The Feasibility of Host Transcriptome Profiling as a Diagnostic Tool for Microbial Etiology in Childhood Cancer Patients with Febrile Neutropenia
Abstract
1. Introduction
2. Results
2.1. Clinical and Microbiological Findings in the Episodes with Sufficient RNA for Sequencing
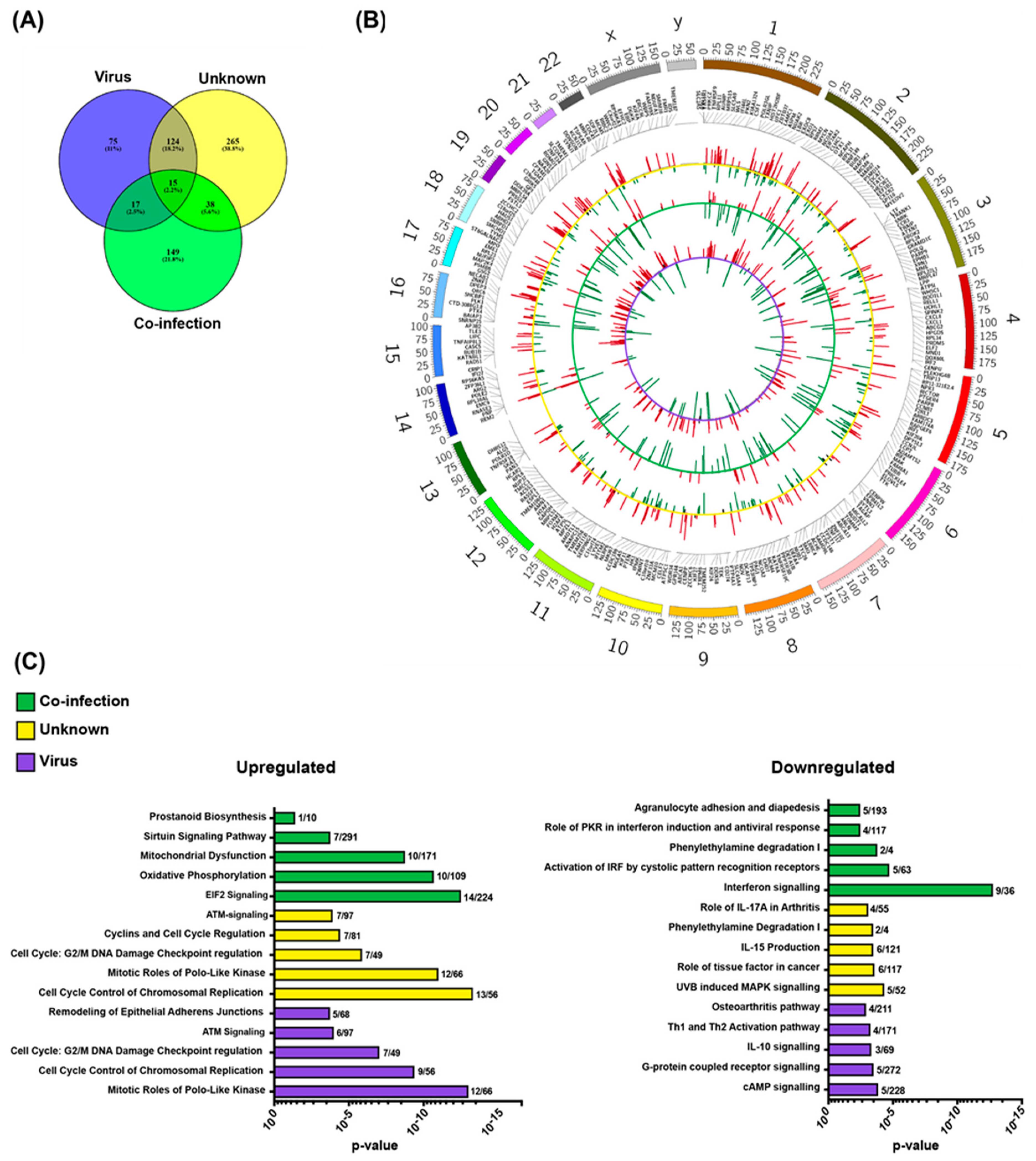
2.2. Pathogen-Related Blood Transcriptomes in Immunosuppressed Children with Neutropenia
3. Discussion
4. Materials and Methods
4.1. Study Cohort
4.2. Blood Sampling, Microbiological Testing and RNA Preparation
4.3. Microbiological Documented Infections and Grouping of Patients
4.4. RNA Preparation, Sequencing and Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| FN | Febrile Neutropenia |
| PAMPs | pathogen-associated molecular patterns |
| PPRs | pattern-recognition receptors |
| INFs | Interferons |
| RNA-seq | RNA sequencing |
| WBC | white blood cell |
| ANC | absolute neutrophil counts |
| CRP | C-reactive protein |
References
- Lehrnbecher, T.; Robinson, P.; Fisher, B.; Alexander, S.; Ammann, R.A.; Beauchemin, M.; Carlesse, F.; Groll, A.H.; Haeusler, G.M.; Santolaya, M.; et al. Guideline for the management of fever and neutropenia in children with cancer and hematopoietic stem-cell transplantation recipients: 2017 update. J. Clin. Oncol. 2017, 35, 2082–2094. [Google Scholar] [CrossRef] [PubMed]
- Agyeman, P.; Kontny, U.; Nadal, D.; Leibundgut, K.; Niggli, F.; Simon, A.; Kronenberg, A.; Frei, R.; Escobar, H.; Kuhne, T.; et al. A prospective multicenter study of microbiologically defined infections in pediatric cancer patients with fever and neutropenia: Swiss Pediatric Oncology Group 2003 fever and neutropenia study. Pediatr. Infect. Dis. J. 2014, 33, e219–e225. [Google Scholar] [CrossRef] [PubMed]
- Lindblom, A.; Bhadri, V.; Soderhall, S.; Ohrmalm, L.; Wong, M.; Norbeck, O.; Lindau, C.; Rotzen-Ostlund, M.; Allander, T.; Catchpoole, D.; et al. Respiratory viruses, a common microbiological finding in neutropenic children with fever. J. Clin. Virol. 2010, 47, 234–237. [Google Scholar] [CrossRef] [PubMed]
- Castagnola, E.; Fontana, V.; Caviglia, I.; Caruso, S.; Faraci, M.; Fioredda, F.; Garre, M.L.; Moroni, C.; Conte, M.; Losurdo, G.; et al. A prospective study on the epidemiology of febrile episodes during chemotherapy-induced neutropenia in children with cancer or after hemopoietic stem cell transplantation. Clin. Infect. Dis. 2007, 45, 1296–1304. [Google Scholar] [CrossRef]
