F.C.; Hofman, A.; Valkenburg, H.A.; Pols, H.A.; Slagboom, P.E.; van Duijn, C.M. A genetic association study of the IGF-1 gene and radiological osteoarthritis in a population-based cohort study (the Rotterdam Study). Ann. Rheum. Dis. 1998, 57, 371–374. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.J.; Keen, L.J.; Billingham, M.J.; Perry, M.J.; Elson, C.J.; Kirwan, J.R.; Sims, J.E.; Doherty, M.; Spector, T.D.; Bidwell, J.L. Extended haplotypes and linkage disequilibrium in the IL1R1-IL1A-IL1B-IL1RN gene cluster: Association with knee osteoarthritis. Genes Immun. 2004, 5, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Moxley, G.; Han, J.; Stern, A.G.; Riley, B.P. Potential influence of IL1B haplotype and IL1A-IL1B-IL1RN extended haplotype on hand osteoarthritis risk. Osteoarthr. Cartil. 2007, 15, 1106–1112. [Google Scholar] [CrossRef] [PubMed]
- Solovieva, S.; Kamarainen, O.P.; Hirvonen, A.; Hamalainen, S.; Laitala, M.; Vehmas, T.; Luoma, K.; Nakki, A.; Riihimaki, H.; Ala-Kokko, L.; et al. Association between interleukin 1 gene cluster polymorphisms and bilateral distal interphalangeal osteoarthritis. J. Rheumatol. 2009, 36, 1977–1986. [Google Scholar] [CrossRef]
- Stern, A.G.; de Carvalho, M.R.; Buck, G.A.; Adler, R.A.; Rao, T.P.; Disler, D.; Moxley, G.; Network, I.N. Association of erosive hand osteoarthritis with a single nucleotide polymorphism on the gene encoding interleukin-1 beta. Osteoarthr. Cartil. 2003, 11, 394–402. [Google Scholar] [CrossRef]
- Kerkhof, H.J.; Doherty, M.; Arden, N.K.; Abramson, S.B.; Attur, M.; Bos, S.D.; Cooper, C.; Dennison, E.M.; Doherty, S.A.; Evangelou, E.; et al. Large-scale meta-analysis of interleukin-1 beta and interleukin-1 receptor antagonist polymorphisms on risk of radiographic hip and knee osteoarthritis and severity of knee osteoarthritis. Osteoarthr. Cartil. 2011, 19, 265–271. [Google Scholar] [CrossRef]
- Attur, M.; Zhou, H.; Samuels, J.; Krasnokutsky, S.; Yau, M.; Scher, J.U.; Doherty, M.; Wilson, A.G.; Bencardino, J.; Hochberg, M.; et al. Interleukin 1 receptor antagonist (IL1RN) gene variants predict radiographic severity of knee osteoarthritis and risk of incident disease. Ann. Rheum. Dis. 2020, 79, 400–407. [Google Scholar] [CrossRef]
- Pola, E.; Papaleo, P.; Pola, R.; Gaetani, E.; Tamburelli, F.C.; Aulisa, L.; Logroscino, C.A. Interleukin-6 gene polymorphism and risk of osteoarthritis of the hip: A case-control study. Osteoarthr. Cartil. 2005, 13, 1025–1028. [Google Scholar] [CrossRef]
- Day-Williams, A.G.; Southam, L.; Panoutsopoulou, K.; Rayner, N.W.; Esko, T.; Estrada, K.; Helgadottir, H.T.; Hofman, A.; Ingvarsson, T.; Jonsson, H.; et al. A variant in MCF2L is associated with osteoarthritis. Am. J. Hum. Genet 2011, 89, 446–450. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, C.; Skelton, A.J.; Rushton, M.D.; Reynard, L.N.; Loughlin, J. Expression analysis of the osteoarthritis genetic susceptibility locus mapping to an intron of the MCF2L gene and marked by the polymorphism rs11842874. BMC Med. Genet 2015, 16, 108. [Google Scholar] [CrossRef] [PubMed]
