Ubiquitination in the ERAD Process
Abstract
1. Introduction
2. Recognition of Target Proteins
3. Retrotranslocation of the Substrate
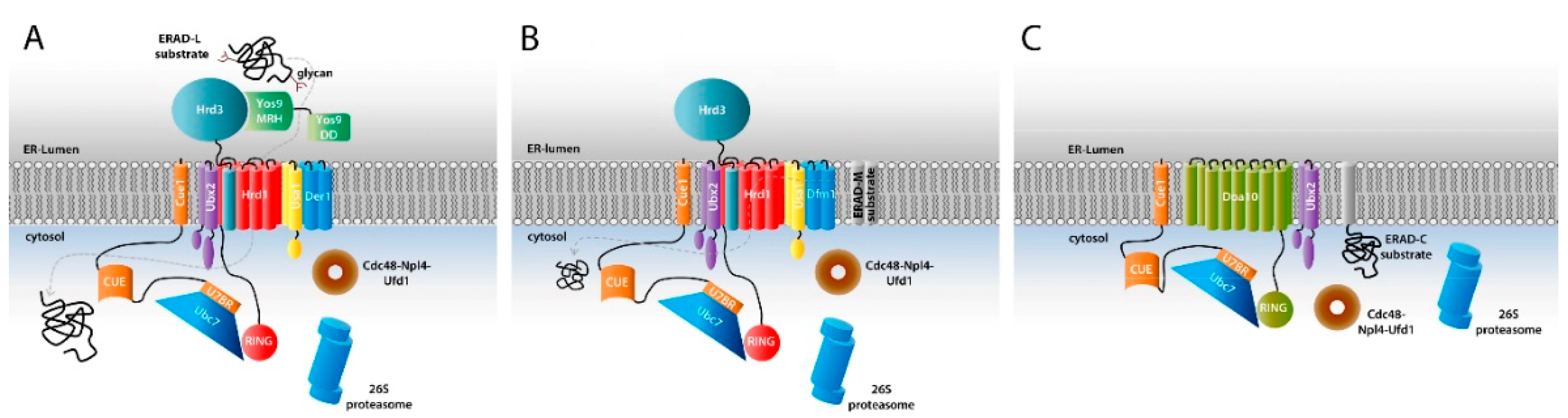
4. Components of the Ubiquitination Machinery
5. High Processivity in ERAD
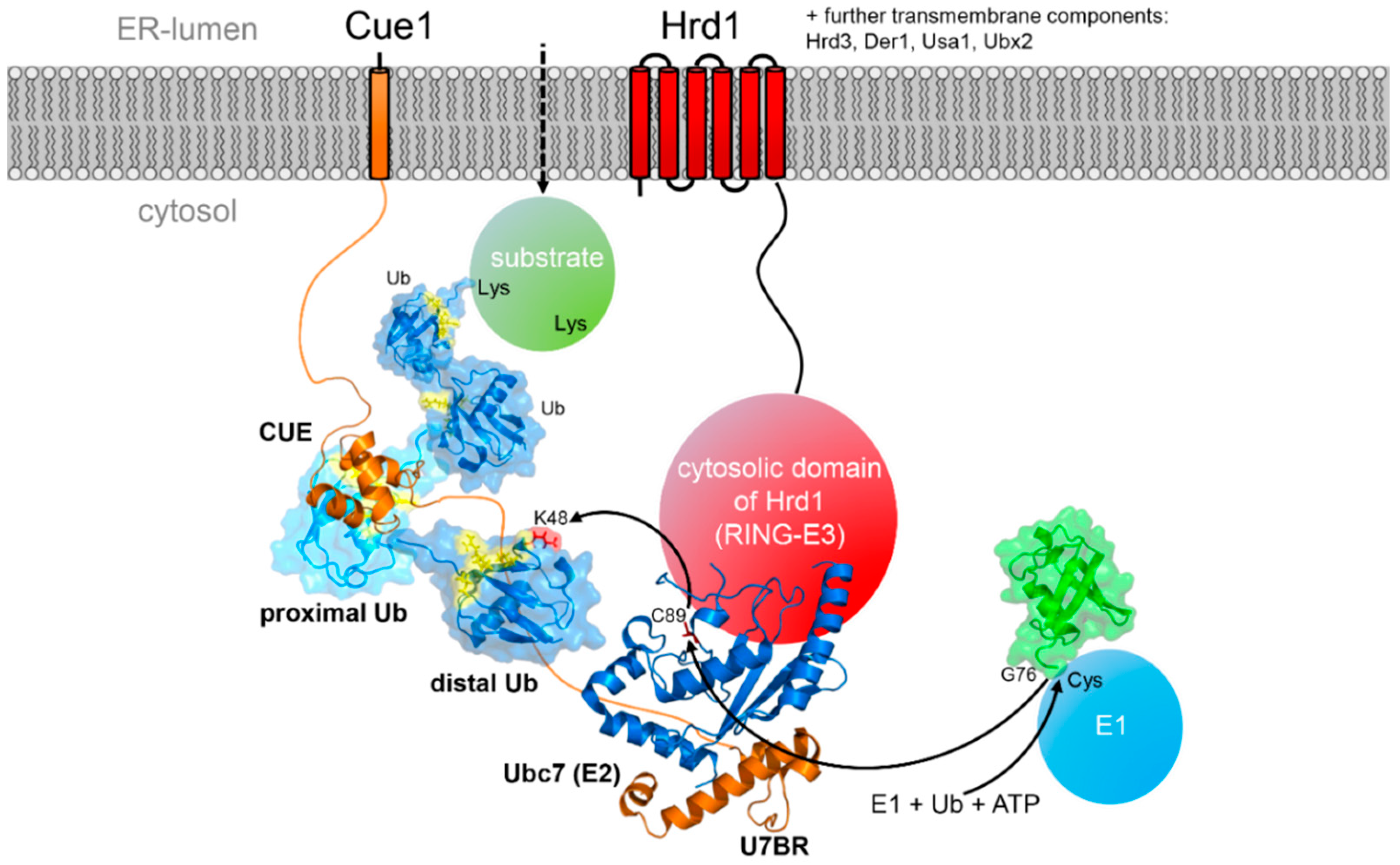
5.1. The Special Role of the E2 Conjugating Enzymes
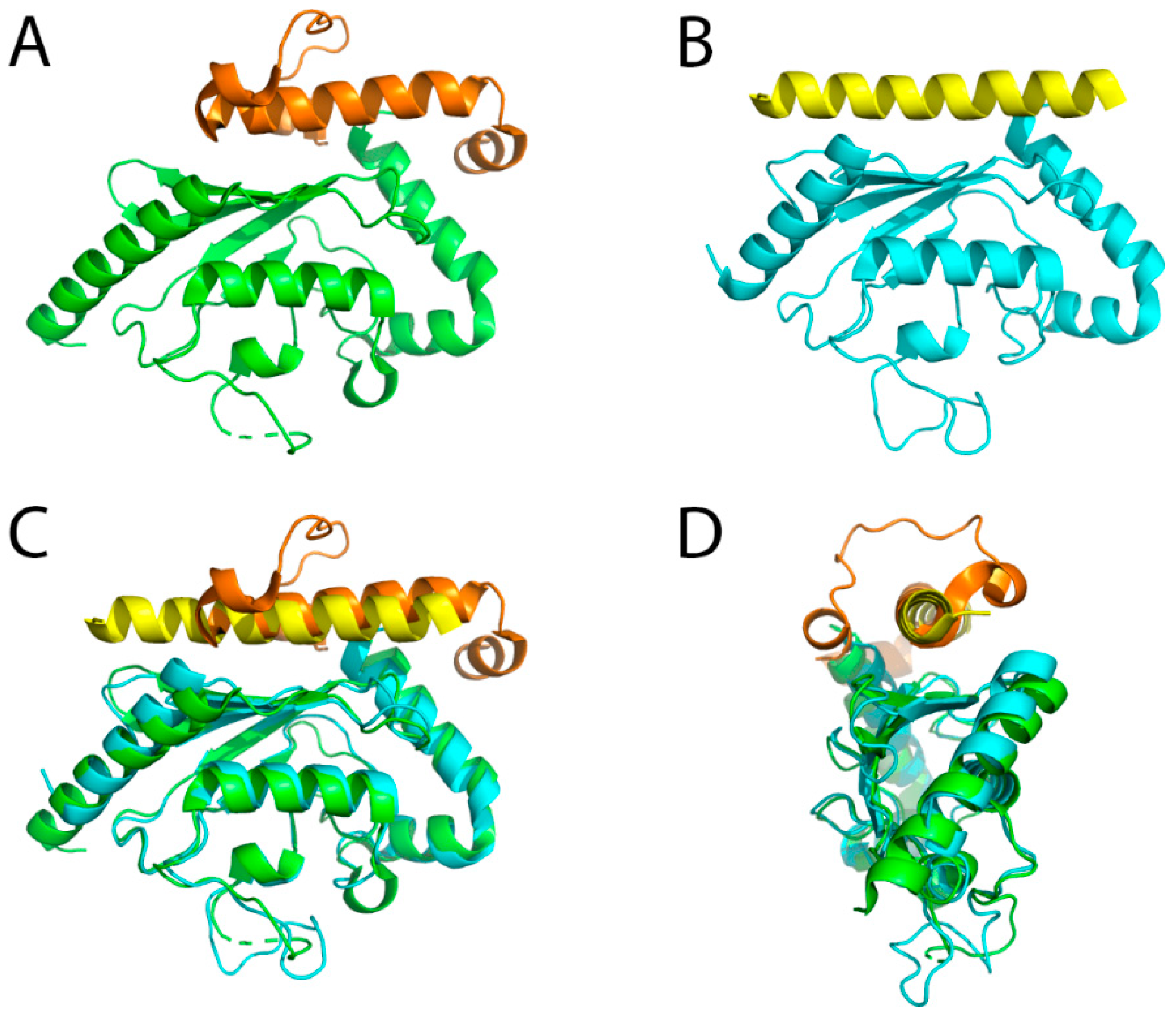
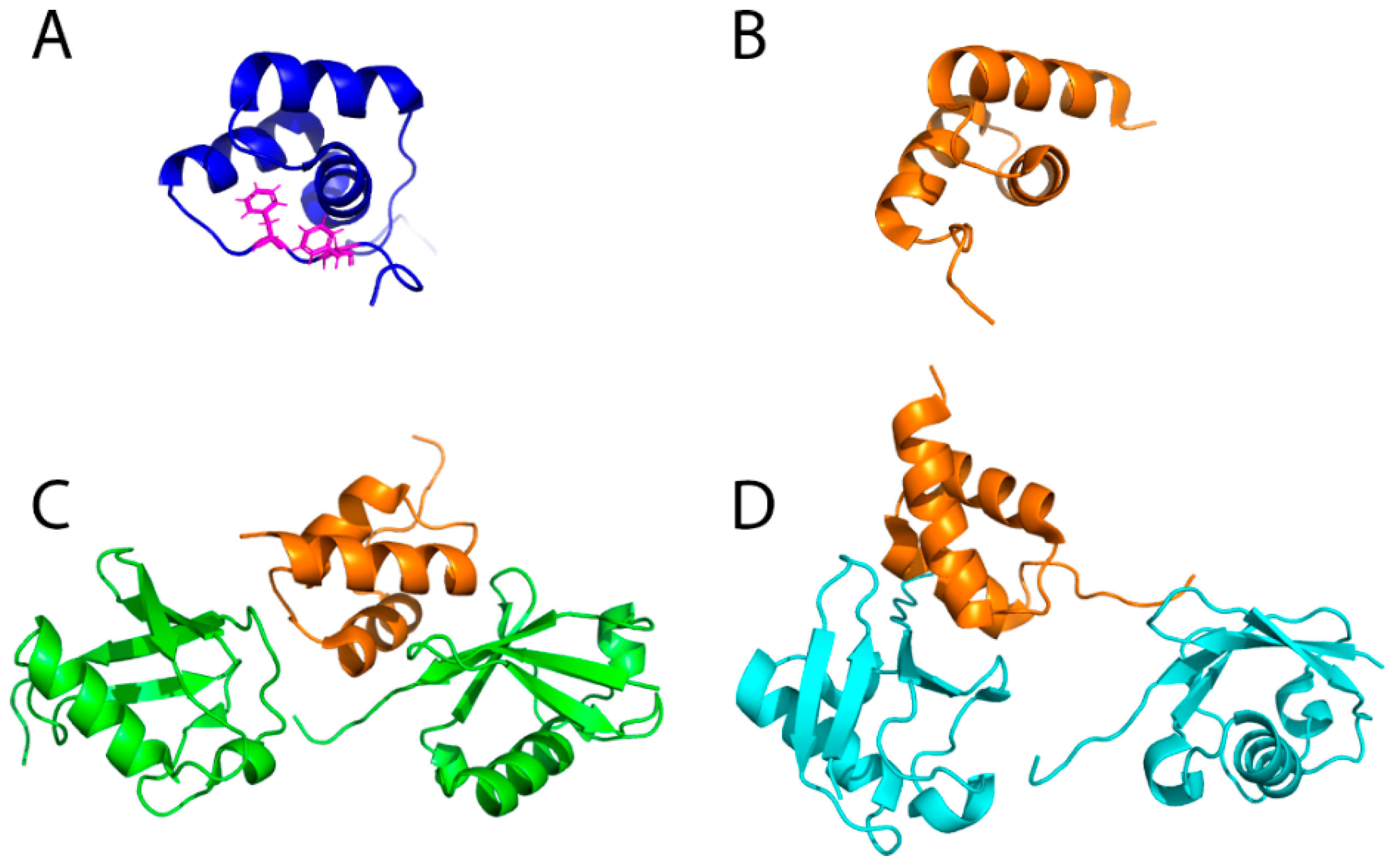
5.2. CUE Domains in the ERAD Process
5.3. Role of E4 Enzymes and Different Chain Linkages
5.4. Degradation of the Ubiquitination Machinery
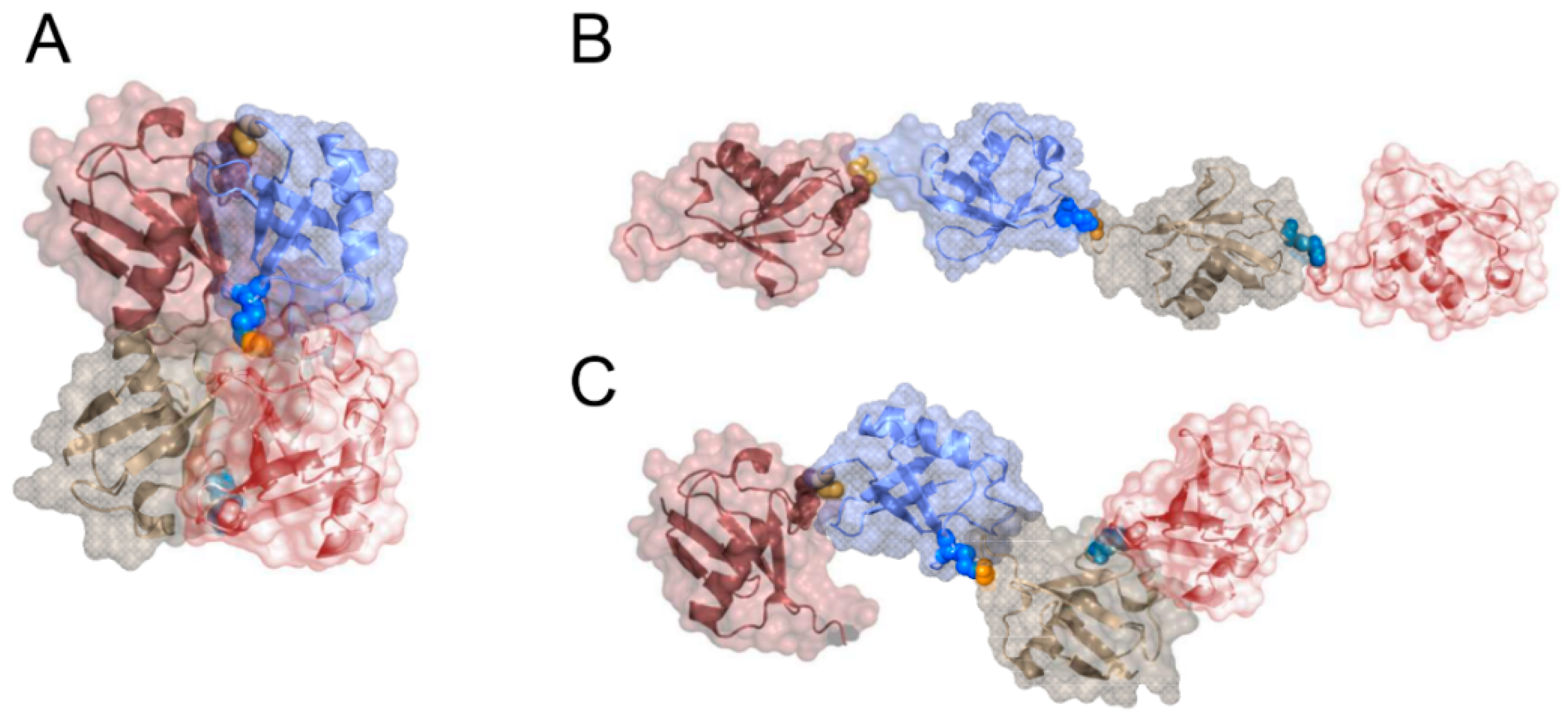
6. The Conformation of the Synthesized Chains
6.1. Binding and Modification of Ubiquitin Chains by Cdc48 and Associated Factors
6.2. Interaction with the Proteasome and the Conformational Space of K48-linked Chains
7. Concluding Remarks
Funding
Conflicts of Interest
References
- Chiti, F.; Dobson, C.M. Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade. Annu. Rev. Biochem. 2017, 86, 27–68. [Google Scholar] [CrossRef] [PubMed]
- Aguzzi, A.; Falsig, J. Prion propagation, toxicity and degradation. Nat. Neurosci. 2012, 15, 936–939. [Google Scholar] [CrossRef] [PubMed]
- Aguzzi, A.; Miele, G. Recent advances in prion biology. Curr. Opin. Neurol. 2004, 17, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Knowles, T.P.; Vendruscolo, M.; Dobson, C.M. The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 2014, 15, 384–396. [Google Scholar] [CrossRef] [PubMed]
- Pohl, C.; Dikic, I. Cellular quality control by the ubiquitin-proteasome system and autophagy. Science 2019, 366, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Gardner, B.M.; Walter, P. Unfolded Proteins Are Ire1-Activating Ligands That Directly Induce the Unfolded Protein Response. Science 2011, 333, 1891–1894. [Google Scholar] [CrossRef] [PubMed]
- Lam, M.; Lawrence, D.A.; Ashkenazi, A.; Walter, P. Confirming a critical role for death receptor 5 and caspase-8 in apoptosis induction by endoplasmic reticulum stress. Cell Death Differ. 2018, 25, 1530–1531. [Google Scholar] [CrossRef]
- Munch, C.; Harper, J.W. Mitochondrial unfolded protein response controls matrix pre-RNA processing and translation. Nature 2016, 534, 710–713. [Google Scholar] [CrossRef]
- Braakman, I.; Hebert, D.N. Protein folding in the endoplasmic reticulum. Cold Spring Harb. Perspect. Biol. 2013, 5, a013201. [Google Scholar] [CrossRef]
- Zhang, S.; Xu, C.C.; Larrimore, K.E.; Ng, D.T.W. Slp1-Emp65: A Guardian Factor that Protects Folding Polypeptides from Promiscuous Degradation. Cell 2017, 171, 346–357. [Google Scholar] [CrossRef]
- Mehrtash, A.B.; Hochstrasser, M. Ubiquitin-dependent protein degradation at the endoplasmic reticulum and nuclear envelope. Semin. Cell Dev. Biol. 2019, 93, 111–124. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.D.; Rapoport, T.A. Mechanistic insights into ER-associated protein degradation. Curr. Opin. Cell Biol. 2018, 53, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Goder, V. Roles of Ubiquitin in Endoplasmic Reticulum-Associated Protein Degradation (ERAD). Curr. Protein Pept. Sci. 2012, 13, 425–435. [Google Scholar] [CrossRef] [PubMed]
- Olzmann, J.A.; Kopito, R.R.; Christianson, J.C. The Mammalian Endoplasmic Reticulum-Associated Degradation System. Cold Spring Harb. Perspect. Biol. 2013, 5, a013185. [Google Scholar] [CrossRef]
- Preston, G.M.; Brodsky, J.L. The evolving role of ubiquitin modification in endoplasmic reticulum-associated degradation. Biochem. J. 2017, 474, 445–469. [Google Scholar] [CrossRef]
- Hirsch, C.; Gauss, R.; Horn, S.C.; Neuber, O.; Sommer, T. The ubiquitylation machinery of the endoplasmic reticulum. Nature 2009, 458, 453–460. [Google Scholar] [CrossRef]
- Aebi, M. N-linked protein glycosylation in the ER. Biochim. Biophys. Acta 2013, 1833, 2430–2437. [Google Scholar] [CrossRef]
- Braakman, I.; Bulleid, N.J. Protein folding and modification in the mammalian endoplasmic reticulum. Annu. Rev. Biochem. 2011, 80, 71–99. [Google Scholar] [CrossRef]
- Clerc, S.; Hirsch, C.; Oggier, D.M.; Deprez, P.; Jakob, C.; Sommer, T.; Aebi, M. Htm1 protein generates the N-glycan signal for glycoprotein degradation in the endoplasmic reticulum. J. Cell Biol. 2009, 184, 159–172. [Google Scholar] [CrossRef]
- Jakob, C.A.; Bodmer, D.; Spirig, U.; Battig, P.; Marcil, A.; Dignard, D.; Bergeron, J.J.; Thomas, D.Y.; Aebi, M. Htm1p, a mannosidase-like protein, is involved in glycoprotein degradation in yeast. EMBO Rep. 2001, 2, 423–430. [Google Scholar] [CrossRef]
- Quan, E.M.; Kamiya, Y.; Kamiya, D.; Denic, V.; Weibezahn, J.; Kato, K.; Weissman, J.S. Defining the glycan destruction signal for endoplasmic reticulum-associated degradation. Mol. Cell 2008, 32, 870–877. [Google Scholar] [CrossRef] [PubMed]
- Hosokawa, N.; Kamiya, Y.; Kamiya, D.; Kato, K.; Nagata, K. Human OS-9, a lectin required for glycoprotein endoplasmic reticulum-associated degradation, recognizes mannose-trimmed N-glycans. J. Biol. Chem. 2009, 284, 17061–17068. [Google Scholar] [CrossRef]