- Torres, J.P.; De la Maza, V.; Kors, L.; Villarroel, M.; Piemonte, P.; Izquierdo, G.; Salgado, C.; Tordecilla, J.; Contardo, V.; Farfan, M.J.; et al. Respiratory viral infections and coinfections in children with cancer, fever and neutropenia: Clinical outcome of infections caused by different respiratory viruses. Pediatr. Infect. Dis. J. 2016, 35, 949–954. [Google Scholar] [CrossRef] [PubMed]
- Suryadevara, M.; Tabarani, C.M.; Bartholoma, N.; Rosenberg, H.F.; Domachowske, J.B. Nasopharyngeal detection of respiratory viruses in febrile neutropenic children. Clin. Pediatr. (Phila) 2012, 51, 1164–1167. [Google Scholar] [CrossRef]
- Rhedin, S.; Lindstrand, A.; Rotzen-Ostlund, M.; Tolfvenstam, T.; Ohrmalm, L.; Rinder, M.R.; Zweygberg-Wirgart, B.; Ortqvist, A.; Henriques-Normark, B.; Broliden, K.; et al. Clinical utility of PCR for common viruses in acute respiratory illness. Pediatrics 2014, 133, e538–e545. [Google Scholar] [CrossRef]
- Soderman, M.; Rhedin, S.; Tolfvenstam, T.; Rotzen-Ostlund, M.; Albert, J.; Broliden, K.; Lindblom, A. Frequent respiratory viral infections in children with febrile neutropenia—A prospective follow-up study. PLoS ONE 2016, 11, e0157398. [Google Scholar] [CrossRef]
- Saito, T.; Gale, M., Jr. Principles of intracellular viral recognition. Curr. Opin. Immunol. 2007, 19, 17–23. [Google Scholar] [CrossRef]
- Zaas, A.K.; Chen, M.; Varkey, J.; Veldman, T.; Hero, A.O., III; Lucas, J.; Huang, Y.; Turner, R.; Gilbert, A.; Lambkin-Williams, R.; et al. Gene expression signatures diagnose influenza and other symptomatic respiratory viral infections in humans. Cell Host Microbe 2009, 6, 207–217. [Google Scholar] [CrossRef]
- Hu, X.; Yu, J.; Crosby, S.D.; Storch, G.A. Gene expression profiles in febrile children with defined viral and bacterial infection. Proc. Natl. Acad. Sci. USA 2013, 110, 12792–12797. [Google Scholar] [CrossRef]
- Herberg, J.A.; Kaforou, M.; Wright, V.J.; Shailes, H.; Eleftherohorinou, H.; Hoggart, C.J.; Cebey-Lopez, M.; Carter, M.J.; Janes, V.A.; Gormley, S.; et al. Diagnostic test accuracy of a 2-transcript host RNA signature for discriminating bacterial vs viral infection in febrile children. JAMA 2016, 316, 835–845. [Google Scholar] [CrossRef]
- Mahajan, P.; Kuppermann, N.; Mejias, A.; Suarez, N.; Chaussabel, D.; Casper, T.C.; Smith, B.; Alpern, E.R.; Anders, J.; Atabaki, S.M.; et al. Association of RNA biosignatures with bacterial infections in febrile infants aged 60 days or younger. JAMA 2016, 316, 846–857. [Google Scholar] [CrossRef] [PubMed]
