Overexpressed BSR1-Mediated Enhancement of Disease Resistance Depends on the MAMP-Recognition System
Abstract
1. Introduction
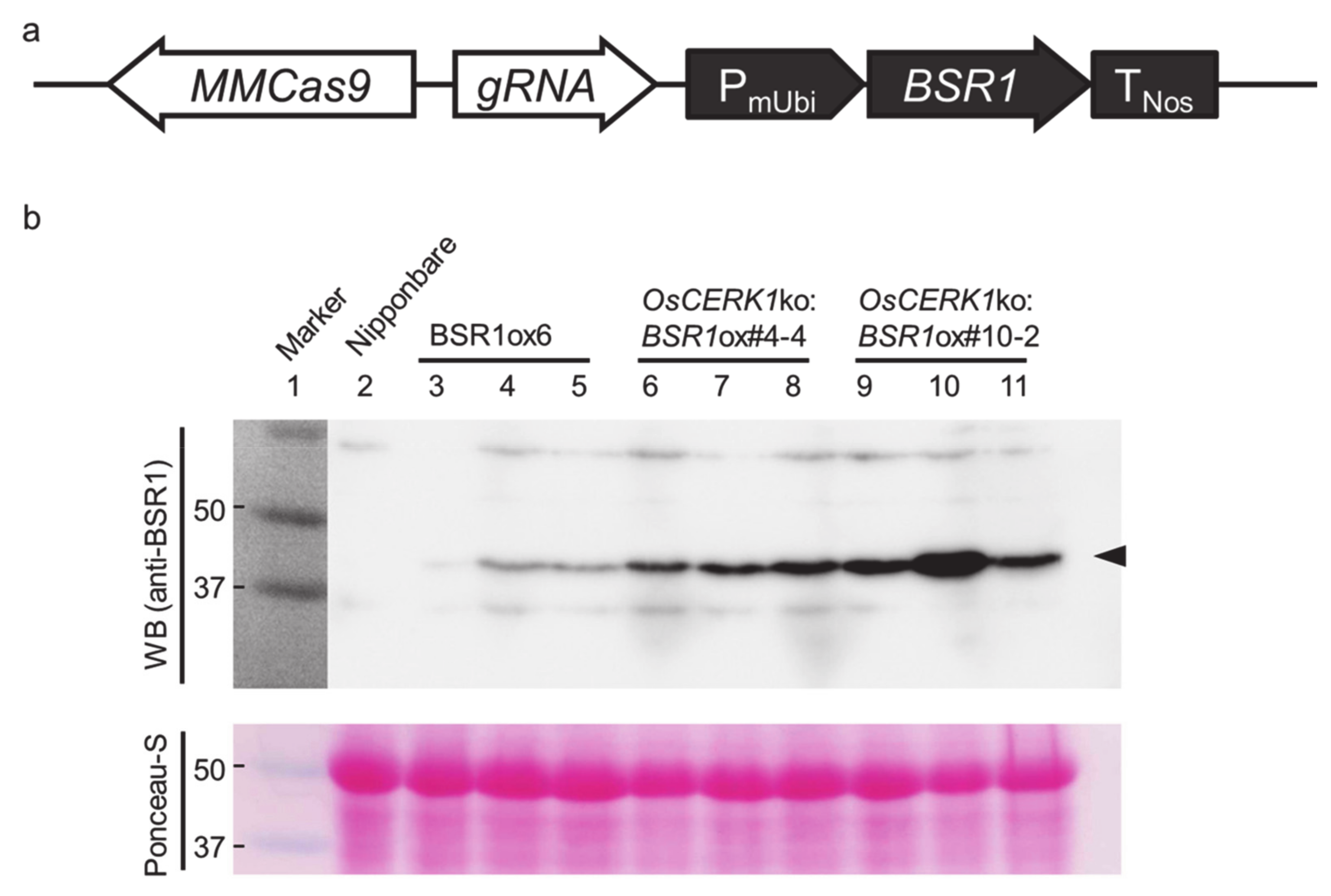
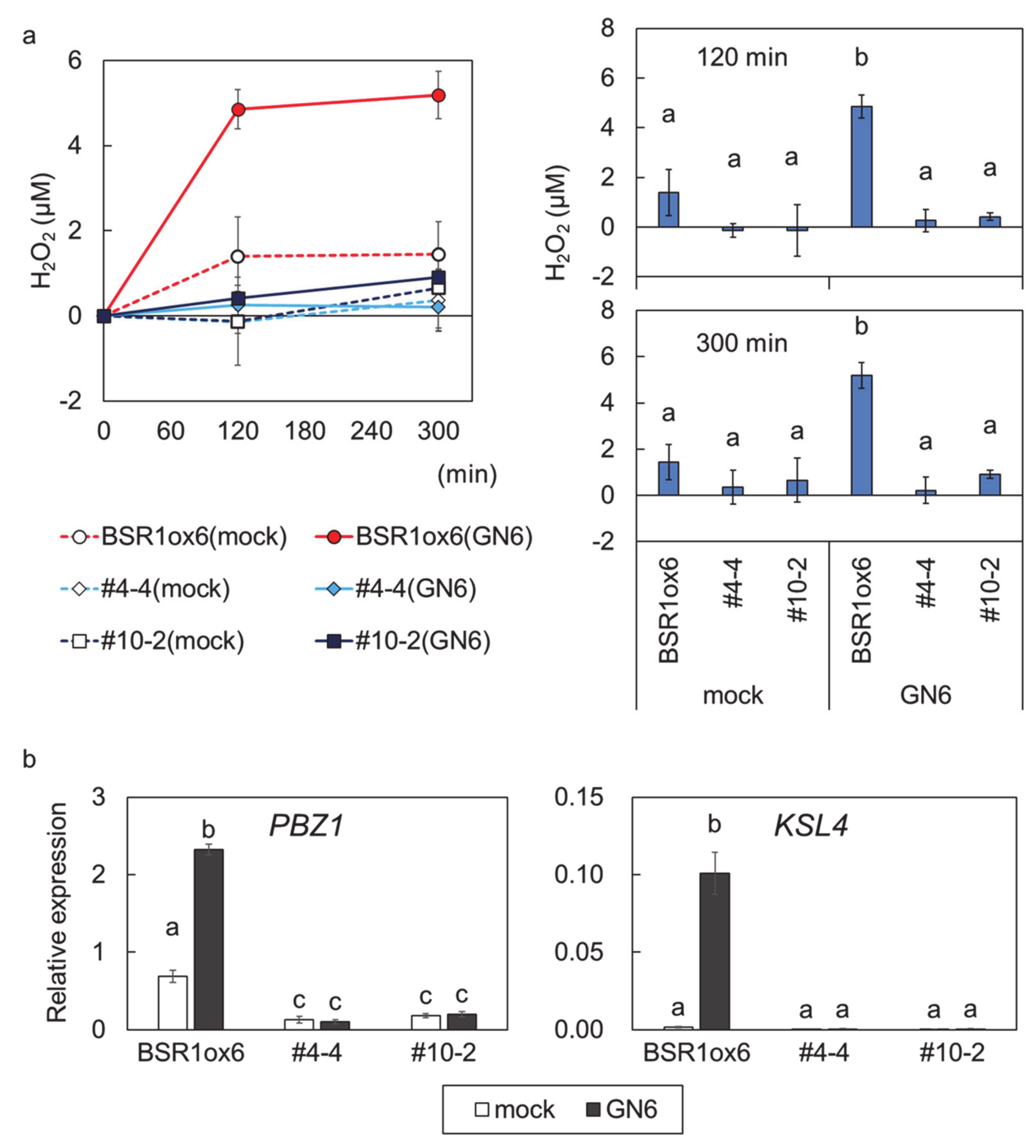
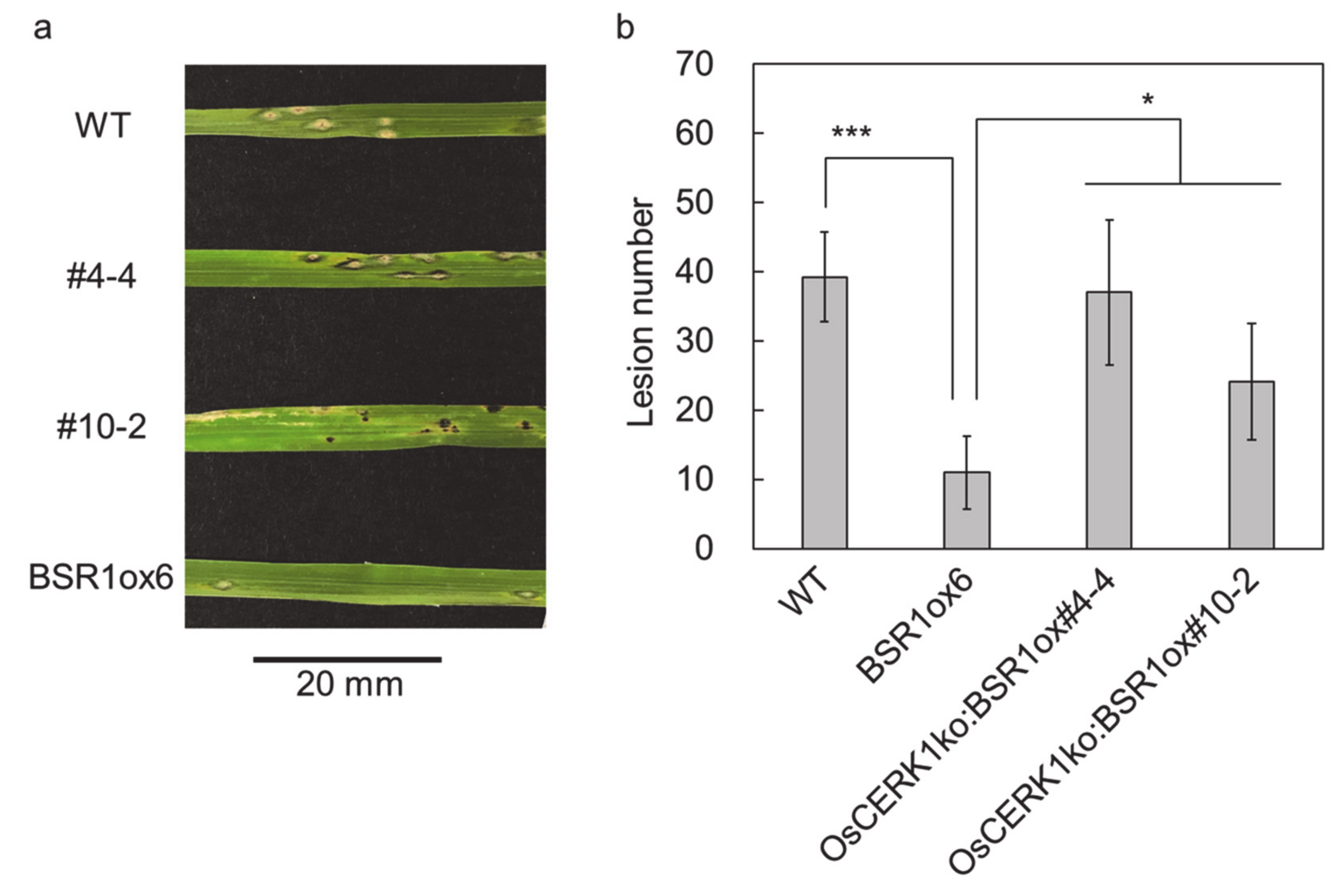
2. Results
3. Discussion
4. Materials and Methods
4.1. Plant and Microbial Materials and Inoculation
4.2. Plasmid Construction and Transformation
4.3. Assay of MAMP-Responsivity
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Boller, T.; Felix, G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 2009, 60, 379–406. [Google Scholar] [CrossRef] [PubMed]
- Monaghan, J.; Zipfel, C. Plant pattern recognition receptor complexes at the plasma membrane. Curr. Opin Plant Biol. 2012, 15, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Macho, A.P.; Zipfel, C. Plant PRRs and the activation of innate immune signaling. Mol. Cell 2014, 54, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, T.; Yamada, K.; Yoshimura, S.; Yamaguchi, K. Chitin receptor-mediated activation of MAP kinases and ROS production in rice and Arabidopsis. Plant. Signal. Behav. 2017, 12, e1361076. [Google Scholar] [CrossRef]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Dubouzet, J.G.; Maeda, S.; Sugano, S.; Ohtake, M.; Hayashi, N.; Ichikawa, T.; Kondou, Y.; Kuroda, H.; Horii, Y.; Matsui, M.; et al. Screening for resistance against Pseudomonas syringae in rice-FOX Arabidopsis lines identified a putative receptor-like cytoplasmic kinase gene that confers resistance to major bacterial and fungal pathogens in Arabidopsis and rice. Plant Biotechnol. J. 2011, 9, 466–485. [Google Scholar] [CrossRef]
