Gut Microbiome Modulation for Preventing and Treating Pediatric Food Allergies
Abstract
1. Introduction
2. Gut Microbiome Dysbiosis and Food Allergy
2.1. Evidence from Human Studies
- -
- -
- Microbiome structure early in life, particularly in the first 6 months of life, is more relevant in FA development [14];
- -
- Dysbiosis could influence not only the occurrence, but also the disease course of FA, as suggested by different gut microbiome features comparing children who outgrow FA with patients with persistent forms of FA [14].
2.2. Evidence from Animal Studies
3. Gut Microbiome: Immune and Nonimmune Mechanisms of Action against Food Allergy
3.1. Mechanisms of Action at the Cellular Level
3.2. Metabolic Level: Immunoregulatory Metabolites
4. Targeting Gut Microbiome in Food Allergy
4.1. Environmental Factors
4.2. Probiotics
4.3. Prebiotics
4.4. Postbiotics
4.5. Synbiotics
4.6. Fecal Microbiota Transplantation
5. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ALDH | Aldehyde dehydrogenases |
| BLG | Beta lactoglobulin |
| CMA | Cow’s milk allergy |
| DCs | Dendritic cells |
| EHCF | Extensively hydrolyzed casein formula |
| FA | Food allergy |
| FoxP3 | Forkhead box P3 |
| FOS | Fructo-oligosaccharides |
| Fbp2 | Fructose-Bisphosphatase 2 |
| GATA-3 | GATA-binding protein 3 |
| GOS | Galacto-oligosaccharides |
| GPCRs | G-protein coupled receptors |
| HDAC | Histone deacetylases |
| IECs | Intestinal epithelial cells |
| IFNγ | Interferon gamma |
| IL | Interleukin |
| ILC | Innate lymphoid cells |
| LGG | Lactobacillus rhamnosus GG |
| NLRP3 | NOD-, LRR- and pyrin domain-containing protein 3 |
| OTUs | Operational taxonomic units |
| PBMCs | Peripheral blood mononuclear cells |
| RA | Retinoic acid |
| RALDH | Retinaldehyde dehydrogenases |
| RORγt | Retinoic acid-related orphan receptor γt |
| Ror2 | Receptor Tyrosine Kinase Like Orphan Receptor 2 |
| SCFAs | Short-chain fatty acids |
| TGFβ | Transforming growth factor-beta |
| Tgfbr3 | Transforming Growth Factor Beta Receptor 3 |
| Th | T helper |
| Treg | T regulatory |
References
- Sicherer, S.H.; Sampson, H.A. Food allergy: A review and update on epidemiology, pathogenesis, diagnosis, prevention, and management. J. Allergy Clin. Immunol. 2018, 141, 41–58. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Holdford, D.; Bilaver, L.; Dyer, A.; Holl, J.L.; Meltzer, D. The economic impact of childhood food allergy in the United States. JAMA Pediatr. 2013, 167, 1026–1031. [Google Scholar] [CrossRef] [PubMed]
- Rook, G.A. 99th Dahlem conference on infection, inflammation and chronic inflammatory disorders: Darwinian medicine and the ‘hygiene’ or ‘old friends’ hypothesis. Clin. Exp. Immunol. 2010, 160, 70–79. [Google Scholar] [PubMed]
- Rook, G.A.; Lowry, C.A.; Raison, C.L. Microbial ‘Old Friends’, immunoregulation and stress resilience. Evol. Med. Public Health 2013, 2013, 46–64. [Google Scholar] [CrossRef]
- Hanski, I.; von Hertzen, L.; Fyhrquist, N.; Koskinen, K.; Torppa, K.; Laatikainen, T.; Karisola, P.; Auvinen, P.; Paulin, L.; Mäkelä, M.J.; et al. Environmental biodiversity, human microbiota, and allergy are interrelated. Proc. Natl. Acad. Sci. USA 2012, 109, 8334–8339. [Google Scholar] [CrossRef]
- Iweala, O.I.; Nagler, C.R. The Microbiome and Food Allergy. Ann. Rev. Immunol. 2019, 37, 377–403. [Google Scholar] [CrossRef]
- Thompson-Chagoyan, O.C.; Vieites, J.M.; Maldonado, J.; Edwards, C.; Gil, A. Changes in faecal microbiota of infants with cow’s milk protein allergy—A Spanish prospective case-control 6-month follow-up study. Pediatr. Allergy Immunol. 2010, 21, e394–e400. [Google Scholar] [CrossRef]
- Bjorksten, B.; Naaber, P.; Sepp, E.; Mikelsaar, M. The intestinal microflora in allergic Estonian and Swedish 2-year-old children. Clin. Exp. Allergy 1999, 29, 342–346. [Google Scholar]
- Bunyavanich, S.; Schadt, E.E. Systems biology of asthma and allergic diseases: A multiscale approach. J. Allergy Clin. Immunol. 2015, 135, 31–42. [Google Scholar] [CrossRef]
- Ling, Z.; Li, Z.; Liu, X.; Cheng, Y.; Luo, Y.; Tong, X.; Yuan, L.; Wang, Y.; Sun, J.; Li, L.; et al. Altered fecal microbiota composition associated with food allergy in infants. Appl. Environ. Microbiol. 2014, 80, 2546–2554. [Google Scholar]