- Kelly, R.S.; Lasky-Su, J.; Yeung, S.J.; Stone, R.M.; Caterino, J.M.; Hagan, S.C.; Lyman, G.H.; Baden, L.R.; Glotzbecker, B.E.; Coyne, C.J.; et al. Integrative omics to detect bacteremia in patients with febrile neutropenia. PLoS ONE 2018, 13, e0197049. [Google Scholar] [CrossRef] [PubMed]
- Bodey, G.P.; Buckley, M.; Sathe, Y.S.; Freireich, E.J. Quantitative relationships between circulating leukocytes and infection in patients with acute leukemia. Ann. Intern. Med. 1966, 64, 328–340. [Google Scholar] [CrossRef]
- Davis, K.; Wilson, S. Febrile neutropenia in paediatric oncology. Paediatr. Child Health 2020, 30, 93–97. [Google Scholar] [CrossRef] [PubMed]
- Hakim, H.; Dallas, R.; Zhou, Y.; Pei, D.; Cheng, C.; Flynn, P.M.; Pui, C.H.; Jeha, S. Acute respiratory infections in children and adolescents with acute lymphoblastic leukemia. Cancer 2016, 122, 798–805. [Google Scholar] [CrossRef] [PubMed]
- Torres, J.P.; Labrana, Y.; Ibanez, C.; Kasaneva, P.; Farfan, M.J.; De la Maza, V.; Villarroel, M.; Vergara, I.; Piemonte, P.; Zubieta, M.; et al. Frequency and clinical outcome of respiratory viral infections and mixed viral-bacterial infections in children with cancer, fever and neutropenia. Pediatr. Infect. Dis. J. 2012, 31, 889–893. [Google Scholar] [CrossRef]
- Heinonen, S.; Jartti, T.; Garcia, C.; Oliva, S.; Smitherman, C.; Anguiano, E.; de Steenhuijsen Piters, W.A.; Vuorinen, T.; Ruuskanen, O.; Dimo, B.; et al. Rhinovirus detection in symptomatic and asymptomatic children: Value of host transcriptome analysis. Am. J. Respir. Crit. Care Med. 2016, 193, 772–782. [Google Scholar] [CrossRef]
- Yu, J.; Peterson, D.R.; Baran, A.M.; Bhattacharya, S.; Wylie, T.N.; Falsey, A.R.; Mariani, T.J.; Storch, G.A. Host gene expression in nose and blood for the diagnosis of viral respiratory infection. J. Infect. Dis. 2019, 219, 1151–1161. [Google Scholar] [CrossRef]
- Barral-Arca, R.; Pardo-Seco, J.; Martinon-Torres, F.; Salas, A. A 2-transcript host cell signature distinguishes viral from bacterial diarrhea and it is influenced by the severity of symptoms. Sci. Rep. 2018, 8, 8043. [Google Scholar] [CrossRef] [PubMed]