- Valdes, A.M.; Doherty, S.; Muir, K.R.; Zhang, W.; Maciewicz, R.A.; Wheeler, M.; Arden, N.; Cooper, C.; Doherty, M. Genetic contribution to radiographic severity in osteoarthritis of the knee. Ann. Rheum. Dis. 2012, 71, 1537–1540. [Google Scholar] [CrossRef] [PubMed]
- Kang, B.; Zhao, F.; Zhang, X.; Deng, X.; He, X. Association between the interaction of SMAD3 polymorphisms with body mass index and osteoarthritis susceptibility. Int. J. Clin. Exp. Pathol. 2015, 8, 7364–7370. [Google Scholar]
- Sharma, A.C.; Srivastava, R.N.; Srivastava, S.R.; Parmar, D.; Singh, A.; Raj, S. Association between Single Nucleotide Polymorphisms of SMAD3 and BMP5 with the Risk of Knee Osteoarthritis. J. Clin. Diagn. Res. 2017, 11, GC01–GC04. [Google Scholar] [CrossRef] [PubMed]
- Valdes, A.M.; Hart, D.J.; Jones, K.A.; Surdulescu, G.; Swarbrick, P.; Doyle, D.V.; Schafer, A.J.; Spector, T.D. Association study of candidate genes for the prevalence and progression of knee osteoarthritis. Arthritis Rheum. 2004, 50, 2497–2507. [Google Scholar] [CrossRef]
- Rodriguez-Lopez, J.; Pombo-Suarez, M.; Loughlin, J.; Tsezou, A.; Blanco, F.J.; Meulenbelt, I.; Slagboom, P.E.; Valdes, A.M.; Spector, T.D.; Gomez-Reino, J.J.; et al. Association of a nsSNP in ADAMTS14 to some osteoarthritis phenotypes. Osteoarthr. Cartil. 2009, 17, 321–327. [Google Scholar] [CrossRef]
- El Khoury, L.; Posthumus, M.; Collins, M.; Handley, C.J.; Cook, J.; Raleigh, S.M. Polymorphic variation within the ADAMTS2, ADAMTS14, ADAMTS5, ADAM12 and TIMP2 genes and the risk of Achilles tendon pathology: A genetic association study. J. Sci. Med. Sport 2013, 16, 493–498. [Google Scholar] [CrossRef]
- Poonpet, T.; Honsawek, S.; Tammachote, N.; Kanitnate, S.; Tammachote, R. ADAMTS14 gene polymorphism associated with knee osteoarthritis in Thai women. Genet Mol. Res. 2013, 12, 5301–5309. [Google Scholar] [CrossRef]
- Wang, D.D.; Gan, Y.H.; Ma, X.C.; Meng, J.H. Association between ADAMTS14 gene polymorphism and the temporomandibular joint osteoarthritis in Chinese Han females. J. Peking Univ. Heal. Sci. 2018, 50, 279–283. [Google Scholar]
- Lv, Z.T.; Liang, S.; Huang, X.J.; Cheng, P.; Zhu, W.T.; Chen, A.M. Association between ADAM12 Single-Nucleotide Polymorphisms and Knee Osteoarthritis: A Meta-Analysis. Biomed. Res. Int. 2017, 2017, 5398181. [Google Scholar] [CrossRef] [PubMed]
- Valdes, A.M.; Doherty, M.; Spector, T.D. The additive effect of individual genes in predicting risk of knee osteoarthritis. Ann. Rheum. Dis. 2008, 67, 124–127. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhou, K.; Chen, Z.; Yang, F.; Zhang, C.; Zhou, Z.; Pei, F. The association between DVWA polymorphisms and osteoarthritis susceptibility: A genetic meta-analysis. Int. J. Clin. Exp. Med. 2015, 8, 12566–12574. [Google Scholar] [PubMed]
