Potential Anti-COVID-19 Therapeutics that Block the Early Stage of the Viral Life Cycle: Structures, Mechanisms, and Clinical Trials
Abstract
1. Introduction
1.1. The Pandemic’s Status and Its Clinical Presentations
1.2. The Life Cycle of SARS-COV-2 and Potential Drug Targets
2. Potential Small Molecule Inhibitors of Early Viral Events in Clinical Trials
2.1. Quinoline-Based Drugs
2.2. Renin-Angiotensin-Aldosterone System (RAAS) Modifiers: Angiotensin Converting Enzyme Inhibitors (ACEIs) and Angiotensin Receptor Blockers (ARBs)
2.3. Hydroxy Methylglutaryl Coenzyme A (HMG-CoA) Reductase Inhibitors: Atorvastatin and Simvastatin
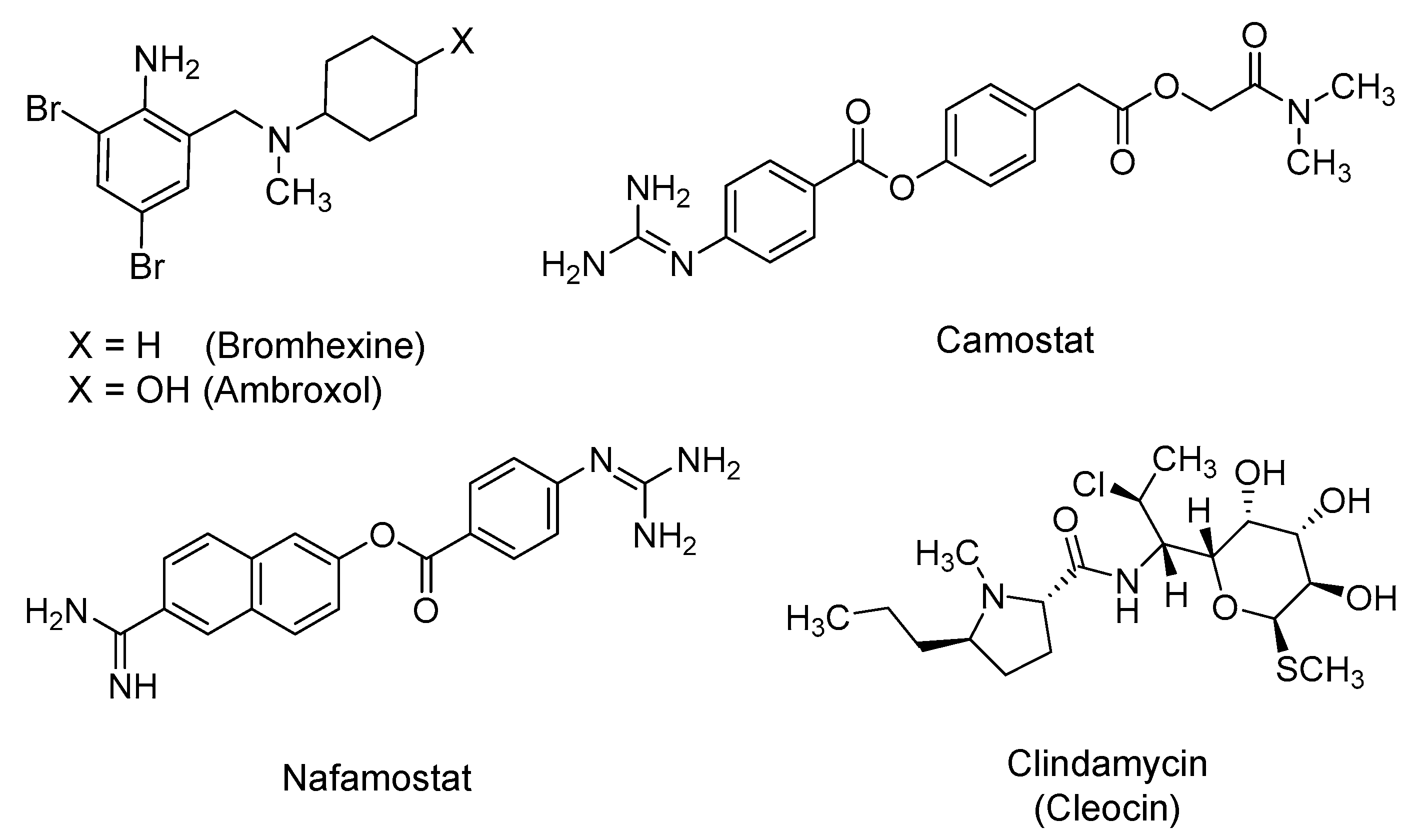
2.4. Bromhexine, Camostat, Nafamostat, and Clindamycin
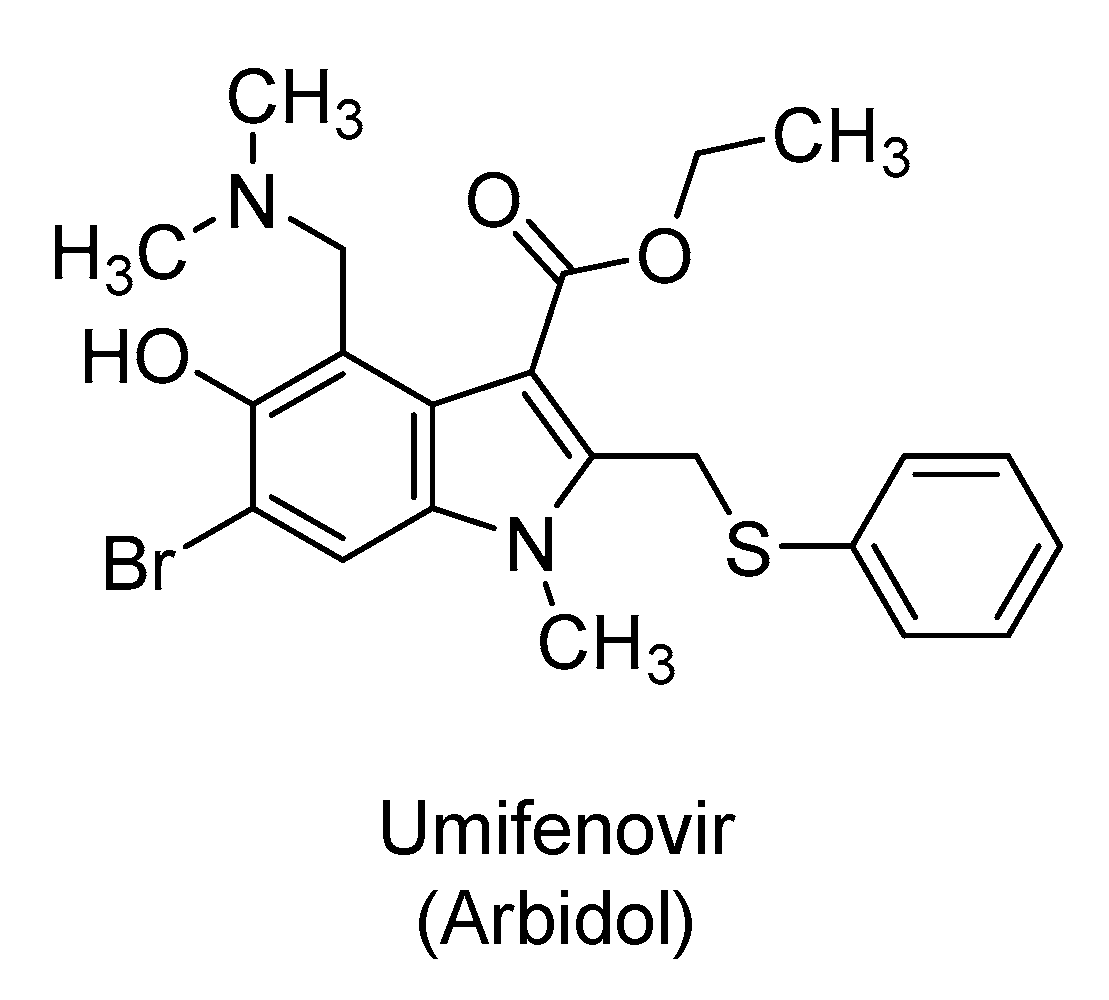
2.5. Umifenovir
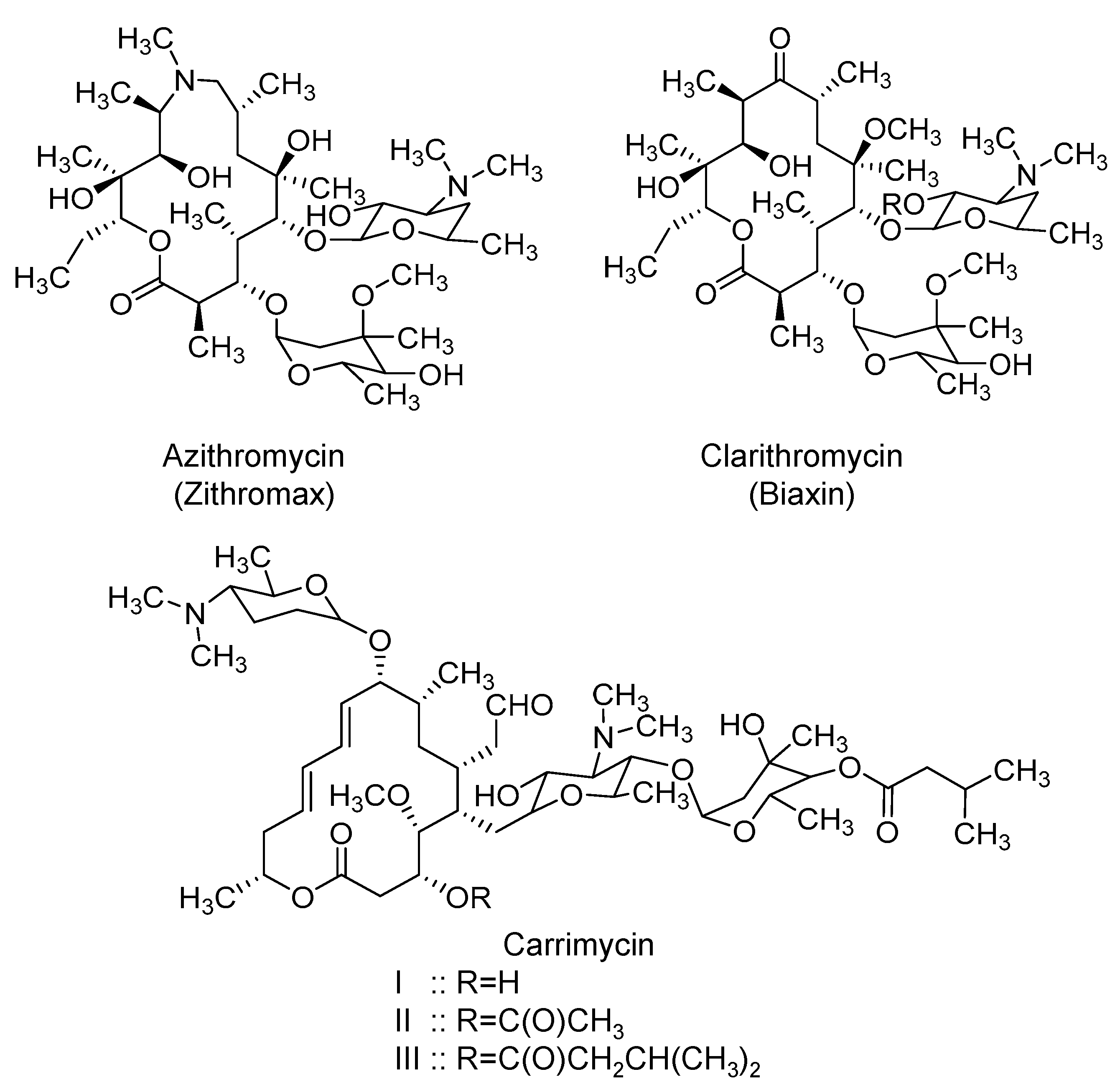
2.6. Macrolides: Azithromycin, Clarithromycin, and Carrimycin
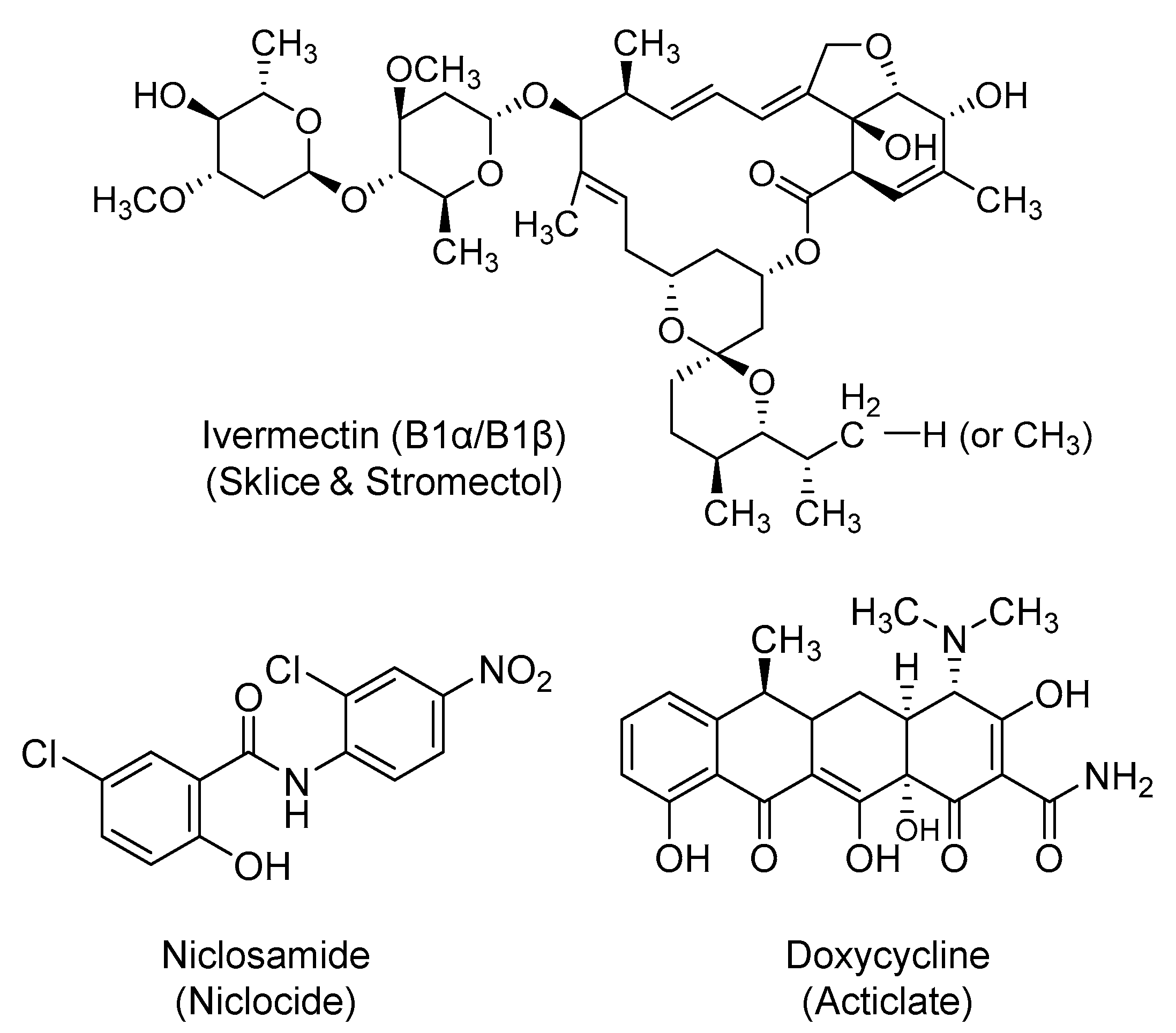
2.7. Ivermectin
2.8. Niclosamide
2.9. Doxycycline
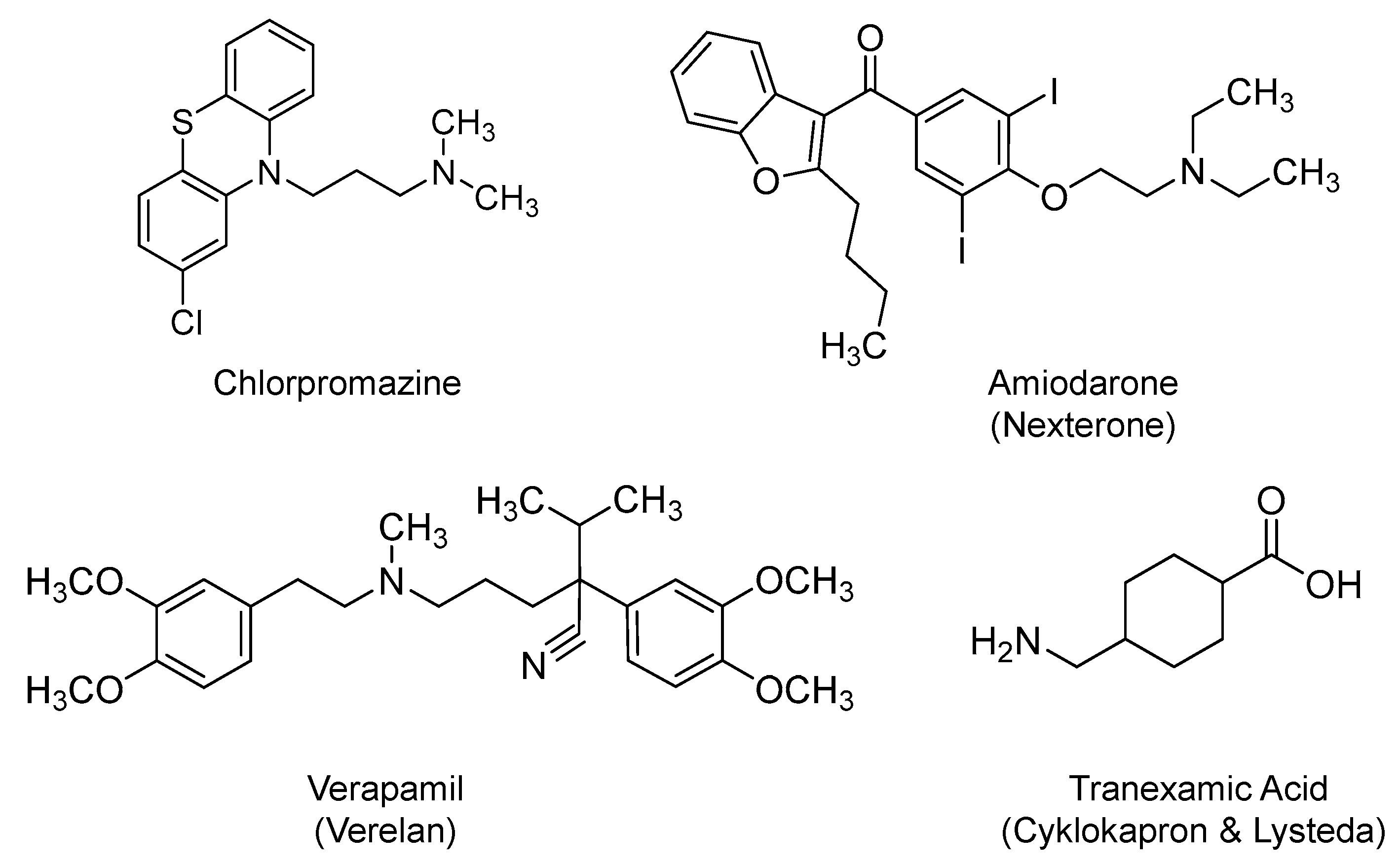
2.10. Chlorpromazine
2.11. Amiodarone and Verapamil
2.12. Tranexamic Acid
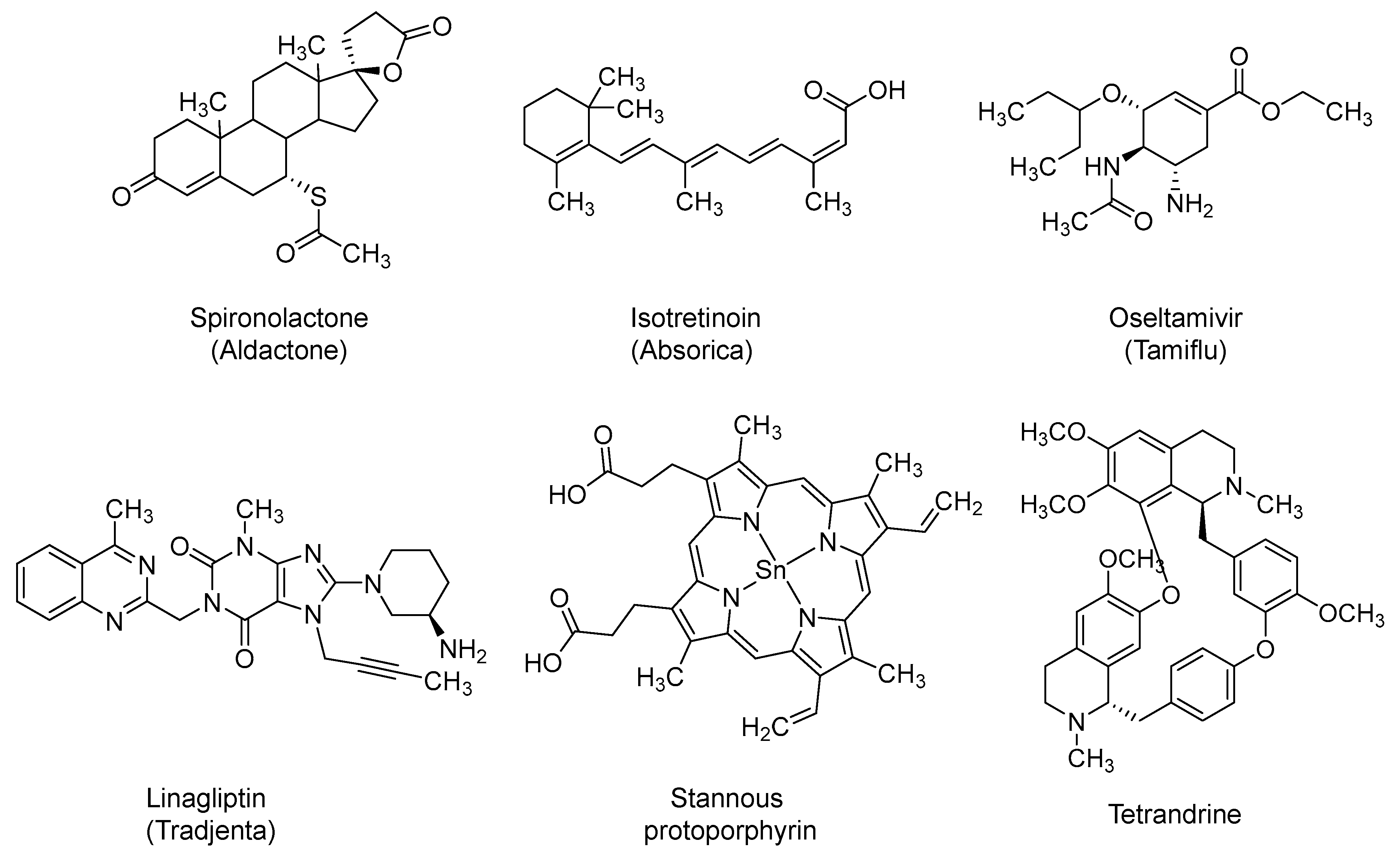
2.13. Spironolactone
2.14. Isotretinoin
2.15. Oseltamivir
2.16. Dipeptidyl Peptidase-4 (DPP-4) Inhibitors: Linagliptin and Sitagliptin
2.17. Stannous Protoporphyrin (also Reported as RBT-9)
2.18. Tetrandrine
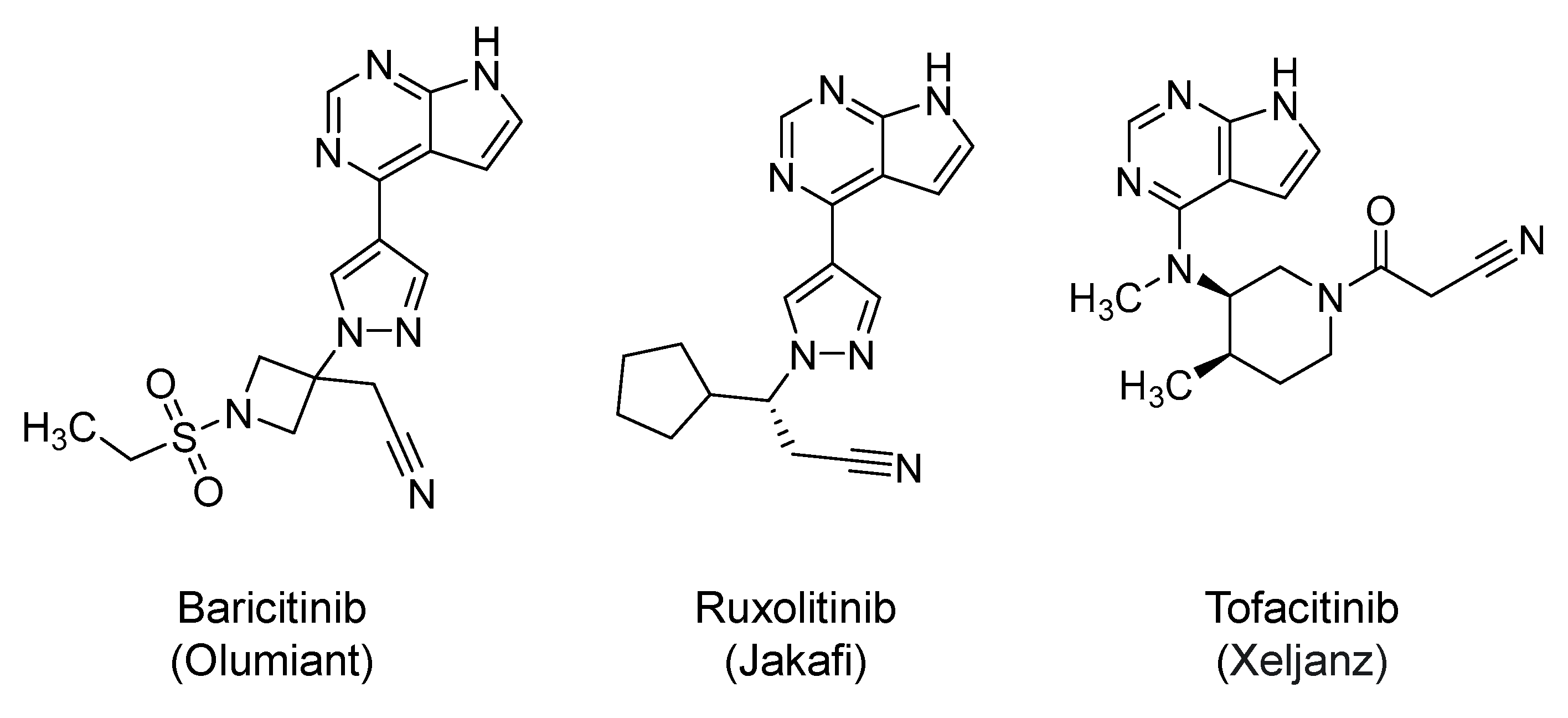
2.19. Janus Kinases (JAKs) Inhibitors: Baricitinib, Ruxolitinib, and Tofacitinib
3. Potential Macromolecular Inhibitors of Early Viral Events in Clinical Trials
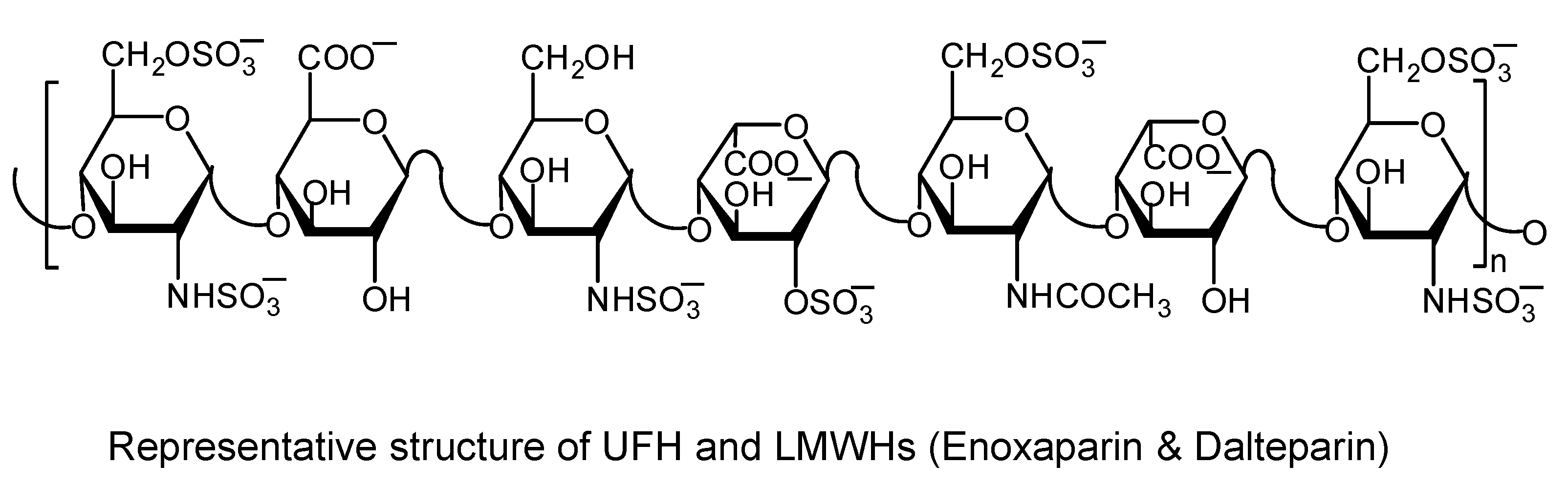
3.1. Heparins
3.2. DAS181
3.3. Recombinant Human ACE2 (rhACE2; also Reported as APN01)
3.4. Combination of REGN10933 and REGN10987
3.5. COVID-19 Convalescent Plasma and Immunoglobulins
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ACE2 | Angiotensin converting enzyme-2 |
| ARDS | Acute respiratory distress syndrome |
| 3CLpro | 3-Chymotrypsin-like protease |
| CC50 | Half maximal cytotoxic concentration |
| COVID-19 | Coronavirus disease-2019 |
| DPP-4 | Dipeptidyl peptidase-4 |
| EC50 | Half maximal effective concentration |
| HIV | Human immunodeficiency virus |
| HMG CoA | Hydroxymethyl glutaryl coenzyme A |
| IC50 | Half maximal inhibitory concentration |
| ICU | Intensive care unit |