- Maeda, S.; Hayashi, N.; Sasaya, T.; Mori, M. Overexpression of BSR1 confers broad-spectrum resistance against two bacterial diseases and two major fungal diseases in rice. Breed. Sci. 2016, 66, 396–406. [Google Scholar] [CrossRef][Green Version]
- Shiu, S.H.; Karlowski, W.M.; Pan, R.; Tzeng, Y.H.; Mayer, K.F.; Li, W.H. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 2004, 16, 1220–1234. [Google Scholar] [CrossRef]
- Vij, S.; Giri, J.; Dansana, P.K.; Kapoor, S.; Tyagi, A.K. The receptor-like cytoplasmic kinase (OsRLCK) gene family in rice: Organization, phylogenetic relationship, and expression during development and stress. Mol. Plant 2008, 1, 732–750. [Google Scholar] [CrossRef]
- Liang, X.; Zhou, J.M. Receptor-Like Cytoplasmic Kinases: Central Players in Plant Receptor Kinase-Mediated Signaling. Annu. Rev. Plant Biol. 2018, 69, 267–299. [Google Scholar] [CrossRef]
- Kanda, Y.; Yokotani, N.; Maeda, S.; Nishizawa, Y.; Kamakura, T.; Mori, M. The receptor-like cytoplasmic kinase BSR1 mediates chitin-induced defense signaling in rice cells. Biosci. Biotechnol. Biochem. 2017, 81, 1497–1502. [Google Scholar] [CrossRef] [PubMed]
- Kanda, Y.; Nakagawa, H.; Nishizawa, Y.; Kamakura, T.; Mori, M. Broad-Spectrum Disease Resistance Conferred by the Overexpression of Rice RLCK BSR1 Results from an Enhanced Immune Response to Multiple MAMPs. Int. J. Mol. Sci. 2019, 20. [Google Scholar] [CrossRef] [PubMed]