- Azad, M.B.; Konya, T.; Guttman, D.S.; Field, C.J.; Sears, M.R.; HayGlass, K.T.; Mandhane, P.J.; Turvey, S.E.; Subbarao, P.; Becker, A.B.; et al. Infant gut microbiota and food sensitization: Associations in the first year of life. Clin. Exp. Allergy 2015, 45, 632–643. [Google Scholar] [CrossRef]
- Chen, C.C.; Chen, K.J.; Kong, M.S.; Chang, H.J.; Huang, J.L. Alterations in the gut microbiota of children with food sensitization in early life. Pediatr. Allergy Immunol. 2016, 27, 254–262. [Google Scholar] [CrossRef]
- Berni Canani, R.; Sangwan, N.; Stefka, A.; Nocerino, R.; Paparo, L.; Aitoro, R.; Calignano, A.; Khan, A.A.; Gilbert, J.A.; Nagler, C.R. Lactobacillus rhamnosus GG-supplemented formula expands butyrate-producing bacterial strains in food allergic infants. ISME J. 2016, 10, 742–750. [Google Scholar] [CrossRef] [PubMed]
- Bunyavanich, S.; Shen, N.; Grishin, A.; Wood, R.; Burks, W.; Dawson, P.; Jones, S.M.; Leung, D.; Sampson, H.; Sicherer, S.; et al. Early–life gut microbiome composition and milk allergy resolution. J. Allergy Clin. Immunol. 2016, 138, 1122–1130. [Google Scholar] [CrossRef] [PubMed]
- Inoue, R.; Sawai, T.; Sawai, C.; Nakatani, M.; Romero-Pérez, G.A.; Ozeki, M.; Nonomura, K.; Tsukahara, T. A preliminary study of gut dysbiosis in children with food allergy. Biosci. Biotechnol. Biochem. 2017, 81, 2396–2399. [Google Scholar] [CrossRef] [PubMed]
- Kourosh, A.; Luna, R.A.; Balderas, M.; Nance, C.; Anagnostou, A.; Devaraj, S.; Davis, C.M. Fecal microbiome signatures are different in food– allergic children compared to siblings and healthy children. Pediatr. Allergy Immunol. 2018, 29, 545–554. [Google Scholar] [CrossRef]
- Fazlollahi, M.; Chun, Y.; Grishin, A.; Wood, R.A.; Burks, A.W.; Dawson, P.; Jones, S.M.; Leung, D.; Sampson, H.A.; Sicherer, S.H.; et al. Early–life gut microbiome and egg allergy. Allergy 2018, 73, 1515–1524. [Google Scholar] [CrossRef]
- Dong, P.; Feng, J.J.; Yan, D.Y.; Lyu, Y.J.; Xu, X. Early–life gut microbiome and cow’s milk allergy—A prospective case—Control 6–month follow–up study. Saudi J. Biol. Sci. 2018, 25, 875–880. [Google Scholar] [CrossRef]
- Savage, J.H.; Lee-Sarwar, K.A.; Sordillo, J.; Bunyavanich, S.; Zhou, Y.; O’Connor, G.; Sandel, M.; Bacharier, L.B.; Zeiger, R.; Sodergren, E.; et al. A prospective microbiome-wide association study of food sensitization and food allergy in early childhood. Allergy 2018, 73, 145–152. [Google Scholar] [CrossRef]
- Díaz, M.; Guadamuro, L.; Espinosa–Martos, I.; Mancabelli, L.; Jiménez, S.; Molinos-Norniella, C.; Pérez-Solis, D.; Milani, C.; Rodríguez, J.M.; Ventura, M.; et al. Microbiota and derived parameters in fecal samples of infants with non–IgE Cow’s milk protein allergy under a restricted diet. Nutrients 2018, 10, 1481. [Google Scholar] [CrossRef]
- Mennini, M.; Fierro, V.; Di Nardo, G.; Pecora, V.; Fiocchi, A. Microbiota in non-IgE-mediated food allergy. Curr. Opin. Allergy Clin. Immunol. 2020, 20, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Berni Canani, R.; De Filippis, F.; Nocerino, R.; Paparo, L.; Di Scala, C.; Cosenza, L.; Della Gatta, G.; Calignano, A.; De Caro, C.; Laiola, M.; et al. Gut microbiota composition and butyrate production in children affected by non-IgE-mediated cow’s milk allergy. Sci. Rep. 2018, 8, 12500. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, J.; Kobayashi, T.; Tanaka, S.; Korenori, Y.; Tateyama, A.; Sakamoto, N.; Kiyohara, C.; Shirakawa, T.; Sonomoto, K. Aberrant structures of fecal bacterial community in allergic infants profiled by 16S rRNA gene pyrosequencing. FEMS Immunol. Med. Microbiol. 2011, 63, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Russell, S.L.; Gold, M.J.; Hartmann, M.; Willing, B.P.; Thorson, L.; Wlodarska, M.; Gill, N.; Blanchet, M.R.; Mohn, W.W.; McNagny, K.M.; et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 2012, 13, 440–447. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, B.; Prioult, G.; Bibiloni, R.; Nicolis, I.; Mercenier, A.; Butel, M.J.; Waligora-Dupriet, A.J. Germ free status and altered caecal subdominant microbiota are associated with a high susceptibility to cow’s milk allergy in mice. FEMS Microbiol. Ecol. 2011, 76, 133–144. [Google Scholar] [CrossRef]
- Bashir, M.E.; Louie, S.; Shi, H.N.; Nagler-Anderson, C. Toll-like receptor 4 signaling by intestinal microbes influences susceptibility to food allergy. J. Immunol. 2004, 172, 6978–6987. [Google Scholar] [CrossRef]