- Szathmary, R.; Bielmann, R.; Nita-Lazar, M.; Burda, P.; Jakob, C.A. Yos9 protein is essential for degradation of misfolded glycoproteins and may function as lectin in ERAD. Mol. Cell 2005, 19, 765–775. [Google Scholar] [CrossRef] [PubMed]
- Gauss, R.; Kanehara, K.; Carvalho, P.; Ng, D.T.; Aebi, M. A complex of Pdi1p and the mannosidase Htm1p initiates clearance of unfolded glycoproteins from the endoplasmic reticulum. Mol. Cell 2011, 42, 782–793. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, A.; Stephanowitz, H.; Krause, E.; Volkwein, C.; Hirsch, C.; Jarosch, E.; Sommer, T. A Complex of Htm1 and the Oxidoreductase Pdi1 Accelerates Degradation of Misfolded Glycoproteins. J. Biol. Chem. 2016, 291, 12195–12207. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.D.; Siggel, M.; Ovchinnikov, S.; Mi, W.; Svetlov, V.; Nudler, E.; Liao, M.F.; Hummer, G.; Rapoport, T.A. Structural basis of ER-associated protein degradation mediated by the Hrd1 ubiquitin ligase complex. Science 2020, 368, eaaz2449. [Google Scholar] [CrossRef]
- Schoebel, S.; Mi, W.; Stein, A.; Ovchinnikov, S.; Pavlovicz, R.; DiMaio, F.; Baker, D.; Chambers, M.G.; Su, H.Y.; Li, D.S.; et al. Cryo-EM structure of the protein-conducting ERAD channel Hrd1 in complex with Hrd3. Nature 2017, 548, 352–355. [Google Scholar] [CrossRef]
- Carvalho, P.; Goder, V.; Rapoport, T.A. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell 2006, 126, 361–373. [Google Scholar] [CrossRef]
- Denic, V.; Quan, E.M.; Weissman, J.S. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell 2006, 126, 349–359. [Google Scholar] [CrossRef]
- Horn, S.C.; Hanna, J.; Hirsch, C.; Volkwein, C.; Schutz, A.; Heinemann, U.; Sommer, T.; Jarosch, E. Usa1 Functions as a Scaffold of the HRD-Ubiquitin Ligase. Mol. Cell 2009, 36, 782–793. [Google Scholar] [CrossRef]
- Neal, S.; Jaeger, P.A.; Duttke, S.H.; Benner, C.K.; Glass, C.; Ideker, T.; Hampton, R. The Dfm1 Derlin Is Required for ERAD Retrotranslocation of Integral Membrane Proteins. Mol. Cell 2018, 69, 306–320. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, P.; Stanley, A.M.; Rapoport, T.A. Retrotranslocation of a Misfolded Luminal ER Protein by the Ubiquitin-Ligase Hrd1p. Cell 2010, 143, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Baldridge, R.D.; Rapoport, T.A. Autoubiquitination of the Hrd1 Ligase Triggers Protein Retrotranslocation in ERAD. Cell 2016, 166, 394–407. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.H.; Rape, M. Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 2009, 10, 755–764. [Google Scholar] [CrossRef]
- Chau, V.; Tobias, J.W.; Bachmair, A.; Marriott, D.; Ecker, D.J.; Gonda, D.K.; Varshavsky, A. A Multiubiquitin Chain Is Confined to Specific Lysine in a Targeted Short-Lived Protein. Science 1989, 243, 1576–1583. [Google Scholar] [CrossRef]
- Gregori, L.; Poosch, M.S.; Cousins, G.; Chau, V. A Uniform Isopeptide-Linked Multiubiquitin Chain Is Sufficient to Target Substrate for Degradation in Ubiquitin-Mediated Proteolysis. J. Biol. Chem. 1990, 265, 8354–8357. [Google Scholar]
- Braten, O.; Livneh, I.; Ziv, T.; Admon, A.; Kehat, I.; Caspi, L.H.; Gonen, H.; Bercovich, B.; Godzik, A.; Jahandideh, S.; et al. Numerous proteins with unique characteristics are degraded by the 26S proteasome following monoubiquitination. Proc. Natl. Acad. Sci. USA 2016, 113, E4639–E4647. [Google Scholar] [CrossRef]
- Shabek, N.; Herman-Bachinsky, Y.; Buchsbaum, S.; Lewinson, O.; Haj-Yahya, M.; Hejjaoui, M.; Lashuel, H.A.; Sommer, T.; Brik, A.; Ciechanover, A. The Size of the Proteasomal Substrate Determines Whether Its Degradation Will Be Mediated by Mono- or Polyubiquitylation. Mol. Cell 2012, 48, 87–97. [Google Scholar] [CrossRef]
- Xu, P.; Duong, D.M.; Seyfried, N.T.; Cheng, D.M.; Xie, Y.; Robert, J.; Rush, J.; Hochstrasser, M.; Finley, D.; Peng, J. Quantitative Proteomics Reveals the Function of Unconventional Ubiquitin Chains in Proteasomal Degradation. Cell 2009, 137, 133–145. [Google Scholar] [CrossRef]
- Chen, B.; Mariano, J.; Tsai, Y.C.; Chan, A.H.; Cohen, M.; Weissman, A.M. The activity of a human endoplasmic reticulum-associated degradation E3, gp78, requires its Cue domain, RING finger, and an E2-binding site. Proc. Natl. Acad. Sci. USA 2006, 103, 341–346. [Google Scholar] [CrossRef]
- Imai, Y.; Soda, M.; Inoue, H.; Hattori, N.; Mizuno, Y.; Takahashi, R. An unfolded putative transmembrane polypeptide, which can lead to endoplasmic reticulum stress, is a substrate of parkin. Cell 2001, 105, 891–902. [Google Scholar] [CrossRef]
- Kikkert, M.; Doolman, R.; Dai, M.; Avner, R.; Hassink, G.; van Voorden, S.; Thanedar, S.; Roitelman, J.; Chau, V.; Wiertz, E. Human HRD1 is an E3 ubiquitin ligase involved in degradation of proteins from the endoplasmic reticulum. J. Biol. Chem. 2004, 279, 3525–3534. [Google Scholar] [CrossRef] [PubMed]
- Lenk, U.; Yu, H.; Walter, J.; Gelman, M.S.; Hartmann, E.; Kopito, R.R.; Sommer, T. A role for mammalian Ubc6 homologues in ER-associated protein degradation. J. Cell Sci. 2002, 115, 3007–3014. [Google Scholar] [PubMed]