- Wang, T.; Liang, Y.; Li, H.; Li, H.; He, Q.; Xue, Y.; Shen, C.; Zhang, C.; Xiang, J.; Ding, J.; et al. Single Nucleotide Polymorphisms and Osteoarthritis: An Overview and a Meta-Analysis. Medicine (Baltim. ) 2016, 95, e2811. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, Y.; Mabuchi, A.; Shi, D.; Kubo, T.; Takatori, Y.; Saito, S.; Fujioka, M.; Sudo, A.; Uchida, A.; Yamamoto, S.; et al. A functional polymorphism in the 5’ UTR of GDF5 is associated with susceptibility to osteoarthritis. Nat. Genet. 2007, 39, 529–533. [Google Scholar] [CrossRef] [PubMed]
- Chapman, K.; Takahashi, A.; Meulenbelt, I.; Watson, C.; Rodriguez-Lopez, J.; Egli, R.; Tsezou, A.; Malizos, K.N.; Kloppenburg, M.; Shi, D.; et al. A meta-analysis of European and Asian cohorts reveals a global role of a functional SNP in the 5’ UTR of GDF5 with osteoarthritis susceptibility. Hum. Mol. Genet. 2008, 17, 1497–1504. [Google Scholar] [CrossRef]
- Williams, F.M.; Popham, M.; Hart, D.J.; de Schepper, E.; Bierma-Zeinstra, S.; Hofman, A.; Uitterlinden, A.G.; Arden, N.K.; Cooper, C.; Spector, T.D.; et al. GDF5 single-nucleotide polymorphism rs143383 is associated with lumbar disc degeneration in Northern European women. Arthritis Rheum. 2011, 63, 708–712. [Google Scholar] [CrossRef]
- Wang, Z.; Li, Y.; Wang, Y.; Wang, X.; Zhang, J.; Tian, J. Association between GDF5 single nucleotide polymorphism rs143383 and lumbar disc degeneration. Exp. Ther. Med. 2018, 16, 1900–1904. [Google Scholar] [CrossRef]
- Southam, L.; Rodriguez-Lopez, J.; Wilkins, J.M.; Pombo-Suarez, M.; Snelling, S.; Gomez-Reino, J.J.; Chapman, K.; Gonzalez, A.; Loughlin, J. An SNP in the 5’-UTR of GDF5 is associated with osteoarthritis susceptibility in Europeans and with in vivo differences in allelic expression in articular cartilage. Hum. Mol. Genet 2007, 16, 2226–2232. [Google Scholar] [CrossRef]
- Rickert, M.; Wang, H.; Wieloch, P.; Lorenz, H.; Steck, E.; Sabo, D.; Richter, W. Adenovirus-mediated gene transfer of growth and differentiation factor-5 into tenocytes and the healing rat Achilles tendon. Connect Tissue Res. 2005, 46, 175–183. [Google Scholar] [CrossRef]
- Liang, H.; Ma, S.Y.; Feng, G.; Shen, F.H.; Joshua Li, X. Therapeutic effects of adenovirus-mediated growth and differentiation factor-5 in a mice disc degeneration model induced by annulus needle puncture. Spine J. 2010, 10, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Chujo, T.; An, H.S.; Akeda, K.; Miyamoto, K.; Muehleman, C.; Attawia, M.; Andersson, G.; Masuda, K. Effects of growth differentiation factor-5 on the intervertebral disc--in vitro bovine study and in vivo rabbit disc degeneration model study. Spine (Phila Pa 1976) 2006, 31, 2909–2917. [Google Scholar] [CrossRef] [PubMed]
- Byers, B.A.; Parrish, W.R.; Geesin, J.; Herzberg, U.; Wadsworth, S.; Bendele, A.M.; Story, B.J. Intra-articular supplementation with recombinant human GDF5 arrests disease progression and stimulates cartilage regeneration in the rat medial meniscus transection (MMT) model of osteoarthritis. Osteoarthr. Cartil. 2014, 22, S471–S472. [Google Scholar] [CrossRef][Green Version]