| LMWHs | Low molecular weight heparins |
| MERS | Middle East respiratory syndrome |
| Mpro | Main protease |
| NSPs | Nonstructural proteins |
| PLpro | Papain-like protease |
| RAAS | Renin-angiotensin-aldosterone system |
| RdRp | RNA-dependent RNA polymerase |
| rhACE2 | Recombinant human ACE2 |
| SARS-CoV-2 | Severe acute respiratory syndrome-coronavirus-2 |
| TMPRSS2 | Transmembrane protease serine 2 |
| UFH | Unfractionated heparin |
References
- World Health Organization. Available online: https://www.who.int/csr/don/05-january-2020-pneumonia-of-unkown-cause-china/en/ (accessed on 31 May 2020).
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Cucinotta, D.; Vanelli, M. WHO Declares COVID-19 a Pandemic. Acta Biomed. 2020, 91, 157–160. [Google Scholar] [PubMed]
- Dong, E.; Du, H.; Gardner, L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020, 20, 533–534. [Google Scholar] [CrossRef]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054e62. [Google Scholar] [CrossRef]
- Kluytmans, M.; Buiting, A.; Pas, S.; Bentvelsen, R.; van den Bijllaardt, W.; van Oudheusden, A.; van Rijen, M.; Verweij, J.; Koopmans, M.; Kluytmans, J. SARS-CoV-2 infection in 86 healthcare workers in two Dutch hospitals in March 2020. medRxiv 2020. [Google Scholar] [CrossRef]
- Liang, W.; Feng, Z.; Rao, S.; Xiao, C.; Xue, X.; Lin, Z.; Zhang, Q.; Qi, W. Diarrhoea may be underestimated: A missing link in 2019 novel coronavirus. Gut 2020, 69, 1141–1143. [Google Scholar] [CrossRef]
- Liang, W.; Feng, Z.; Rao, S.; Xiao, C.; Xue, X.; Lin, Z.; Zhang, Q.; Qi, W. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy 2020. [Google Scholar] [CrossRef]
- Mao, L.; Jin, H.; Wang, M.; Hu, Y.; Chen, S.; He, Q.; Chang, J.; Hong, C.; Zhou, Y.; Wang, D.; et al. Neurologic manifestations of hospitalized patients with coronavirus disease 2019 in Wuhan, China. JAMA Neurol. 2020, 77, 1–9. [Google Scholar] [CrossRef]
- Giacomelli, A.; Pezzati, L.; Conti, F.; Bernacchia, D.; Siano, M.; Oreni, L.; Rusconi, S.; Gervasoni, C.; Ridolfo, A.L.; Rizzardini, G.; et al. Self-reported olfactory and taste disorders in SARS-CoV-2 patients: A cross-sectional study. Clin. Infect. Dis. 2020, ciaa330. [Google Scholar] [CrossRef]