- Hazebrouck, S.; Przybylski-Nicaise, L.; Ah-Leung, S.; Adel-Patient, K.; Corthier, G.; Wal, J.M.; Rabot, S. Allergic sensitization to bovine beta-lactoglobulin: Comparison between germ-free and conventional BALB/c mice. Int. Arch. Allergy Immunol. 2009, 148, 65–72. [Google Scholar] [CrossRef]
- Winkler, P.; Ghadimi, D.; Schrezenmeir, J.; Kraehenbuhl, J.P. Molecular and cellular basis of microflora-host interactions. J. Nutr. 2007, 137, 756S–772S. [Google Scholar] [CrossRef]
- Noval Rivas, M.; Burton, O.T.; Wise, P.; Zhang, Y.Q.; Hobson, S.A.; Garcia Lloret, M.; Chehoud, C.; Kuczynski, J.; DeSantis, T.; Warrington, J.; et al. A microbiota signature associated with experimental food allergy promotes allergic sensitization and anaphylaxis. J. Allergy Clin. Immunol. 2013, 131, 201–212. [Google Scholar] [CrossRef]
- Stefka, A.T.; Feehley, T.; Tripathi, P.; Qiu, J.; McCoy, K.; Mazmanian, S.K.; Tjota, M.Y.; Seo, G.Y.; Cao, S.; Theriault, B.R.; et al. Commensal bacteria protect against food allergen sensitization. Proc. Natl. Acad. Sci. USA 2014, 111, 13145–13150. [Google Scholar] [CrossRef]
- Feehley, T.; Plunkett, C.H.; Bao, R.; Choi Hong, S.M.; Culleen, E.; Belda-Ferre, P.; Campbell, E.; Aitoro, R.; Nocerino, R.; Paparo, L.; et al. Healthy infants harbor intestinal bacteria that protect against food allergy. Nat. Med. 2019, 25, 448–453. [Google Scholar] [CrossRef]
- Atarashi, K.; Tanoue, T.; Oshima, K.; Suda, W.; Nagano, Y.; Nishikawa, H.; Fukuda, S.; Saito, T.; Narushima, S.; Hase, K.; et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013, 500, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Atarashi, K.; Tanoue, T.; Shima, T.; Imaoka, A.; Kuwahara, T.; Momose, Y.; Cheng, G.; Yamasaki, S.; Saito, T.; Ohba, Y.; et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011, 331, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Ohnmacht, C.; Park, J.; Cording, S.; Wing, J.B.; Atarashi, K.; Obata, Y.; Gaboriau-Routhiau, V.; Marques, R.; Dulauroy, S.; Fedoseeva, M.; et al. The microbiota regulates type 2 immunity through RORgt+ T cells. Science 2015, 349, 989–993. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Gadir, A.; Stephen-Victor, E.; Gerber, G.K.; Noval Rivas, M.; Wang, S.; Harb, H.; Wang, L.; Li, N.; Crestani, E.; Spielman, S.; et al. Microbiota therapy acts via a regulatory T cell MyD88/RORγt pathway to suppress food allergy. Nat. Med. 2019, 25, 1164–1174. [Google Scholar] [CrossRef]
- Wang, S.; Charbonnier, L.M.; Noval Rivas, M.; Georgiev, P.; Li, N.; Gerber, G.; Bry, L.; Chatila, T.A. MyD88 adaptor-dependent microbial sensing by regulatory T cells promotes mucosal tolerance and enforces commensalism. Immunity 2015, 43, 289–303. [Google Scholar] [CrossRef] [PubMed]
- Kubinak, J.L.; Petersen, C.; Stephens, W.Z.; Soto, R.; Bake, E.; O’Connell, R.M.; Round, J.L. MyD88 signaling in T cells directs IgAmediated of the microbiota to promote health. Cell Host Microbe 2015, 17, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, G.P.; Ladinsky, M.S.; Yu, K.B.; Sanders, J.G.; Yoo, B.B.; Chou, W.C.; Conner, M.E.; Earl, A.M.; Knight, R.; Bjorkman, P.J.; et al. Gut microbiota utilize immunoglobulin A for mucosal colonization. Science 2018, 360, 795–800. [Google Scholar] [CrossRef]
- Tait Wojno, E.D.; Artis, D. Emerging concepts and future challenges in innate lymphoid cell biology. J. Exp. Med. 2016, 213, 2229–2248. [Google Scholar] [CrossRef]
- Eyerich, K.; Dimartino, V. IL-17 and IL-22 in immunity: Driving protection and pathology. Eur. J. Immunol. 2017, 47, 607–614. [Google Scholar] [CrossRef]
- Wesemann, D.R.; Portuguese, A.J. Microbial colonization influences early B-lineage development in the gut lamina propria. Nature 2013, 501, 112–115. [Google Scholar] [CrossRef] [PubMed]
- Rosser, E.C.; Mauri, C. Regulatory B cells: Origin, phenotype, and function. Immunity 2015, 42, 607–612. [Google Scholar] [CrossRef] [PubMed]
- Van de Veen, W.; Stanic, B. IgG4 production is confined to human IL-10-producing regulatory B cells that suppress antigen-specific immune responses. J. Allergy Clin. Immunol. 2013, 131, 1204–1212. [Google Scholar] [CrossRef]
- McKenzie, C.; Tan, J.; Macia, L.; Mackay, C.R. The nutrition–gut microbiome–physiology axis and allergic diseases. Immunol. Rev. 2017, 278, 277–295. [Google Scholar] [CrossRef] [PubMed]