- Parrish, W.R.; Byers, B.A.; Su, D.; Geesin, J.; Herzberg, U.; Wadsworth, S.; Bendele, A.; Story, B. Intra-articular therapy with recombinant human GDF5 arrests disease progression and stimulates cartilage repair in the rat medial meniscus transection (MMT) model of osteoarthritis. Osteoarthr. Cartil. 2017, 25, 554–560. [Google Scholar] [CrossRef] [PubMed]
- den Hollander, W.; Ramos, Y.F.; Bomer, N.; Elzinga, S.; van der Breggen, R.; Lakenberg, N.; de Dijcker, W.J.; Suchiman, H.E.; Duijnisveld, B.J.; Houwing-Duistermaat, J.J.; et al. Transcriptional associations of osteoarthritis-mediated loss of epigenetic control in articular cartilage. Arthritis Rheumatol. 2015, 67, 2108–2116. [Google Scholar] [CrossRef]
- Im, G.I.; Choi, Y.J. Epigenetics in osteoarthritis and its implication for future therapeutics. Expert Opin. Biol. 2013, 13, 713–721. [Google Scholar] [CrossRef]
- Kim, H.; Kang, D.; Cho, Y.; Kim, J.H. Epigenetic Regulation of Chondrocyte Catabolism and Anabolism in Osteoarthritis. Mol. Cells 2015, 38, 677–684. [Google Scholar] [CrossRef]
- Loughlin, J.; Reynard, L.N. Osteoarthritis: Epigenetics of articular cartilage in knee and hip OA. Nat. Rev. Rheumatol. 2015, 11, 6–7. [Google Scholar] [CrossRef]
- Raman, S.; FitzGerald, U.; Murphy, J.M. Interplay of Inflammatory Mediators with Epigenetics and Cartilage Modifications in Osteoarthritis. Front Bioeng. Biotechnol. 2018, 6, 22. [Google Scholar] [CrossRef]
- Reynard, L.N.; Loughlin, J. Genetics and epigenetics of osteoarthritis. Maturitas 2012, 71, 200–204. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, J. Epigenetics and Osteoarthritis. Genes Dis. 2015, 2, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Szczepanek, J. Role of microRNA dysregulation in childhood acute leukemias: Diagnostics, monitoring and therapeutics: A comprehensive review. World J. Clin. Oncol. 2020, 11, 348–369. [Google Scholar] [CrossRef]
- Endisha, H.; Rockel, J.; Jurisica, I.; Kapoor, M. The complex landscape of microRNAs in articular cartilage: Biology, pathology, and therapeutic targets. JCI Insight 2018, 3. [Google Scholar] [CrossRef] [PubMed]
- Jackson, A.; Linsley, P.S. The therapeutic potential of microRNA modulation. Discov. Med. 2010, 9, 311–318. [Google Scholar] [PubMed]
- Jackson, A.L.; Levin, A.A. Developing microRNA therapeutics: Approaching the unique complexities. Nucleic Acid 2012, 22, 213–225. [Google Scholar] [CrossRef] [PubMed]
- Sondag, G.R.; Haqqi, T.M. The Role of MicroRNAs and Their Targets in Osteoarthritis. Curr. Rheumatol. Rep. 2016, 18. [Google Scholar] [CrossRef]
- Genemaras, A.; Charles Huang, C.Y.D.; Kaplan, L. MicroRNA Therapies for Osteoarthritis and its Symptoms. Curr. Tissue Eng. 2015, 4, 86–94. [Google Scholar] [CrossRef]
- Huang, J.; Zhao, L.; Fan, Y.; Liao, L.; Ma, P.X.; Xiao, G.; Chen, D. The microRNAs miR-204 and miR-211 maintain joint homeostasis and protect against osteoarthritis progression. Nat. Commun. 2019, 10, 2876. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, H.; Sun, Q.; Wang, Y.; Yang, J.; Yang, J.; Zhang, T.; Luo, S.; Wang, L.; Jiang, Y.; et al. Intra-articular Delivery of Antago-miR-483-5p Inhibits Osteoarthritis by Modulating Matrilin 3 and Tissue Inhibitor of Metalloproteinase 2. Mol. Ther. 2017, 25, 715–727. [Google Scholar] [CrossRef]