- Wu, P.; Duan, F.; Luo, C.; Liu, Q.; Qu, X.; Liang, L.; Wu, K. Characteristics of ocular findings of patients with coronavirus disease 2019 (COVID-19) in Hubei Province, China. JAMA Ophthalmol. 2020, 138, 575–578. [Google Scholar] [CrossRef]
- Recalcati, S. Cutaneous manifestations in COVID-19: A first perspective. J. Eur. Acad. Dermatol. Venereol. 2020, 34, e212–e213. [Google Scholar] [CrossRef] [PubMed]
- Epidemiology Working Group for NCIP Epidemic Response; Chinese Center for Disease Control and Prevention. The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China. Zhonghua Liu Xing Bing Xue Za Zhi 2020, 41, 145–151. [Google Scholar]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients with 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA 2020, 323, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.Y.; Ma, Y.T.; Zhang, J.Y.; Xie, X. COVID-19 and the cardiovascular system. Nat. Rev. Cardiol. 2020, 17, 259–260. [Google Scholar] [CrossRef]
- Qin, C.; Zhou, L.; Hu, Z.; Zhang, S.; Yang, S.; Tao, Y.; Xie, C.; Ma, K.; Shang, K.; Wang, W.; et al. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin. Infect. Dis. 2020, ciaa248. [Google Scholar] [CrossRef]
- Zheng, M.; Gao, Y.; Wang, G.; Song, G.; Liu, S.; Sun, D.; Xu, Y.; Tian, Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell Mol. Immunol. 2020, 17, 533–535. [Google Scholar] [CrossRef]
- Mehta, P.; McAuley, D.F.; Brown, M.; Sanchez, E.; Tattersall, R.S.; Manson, J.J.; HLH Across Speciality Collaboration. COVID-19: Consider cytokine storm syndromes and immunosuppression. Lancet 2020, 395, 1033–1034. [Google Scholar] [CrossRef]
- Demelo-Rodríguez, P.; Cervilla-Muñoz, E.; Ordieres-Ortega, L.; Parra-Virto, A.; Toledano-Macías, M.; Toledo-Samaniego, N.; García-García, A.; García-Fernández-Bravo, I.; Ji, Z.; de-Miguel-Diez, J.; et al. Incidence of asymptomatic deep vein thrombosis in patients with COVID-19 pneumonia and elevated D-dimer levels. Thromb. Res. 2020, 192, 23–26. [Google Scholar] [CrossRef]
- Oxley, T.J.; Mocco, J.; Majidi, S.; Kellner, C.P.; Shoirah, H.; Singh, I.P.; De Leacy, R.A.; Shigematsu, T.; Ladner, T.R.; Yaeger, K.A.; et al. Large-vessel stroke as a presenting feature of Covid-19 in the young. N. Engl. J. Med. 2020, 382, e60. [Google Scholar] [CrossRef]