- Dhondalay, G.K.; Rael, E.; Acharya, S.; Zhang, W.; Sampath, V.; Galli, S.J.; Tibshirani, R.; Boyd, S.D.; Maecker, H.; Nadeau, K.C.; et al. Food allergy and omics. J. Allergy Clin. Immunol. 2018, 141, 20–29. [Google Scholar] [CrossRef]
- Berni Canani, R.; Di Costanzo, M.; Leone, L.; Pedata, M.; Meli, R.; Calignano, A. Potential beneficial effects of butyrate in intestinal and extraintestinal diseases. World J. Gastroenterol. 2011, 17, 1519–1528. [Google Scholar] [CrossRef]
- Hamer, H.M.; Jonkers, D.; Venema, K.; Vanhoutvin, S.; Troost, F.J.; Brummer, R.J. Review article: The role of butyrate on colonic function. Aliment. Pharmacol. Ther. 2008, 27, 104–119. [Google Scholar] [CrossRef]
- Tan, J.; McKenzie, C.; Vuillermin, P.J.; Goverse, G.; Vinuesa, C.G.; Mebius, R.E.; Macia, L.; Mackay, C.R. Dietary fiber and bacterial SCFA enhance oral tolerance and protect against food allergy through diverse cellular pathways. Cell Rep. 2016, 15, 2809–2824. [Google Scholar] [CrossRef] [PubMed]
- Macia, L.; Tan, J.; Vieira, A.T.; Leach, K.; Stanley, D.; Luong, S.; Maruya, M.; Ian McKenzie, C.; Hijikata, A.; Wong, C.; et al. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat. Commun. 2015, 6, 6734. [Google Scholar] [CrossRef]
- Rooks, M.G.; Garrett, W.S. Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 2016, 16, 341–352. [Google Scholar] [CrossRef]
- Feng, Y.; Wang, Y.; Wang, P.; Huang, Y.; Wang, F. Short-Chain Fatty Acids Manifest Stimulative and Protective Effects on Intestinal Barrier Function Through the Inhibition of NLRP3 Inflammasome and Autophagy. Cell Physiol. Biochem. 2018, 49, 190–205. [Google Scholar] [CrossRef] [PubMed]
- Furusawa, Y.; Obata, Y. Commensal microbe-derived butyrate induces differentiation of colonic regulatory T cells. Nature 2013, 504, 446–450. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly-Y, M.; Glickman, J.N.; Garrett, W.S. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; van der Veeken, J.; deRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T–cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef]
- Goverse, G.; Molenaar, R.; Macia, L.; Tan, J.; Erkelens, M.N.; Konijn, T.; Knippenberg, M.; Cook, E.C.; Hanekamp, D.; Veldhoen, M.; et al. Diet-derived short chain fatty acids stimulate intestinal epithelial cells to induce mucosal tolerogenic dendritic cells. J. Immunol. 2017, 198, 2172–2181. [Google Scholar] [CrossRef]
- Kim, M.; Qie, Y.; Park, J.; Kim, C.H. Gut microbial metabolites fuel host antibody responses. Cell Host Microbe 2016, 20, 202–214. [Google Scholar] [CrossRef]
- Tao, R.; de Zoeten, E.F.; Ozkaynak, E.; Chen, C.; Wang, L.; Porrett, P.M.; Li, B.; Turka, L.A.; Olson, E.N.; Greene, M.I.; et al. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat. Med. 2007, 13, 1299–1307. [Google Scholar] [CrossRef]
- Louis, P.; Flint, H.J. Diversity, metabolism and microbial ecology of butyrate–producing bacteria from the human large intestine. FEMS Microbiol. Lett. 2009, 294, 1–8. [Google Scholar] [CrossRef]
- Di Costanzo, M.; Paparo, L.; Aitoro, R.; Cosenza, L.; Nocerino, R.; Cozzolino, T.; Pezzella, V.; Vallone, G.; Berni Canani, R. Potential Beneficial Effects of Butyrate against Food Allergy. In Butyrate: Food Sources, Functions and Health Benefits; Li, C.-J., Ed.; Biochemistry Research Trends: New York, NY, USA, 2014; pp. 81–90. [Google Scholar]
- Aitoro, R.; Paparo, L.; Amoroso, A.; Di Costanzo, M.; Cosenza, L.; Granata, V.; Di Scala, C.; Nocerino, R.; Trinchese, G.; Montella, M.; et al. Gut microbiota as a target for preventive and therapeutic intervention against food allergy. Nutrients 2017, 9, 672. [Google Scholar] [CrossRef]
- Vonk, M.M.; Blokhuis, B.R.J.; Diks, M.A.P.; Wagenaar, L.; Smit, J.J.; Pieters, R.; Garssen, J.; Knippels, L.; van Esch, B. Butyrate Enhances Desensitization Induced by Oral Immunotherapy in Cow’s Milk Allergic Mice. Mediat. Inflamm. 2019, 2019, 1–12. [Google Scholar] [CrossRef]
- Crestani, E.; Harb, H.; Charbonnier, L.M.; Leirer, J.; Motsinger-Reif, A.; Rachid, R.; Phipatanakul, W.; Kaddurah-Daouk, R.; Chatila, T.A. Untargeted metabolomic profiling identifies disease-specific signatures in food allergy and asthma. J. Allergy Clin. Immunol. 2020, 145, 897–906. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.H.; Song, Y.; Wu, W.; Yu, K.; Zhang, G. The gut microbiota, environmental factors, and links to the development of food allergy. Clin. Mol. Allergy 2020, 18, 5. [Google Scholar] [CrossRef] [PubMed]