- Nagata, Y.; Nakasa, T.; Mochizuki, Y.; Ishikawa, M.; Miyaki, S.; Shibuya, H.; Yamasaki, K.; Adachi, N.; Asahara, H.; Ochi, M. Induction of apoptosis in the synovium of mice with autoantibody-mediated arthritis by the intraarticular injection of double-stranded MicroRNA-15a. Arthritis Rheum. 2009, 60, 2677–2683. [Google Scholar] [CrossRef]
- Ko, J.Y.; Lee, M.S.; Lian, W.S.; Weng, W.T.; Sun, Y.C.; Chen, Y.S.; Wang, F.S. MicroRNA-29a Counteracts Synovitis in Knee Osteoarthritis Pathogenesis by Targeting VEGF. Sci. Rep. 2017, 7, 3584. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, K.; Oreffo, R.O.; Gibson, M.B.; Goldring, M.B.; Roach, H.I. DNA demethylation at specific CpG sites in the IL1B promoter in response to inflammatory cytokines in human articular chondrocytes. Arthritis Rheum. 2009, 60, 3303–3313. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, K.; Nakasa, T.; Miyaki, S.; Ishikawa, M.; Deie, M.; Adachi, N.; Yasunaga, Y.; Asahara, H.; Ochi, M. Expression of MicroRNA-146a in osteoarthritis cartilage. Arthritis Rheum. 2009, 60, 1035–1041. [Google Scholar] [CrossRef]
- Huh, Y.H.; Ryu, J.H.; Chun, J.S. Regulation of type II collagen expression by histone deacetylase in articular chondrocytes. J. Biol. Chem. 2007, 282, 17123–17131. [Google Scholar] [CrossRef] [PubMed]
- Martinek, V.; Ueblacker, P.; Imhoff, A.B. Current Concepts of Gene Therapy and Cartilage Repair. J. Bone Jt Surg. Br. Vol. 2003, 85-B, 782–788. [Google Scholar] [CrossRef]
- Venkatesan, J.K.; Rey-Rico, A.; Cucchiarini, M. Current Trends in Viral Gene Therapy for Human Orthopaedic Regenerative Medicine. Tissue Eng. Regen Med. 2019, 16, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Evans, C.H.; Huard, J. Gene therapy approaches to regenerating the musculoskeletal system. Nat. Rev. Rheumatol. 2015, 11, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Pascher, A.; Palmer, G.D.; Steinert, A.; Oligino, T.; Gouze, E.; Gouze, J.N.; Betz, O.; Spector, M.; Robbins, P.D.; Evans, C.H.; et al. Gene delivery to cartilage defects using coagulated bone marrow aspirate. Gene 2004, 11, 133–141. [Google Scholar] [CrossRef]
- Sieker, J.T.; Kunz, M.; Weissenberger, M.; Gilbert, F.; Frey, S.; Rudert, M.; Steinert, A.F. Direct bone morphogenetic protein 2 and Indian hedgehog gene transfer for articular cartilage repair using bone marrow coagulates. Osteoarthr. Cartil. 2015, 23, 433–442. [Google Scholar] [CrossRef]
- Ivkovic, A.; Pascher, A.; Hudetz, D.; Maticic, D.; Jelic, M.; Dickinson, S.; Loparic, M.; Haspl, M.; Windhager, R.; Pecina, M. Articular cartilage repair by genetically modified bone marrow aspirate in sheep. Gene 2010, 17, 779–789. [Google Scholar] [CrossRef]
- Cucchiarini, M.; Madry, H. Overexpression of human IGF-I via direct rAAV-mediated gene transfer improves the early repair of articular cartilage defects in vivo. Gene 2014, 21, 811–819. [Google Scholar] [CrossRef] [PubMed]