- Bangalore, S.; Sharma, A.; Slotwiner, A.; Yatskar, L.; Harari, R.; Shah, B.; Ibrahim, H.; Friedman, G.H.; Thompson, C.; Alviar, C.L.; et al. ST-segment elevation in patients with Covid-19-A case series. N. Engl. J. Med. 2020, 382, 2478–2480. [Google Scholar] [CrossRef]
- Bellosta, R.; Luzzani, L.; Natalini, G.; Pegorer, M.A.; Attisani, L.; Cossu, L.G.; Ferrandina, C.; Fossati, A.; Conti, E.; Bush, R.L.; et al. Acute limb ischemia in patients with COVID-19 pneumonia. J. Vasc. Surg. 2020. [Google Scholar] [CrossRef] [PubMed]
- Bompard, F.; Monnier, H.; Saab, I.; Tordjman, M.; Abdoul, H.; Fournier, L.; Sanchez, O.; Lorut, C.; Chassagnon, G.; Revel, M.P. Pulmonary embolism in patients with Covid-19 pneumonia. Eur. Respir. J. 2020, 2001365. [Google Scholar] [CrossRef] [PubMed]
- Tang, N.; Li, D.; Wang, X.; Sun, Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J. Thromb. Haemost. 2020, 18, 844–847. [Google Scholar] [CrossRef] [PubMed]
- Bikdeli, B.; Madhavan, M.V.; Jimenez, D.; Chuich, T.; Dreyfus, I.; Driggin, E.; Nigoghossian, C.; Ageno, W.; Madjid, M.; Guo, Y.; et al. COVID-19 and Thrombotic or Thromboembolic Disease: Implications for Prevention, Antithrombotic Therapy, and Follow-Up: JACC State-of-the-Art Review. J. Am. Coll. Cardiol. 2020, 75, 2950–2973. [Google Scholar] [CrossRef] [PubMed]
- Cui, S.; Chen, S.; Li, X.; Liu, S.; Wang, F. Prevalence of venous thromboembolism in patients with severe novel coronavirus pneumonia. J. Thromb. Haemost. 2020, 18, 1421–1424. [Google Scholar] [CrossRef]
- Klok, F.A.; Kruip, M.; van der Meer, N.; Arbous, M.S.; Gommers, D.; Kant, K.M.; Kaptein, F.; van Paassen, J.; Stals, M.; Huisman, M.V.; et al. Incidence of thrombotic complications in critically ill ICU patients with COVID-19. Thromb. Res. 2020, 191, 145–147. [Google Scholar] [CrossRef]
- Helms, J.; Tacquard, C.; Severac, F.; Leonard-Lorant, I.; Ohana, M.; Delabranche, X.; Merdji, H.; Clere-Jehl, R.; Schenck, M.; Schneider, F.; et al. High risk of thrombosis in patients with severe SARS-CoV-2 infection: A multicenter prospective cohort study. Intensive Care Med. 2020, 46, 1089–1098. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef]
- Zou, X.; Chen, K.; Zou, J.; Han, P.; Hao, J.; Han, Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020, 14, 185–192. [Google Scholar] [CrossRef]
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef] [PubMed]