- Sugano, S.; Maeda, S.; Hayashi, N.; Kajiwara, H.; Inoue, H.; Jiang, C.J.; Takatsuji, H.; Mori, M. Tyrosine phosphorylation of a receptor-like cytoplasmic kinase, BSR1, plays a crucial role in resistance to multiple pathogens in rice. Plant J. 2018, 96, 1137–1147. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Nakano, T.; Takamizawa, D.; Desaki, Y.; Ishii-Minami, N.; Nishizawa, Y.; Minami, E.; Okada, K.; Yamane, H.; Kaku, H.; et al. Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 2010, 64, 204–214. [Google Scholar] [CrossRef] [PubMed]
- Ao, Y.; Li, Z.; Feng, D.; Xiong, F.; Liu, J.; Li, J.F.; Wang, M.; Wang, J.; Liu, B.; Wang, H.B. OsCERK1 and OsRLCK176 play important roles in peptidoglycan and chitin signaling in rice innate immunity. Plant J. 2014, 80, 1072–1084. [Google Scholar] [CrossRef] [PubMed]
- Kouzai, Y.; Mochizuki, S.; Nakajima, K.; Desaki, Y.; Hayafune, M.; Miyazaki, H.; Yokotani, N.; Ozawa, K.; Minami, E.; Kaku, H.; et al. Targeted gene disruption of OsCERK1 reveals its indispensable role in chitin perception and involvement in the peptidoglycan response and immunity in rice. Mol. Plant Microbe Interact. 2014, 27, 975–982. [Google Scholar] [CrossRef] [PubMed]
- Desaki, Y.; Kouzai, Y.; Ninomiya, Y.; Iwase, R.; Shimizu, Y.; Seko, K.; Molinaro, A.; Minami, E.; Shibuya, N.; Kaku, H.; et al. OsCERK1 plays a crucial role in the lipopolysaccharide-induced immune response of rice. New Phytol. 2018, 217, 1042–1049. [Google Scholar] [CrossRef]