- Cucchiarini, M.; Orth, P.; Madry, H. Direct rAAV SOX9 administration for durable articular cartilage repair with delayed terminal differentiation and hypertrophy in vivo. J. Mol. Med. (Berl.) 2013, 91, 625–636. [Google Scholar] [CrossRef] [PubMed]
- Cucchiarini, M.; Madry, H.; Ma, C.; Thurn, T.; Zurakowski, D.; Menger, M.D.; Kohn, D.; Trippel, S.B.; Terwilliger, E.F. Improved tissue repair in articular cartilage defects in vivo by rAAV-mediated overexpression of human fibroblast growth factor 2. Mol. Ther. 2005, 12, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Gobbi, A.; Kon, E.; Filardo, G.; Delcogliano, M.; Montaperto, C.; Boldrini, L.; Bathan, L.; Marcacci, M., 2nd. Gen ACI Vs Microfracture in Knee Chondral Defect Treatment: Comparative study at 5 years (SS-61). Arthrosc. J. Arthrosc. Relat. Surg. 2009, 25, e33–e34. [Google Scholar] [CrossRef]
- Brittberg, M.; Lindahl, A.; Nilsson, A.; Ohlsson, C.; Isaksson, O.; Peterson, L. Treatment of deep cartilage defects in the knee with autologous chondrocyte transplantation. N. Engl. J. Med. 1994, 331, 889–895. [Google Scholar] [CrossRef]
- Peterson, L.; Brittberg, M.; Kiviranta, I.; Akerlund, E.L.; Lindahl, A. Autologous chondrocyte transplantation. Biomechanics and long-term durability. Am. J. Sports Med. 2002, 30, 2–12. [Google Scholar] [CrossRef]
- Kang, R.; Marui, T.; Ghivizzani, S.C.; Nita, I.M.; Georgescu, H.I.; Suh, J.K.; Robbins, P.D.; Evans, C.H. Ex vivo gene transfer to chondrocytes in full-thickness articular cartilage defects: A feasibility study. Osteoarthr. Cartil. 1997, 5, 139–143. [Google Scholar] [CrossRef]
- Orth, P.; Kaul, G.; Cucchiarini, M.; Zurakowski, D.; Menger, M.D.; Kohn, D.; Madry, H. Transplanted articular chondrocytes co-overexpressing IGF-I and FGF-2 stimulate cartilage repair in vivo. Knee Surg. Sports Traumatol. Arthrosc. 2011, 19, 2119–2130. [Google Scholar] [CrossRef]
- Ortved, K.F.; Begum, L.; Mohammed, H.O.; Nixon, A.J. Implantation of rAAV5-IGF-I transduced autologous chondrocytes improves cartilage repair in full-thickness defects in the equine model. Mol. Ther. 2015, 23, 363–373. [Google Scholar] [CrossRef]
- Ha, C.W.; Noh, M.J.; Choi, K.B.; Lee, K.H. Initial phase I safety of retrovirally transduced human chondrocytes expressing transforming growth factor-beta-1 in degenerative arthritis patients. Cytotherapy 2012, 14, 247–256. [Google Scholar] [CrossRef]
- Evans, C.H.; Liu, F.J.; Glatt, V.; Hoyland, J.A.; Kirker-Head, C.; Walsh, A.; Betz, O.; Wells, J.W.; Betz, V.; Porter, R.M.; et al. Use of genetically modified muscle and fat grafts to repair defects in bone and cartilage. Eur. Cell Mater. 2009, 18, 96–111. [Google Scholar] [CrossRef] [PubMed]
- Rowland, C.R.; Glass, K.A.; Ettyreddy, A.R.; Gloss, C.C.; Matthews, J.R.L.; Huynh, N.P.T.; Guilak, F. Regulation of decellularized tissue remodeling via scaffold-mediated lentiviral delivery in anatomically-shaped osteochondral constructs. Biomaterials 2018, 177, 161–175. [Google Scholar] [CrossRef] [PubMed]