- Renz, H.; Allen, K.J.; Sicherer, S.H.; Sampson, H.A.; Lack, G.; Beyer, K.; Oettgen, H.C. Food allergy. Nat. Rev. Dis. Primers 2018, 4, 17098. [Google Scholar] [CrossRef]
- Lyons, K.E.; Ryan, C.A.; Dempsey, E.M.; Ross, R.P.; Stanton, C. Breast Milk, a Source of Beneficial Microbes and Associated Benefits for Infant Health. Nutrients 2020, 12, 1039. [Google Scholar] [CrossRef] [PubMed]
- Du Toit, G.; Roberts, G.; Sayre, P.H.; Plaut, M.; Bahnson, H.T.; Mitchell, H.; Radulovic, S.; Chan, S.; Fox, A.; Turcanu, V.; et al. Identifying infants at high risk of peanut allergy: The Learning Early About Peanut Allergy (LEAP) screening study. J. Allergy Clin. Immunol. 2013, 131, 135–143.e1–12. [Google Scholar] [CrossRef]
- Mitselou, N.; Hallberg, J.; Stephansson, O.; Almqvist, C.; Melén, E.; Ludvigsson, J.F. Cesarean delivery, preterm birth, and risk of food allergy: Nationwide Swedish cohort study of more than 1 million children. J. Allergy Clin. Immunol. 2018, 142, 1510–1514. [Google Scholar] [CrossRef]
- Biasucci, G.; Rubini, M.; Riboni, S.; Morelli, L.; Bessi, E.; Retetangos, C. Mode of delivery affects the bacterial community in the newborn gut. Early Hum. Dev. 2010, 86, 13–15. [Google Scholar] [CrossRef]
- Guibas, G.V.; Moschonis, G.; Xepapadaki, P.; Roumpedaki, E.; Androutsos, O.; Manios, Y.; Papadopoulos, N.G. Conception via in vitro fertilization and delivery by Caesarean section are associated with paediatric asthma incidence. Clin. Exp. Allergy 2013, 43, 1058–1066. [Google Scholar] [CrossRef]
- Greenwood, C.; Morrow, A.L.; Lagomarcino, A.J.; Altaye, M.; Taft, D.H.; Yu, Z.; Newburg, D.S.; Ward, D.V.; Schibler, K.R. Early empiric antibiotic use in preterm infants is associated with lower bacterial diversity and higher relative abundance of Enterobacter. J. Pediatr. 2014, 165, 23–29. [Google Scholar] [CrossRef]
- Arboleya, S.; Sánchez, B.; Milani, C.; Duranti, S.; Solís, G.; Fernández, N.; de los Reyes-Gavilán, C.G.; Ventura, M.; Margolles, A.; Gueimonde, M. Intestinal microbiota development in preterm neonates and effect of perinatal antibiotics. J. Pediatr. 2015, 166, 538–544. [Google Scholar] [CrossRef]
- Fouhy, F.; Guinane, C.M.; Hussey, S.; Wall, R.; Ryan, C.A.; Dempsey, E.M.; Murphy, B.; Ross, R.P.; Fitzgerald, G.F.; Stanton, C.; et al. High–throughput sequencing reveals the incomplete, short–term recovery of infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamicin. Antimicrob. Agents Chemother. 2012, 56, 5811–5820. [Google Scholar] [CrossRef] [PubMed]
- Biasucci, G. Gut perturbation and probiotics in neonatology. J. Pediatr. Neonat. Ind. Med. 2018, 7, e070202. [Google Scholar]
- Silvers, K.M.; Frampton, C.M.; Wickens, K.; Pattemore, P.K.; Ingham, T.; Fishwick, D.; Crane, J.; Town, G.I.; Epton, M.J.; New Zealand Asthma and Allergy Cohort Study Group. Breastfeeding protects against current asthma up to 6 years of age. J. Pediatr. 2012, 160, 991–996. [Google Scholar] [CrossRef]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking long–term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef] [PubMed]
- Trompette, A.; Gollwitzer, E.S.; Yadava, K.; Sichelstiel, A.K.; Sprenger, N.; Ngom-Bru, C.; Blanchard, C.; Junt, T.; Nicod, L.P.; Harris, N.L.; et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 2014, 20, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Jordakieva, G.; Kundi, M.; Untersmayr, E.; Pali-Schöll, I.; Reichardt, B.; Jensen-Jarolim, E. Country-wide medical records infer increased allergy risk of gastric acid inhibition. Nat. Commun. 2019, 10, 3298. [Google Scholar] [CrossRef] [PubMed]
- Schöll, I.; Ackermann, U.; Ozdemir, C.; Blümer, N.; Dicke, T.; Sel, S.; Sel, S.; Wegmann, M.; Szalai, K.; Knittelfelder, R.; et al. Anti-ulcer treatment during pregnancy induces food allergy in mouse mothers and a Th2-bias in their offspring. FASEB J. 2007, 21, 1264–1270. [Google Scholar]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Berni Canani, R.; Flint, H.J.; Salminen, S.; et al. The International scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotics. Nat. Rev. Gastrol. Hepat. 2014, 11, 506–514. [Google Scholar] [CrossRef]