- Mikami, M.; Toki, S.; Endo, M. Comparison of CRISPR/Cas9 expression constructs for efficient targeted mutagenesis in rice. Plant Mol. Biol. 2015, 88, 561–572. [Google Scholar] [CrossRef]
- Zhang, X.; Dong, W.; Sun, J.; Feng, F.; Deng, Y.; He, Z.; Oldroyd, G.E.; Wang, E. The receptor kinase CERK1 has dual functions in symbiosis and immunity signalling. Plant J. 2015, 81, 258–267. [Google Scholar] [CrossRef]
- Lei, Y.; Lu, L.; Liu, H.Y.; Li, S.; Xing, F.; Chen, L.L. CRISPR-P: A web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol. Plant 2014, 7, 1494–1496. [Google Scholar] [CrossRef]
- Toki, S.; Hara, N.; Ono, K.; Onodera, H.; Tagiri, A.; Oka, S.; Tanaka, H. Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J. 2006, 47, 969–976. [Google Scholar] [CrossRef]
- Desaki, Y.; Shimada, H.; Takahashi, S.; Sakurayama, C.; Kawai, M.; Kaku, H.; Shibuya, N. Handmade leaf cutter for efficient and reliable ROS assay. Plant Biotechnol. (Tokyo) 2019, 36, 275–278. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.J.; Shimono, M.; Sugano, S.; Kojima, M.; Yazawa, K.; Yoshida, R.; Inoue, H.; Hayashi, N.; Sakakibara, H.; Takatsuji, H. Abscisic acid interacts antagonistically with salicylic acid signaling pathway in rice-Magnaporthe grisea interaction. Mol. Plant Microbe Interact. 2010, 23, 791–798. [Google Scholar] [CrossRef] [PubMed]
| Target | Line | Cleavage Site Sequence | Frameshift |
|---|---|---|---|
| (Site #4) | WT | -GCTGGCTTCCTTCTACGTGACGCCGAACCAGAACGTCAC- | |
| Transformant#4-2 | -GCTGGCTTCCTTCTaACGTGACGCCGAACCAGAACGTCAC- | +1 | |
| -GCTGGCTTCCTTCT-CGTGACGCCGAACCAGAACGTCAC- | −1 | ||