- Belouzard, S.; Chu, V.C.; Whittaker, G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Version 2. Proc. Natl. Acad. Sci. USA 2009, 106, 5871–5876. [Google Scholar] [CrossRef] [PubMed]
- Raj, V.S.; Mou, H.; Smits, S.L.; Dekkers, D.H.; Müller, M.A.; Dijkman, R.; Muth, D.; Demmers, J.A.; Zaki, A.; Fouchier, R.A. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 2013, 495, 251–254. [Google Scholar] [CrossRef] [PubMed]
- Wentworth, D.E.; Holmes, K.V. Molecular determinants of species specificity in the coronavirus receptor aminopeptidase N (CD13): Influence of N-linked glycosylation. J. Virol. 2001, 75, 9741–9752. [Google Scholar] [CrossRef]
- Yang, N.; Shen, H.M. Targeting the Endocytic Pathway and Autophagy Process as a Novel Therapeutic Strategy in COVID-19. Int. J. Biol. Sci. 2020, 16, 1724–1731. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Pöhlmann, S. A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol. Cell. 2020, 78, 779–784.e5. [Google Scholar] [CrossRef]
- Liu, T.; Luo, S.; Libby, P.; Shi, G.P. Cathepsin L-selective inhibitors: A potentially promising treatment for COVID-19 patients. Pharmacol. Ther. 2020, 213, 107587. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Ou, X.; Liu, Y.; Lei, X.; Li, P.; Mi, D.; Ren, L.; Guo, L.; Guo, R.; Chen, T.; Hu, J.; et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020, 11, 1620. [Google Scholar] [CrossRef]
- Ji, H.L.; Zhao, R.; Matalon, S.; Matthay, M.A. Elevated plasmin(ogen) as a common risk factor for COVID-19 susceptibility. Physiol. Rev. 2020, 100, 1065–1075. [Google Scholar] [CrossRef]
- Du, L.; Kao, R.Y.; Zhou, Y.; He, Y.; Zhao, G.; Wong, C.; Jiang, S.; Yuen, K.Y.; Jin, D.Y.; Zheng, B.J. Cleavage of spike protein of SARS coronavirus by protease factor Xa is associated with viral infectivity. Biochem. Biophys. Res. Commun. 2007, 359, 174–179. [Google Scholar] [CrossRef] [PubMed]
- Khailany, R.A.; Safdar, M.; Ozaslan, M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020, 19, 100682. [Google Scholar] [CrossRef]
- Astuti, I.; Ysrafil. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): An overview of viral structure and host response. Diabetes Metab. Syndr. 2020, 14, 407–412. [Google Scholar] [CrossRef]