- Paparo, L.; Aitoro, R.; Nocerino, R.; Fierro, C.; Bruno, C.; Berni Canani, R. Direct effects of fermented cow’s milk product with Lactobacillus paracasei CBA L74 on human enterocytes. Benef. Microbes 2018, 9, 165–172. [Google Scholar] [CrossRef]
- Tulyeu, J.; Kumagai, H.; Jimbo, E.; Watanabe, S.; Yokoyama, K.; Cui, L.; Osaka, H.; Mieno, M.; Yamagata, T. Probiotics Prevents Sensitization to Oral Antigen and Subsequent Increases in Intestinal Tight Junction Permeability in Juvenile-Young Adult Rats. Microorganisms 2019, 7, 463. [Google Scholar] [CrossRef]
- Hardy, H.; Harris, J.; Lyon, E.; Beal, J.; Foey, A.D. Probiotics, Prebiotics and Immunomodulation of gut mucosal defences: Homeostatis and immunopathology. Nutrients 2013, 5, 1869–1912. [Google Scholar] [CrossRef] [PubMed]
- Torii, A.; Torii, S.; Fujiwara, S.; Tanaka, H.; Inagaki, N.; Nagai, H. Lactobacillus acidophilus strain L−92 regulates theproduction of Th1 cytokine as well as Th2 cytokines. Allergol. Int. 2007, 56, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Niers, L.E.; Timmerman, H.M.; Rijkers, G.T.; van Bleek, G.M.; van Uden, N.O.; Knol, E.F.; Kapsenberg, M.L.; Kimpen, J.L.; Hoekstra, M.O. Identification of strong interleukin−10 inducinglactic acid bacteria which down–regulate T helper type 2 cytokines. Clin. Exp. Allergy 2005, 35, 1481–1489. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Kitazawa, H.; Iwabuchi, N.; Xiao, J.Z.; Miyaji, K.; Iwatsuki, K.; Saito, T. Oral administration of an immunostimulatory DNA sequence from Bifidobacterium longum improves Th1/Th2 balance in a murine model. Biosci. Biotechnol. Biochem. 2006, 70, 2013–2017. [Google Scholar] [CrossRef][Green Version]
- Huang, C.H.; Lin, Y.C.; Jan, T.R. Lactobacillus reuteri induces intestinal immune tolerance against food allergy in mice. J. Funct. Foods 2017, 31, 44–51. [Google Scholar] [CrossRef]
- Song, J.; Li, Y.; Li, J.; Wang, H.; Zhang, Y.; Suo, H. Lactobacillus rhamnosus 2016SWU.05.0601 regulates immune balance in ovalbumin-sensitized mice by modulating the immune-related transcription factors expression and gut microbiota. J. Sci. Food Agric. 2020. [Google Scholar] [CrossRef]
- Fu, L.; Song, J.; Wang, C.; Fu, S.; Wang, Y. Bifidobacterium infantis Potentially Alleviates Shrimp Tropomyosin-Induced Allergy by Tolerogenic Dendritic Cell-Dependent Induction of Regulatory T Cells and Alterations in Gut Microbiota. Front. Immunol. 2017, 8, 1536. [Google Scholar] [CrossRef]
- Maiga, M.A.; Morin, S.; Bernard, H.; Rabot, S.; Adel-Patient, K.; Hazebrouck, S. Neonatal mono–colonization of germ–free mice with Lactobacillus casei enhances casein immunogenicity after oral sensitization to cow’s milk. Mol. Nutr. Food Res. 2017, 61, 1600862. [Google Scholar] [CrossRef]
- Liu, M.Y.; Yang, Z.Y.; Dai, W.K.; Huang, J.Q.; Li, Y.H.; Zhang, J.; Qiu, C.Z.; Wei, C.; Zhou, Q.; Sun, X.; et al. Protective effect of Bifidobacterium infantis CGMCC313–2 on ovalbumin–induced airway asthma and b–lactoglobulin induced intestinal food allergy mouse models. World J. Gastroenterol. 2017, 23, 2149–2158. [Google Scholar] [CrossRef]
- Berni Canani, R.; Nocerino, R.; Terrin, G.; Coruzzo, A.; Cosenza, L.; Leone, L.; Troncone, R. Effect of Lactobacillus GG on tolerance acquisition in infants with cow’s milk allergy a randomized trial. J. Allergy Clin. Immunol. 2012, 129, 580–582. [Google Scholar] [CrossRef]
- Berni Canani, R.; Nocerino, R.; Terrin, G.; Frediani, T.; Lucarelli, S.; Cosenza, L.; Passariello, A.; Leone, L.; Granata, V.; Di Costanzo, M.; et al. Formula selection for management of children with cow milk allergy influences the rate of acquisition of tolerance: A prospective multicenter study. J. Pediatr. 2013, 163, 771–777. [Google Scholar] [CrossRef] [PubMed]
- Berni Canani, R.; Di Costanzo, M.; Bedogni, G.; Amoroso, A.; Cosenza, L.; Di Scala, C.; Granata, V.; Nocerino, R. Extensively hydrolyzed casein formula containing Lactobacillus rhamnosus GG reduces the occurrence of other allergic manifestation sin children with cow’s milk allergy: 3–year randomized controlled trial. J. Allergy Clin. Immunol. 2017, 139, 1906–1913. [Google Scholar] [CrossRef] [PubMed]
- Fox, A.; Bird, J.A.; Fiocchi, A.; Knol, J.; Meyer, R.; Salminen, S.; Sitang, G.; Szajewska, H.; Papadopoulos, N. The potential for pre-, pro- and synbiotics in the management of infants at risk of cow’s milk allergy or with cow’s milk allergy: An exploration of the rationale, available evidence and remaining questions. World Allergy Organ. J. 2019, 12, 100034. [Google Scholar] [CrossRef] [PubMed]