- Arteaga, M.F.; Wang, L.; Ravid, T.; Hochstrasser, M.; Canessa, C.M. An amphipathic helix targets serum and glucocorticoid-induced kinase 1 to the endoplasmic reticulum-associated ubiquitin-conjugation machinery. Proc. Natl. Acad. Sci. USA 2006, 103, 11178–11183. [Google Scholar] [CrossRef]
- Younger, J.M.; Chen, L.L.; Ren, H.Y.; Rosser, M.F.N.; Turnbull, E.L.; Fan, C.Y.; Patterson, C.; Cyr, D.M. Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell 2006, 126, 571–582. [Google Scholar] [CrossRef]
- Kreft, S.G.; Wang, L.; Hochstrasser, M. Membrane topology of the yeast endoplasmic reticulum-localized ubiquitin ligase Doa10 and comparison with its human ortholog TEB4 (MARCH-VI). J. Biol. Chem. 2006, 281, 4646–4653. [Google Scholar] [CrossRef] [PubMed]
- Foresti, O.; Rodriguez-Vaello, V.; Funaya, C.; Carvalho, P. Quality control of inner nuclear membrane proteins by the Asi complex. Science 2014, 346, 751–755. [Google Scholar] [CrossRef]
- Khmelinskii, A.; Blaszczak, E.; Pantazopoulou, M.; Fischer, B.; Omnus, D.J.; Le Dez, G.; Brossard, A.; Gunnarsson, A.; Barry, J.D.; Meurer, M.; et al. Protein quality control at the inner nuclear membrane. Nature 2014, 516, 410–413. [Google Scholar] [CrossRef]
- Fang, S.Y.; Ferrone, M.; Yang, C.H.; Jensen, J.P.; Tiwari, S.; Weissman, A.M. The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 2001, 98, 14422–14427. [Google Scholar] [CrossRef]
- Nadav, E.; Shmueli, A.; Barr, H.; Gonen, H.; Ciechanover, A.; Reiss, Y. A novel mammalian endoplasmic reticulum ubiquitin ligase homologous to the yeast Hrd1. Biochem. Biophys. Res. Commun. 2003, 303, 91–97. [Google Scholar] [CrossRef]
- Hassink, G.; Kikkert, M.; van Voorden, S.; Lee, S.J.; Spaapen, R.; van Laar, T.; Coleman, C.S.; Bartee, E.; Fruh, K.; Chau, V.; et al. TEB4 is a C4HC3 RING finger-containing ubiquitin ligase of the endoplasmic reticulum. Biochem. J. 2005, 388, 647–655. [Google Scholar] [CrossRef] [PubMed]
- Araki, K.; Nagata, K. Protein folding and quality control in the ER. Cold Spring Harb. Perspect. Biol. 2011, 3, a007526. [Google Scholar] [CrossRef] [PubMed]
- Paul, I.; Ghosh, M.K. The E3 Ligase CHIP: Insights into Its Structure and Regulation. Biomed. Res. Int. 2014, 2014, 918183. [Google Scholar] [CrossRef] [PubMed]
- Oda, Y.; Hosokawa, N.; Wada, I.; Nagata, K. EDEM as an acceptor of terminally misfolded glycoproteins released from calnexin. Science 2003, 299, 1394–1397. [Google Scholar] [CrossRef] [PubMed]
- Christianson, J.C.; Shaler, T.A.; Tyler, R.E.; Kopito, R.R. OS-9 and GRP94 deliver mutant alpha 1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat. Cell Biol. 2008, 10, 272–282. [Google Scholar] [CrossRef] [PubMed]
- Apperizeller-Herzog, C.; Ellgaard, L. The human PDI family: Versatility packed into a single fold. Biochim. Biophys. Acta 2008, 1783, 535–548. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, M.; Yasui, S.; Niinuma, Y.; Arai, K.; Omura, T.; Okuma, Y.; Nomura, Y. A different pathway in the endoplasmic reticulum stress-induced expression of human HRD1 and SEL1 genes. FEBS Lett. 2007, 581, 5355–5360. [Google Scholar] [CrossRef]
- Lilley, B.N.; Ploegh, H.L. A membrane protein required for dislocation of misfolded proteins from the ER. Nature 2004, 429, 834–840. [Google Scholar] [CrossRef]
- Schulze, A.; Standera, S.; Buerger, E.; Kikkert, M.; van Voorden, S.; Wiertz, E.; Koning, F.; Kloetzel, P.M.; Seeger, M. The ubiquitin-domain protein HERP forms a complex with components of the endoplasmic reticulum associated degradation pathway. J. Mol. Biol. 2005, 354, 1021–1027. [Google Scholar] [CrossRef]
- Rabinovich, E.; Kerem, A.; Frohlich, K.U.; Diamant, N.; Bar-Nun, S. AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol. Cell. Biol. 2002, 22, 626–634. [Google Scholar] [CrossRef]
- Nowis, D.; McConnell, E.; Wojcik, C. Destabilization of the VCP-Ufd1-Npl4 complex is associated with decreased levels of ERAD substrates. Exp. Cell Res. 2006, 312, 2921–2932. [Google Scholar] [CrossRef] [PubMed]
- Bays, N.W.; Wilhovsky, S.K.; Goradia, A.; Hodgkiss-Harlow, K.; Hampton, R.Y. HRD4/NPL4 is required for the proteasomal processing of ubiquitinated ER proteins. Mol. Biol. Cell 2001, 12, 4114–4128. [Google Scholar] [CrossRef] [PubMed]
- Zehmer, J.K.; Bartz, R.; Bisel, B.; Liu, P.S.; Seemann, J.; Anderson, R.G.W. Targeting sequences of UBXD8 and AAM-B reveal that the ER has a direct role in the emergence and regression of lipid droplets. J. Cell Sci. 2009, 122, 3694–3702. [Google Scholar] [CrossRef] [PubMed]
- Kalchman, M.A.; Graham, R.K.; Xia, G.; Koide, H.B.; Hodgson, J.G.; Graham, K.C.; Goldberg, Y.P.; Gietz, R.D.; Pickart, C.M.; Hayden, M.R. Huntingtin is ubiquitinated and interacts with a specific ubiquitin-conjugating enzyme. J. Biol. Chem. 1996, 271, 19385–19394. [Google Scholar] [CrossRef]
- Scheffner, M.; Huibregtse, J.M.; Howley, P.M. Identification of a Human Ubiquitin-Conjugating Enzyme That Mediates the E6-Ap-Dependent Ubiquitination of P53. Proc. Natl. Acad. Sci. USA 1994, 91, 8797–8801. [Google Scholar] [CrossRef]