- Zhang, L.; Lin, D.; Sun, X.; Curth, U.; Drosten, C.; Sauerhering, L.; Becker, S.; Rox, K.; Hilgenfeld, R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science 2020, 368, 409–412. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Yan, L.; Huang, Y.; Liu, F.; Zhao, Y.; Cao, L.; Wang, T.; Sun, Q.; Ming, Z.; Zhang, L.; et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science 2020, 368, 779–782. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Hillyer, C.; Du, L. Neutralizing Antibodies against SARS-CoV-2 and Other Human Coronaviruses. Trends Immunol. 2020, 41, 355–359. [Google Scholar] [CrossRef] [PubMed]
- Atri, D.; Siddiqi, H.K.; Lang, J.; Nauffal, V.; Morrow, D.A.; Bohula, E.A. COVID-19 for the Cardiologist: A Current Review of the Virology, Clinical Epidemiology, Cardiac and Other Clinical Manifestations and Potential Therapeutic Strategies. JACC Basic Transl. Sci. 2020, 5, 518–536. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; He, Y.; Zhou, Y.; Liu, S.; Zheng, B.J.; Jiang, S. The spike protein of SARS-CoV-a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009, 7, 226–236. [Google Scholar] [CrossRef]
- Barber, B.E. Chloroquine and Hydroxychloroquine. In Kucers’ the Use of Antibiotics: A Clinical Review of Antibacterial, Antifungal, Antiparasitic, and Antiviral Drugs, 7th ed.; Grayson, M.L., Ed.; CRC Press: Boca Raton, FL, USA, 2018; pp. 3030–3048. [Google Scholar]
- Rolain, M.J.; Colson Raoult, D. Recycling of chloroquine and its hydroxyl analogue to face bacterial, fungal and viral infections in the 21st century. Int. J. Antimicrob. Agents 2007, 30, 297–308. [Google Scholar] [CrossRef]
- Ashley, E.A.; Phyo, A.P. Drugs in development for malaria. Drugs 2018, 78, 861–879. [Google Scholar] [CrossRef]
- Fong, K.Y.; Wright, D.W. Hemozoin and antimalarial drug discovery. Future Med. Chem. 2013, 5, 1437–1450. [Google Scholar] [CrossRef] [PubMed]
- Camarda, G.; Jirawatcharadech, P.; Priestley, R.S.; Saif, A.; March, S.; Wong, M.H.L.; Leung, S.; Miller, A.B.; Baker, D.A.; Alano, P.; et al. Antimalarial activity of primaquine operates via a two-step biochemical relay. Nat. Commun. 2019, 10, 3226. [Google Scholar] [CrossRef] [PubMed]
- U.S. Food and Drug Administration. Emergency Use Authorization Information. Available online: https://www.fda.gov/emergency-preparedness-and-response/mcm-legal-regulatory-and-policy-framework/emergency-use-authorization (accessed on 27 June 2020).