- Fiocchi, A.; Brozek, J.; Schunemann, H.; Bahna, S.L.; von Berg, A.; Beyer, K.; Bozzola, M.; Bradsher, J.; Compalati, E.; Ebisawa, M.; et al. World allergy organization (WAO) diagnosis and rationale for action against cow’s milk allergy (DRACMA) guidelines. World Allergy Organ. J. 2010, 21, 57–161. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.L.; Ponsonby, A.L.; Orsini, F.; Tey, D.; Robinson, M.; Su, E.L.; Licciardi, P.; Burks, W.; Donath, S. Administration of a probiotic with peanut oral immunotherapy: A randomized trial. J. Allergy Clin. Immunol. 2015, 135, 737–744.e8. [Google Scholar] [CrossRef]
- Hsiao, K.C.; Ponsonby, A.L.; Axelrad, C.; Pitkin, S.; Tang, M.; PPOIT Study Team. Long-term clinical and immunological effects of probiotic and peanut oral immunotherapy after treatment cessation: 4-year follow-up of a randomised, double-blind, placebo-controlled trial. Lancet Child. Adolesc. Health 2017, 1, 97–105. [Google Scholar] [CrossRef]
- Gibson, G.R.; Hutkins, R.; Sanders, M.E.; Prescott, S.L.; Reimer, R.A.; Salminen, S.J.; Scott, K.; Stanton, C.; Swanson, K.S.; Cani, P.; et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 491–502. [Google Scholar] [CrossRef]
- Miqdady, M.; Al Mistarihi, J.; Azaz, A.; Rawat, D. Prebiotics in the Infant Microbiome: The Past, Present, and Future. Pediatr Gastroenterol. Hepatol. Nutr. 2020, 23, 1–14. [Google Scholar] [CrossRef]
- Tang, M.L.; Lahtinen, S.J.; Boyle, R.J. Probiotics and prebiotics: Clinical effects in allergic disease. Curr. Opin. Pediatr. 2010, 22, 626–634. [Google Scholar] [CrossRef]
- Osborn, D.A.; Sinn, J.K. Prebiotics in infants for prevention of allergy. Cochrane Database Syst. Rev. 2013, 3, CD006474. [Google Scholar] [CrossRef]
- Braegger, C.; Chmielewska, A.; Decsi, T.; Kolacek, S.; Mihatsch, W.; Moreno, L.; Pieścik, M.; Puntis, J.; Shamir, R.; Szajewska, H.; et al. Supplementation of infant formula with probiotics and/or prebiotics: A systematic review and comment by the ESPGHAN committee on nutrition. J. Pediatr Gastroenterol. Nutr. 2011, 52, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Homayouni Rad, A.; Aghebati Maleki, L.; Samadi Kafil, H.; Abbasi, A. Postbiotics: A novel strategy in food allergy treatment. Crit. Rev. Food Sci. Nutr. 2020, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Rad, A.H.; Maleki, L.A.; Kafil, H.S.; Zavoshti, H.F.; Abbasi, A. Postbiotics as novel health-promoting ingredients in functional foods. Health Promot. Perspect. 2020, 10, 3–4. [Google Scholar]
- Roduit, C.; Frei, R.; Ferstl, R.; Loeliger, S.; Westermann, P.; Rhyner, C.; Schiavi, E.; Barcik, W.; Rodriguez-Perez, N.; Wawrzyniak, M.; et al. High levels of butyrate and propionate in early life are associated with protection against atopy. Allergy 2019, 74, 799–809. [Google Scholar] [CrossRef]
- Sandin, A.; Bråbäck, L.; Norin, E.; Björkstén, B. Faecal short chain fatty acid pattern and allergy in early childhood. Acta Paediatr. 2009, 98, 823–827. [Google Scholar] [CrossRef]
- Cait, A.; Cardenas, E.; Dimitriu, P.A.; Amenyogbe, N.; Dai, D.; Cait, J.; Sbihi, H.; Stiemsma, L.; Subbarao, P.; Mandhane, P.J.; et al. Reduced genetic potential for butyrate fermentation in the gut microbiome of infants who develop allergic sensitization. J. Allergy Clin. Immunol. 2019, 144, 1438–1647. [Google Scholar] [CrossRef]
- Markowiak, P.; Slizewska, K. Effects of probiotics, prebiotics, and synbiotics on human health. Nutrients 2017, 9, 1021. [Google Scholar] [CrossRef]
- Candy, D.C.A.; Van Ampting, M.T.J.; Oude Nijhuis, M.M.; Wopereis, H.; Butt, A.M.; Peroni, D.G.; Vandenplas, Y.; Fox, A.T.; Shah, N.; West, C.E.; et al. A synbiotic-containing amino-acid-based formula improves gut microbiota in non-IgE-mediated allergic infants. Pediatr. Res. 2018, 83, 677–686. [Google Scholar] [CrossRef]
- Li, N.; Yu, Y.; Chen, X.; Gao, S.; Zhang, Q.; Xu, C. Bifidobacterium breve M-16V alters the gut microbiota to alleviate OVA-induced food allergy through IL-33/ST2 signal pathway. J. Cell. Physiol. 2020. [Google Scholar] [CrossRef]
- Borody, T.J.; Khoruts, A. Fecal microbiota transplantation and emerging applications. Nat. Rev. Gastroenterol. Hepatol. 2011, 9, 88–96. [Google Scholar] [CrossRef]