- Zaslav, K.; McAdams, T.; Scopp, J.; Theosadakis, J.; Mahajan, V.; Gobbi, A. New Frontiers for Cartilage Repair and Protection. Cartilage 2012, 3, 77S–86S. [Google Scholar] [CrossRef] [PubMed]
- Cucchiarini, M.; Madry, H. Biomaterial-guided delivery of gene vectors for targeted articular cartilage repair. Nat. Rev. Rheumatol. 2019, 15, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Moutos, F.T.; Glass, K.A.; Compton, S.A.; Ross, A.K.; Gersbach, C.A.; Guilak, F.; Estes, B.T. Anatomically shaped tissue-engineered cartilage with tunable and inducible anticytokine delivery for biological joint resurfacing. Proc. Natl. Acad. Sci. USA 2016, 113, E4513–E4522. [Google Scholar] [CrossRef] [PubMed]
- Lam, J.; Lu, S.; Kasper, F.K.; Mikos, A.G. Strategies for controlled delivery of biologics for cartilage repair. Adv. Drug Deliv. Rev. 2015, 84, 123–134. [Google Scholar] [CrossRef]
- Rey-Rico, A.; Frisch, J.; Venkatesan, J.K.; Schmitt, G.; Rial-Hermida, I.; Taboada, P.; Concheiro, A.; Madry, H.; Alvarez-Lorenzo, C.; Cucchiarini, M. PEO-PPO-PEO Carriers for rAAV-Mediated Transduction of Human Articular Chondrocytes in Vitro and in a Human Osteochondral Defect Model. ACS Appl. Mater. Interfaces 2016, 8, 20600–20613. [Google Scholar] [CrossRef]
- Pannier, A.K.; Shea, L.D. Controlled release systems for DNA delivery. Mol. Ther. 2004, 10, 19–26. [Google Scholar] [CrossRef]
- Glass, K.A.; Link, J.M.; Brunger, J.M.; Moutos, F.T.; Gersbach, C.A.; Guilak, F. Tissue-engineered cartilage with inducible and tunable immunomodulatory properties. Biomaterials 2014, 35, 5921–5931. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Chen, Y.R.; Song, Y.F.; Yang, M.; Ye, J.; Zhou, G.; Yu, J.K. Scaffold-Based Gene Therapeutics for Osteochondral Tissue Engineering. Front Pharm. 2019, 10, 1534. [Google Scholar] [CrossRef]
- Rey-Rico, A.; Venkatesan, J.K.; Schmitt, G.; Speicher-Mentges, S.; Madry, H.; Cucchiarini, M. Effective Remodelling of Human Osteoarthritic Cartilage by sox9 Gene Transfer and Overexpression upon Delivery of rAAV Vectors in Polymeric Micelles. Mol. Pharm. 2018, 15, 2816–2826. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Rodriguez, P.; Rey-Rico, A.; Madry, H.; Landin, M.; Cucchiarini, M. Effective genetic modification and differentiation of hMSCs upon controlled release of rAAV vectors using alginate/poloxamer composite systems. Int. J. Pharm. 2015, 496, 614–626. [Google Scholar] [CrossRef] [PubMed]
- Rey-Rico, A.; Babicz, H.; Madry, H.; Concheiro, A.; Alvarez-Lorenzo, C.; Cucchiarini, M. Supramolecular polypseudorotaxane gels for controlled delivery of rAAV vectors in human mesenchymal stem cells for regenerative medicine. Int. J. Pharm. 2017, 531, 492–503. [Google Scholar] [CrossRef] [PubMed]
- Rey-Rico, A.; Venkatesan, J.K.; Schmitt, G.; Concheiro, A.; Madry, H.; Alvarez-Lorenzo, C.; Cucchiarini, M. rAAV-mediated overexpression of TGF-beta via vector delivery in polymeric micelles stimulates the biological and reparative activities of human articular chondrocytes in vitro and in a human osteochondral defect model. Int. J. Nanomed. 2017, 12, 6985–6996. [Google Scholar] [CrossRef]