- Katsanis, N.; Fisher, E.M.C. Identification, expression, and chromosomal localization of ubiquitin conjugating enzyme 7 (UBE2G2), a human homologue of the Saccharomyces cerevisiae Ubc7 gene. Genomics 1998, 51, 128–131. [Google Scholar] [CrossRef]
- Matsumura, Y.; Sakai, J.; Skach, W.R. Endoplasmic Reticulum Protein Quality Control Is Determined by Cooperative Interactions between Hsp/c70 Protein and the CHIP E3 Ligase. J. Biol. Chem. 2013, 288, 31069–31079. [Google Scholar] [CrossRef]
- Matsuda, N.; Suzuki, T.; Tanaka, K.; Nakano, A. Rma1, a novel type of RING finger protein conserved from Arabidopsis to human, is a membrane-bound ubiquitin ligase. J. Cell Sci. 2001, 114, 1949–1957. [Google Scholar]
- Mahoney, J.A.; Odin, J.A.; White, S.M.; Shaffer, D.; Koff, A.; Casciola-Rosen, L.; Rosen, A. The human homologue of the yeast polyubiquitination factor Ufd2p is cleaved by caspase 6 and granzyme B during apoptosis. Biochem. J. 2002, 361, 587–595. [Google Scholar] [CrossRef]
- Bohm, S.; Lamberti, G.; Fernandez-Saiz, V.; Stapf, C.; Buchberger, A. Cellular Functions of Ufd2 and Ufd3 in Proteasomal Protein Degradation Depend on Cdc48 Binding. Mol. Cell. Biol. 2011, 31, 1528–1539. [Google Scholar] [CrossRef]
- Spandl, J.; Lohmann, D.; Kuerschner, L.; Moessinger, C.; Thiele, C. Ancient Ubiquitous Protein 1 (AUP1) Localizes to Lipid Droplets and Binds the E2 Ubiquitin Conjugase G2 (Ube2g2) via Its G2 Binding Region. J. Biol. Chem. 2011, 286, 5599–5606. [Google Scholar] [CrossRef] [PubMed]
- Ernst, R.; Mueller, B.; Ploegh, H.L.; Schlieker, C. The Otubain YOD1 Is a Deubiquitinating Enzyme that Associates with p97 to Facilitate Protein Dislocation from the ER. Mol. Cell 2009, 36, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Ng, J.M.Y.; Vermeulen, W.; van der Horst, G.T.J.; Bergink, S.; Sugasawa, K.; Vrieling, H.; Hoeijmakers, J.H.J. A novel regulation mechanism of DNA repair by damage-induced and RAD23-dependent stabilization of xeroderma pigmentosum group C protein. Gene. Dev. 2003, 17, 1630–1645. [Google Scholar] [CrossRef] [PubMed]
- Chuang, K.H.; Liang, F.S.; Higgins, R.; Wang, Y.C. Ubiquilin/Dsk2 promotes inclusion body formation and vacuole (lysosome)-mediated disposal of mutated huntingtin. Mol. Biol. Cell 2016, 27, 2025–2036. [Google Scholar] [CrossRef] [PubMed]
- Weber, A.; Cohen, I.; Popp, O.; Dittmar, G.; Reiss, Y.; Sommer, T.; Ravid, T.; Jarosch, E. Sequential Poly-ubiquitylation by Specialized Conjugating Enzymes Expands the Versatility of a Quality Control Ubiquitin Ligase. Mol. Cell 2016, 63, 827–839. [Google Scholar] [CrossRef]
- Wang, X.L.; Herr, R.A.; Rabelink, M.; Hoeben, R.C.; Wiertz, E.J.H.J.; Hansen, T.H. Ube2j2 ubiquitinates hydroxylated amino acids on ER-associated degradation substrates. J. Cell Biol. 2009, 187, 655–668. [Google Scholar] [CrossRef]
- Shimizu, Y.; Okuda-Shimizu, Y.; Hendershot, L.M. Ubiquitylation of an ERAD Substrate Occurs on Multiple Types of Amino Acids. Mol. Cell 2010, 40, 917–926. [Google Scholar] [CrossRef]
- Ishikura, S.; Weissman, A.M.; Bonifacino, J.S. Serine Residues in the Cytosolic Tail of the T-cell Antigen Receptor alpha-Chain Mediate Ubiquitination and Endoplasmic Reticulum-associated Degradation of the Unassembled Protein. J. Biol. Chem. 2010, 285, 23916–23924. [Google Scholar] [CrossRef]
- Rapoport, T.A. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 2007, 450, 663–669. [Google Scholar] [CrossRef]
- Ravid, T.; Hochstrasser, M. Autoregulation of an E2 enzyme by ubiquitin-chain assembly on its catalytic residue. Nat. Cell Biol. 2007, 9, 422–427. [Google Scholar] [CrossRef]
- Li, W.; Tu, D.; Li, L.Y.; Wollert, T.; Ghirlando, R.; Brunger, A.T.; Ye, Y.H. Mechanistic insights into active site-associated polyubiquitination by the ubiquitin-conjugating enzyme Ube2g2. Proc. Natl. Acad. Sci. USA 2009, 106, 3722–3727. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Tu, D.Q.; Brunger, A.T.; Ye, Y.H. A ubiquitin ligase transfers preformed polyubiquitin chains from a conjugating enzyme to a substrate. Nature 2007, 446, 333–337. [Google Scholar] [CrossRef] [PubMed]
- Bazirgan, O.A.; Hampton, R.Y. Cue1p is an activator of Ubc7p E2 activity in vitro and in vivo. J. Biol. Chem. 2008, 283, 12797–12810. [Google Scholar] [CrossRef] [PubMed]
- Kostova, Z.; Mariano, J.; Scholz, S.; Koenig, C.; Weissman, A.M. A Ubc7p-binding domain in Cue1p activates ER-associated protein degradation. J. Cell Sci. 2009, 122, 1374–1381. [Google Scholar] [CrossRef]
- Metzger, M.B.; Liang, Y.H.; Das, R.; Mariano, J.; Li, S.J.; Li, J.; Kostova, Z.; Byrd, R.A.; Ji, X.H.; Weissman, A.M. A Structurally Unique E2-Binding Domain Activates Ubiquitination by the ERAD E2, Ubc7p, through Multiple Mechanisms. Mol. Cell 2013, 50, 516–527. [Google Scholar] [CrossRef]