- Savarino, A.; Di Trani, L.; Donatelli, I.; Cauda, R.; Cassone, A. New insights into the antiviral effects of chloroquine. Lancet Infect. Dis. 2006, 6, 67–69. [Google Scholar] [CrossRef]
- Wang, M.; Cao, R.; Zhang, L.; Yang, X.; Liu, J.; Xu, M.; Shi, Z.; Hu, Z.; Zhong, W.; Xiao, G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020, 30, 269–271. [Google Scholar] [CrossRef]
- Liu, J.; Cao, R.; Xu, M.; Wang, X.; Zhang, H.; Hu, H.; Li, Y.; Hu, Z.; Zhong, W.; Wang, M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020, 6, 16. [Google Scholar] [CrossRef]
- Vincent, M.J.; Bergeron, E.; Benjannet, S.; Erickson, B.R.; Rollin, P.E.; Ksiazek, T.G.; Seidah, N.G.; Nichol, S.T. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. 2005, 2, 69. [Google Scholar] [CrossRef]
- Browning, D.J. Pharmacology of chloroquine and hydroxychloroquine. In Hydroxychloroquine and Chloroquine Retinopathy; Springer: New York, NY, USA, 2014; pp. 35–63. [Google Scholar]
- Keyaerts, E.; Vijgen, L.; Maes, P.; Neyts, J.; Van Ranst, M. In vitro inhibition of severe acute respiratory syndrome coronavirus by chloroquine. Biochem. Biophys. Res. Commun. 2004, 323, 264–268. [Google Scholar] [CrossRef]
- Devaux, C.A.; Rolain, J.M.; Colson, P.; Raoult, D. New insights on the antiviral effects of chloroquine against coronavirus: What to expect for COVID-19? Int. J. Antimicrob. Agents 2020, 55, 105938. [Google Scholar] [CrossRef]
- Sahraei, Z.; Shabani, M.; Shokouhi, S.; Saffaei, A. Aminoquinolines against coronavirus disease 2019 (COVID-19): Chloroquine or hydroxychloroquine. Int. J. Antimicrob. Agents 2020, 55, 105945. [Google Scholar] [CrossRef]
- Zhou, D.; Dai, S.M.; Tong, Q. COVID-19: A recommendation to examine the effect of hydroxychloroquine in preventing infection and progression. J. Antimicrob. Chemother. 2020, 75, 1667–1670. [Google Scholar] [CrossRef]
- Huang, M.; Tang, T.; Pang, P.; Li, M.; Ma, R.; Lu, J.; Shu, J.; You, Y.; Chen, B.; Liang, J.; et al. Treating COVID-19 with chloroquine. J. Mol. Cell Biol. 2020, 12, 322–325. [Google Scholar] [CrossRef] [PubMed]
- Borba, M.; Val, F.; Sampaio, V.S.; Alexandre, M.; Melo, G.C.; Brito, M.; Mourão, M.; Brito-Sousa, J.D.; Baía-da-Silva, D.; Guerra, M.; et al. Effect of high vs low doses of chloroquine diphosphate as adjunctive therapy for patients hospitalized with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection: A randomized clinical trial. JAMA Netw. Open. 2020, 3, e208857. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Hu, J.; Zhang, Z.; Jiang, S.; Han, S.; Yan, D.; Zhuang, R.; Hu, B.; Zhang, Z. Efficacy of hydroxychloroquine in patients with COVID-19: Results of a randomized clinical trial. medRxiv 2020. [Google Scholar] [CrossRef]
- Tang, W.; Cao, Z.; Han, M.; Wang, Z.; Chen, J.; Sun, W.; Wu, Y.; Xiao, W.; Liu, S.; Chen, E.; et al. Hydroxychloroquine in patients with mainly mild to moderate coronavirus disease 2019: Open label, randomised controlled trial. BMJ 2020, 369, m1849. [Google Scholar] [CrossRef]
- Molina, J.M.; Delaugerre, C.; Le Goff, J.; Mela-Lima, B.; Ponscarme, D.; Goldwirt, L.; de Castro, N. No evidence of rapid antiviral clearance or clinical benefit with the combination of hydroxychloroquine and azithromycin in patients with severe COVID-19 infection. Med. Mal. Infect. 2020, 50, 384. [Google Scholar] [CrossRef]
- Magagnoli, J.; Narendran, S.; Pereira, F.; Cummings, T.; Hardin, J.W.; Sutton, S.S.; Ambati, J. Outcomes of hydroxychloroquine usage in United States veterans hospitalized with Covid-19. medRxiv 2020. [Google Scholar] [CrossRef]
- Rosenberg, E.S.; Dufort, E.M.; Udo, T.; Wilberschied, L.A.; Kumar, J.; Tesoriero, J.; Weinberg, P.; Kirkwood, J.; Muse, A.; DeHovitz, J.; et al. Association of Treatment with Hydroxychloroquine or Azithromycin with In-Hospital Mortality in Patients with COVID-19 in New York State. JAMA 2020, 323, 2493–2502. [Google Scholar] [CrossRef]
- Geleris, J.; Sun, Y.; Platt, J.; Zucker, J.; Baldwin, M.; Hripcsak, G.; Labella, A.; Manson, D.K.; Kubin, C.; Barr, R.G.; et al. Observational Study of Hydroxychloroquine in Hospitalized Patients with Covid-19. N. Engl. J. Med. 2020, 382, 2411–2418. [Google Scholar] [CrossRef]
- Horby, P.; Landray, M. Statement from the Chief Investigators of the Randomised Evalution of COVID-19 Therapy (RECOVERY) Trial on Hydroxychloroquine. 5 June 2020. Available online: https://www.recoverytrial.net/news/statement-from-the-chief-investigators-of-the-randomised-evaluation-of-covid-19-therapy-recovery-trial-on-hydroxychloroquine-5-june-2020-no-clinical-benefit-from-use-of-hydroxychloroquine-in-hospitalised-patients-with-covid-19 (accessed on 5 June 2020).