- Holcomb, Z.E.; Tsalik, E.L.; Woods, C.W.; McClain, M.T. Host-based peripheral blood gene expression analysis for diagnosis of infectious diseases. J. Clin. Microbiol. 2017, 55, 360–368. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Carballa, A.; Cebey-Lopez, M.; Pardo-Seco, J.; Barral-Arca, R.; Rivero-Calle, I.; Pischedda, S.; Curras-Tuala, M.J.; Gomez-Rial, J.; Barros, F.; Martinon-Torres, F.; et al. A qPCR expression assay of IFI44L gene differentiates viral from bacterial infections in febrile children. Sci. Rep. 2019, 9, 11780. [Google Scholar] [CrossRef] [PubMed]
- Marotta, C.; Di Gennaro, F.; Pizzol, D.; Madeira, G.; Monno, L.; Saracino, A.; Putoto, G.; Casuccio, A.; Mazzucco, W. The at risk child clinic (ARCC): 3 years of health activities in support of the most vulnerable children in Beira, Mozambique. Int. J. Environ. Res. Public Health 2018, 15, 1350. [Google Scholar] [CrossRef] [PubMed]
- Tiveljung-Lindell, A.; Rotzen-Ostlund, M.; Gupta, S.; Ullstrand, R.; Grillner, L.; Zweygberg-Wirgart, B.; Allander, T. Development and implementation of a molecular diagnostic platform for daily rapid detection of 15 respiratory viruses. J. Med. Virol. 2009, 81, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]

| Sufficient RNA for RNA-seq N = 43 | Insufficient RNA for RNA-seq N = 20 | p-Value | |
|---|---|---|---|
| Age, median (range) | 7.6, (0.8–16.1) | 8.25, (0.9–12.3) | 0.83 |
| Gender (n girl) (%) | 23 (53) | 10 (50) | 1 |
| Hematological malignancy, n (%) | 16 (37) | 13 (65) | 0.06 |
| WBC a (109/L), median (range) | 1.6 (0.2–14.6) | 0.3 (0.1–0.7) | <0.001 |
| ANC b (109/L), median (range) | 0.1 (<0.1 c–10.9) | <0.1 c (<0.1–<0.1) | <0.001 |
| Temp max, median (range) | 39.2 (38.1–40.5) | 39.0 (38.1–40.2) | 0.67 |
| CRP max, median (range) | 73 (4–412) | 117 (41–341) | <0.01 |
| Days with antibiotics, median (range) | 7 (0–30) | 8.5 (5–17) | <0.05 |
| Respiratory virus infection, n (%) | 15 (35) | 3 (15) | 0.14 |
| Unknown etiology, n (%) | 22 (51) | 13 (65) | 0.42 |
| Co-infection, n (%) | 4 (9) | 1 (5) | 1.0 |
| Bacterial infection, n (%) | 2 (5) | 3 (15) | 0.32 |
| Viral Infection *,a n = 15 | Unknown Etiology *,b n = 22 | Co-Infection * n = 4 | Bacterial Infection *,c n = 2 | Control * n = 12 | |
|---|---|---|---|---|---|
| Age (Median, range; IQR) | 9.9 (0.8–16.1) | 7.5 (0.5–16.0) | 7.1 (3.4–10.1) | 3.0 (1.5–4.5) | 9.7 (0.6–15.8) |
| Gender (n girl) (%) | 10 (67) | 11 (50) | 1 (25) | 1 (50) | 8 (67) |
| Hematological d, n (%) | 6 (40) | 8 (36) | 0 | 2 (100) | 5 (42) |
| Hight intensity treatment n (%) | 6 (40) | 9 (41) | 2 (50) | 2 (100) | 6 (50) |
| Medium intensity treatment, n (%) | 9 (60) | 13 (59) | 2 (50) | 0 | 6 (50) |