- Das, R.; Mariano, J.; Tsai, Y.C.; Kalathur, R.C.; Kostova, Z.; Li, J.; Tarasov, S.G.; McFeeters, R.L.; Altieri, A.S.; Ji, X.H.; et al. Allosteric Activation of E2-RING Finger-Mediated Ubiquitylation by a Structurally Defined Specific E2-Binding Region of gp78. Mol. Cell 2009, 34, 674–685. [Google Scholar] [CrossRef]
- Ponting, C.P. Proteins of the endoplasmic-reticulum-associated degradation pathway: Domain detection and function prediction. Biochem. J. 2000, 351, 527–535. [Google Scholar] [CrossRef]
- Biederer, T.; Volkwein, C.; Sommer, T. Role of Cue1p in ubiquitination and degradation at the ER surface. Science 1997, 278, 1806–1809. [Google Scholar] [CrossRef]
- Klemm, E.J.; Spooner, E.; Ploegh, H.L. Dual Role of Ancient Ubiquitous Protein 1 (AUP1) in Lipid Droplet Accumulation and Endoplasmic Reticulum (ER) Protein Quality Control. J. Biol. Chem. 2011, 286, 37602–37614. [Google Scholar] [CrossRef]
- Prag, G.; Misra, S.; Jones, E.A.; Ghirlando, R.; Davies, B.A.; Horazdovsky, B.F.; Hurley, J.H. Mechanism of ubiquitin recognition by the CUE domain of Vps9p. Cell 2003, 113, 609–620. [Google Scholar] [CrossRef]
- Mitra, S.; Traughber, C.A.; Brannon, M.K.; Gomez, S.; Capelluto, D.G.S. Ubiquitin Interacts with the Tollip C2 and CUE Domains and Inhibits Binding of Tollip to Phosphoinositides. J. Biol. Chem. 2013, 288, 25780–25791. [Google Scholar] [CrossRef] [PubMed]
- Bagola, K.; von Delbruck, M.; Dittmar, G.; Scheffner, M.; Ziv, I.; Glickman, M.H.; Ciechanover, A.; Sommer, T. Ubiquitin binding by a CUE domain regulates ubiquitin chain formation by ERAD E3 ligases. Mol. Cell 2013, 50, 528–539. [Google Scholar] [CrossRef] [PubMed]
- Shih, S.C.; Prag, G.; Francis, S.A.; Sutanto, M.A.; Hurley, J.H.; Hicke, L. A ubiquitin-binding motif required for intramolecular monoubiquitylation, the CUE domain. EMBO J. 2003, 22, 1273–1281. [Google Scholar] [CrossRef] [PubMed]
- Kang, R.S.; Daniels, C.M.; Francis, S.A.; Shih, S.C.; Salerno, W.J.; Hicke, L.; Radhakrishnan, I. Solution structure of a CUE-ubiquitin complex reveals a conserved mode of ubiquitin binding. Cell 2003, 113, 621–630. [Google Scholar] [CrossRef]
- Liu, S.; Chen, Y.H.; Li, J.; Huang, T.; Tarasov, S.; King, A.; Weissman, A.M.; Byrd, R.A.; Das, R. Promiscuous Interactions of gp78 E3 Ligase CUE Domain with Polyubiquitin Chains. Structure 2012, 20, 2138–2150. [Google Scholar] [CrossRef] [PubMed]
- von Delbruck, M.; Kniss, A.; Rogov, V.V.; Pluska, L.; Bagola, K.; Lohr, F.; Guntert, P.; Sommer, T.; Dotsch, V. The CUE Domain of Cue1 Aligns Growing Ubiquitin Chains with Ubc7 for Rapid Elongation. Mol. Cell 2016, 62, 918–928. [Google Scholar] [CrossRef]
- Choi, Y.S.; Lee, Y.J.; Lee, S.Y.; Shi, L.; Ha, J.H.; Cheong, H.K.; Cheong, C.; Cohen, R.E.; Ryu, K.S. Differential ubiquitin binding by the acidic loops of Ube2g1 and Ube2r1 enzymes distinguishes their Lys-48-ubiquitylation activities. J. Biol. Chem. 2015, 290, 2251–2263. [Google Scholar] [CrossRef]
- Meyer, H.J.; Rape, M. Enhanced Protein Degradation by Branched Ubiquitin Chains. Cell 2014, 157, 910–921. [Google Scholar] [CrossRef]
- Wickliffe, K.E.; Lorenz, S.; Wemmer, D.E.; Kuriyan, J.; Rape, M. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell 2011, 144, 769–781. [Google Scholar] [CrossRef]
- Liu, W.X.; Shang, Y.L.; Li, W. gp78 elongates of polyubiquitin chains from the distal end through the cooperation of its G2BR and CUE domains. Sci. Rep. 2014, 4, 7138. [Google Scholar] [CrossRef]
- Gao, F.Y.; Shang, Y.L.; Liu, W.X.; Li, W. The linkage specificity determination of Ube2g2-gp78 mediated polyubiquitination. Biochem. Biophys. Res. Commun. 2016, 473, 1139–1143. [Google Scholar] [CrossRef]
- Brzovic, P.S.; Lissounov, A.; Christensen, D.E.; Hoyt, D.W.; Klevit, R.E. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol. Cell 2006, 21, 873–880. [Google Scholar] [CrossRef] [PubMed]
- Buetow, L.; Gabrielsen, M.; Anthony, N.G.; Dou, H.; Patel, A.; Aitkenhead, H.; Sibbet, G.J.; Smith, B.O.; Huang, D.T. Activation of a Primed RING E3-E2-Ubiquitin Complex by Non-Covalent Ubiquitin. Mol. Cell 2015, 58, 297–310. [Google Scholar] [CrossRef] [PubMed]
- Brown, N.G.; Watson, E.R.; Weissmann, F.; Jarvis, M.A.; VanderLinden, R.; Grace, C.R.R.; Frye, J.J.; Qiao, R.P.; Dube, P.; Petzold, G.; et al. Mechanism of Polyubiquitination by Human Anaphase-Promoting Complex: RING Repurposing for Ubiquitin Chain Assembly. Mol. Cell 2014, 56, 246–260. [Google Scholar] [CrossRef] [PubMed]
- Wright, J.D.; Mace, P.D.; Day, C.L. Secondary ubiquitin-RING docking enhances Arkadia and Ark2C E3 ligase activity. Nat. Struct. Mol. Biol. 2016, 23, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Koegl, M.; Hoppe, T.; Schlenker, S.; Ulrich, H.D.; Mayer, T.U.; Jentsch, S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 1999, 96, 635–644. [Google Scholar] [CrossRef]
- Liu, C.; Liu, W.X.; Ye, Y.H.; Li, W. Ufd2p synthesizes branched ubiquitin chains to promote the degradation of substrates modified with atypical chains. Nat. Commun. 2017, 8, 14274. [Google Scholar] [CrossRef]