| Transformant#4-4 | -GCTGGCTTCCTTCTaACGTGACGCCGAACCAGAACGTCAC- | +1 | |
| -GCTGGCTTCCTTCTaACGTGACGCCGAACCAGAACGTCAC- | +1 | ||
| (Site #10) | WT | -GTGCAGCGCCGGGTGCGACCTCGCGCTGGCTTCCTTCTA- | |
| Transformant#10-2 | -GTGCAGCGCCGGGTtGCGACCTCGCGCTGGCTTCCTTCTA- | +1 | |
| -GTGCAGCGCCGGGTtGCGACCTCGCGCTGGCTTCCTTCTA- | +1 | ||
| Transformant#10-6 | -GTGCAGCGCCGGGT--GACCTCGCGCTGGCTTCCTTCTA- | −2 | |
| -GTGCAGCGCCGGGTaGCGACCTCGCGCTGGCTTCCTTCTA- | +1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kanda, Y.; Nishizawa, Y.; Kamakura, T.; Mori, M. Overexpressed BSR1-Mediated Enhancement of Disease Resistance Depends on the MAMP-Recognition System. Int. J. Mol. Sci. 2020, 21, 5397. https://doi.org/10.3390/ijms21155397
Kanda Y, Nishizawa Y, Kamakura T, Mori M. Overexpressed BSR1-Mediated Enhancement of Disease Resistance Depends on the MAMP-Recognition System. International Journal of Molecular Sciences. 2020; 21(15):5397. https://doi.org/10.3390/ijms21155397
Chicago/Turabian StyleKanda, Yasukazu, Yoko Nishizawa, Takashi Kamakura, and Masaki Mori. 2020. "Overexpressed BSR1-Mediated Enhancement of Disease Resistance Depends on the MAMP-Recognition System" International Journal of Molecular Sciences 21, no. 15: 5397. https://doi.org/10.3390/ijms21155397
APA StyleKanda, Y., Nishizawa, Y., Kamakura, T., & Mori, M. (2020). Overexpressed BSR1-Mediated Enhancement of Disease Resistance Depends on the MAMP-Recognition System. International Journal of Molecular Sciences, 21(15), 5397. https://doi.org/10.3390/ijms21155397