| WBC e count (109/L) (Median, range) | 1.3 (0.2–3.8) | 1.7 (0.2–14.6) | 3.1 (1.3–4.5) | 1.0 (0.3–1.6) | 3.7 (0.7–0.6) |
| ANC e,f (109/L) (Median, range) | 0.1 (<0.1–1.3) | 0.2 (<0.1–10.9) | <0.1 (<0.1–0.4) | <0.1 (<0.1–<0.1) | 2.1 (0.1–5.6) |
| Temp max (median, range) | 39.1 (38.1–40.5) | 39.1 (38.1–40.2) | 39.2 (38.6–39.8) | 39.2 (39.4–40.3) | N/A |
| CRP max (median, range) | 66 (6–240) | 67 (4–412) | 86 (22–257) | 174 (141–207) | N/A |
| Days with neutropenia g (median range) | 6 (2–37) | 9 (1–>30) | 5.5 (4–13) | 255 (20–>30) | N/A |
| Days with fever (median, range) | 2 (1–6) | 2 (1–16) | 1.5 (1–3) | 3 (2–4) | N/A |
| Days with antibiotics (median range) | 7 (0–10) | 7 (0–30) | 8 (7–13) | 15.5 (14–17) | N/A |
| Days at hospital (median, range) | 5 (0–9) | 5 (2–30) | 5 (3–17) | 9.5 (8–11) | N/A |
| Respiratory symptoms, n (%) | 13 (87) | 14 (64) | 4 (100) | 1 (50) | N/A |
| Gastrointestinal symptoms, n (%) | 3 (20) | 3 (14) | 2 (50) | 0 (0) | N/A |
| Local symptoms, n (%) | 1 (7) | 2 (9) | 2 (50) | 1 (50) | N/A |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wahlund, M.; Sinha, I.; Broliden, K.; Saghafian-Hedengren, S.; Nilsson, A.; Berggren, A. The Feasibility of Host Transcriptome Profiling as a Diagnostic Tool for Microbial Etiology in Childhood Cancer Patients with Febrile Neutropenia. Int. J. Mol. Sci. 2020, 21, 5305. https://doi.org/10.3390/ijms21155305
Wahlund M, Sinha I, Broliden K, Saghafian-Hedengren S, Nilsson A, Berggren A. The Feasibility of Host Transcriptome Profiling as a Diagnostic Tool for Microbial Etiology in Childhood Cancer Patients with Febrile Neutropenia. International Journal of Molecular Sciences. 2020; 21(15):5305. https://doi.org/10.3390/ijms21155305
Chicago/Turabian StyleWahlund, Martina, Indranil Sinha, Kristina Broliden, Shanie Saghafian-Hedengren, Anna Nilsson, and Anna Berggren. 2020. "The Feasibility of Host Transcriptome Profiling as a Diagnostic Tool for Microbial Etiology in Childhood Cancer Patients with Febrile Neutropenia" International Journal of Molecular Sciences 21, no. 15: 5305. https://doi.org/10.3390/ijms21155305
APA StyleWahlund, M., Sinha, I., Broliden, K., Saghafian-Hedengren, S., Nilsson, A., & Berggren, A. (2020). The Feasibility of Host Transcriptome Profiling as a Diagnostic Tool for Microbial Etiology in Childhood Cancer Patients with Febrile Neutropenia. International Journal of Molecular Sciences, 21(15), 5305. https://doi.org/10.3390/ijms21155305