- Kim, W.; Bennett, E.J.; Huttlin, E.L.; Guo, A.; Li, J.; Possemato, A.; Sowa, M.E.; Rad, R.; Rush, J.; Comb, M.J.; et al. Systematic and Quantitative Assessment of the Ubiquitin-Modified Proteome. Mol. Cell 2011, 44, 325–340. [Google Scholar] [CrossRef]
- Gardner, R.G.; Swarbrick, G.M.; Bays, N.W.; Cronin, S.R.; Wilhovsky, S.; Seelig, L.; Kim, C.; Hampton, R. Endoplasmic reticulum degradation requires lumen to cytosol signaling: Transmembrane control of Hrd1p by Hrd3p. J. Cell Biol. 2000, 151, 69–82. [Google Scholar] [CrossRef]
- Carroll, S.M.; Hampton, R.Y. Usa1p Is Required for Optimal Function and Regulation of the Hrd1p Endoplasmic Reticulum-associated Degradation Ubiquitin Ligase. J. Biol. Chem. 2010, 285, 5146–5156. [Google Scholar] [CrossRef]
- Plemper, R.K.; Bordallo, J.; Deak, P.M.; Taxis, C.; Hitt, R.; Wolf, D.H. Genetic interactions of Hrd3p and Der3p/Hrd1p with Sec61p suggest a retro-translocation complex mediating protein transport for ER degradation. J. Cell Sci. 1999, 112, 4123–4134. [Google Scholar] [PubMed]
- Nakatsukasa, K.; Brodsky, J.L.; Kamura, T. A stalled retrotranslocation complex reveals physical linkage between substrate recognition and proteasomal degradation during ER-associated degradation. Mol. Biol. Cell 2013, 24, 1765–1775. [Google Scholar] [CrossRef] [PubMed]
- Gauss, R.; Sommer, T.; Jarosch, E. The Hrd1p ligase complex forms a linchpin between ER-lumenal substrate selection and Cdc48p recruitment. EMBO J. 2006, 25, 1827–1835. [Google Scholar] [CrossRef] [PubMed]
- Walter, J.; Urban, J.; Volkwein, C.; Sommer, T. Sec61p-independent degradation of the tail-anchored ER membrane protein Ubc6p. EMBO J. 2001, 20, 3124–3131. [Google Scholar] [CrossRef]
- Kreft, S.G.; Hochstrasser, M. An Unusual Transmembrane Helix in the Endoplasmic Reticulum Ubiquitin Ligase Doa10 Modulates Degradation of Its Cognate E2 Enzyme. J. Biol. Chem. 2011, 286, 20163–20174. [Google Scholar] [CrossRef] [PubMed]
- Claessen, J.H.L.; Mueller, B.; Spooner, E.; Pivorunas, V.L.; Ploegh, H.L. The Transmembrane Segment of a Tail-anchored Protein Determines Its Degradative Fate through Dislocation from the Endoplasmic Reticulum. J. Biol. Chem. 2010, 285, 20732–20739. [Google Scholar] [CrossRef]
- Lam, S.Y.; Murphy, C.; Foley, L.A.; Ross, S.A.; Wang, T.C.; Fleming, J.V. The human ubiquitin conjugating enzyme UBE2J2 (Ubc6) is a substrate for proteasomal degradation. Biochem. Biophys. Res. Commun. 2014, 451, 361–366. [Google Scholar] [CrossRef]
- Varshavsky, A. N-degron and C-degron pathways of protein degradation. Proc. Natl. Acad. Sci. USA 2019, 116, 358–366. [Google Scholar] [CrossRef]
- Flierman, D.; Ye, Y.H.; Dai, M.; Chau, V.; Rapoport, T.A. Polyubiquitin serves as a recognition signal, rather than a ratcheting molecule, during retrotranslocation of proteins across the endoplasmic reticulum membrane. J. Biol. Chem. 2003, 278, 34774–34782. [Google Scholar] [CrossRef]
- Meyer, H.H.; Wang, Y.Z.; Warren, G. Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and Ufd1-Npl4. EMBO J. 2002, 21, 5645–5652. [Google Scholar] [CrossRef]
- Meyer, H.H.; Shorter, J.G.; Seemann, J.; Pappin, D.; Warren, G. A complex of mammalian Ufd1 and Npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. EMBO J. 2000, 19, 2181–2192. [Google Scholar] [CrossRef] [PubMed]
- Neuber, O.; Jarosch, E.; Volkwein, C.; Walter, J.; Sommer, T. Ubx2 links the Cdc48 complex to ER-associated protein degradation. Nat. Cell Biol. 2005, 7, 993–998. [Google Scholar] [CrossRef]
- Stein, A.; Ruggiano, A.; Carvalho, P.; Rapoport, T.A. Key Steps in ERAD of Luminal ER Proteins Reconstituted with Purified Components. Cell 2014, 158, 1375–1388. [Google Scholar] [CrossRef]
- Schuberth, C.; Buchberger, A. Membrane-bound Ubx2 recruits Cdc48 to ubiquitin ligases and their substrates to ensure efficient ER-associated protein degradation. Nat. Cell Biol. 2005, 7, 999–1006. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Isaacson, R.; Kim, H.T.; Silver, P.A.; Wagner, G. Ufd1 exhibits the AAA-ATPase fold with two distinct ubiquitin interaction sites. Structure 2005, 13, 995–1005. [Google Scholar] [CrossRef] [PubMed]
- Rape, M.; Hoppe, T.; Gorr, I.; Kalocay, M.; Richly, H.; Jentsch, S. Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell 2001, 107, 667–677. [Google Scholar] [CrossRef]
- Ye, Y.H.; Meyer, H.H.; Rapoport, T.A. Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: Dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J. Cell Biol. 2003, 162, 71–84. [Google Scholar] [CrossRef]
- Nakatsukasa, K.; Huyer, G.; Michaelis, S.; Brodsky, J.L. Dissecting the ER-Associated degradation of a misfolded polytopic membrane protein. Cell 2008, 132, 101–112. [Google Scholar] [CrossRef]
- Liu, C.; van Dyk, D.; Xu, P.; Choe, V.; Pan, H.H.; Peng, J.M.; Andrews, B.; Rao, H. Ubiquitin Chain Elongation Enzyme Ufd2 Regulates a Subset of Doa10 Substrates. J. Biol. Chem. 2010, 285, 10265–10272. [Google Scholar] [CrossRef]
- Rumpf, S.; Jentsch, S. Functional division of substrate processing cofactors of the ubiquitin-selective Cdc48 chaperone. Mol. Cell 2006, 21, 261–269. [Google Scholar] [CrossRef]