- Gupta, S.; Allen-Vercoe, E.; Petrof, E.O. Fecal microbiota transplantation: In perspective. Ther. Adv. Gastroenterol. 2016, 9, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.-X.; Li, Y.-H.; Dai, W.-K.; Li, X.S.; Qiu, C.Z.; Ruan, M.L.; Zou, B.; Dong, C.; Liu, Y.H.; He, J.Y.; et al. Fecal microbiota transplantation induces remission of infantile allergic colitis through gut microbiota re-establishment. World J. Gastroenterol. 2017, 23, 8570–8581. [Google Scholar] [CrossRef]
- Bunyavanich, S.; Berin, M.C. Food allergy and the microbiome: Current understandings and future directions. J. Allergy Clin. Immunol. 2019, 144, 1468–1477. [Google Scholar] [CrossRef] [PubMed]
- Blázquez, A.B.; Berin, M.C. Microbiome and food allergy. Transl. Res. 2017, 179, 199–203. [Google Scholar] [CrossRef] [PubMed]
- Shu, S.A.; Yuen, A.W.T.; Woo, E.; Chu, K.H.; Kwan, H.S.; Yang, G.X.; Yang, Y.; Leung, P. Microbiota and Food Allergy. Clin. Rev. Allergy Immunol. 2019, 57, 83–97. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Ho, H.E.; Bunyavanich, S. The gut microbiome in food allergy. Ann. Allergy Asthma Immunol. 2019, 122, 276–282. [Google Scholar] [CrossRef]
- Berni Canani, R.; Paparo, L.; Nocerino, R.; Di Scala, C.; Della Gatta, G.; Maddalena, Y.; Buono, A.; Bruno, C.; Voto, L.; Ercolini, D. Gut microbiome as target for innovative strategies against food allergy. Front. Immunol. 2019, 10, 191. [Google Scholar] [CrossRef]
- Stephen-Victor, E.; Chatila, T.A. Regulation of oral immune tolerance by the microbiome in food allergy. Curr. Opin. Immunol. 2019, 60, 141–147. [Google Scholar] [CrossRef]
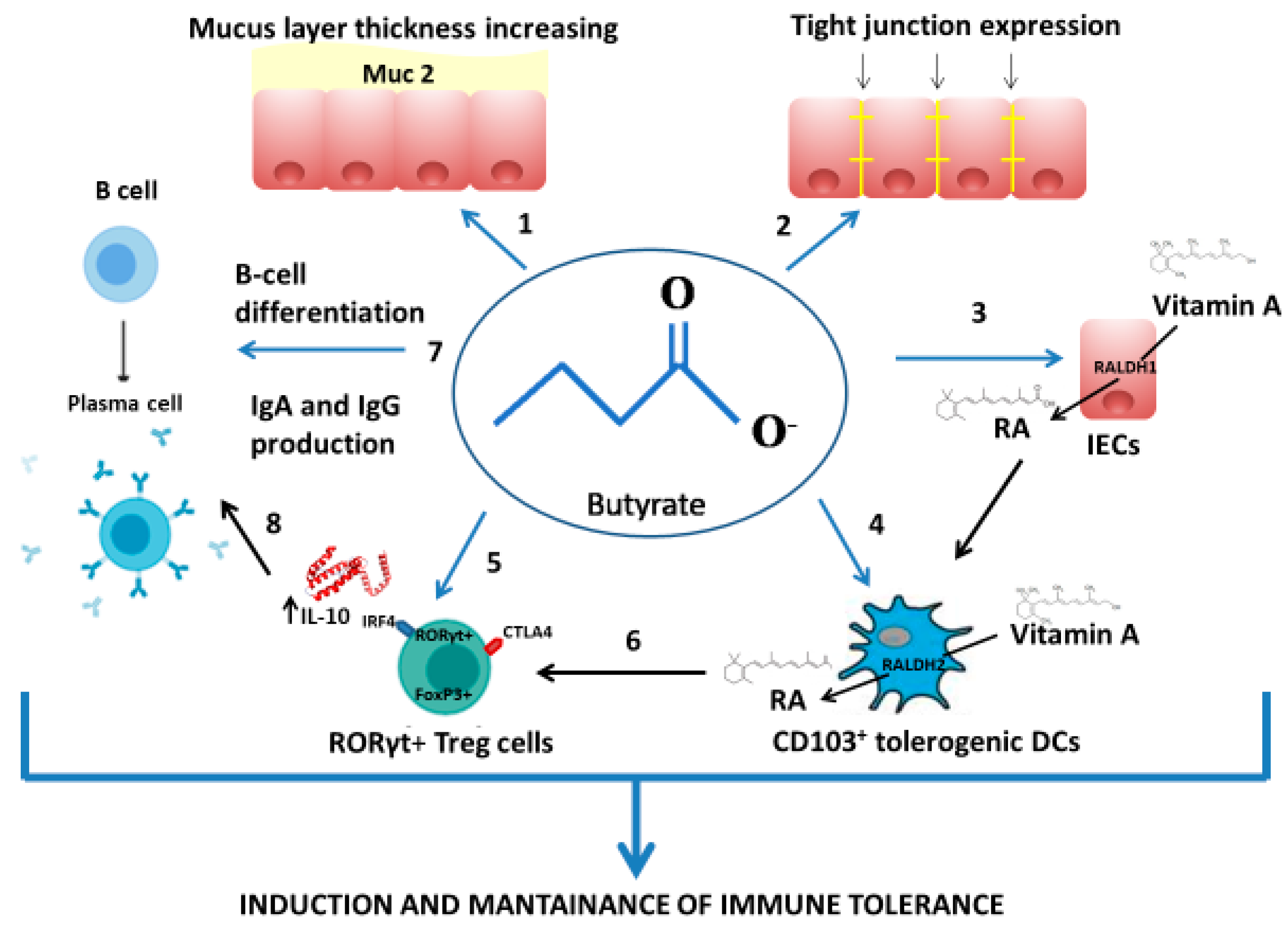
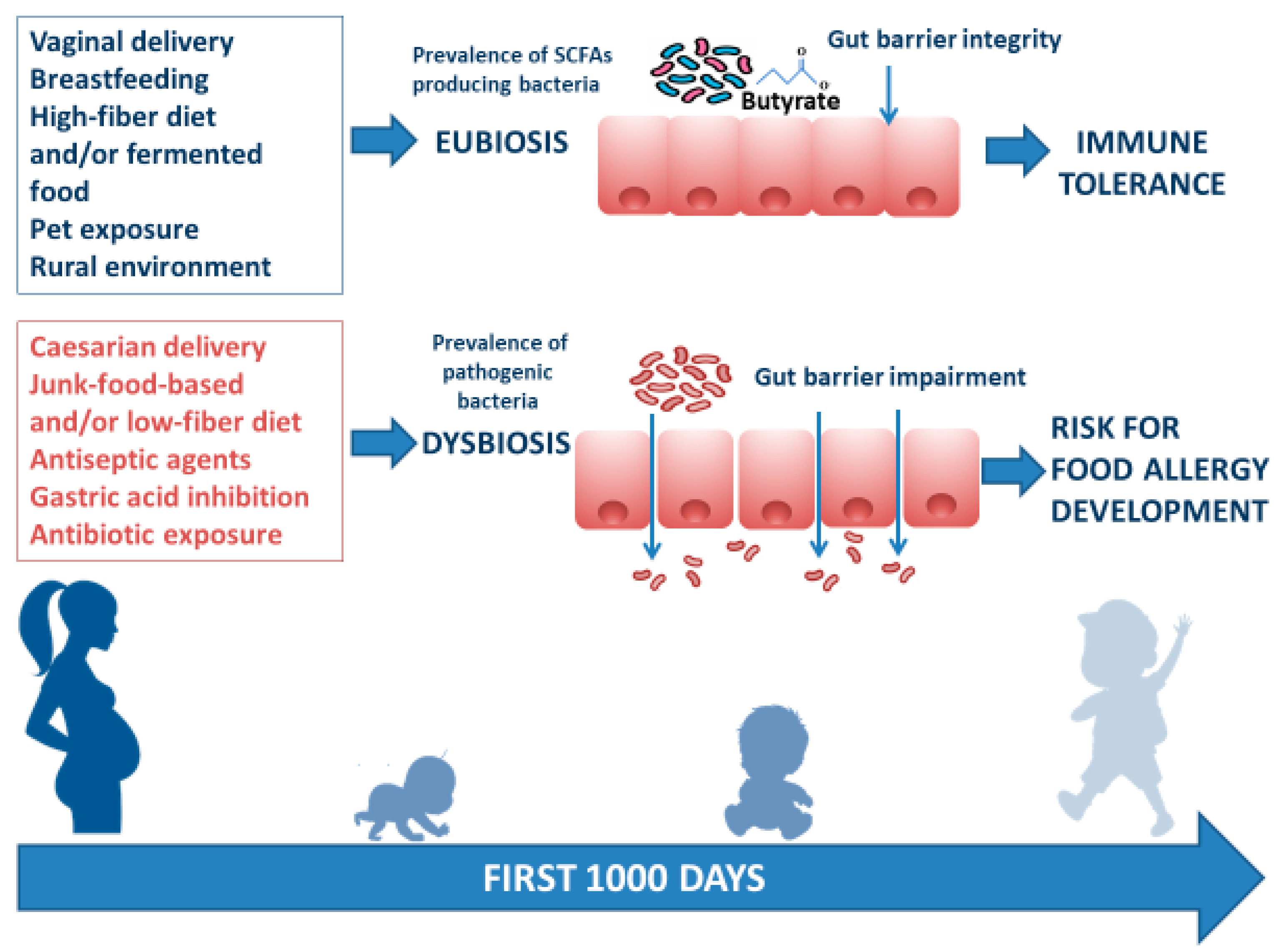
| Food Allergy | OTUs | Diversity | Main Features Associated with Food Allergy | |
|---|---|---|---|---|
| Ling et al. 2014 [10] (n = 34; FA) | Cow’s milk, egg, wheat, nut, peanuts, fish, shrimp, soybean | ↓ | = | ↑ Bacteroidetes ↑ Proteobacteria ↑Actinobacteria ↓ Firmicutes |
| Azad et al. 2015 [11] (n = 12; FS) | Cow’s milk, egg, peanut | ↓ | = | ↓ Enterobacteriaceae ↓ Bacteroidaceae |
| Chen et al. 2015 [12] (n = 23; FS) | Egg white, cow’s milk, wheat, peanut, soybean | N.R. | ↓ | ↑ Firmicutes ↑ Proteobacteria ↑ Actinobacteria ↓ Veillonella |
| Berni Canani et al. 2016 [13] (n = 39; FA) | Cow’s milk | ↑ | N.R. | ↑Ruminococcaceae ↑ Lachnospiraceae ↓Bifidobacteriaceae ↓Streptococcaceae ↓Enterobacteriaceae |
| Bunyavanich et al. 2016 [14] (n = 226; FA) | Cow’s milk | ↑ | N.R. | ↑ Bacteroidetes ↑Enterobacter |
| Inoue et al. 2017 [15] (n = 4; FA) | Egg, wheat, soybean, sesame, cow’s milk, peanut, shrimp, crab | N.R. | N.R. | ↑ Lachnospira ↑ Veillonella ↑ Sutterella ↓ Dorea ↓ Akkermansia |
| Kourosh et al. 2018 [16] (n = 68; FA) | Tree nuts, fish, milk, egg, sesame, soy | ↑ | N.R. | ↑Oscillobacter valericigenes ↑Lachnocrostidium bolteae ↑ Faecalibacterium sp. |
| Fazlollahi et al. 2018 [17] (n = 141; FA) | Egg | N.R. | N.R. | ↑ Lachnospiraceae ↑ Streptococcaceae ↑ Leuconostocaceae |
| Dong et al. 2018 [18] (n = 60; FA) | Cow’s milk | N.R. | ↓ | ↑ Lactobacillaceae ↓ Bifidocacteriaceae ↓ Ruminococcaceae |
| Savage et al. 2018 [19] (n = 14; FA) | Cow’s milk, egg, wheat, soy, walnut, peanut | = | = | ↓ Citrobacter ↓ Oscillospira ↓ Lactococcus ↓ Dorea |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Costanzo, M.; Carucci, L.; Berni Canani, R.; Biasucci, G. Gut Microbiome Modulation for Preventing and Treating Pediatric Food Allergies. Int. J. Mol. Sci. 2020, 21, 5275. https://doi.org/10.3390/ijms21155275
Di Costanzo M, Carucci L, Berni Canani R, Biasucci G. Gut Microbiome Modulation for Preventing and Treating Pediatric Food Allergies. International Journal of Molecular Sciences. 2020; 21(15):5275. https://doi.org/10.3390/ijms21155275
Chicago/Turabian StyleDi Costanzo, Margherita, Laura Carucci, Roberto Berni Canani, and Giacomo Biasucci. 2020. "Gut Microbiome Modulation for Preventing and Treating Pediatric Food Allergies" International Journal of Molecular Sciences 21, no. 15: 5275. https://doi.org/10.3390/ijms21155275
APA StyleDi Costanzo, M., Carucci, L., Berni Canani, R., & Biasucci, G. (2020). Gut Microbiome Modulation for Preventing and Treating Pediatric Food Allergies. International Journal of Molecular Sciences, 21(15), 5275. https://doi.org/10.3390/ijms21155275