- Brunger, J.M.; Huynh, N.P.; Guenther, C.M.; Perez-Pinera, P.; Moutos, F.T.; Sanchez-Adams, J.; Gersbach, C.A.; Guilak, F. Scaffold-mediated lentiviral transduction for functional tissue engineering of cartilage. Proc. Natl. Acad. Sci. USA 2014, 111, E798–E806. [Google Scholar] [CrossRef]
- Zhao, R.; Peng, X.; Li, Q.; Song, W. Effects of phosphorylatable short peptide-conjugated chitosan-mediated IL-1Ra and igf-1 gene transfer on articular cartilage defects in rabbits. PLoS ONE 2014, 9, e112284. [Google Scholar] [CrossRef]
- Rey-Rico, A.; Venkatesan, J.K.; Frisch, J.; Schmitt, G.; Monge-Marcet, A.; Lopez-Chicon, P.; Mata, A.; Semino, C.; Madry, H.; Cucchiarini, M. Effective and durable genetic modification of human mesenchymal stem cells via controlled release of rAAV vectors from self-assembling peptide hydrogels with a maintained differentiation potency. Acta Biomater. 2015, 18, 118–127. [Google Scholar] [CrossRef]
- Rey-Rico, A.; Venkatesan, J.K.; Frisch, J.; Rial-Hermida, I.; Schmitt, G.; Concheiro, A.; Madry, H.; Alvarez-Lorenzo, C.; Cucchiarini, M. PEO-PPO-PEO micelles as effective rAAV-mediated gene delivery systems to target human mesenchymal stem cells without altering their differentiation potency. Acta Biomater. 2015, 27, 42–52. [Google Scholar] [CrossRef]
- Meng, W.; Rey-Rico, A.; Claudel, M.; Schmitt, G.; Speicher-Mentges, S.; Pons, F.; Lebeau, L.; Venkatesan, J.K.; Cucchiarini, M. rAAV-Mediated Overexpression of SOX9 and TGF-beta via Carbon Dot-Guided Vector Delivery Enhances the Biological Activities in Human Bone Marrow-Derived Mesenchymal Stromal Cells. Nanomaterials 2020, 10. [Google Scholar] [CrossRef]
- Leng, P.; Ding, C.R.; Zhang, H.N.; Wang, Y.Z. Reconstruct large osteochondral defects of the knee with hIGF-1 gene enhanced Mosaicplasty. Knee 2012, 19, 804–811. [Google Scholar] [CrossRef]
- Yang, S.; Qian, Z.; Liu, D.; Wen, N.; Xu, J.; Guo, X. Integration of C-type natriuretic peptide gene-modified bone marrow mesenchymal stem cells with chitosan/silk fibroin scaffolds as a promising strategy for articular cartilage regeneration. Cell Tissue Bank 2019, 20, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Venkatesan, J.K.; Gardner, O.; Rey-Rico, A.; Eglin, D.; Alini, M.; Stoddart, M.J.; Cucchiarini, M.; Madry, H. Improved Chondrogenic Differentiation of rAAV SOX9-Modified Human MSCs Seeded in Fibrin-Polyurethane Scaffolds in a Hydrodynamic Environment. Int. J. Mol. Sci. 2018, 19. [Google Scholar] [CrossRef] [PubMed]
- Pi, Y.; Zhang, X.; Shi, J.; Zhu, J.; Chen, W.; Zhang, C.; Gao, W.; Zhou, C.; Ao, Y. Targeted delivery of non-viral vectors to cartilage in vivo using a chondrocyte-homing peptide identified by phage display. Biomaterials 2011, 32, 6324–6332. [Google Scholar] [CrossRef] [PubMed]
- Evans, C.H. The vicissitudes of gene therapy. Bone Jt. Re 2019, 8, 469–471. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).