- Richly, H.; Rape, M.; Braun, S.; Rumpf, S.; Hoege, C.; Jentsch, S. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell 2005, 120, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Bodnar, N.O.; Rapoport, T.A. Molecular Mechanism of Substrate Processing by the Cdc48 ATPase Complex. Cell 2017, 169, 722–735. [Google Scholar] [CrossRef] [PubMed]
- Medicherla, B.; Kostova, Z.; Schaefer, A.; Wolf, D.H. A genomic screen identifies Dsk2p and Rad23p as essential components of ER-associated degradation. EMBO Rep. 2004, 5, 692–697. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.; Mi, K.X.; Rao, H. Multiple interactions of Rad23 suggest a mechanism for ubiquitylated substrate delivery important in proteolysis. Mol. Biol. Cell 2004, 15, 3357–3365. [Google Scholar] [CrossRef]
- Elsasser, S.; Gali, R.R.; Schwickart, M.; Larsen, C.N.; Leggett, D.S.; Muller, B.; Feng, M.T.; Tubing, F.; Dittmar, G.A.; Finley, D. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat. Cell Biol. 2002, 4, 725–730. [Google Scholar] [CrossRef]
- Elsasser, S.; Chandler-Militello, D.; Muller, B.; Hanna, J.; Finley, D. Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J. Biol. Chem. 2004, 279, 26817–26822. [Google Scholar] [CrossRef]
- Husnjak, K.; Elsasser, S.; Zhang, N.X.; Chen, X.; Randles, L.; Shi, Y.; Hofmann, K.; Walters, K.J.; Finley, D.; Dikic, I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 2008, 453, 481–488. [Google Scholar] [CrossRef]
- Schreiner, P.; Chen, X.; Husnjak, K.; Randles, L.; Zhang, N.X.; Elsasser, S.; Finley, D.; Dikic, I.; Walters, K.J.; Groll, M. Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature 2008, 453, 548–552. [Google Scholar] [CrossRef]
- Chen, L.; Shinde, U.; Ortolan, T.G.; Madura, K. Ubiquitin-associated (UBA) domains in Rad23 bind ubiquitin and promote inhibition of multi-ubiquitin chain assembly. EMBO Rep. 2001, 2, 933–938. [Google Scholar] [CrossRef]
- Lam, Y.A.; Lawson, T.G.; Velayutham, M.; Zweier, J.L.; Pickart, C.M. A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature 2002, 416, 763–767. [Google Scholar] [CrossRef]
- Thrower, J.S.; Hoffman, L.; Rechsteiner, M.; Pickart, C.M. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000, 19, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Yau, R.; Rape, M. The increasing complexity of the ubiquitin code. Nat. Cell Biol. 2016, 18, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.T.; Ciechanover, A. The Ubiquitin Code in the Ubiquitin-Proteasome System and Autophagy. Trends Biochem. Sci. 2017, 42, 873–886. [Google Scholar] [CrossRef] [PubMed]
- Varadan, R.; Walker, O.; Pickart, C.; Fushman, D. Structural properties of polyubiquitin chains in solution. J. Mol. Biol. 2002, 324, 637–647. [Google Scholar] [CrossRef]
- Ryabov, Y.; Fushman, D. Interdomain mobility in di-ubiquitin revealed by NMR. Proteins 2006, 63, 787–796. [Google Scholar] [CrossRef]
- Cook, W.J.; Jeffrey, L.C.; Carson, M.; Chen, Z.; Pickart, C.M. Structure of a diubiquitin conjugate and a model for interaction with ubiquitin conjugating enzyme (E2). J. Biol. Chem. 1992, 267, 16467–16471. [Google Scholar] [CrossRef]
- Eddins, M.J.; Varadan, R.; Flushman, D.; Pickart, C.M.; Wolberger, C. Crystal structure and solution NMR studies of Lys48-linked tetraubiquitin at neutral pH. J. Mol. Biol. 2007, 367, 204–211. [Google Scholar] [CrossRef]
- Ye, Y.; Blaser, G.; Horrocks, M.H.; Ruedas-Rama, M.J.; Ibrahim, S.; Zhukov, A.A.; Orte, A.; Klenerman, D.; Jackson, S.E.; Komander, D. Ubiquitin chain conformation regulates recognition and activity of interacting proteins. Nature 2012, 492, 266–270. [Google Scholar] [CrossRef]
- Hirano, T.; Serve, O.; Yagi-Utsumi, M.; Takemoto, E.; Hiromoto, T.; Satoh, T.; Mizushima, T.; Kato, K. Conformational Dynamics of Wild-type Lys-48-linked Diubiquitin in Solution. J. Biol. Chem. 2011, 286, 37496–37502. [Google Scholar] [CrossRef]
- Fushman, D.; Walker, O. Exploring the Linkage Dependence of Polyubiquitin Conformations Using Molecular Modeling. J. Mol. Biol. 2010, 395, 803–814. [Google Scholar] [CrossRef][Green Version]
- Weeks, S.D.; Grasty, K.C.; Hernandez-Cuebas, L.; Loll, P.J. Crystal structures of Lys-63-linked tri- and di-ubiquitin reveal a highly extended chain architecture. Proteins-Struct. Funct. Bioinformatics 2009, 77, 753–759. [Google Scholar] [CrossRef] [PubMed]
- Datta, A.B.; Hura, G.L.; Wolberger, C. The Structure and Conformation of Lys63-Linked Tetraubiquitin. J. Mol. Biol. 2009, 392, 1117–1124. [Google Scholar] [CrossRef] [PubMed]
- Komander, D.; Reyes-Turcu, F.; Licchesi, J.D.F.; Odenwaelder, P.; Wilkinson, K.D.; Barford, D. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 2009, 10, 466–473. [Google Scholar] [CrossRef] [PubMed]
- Varadan, R.; Assfalg, M.; Haririnia, A.; Raasi, S.; Pickart, C.; Fushman, D. Solution conformation of Lys63-linked di-ubiquitin chain provides clues to functional diversity of polyubiquitin signaling. J. Biol. Chem. 2004, 279, 7055–7063. [Google Scholar] [CrossRef] [PubMed]
- Tenno, T.; Fujiwara, K.; Tochio, H.; Iwai, K.; Morita, E.H.; Hayashi, H.; Murata, S.; Hiroaki, H.; Sato, M.; Tanaka, K.; et al. Structural basis for distinct roles of Lys63- and Lys48-linked polyubiquitin chains. Genes Cells 2004, 9, 865–875. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, M.L.; Wickliffe, K.E.; Dong, K.C.; Yu, C.; Bosanac, I.; Bustos, D.; Phu, L.; Kirkpatrick, D.S.; Hymowitz, S.G.; Rape, M.; et al. K11-Linked Polyubiquitination in Cell Cycle Control Revealed by a K11 Linkage-Specific Antibody. Mol. Cell 2010, 39, 477–484. [Google Scholar] [CrossRef]
- Bremm, A.; Freund, S.M.V.; Komander, D. Lys11-linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne. Nat. Struct. Mol. Biol. 2010, 17, 939–947. [Google Scholar] [CrossRef]
- Castaneda, C.A.; Kashyap, T.R.; Nakasone, M.A.; Krueger, S.; Fushman, D. Unique Structural, Dynamical, and Functional Properties of K11-Linked Polyubiquitin Chains. Structure 2013, 21, 1168–1181. [Google Scholar] [CrossRef]
- Zhang, Y.; Vukovic, L.; Rudack, T.; Han, W.; Schulten, K. Recognition of Poly-Ubiquitins by the Proteasome through Protein Refolding Guided by Electrostatic and Hydrophobic Interactions. J. Phys. Chem. B 2016, 120, 8137–8146. [Google Scholar] [CrossRef]
- Kravtsova-Ivantsiv, Y.; Cohen, S.; Ciechanover, A. Modification by Single Ubiquitin Moieties Rather Than Polyubiquitination Is Sufficient for Proteasomal Processing of the p105 NF-kappa B Precursor. Mol. Cell 2009, 33, 496–504. [Google Scholar] [CrossRef]
- Cook, B.W.; Lacoursiere, R.E.; Shaw, G.S. Recruitment of Ubiquitin within an E2 Chain Elongation Complex. Biophys. J. 2020, 118, 1679–1689. [Google Scholar] [CrossRef] [PubMed]
- Phillips, C.L.; Thrower, J.; Pickart, C.M.; Hill, C.P. Structure of a new crystal form of tetraubiquitin. Acta Crystallogr. D 2001, 57, 341–344. [Google Scholar] [CrossRef] [PubMed]
- Kniss, A.; Schuetz, D.; Kazemi, S.; Pluska, L.; Spindler, P.E.; Rogov, V.V.; Husnjak, K.; Dikic, I.; Guntert, P.; Sommer, T.; et al. Chain Assembly and Disassembly Processes Differently Affect the Conformational Space of Ubiquitin Chains. Structure 2018, 26, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Berg, A.; Franke, L.; Scheffner, M.; Peter, C. Machine Learning Driven Analysis of Large Scale Simulations Reveals Conformational Characteristics of Ubiquitin Chains. J. Chem. Theory Comput. 2020, 16, 3205–3220. [Google Scholar] [CrossRef] [PubMed]
- Younger, J.M.; Ren, H.Y.; Chen, L.L.; Fan, C.Y.; Fields, A.; Patterson, C.; Cyr, D.M. A foldable CFTR Delta F508 biogenic intermediate accumulates upon inhibition of the Hsc70-CHIP E3 ubiquitin ligase. J. Cell Biol. 2004, 167, 1075–1085. [Google Scholar] [CrossRef] [PubMed]
- Grove, D.E.; Fan, C.Y.; Ren, H.Y.; Cyr, D.M. The endoplasmic reticulum-associated Hsp40 DNAJB12 and Hsc70 cooperate to facilitate RMA1 E3-dependent degradation of nascent CFTR Delta F508. Mol. Biol. Cell 2011, 22, 301–314. [Google Scholar] [CrossRef]
- Morito, D.; Hirao, K.; Oda, Y.; Hosokawa, N.; Tokunaga, F.; Cyr, D.M.; Tanaka, K.; Iwai, K.; Nagata, K. Gp78 cooperates with RMA1 in endoplasmic reticulum-associated degradation of CFTR Delta F508. Mol. Biol. Cell 2008, 19, 1328–1336. [Google Scholar] [CrossRef]

| Yeast | Human Homologue | Reference |
|---|---|---|
| Target recognition | ||
| Htm1/Mnl1 | EDEM1-3 | [54] |
| Yos9 | OS-9 | [55] |
| Pdi1 | Pdi | [56] |
| Hrd3 | Sel1L | [57] |
| Retrotranslocation | ||
| Der1 | Derlin1 | [58] |
| Dfm1 | Derlin1 | [58] |
| Usa1 | HERP | [59] |
| Cdc48 | p97 | [60] |
| Ufd1 | Ufd1 | [61] |
| Npl4 | Npl4 | [62] |
| Ubx2 | Ubxd8 | [63] |
| E2 ubiquitin conjugating enzymes | ||
| Ubc1 | Ube2k | [64] |
| Ubc4 | Ube2d1 | [65] |
| Ubc6 | Ube2j1-2 | [43] |
| Ubc7 | Ube2g1-2 | [66] |
| E3 ubiquitin ligase enzymes | ||
| Hrd1 | Hrd1 | [57] |
| Doa10 | TEB4 | [51] |
| Asi | ||
| gp78 | [49] | |
| CHIP | [67] | |
| RMA1 | [68] | |
| E4 ubiquitin chain elongation factors | ||
| Ufd2 | Ufd2 | [69] |
| Doa1 | Ufd3 | [70] |
| Cue1 | ||
| AUP1 | [71] | |
| Deubiquitinase | ||
| Otu1 | Yod1 | [72] |
| Escort factors | ||
| Rad23 | Hr23a-b | [73] |
| Dsk2 | Ubiquilin-1 | [74] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lopata, A.; Kniss, A.; Löhr, F.; Rogov, V.V.; Dötsch, V. Ubiquitination in the ERAD Process. Int. J. Mol. Sci. 2020, 21, 5369. https://doi.org/10.3390/ijms21155369
Lopata A, Kniss A, Löhr F, Rogov VV, Dötsch V. Ubiquitination in the ERAD Process. International Journal of Molecular Sciences. 2020; 21(15):5369. https://doi.org/10.3390/ijms21155369
Chicago/Turabian StyleLopata, Anna, Andreas Kniss, Frank Löhr, Vladimir V. Rogov, and Volker Dötsch. 2020. "Ubiquitination in the ERAD Process" International Journal of Molecular Sciences 21, no. 15: 5369. https://doi.org/10.3390/ijms21155369
APA StyleLopata, A., Kniss, A., Löhr, F., Rogov, V. V., & Dötsch, V. (2020). Ubiquitination in the ERAD Process. International Journal of Molecular Sciences, 21(15), 5369. https://doi.org/10.3390/ijms21155369