- Nuffield Department of Population Health. Study protocol for randomized evaluation of Covid-19 therapy (RECOVERY). Available online: https://www.recoverytrial.net/files/recovery-protocol-v6-0-2020-05-14.pdf (accessed on 30 June 2020).
- Mahevas, M.; Tran, V.; Roumier, M.; Chabrol, A.; Paule, R.; Guillaud, C.; Gallien, S.; Lepeule, R.; Szwebel, T.-A.; Perrodeau, E.; et al. No evidence of clinical efficacy of hydroxychloroquine in patients hospitalized for COVID-19 infection with oxygen requirement: Results of a study using routinely collected data to emulate a target trial. medRxiv 2020. [Google Scholar] [CrossRef]
- Million, M.; Lagier, J.C.; Gautret, P.; Colson, P.; Fournier, P.E.; Amrane, S.; Hocquart, M.; Mailhe, M.; Esteves-Vieira, V.; Doudier, B.; et al. Early treatment of COVID-19 patients with hydroxychloroquine and azithromycin: A retrospective analysis of 1061 cases in Marseille, France. Travel Med. Infect. Dis. 2020, 35, 101738. [Google Scholar] [CrossRef]
- Machiels, J.D.; Bleeker-Rovers, C.P.; Ter Heine, R.; Rahamat-Langendoen, J.; de Mast, Q.; Ten Oever, J.; Bousema, T.; van Crevel, R.; Wertheim, H.F. Reply to Gautret et al: Hydroxychloroquine sulfate and azithromycin for COVID-19: What is the evidence and what are the risks? Int. J. Antimicrob. Agents 2020, 106056. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Liu, D.; Liu, L.; Liu, P.; Xu, Q.; Xia, L.; Ling, Y.; Huang, D.; Song, S.; Zhang, D.; et al. A pilot study of hydroxychloroquine in treatment of patients with moderate COVID-19. J. Zhejiang Univ. 2020, 49, 215–219. [Google Scholar]
- Gautret, P.; Lagier, J.C.; Parola, P.; Hoang, V.T.; Meddeb, L.; Sevestre, J.; Mailhe, M.; Doudier, B.; Aubry, C.; Amrane, S.; et al. Clinical and microbiological effect of a combination of hydroxychloroquine and azithromycin in 80 COVID-19 patients with at least a six-day follow up: A pilot observational study. Travel Med. Infect. Dis. 2020, 34, 101663. [Google Scholar] [CrossRef]
- Boulware, D.R.; Pullen, M.F.; Bangdiwala, A.S.; Pastick, K.A.; Lofgren, S.M.; Okafor, E.C.; Skipper, C.P.; Nascene, A.A.; Nicol, M.R.; Abassi, M.; et al. A Randomized Trial of Hydroxychloroquine as Postexposure Prophylaxis for Covid-19. N. Engl. J. Med. 2020, NEJMoa2016638. [Google Scholar] [CrossRef] [PubMed]
- Chary, M.A.; Barbuto, A.F.; Izadmehr, S.; Hayes, B.D.; Burns, M.M. COVID-19: Therapeutics and Their Toxicities. J. Med. Toxicol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Marmor, M.F.; Kellner, U.; Lai, T.Y.; Melles, R.B.; Mieler, W.F. American Academy of Ophthalmology. Recommendations on Screening for Chloroquine and Hydroxychloroquine Retinopathy (2016 Revision). Ophthalmology 2016, 123, 1386–1394. [Google Scholar] [CrossRef]
- Shippey, E.A.; Wagler, V.D.; Collamer, A.N. Hydroxychloroquine: An old drug with new relevance. Cleve. Clin. J. Med. 2018, 85, 459–467. [Google Scholar] [CrossRef]
- Nevin, R.L. A serious nightmare: Psychiatric and neurologic adverse reactions to mefloquine are serious adverse reactions. Pharmacol. Res. Perspect. 2017, 5, e00328. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration. Primaquine. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2017/008316s023lbl.pdf (accessed on 15 May 2020).
- National Institutes of Health. Coronavirus Disease 2019 (COVID-19) Treatment Guidelines. Available online: https://www.covid19treatmentguidelines.nih.gov/antiviral-therapy/chloroquine-or-hydroxychloroquine/ (accessed on 27 June 2020).
- U.S. Food and Drug Administration. FDA Drug Safety Communication: FDA Cautions against Use of Hydroxychloroquine or Chloroquine for COVID-19 outside of the Hospital Setting or a Clinical Trial due to Risk of Heart Rhythm Problems. Available online: https://www.fda.gov/media/137250/download (accessed on 27 June 2020).
- National Health Commission (NHC); State Administration of Traditional Chinese Medicine (Trial Version 7). Diagnosis and Treatment Protocol for Novel Coronavirus Pneumonia. Available online: http://busan.china-consulate.org/chn/zt/4/P020200310548447287942.pdf (accessed on 27 June 2020).
- U.S. Food and Drug Administration. Letter of authorization: Emergency Use Authorization for Use of Chloroquine Phosphate or Hydroxychloroquine Sulfate Supplied from the Strategic National Stockpile for Treatment of 2019 Coronavirus Disease. 28 March 2020. Available online: https://www.fda.gov/media/136534/download (accessed on 27 June 2020).
- U.S. Food and Drug Administration. Letter Regarding Revocation of Emergency Use Authorization (EUA) for Emergency Use of Chloroquine Phosphate and Hydroxychloroquine Sulfate Supplied from the Strategic National Stockpile for Treatment of Coronavirus Disease 2019. 15 June 2020. Available online: https://www.fda.gov/media/138945/download (accessed on 27 June 2020).
- Benowitz, N.L. Section III: Cardiovascular-renal drugs. In Basic & Clinical Pharmacology; Katzung, B.G., Ed.; Lange Medical Books/McGraw Hill: New York, NY, USA, 2017. [Google Scholar]
- Fang, L.; Karakiulakis, G.; Roth, M. Are patients with hypertension and diabetes mellitus at increased risk for COVID-19 infection? Lancet Respir. Med. 2020, 8, e21. [Google Scholar] [CrossRef]
- Ingraham, N.E.; Barakat, A.G.; Reilkoff, R.; Bezdicek, T.; Schacker, T.; Chipman, J.G.; Tignanelli, C.J.; Puskarich, M.A. Understanding the Renin-Angiotensin-Aldosterone-SARS-CoV-Axis: A Comprehensive Review. Eur. Respir. J. 2020. [Google Scholar] [CrossRef]
- Gurwitz, D. Angiotensin receptor blockers as tentative SARS-CoV-2 therapeutics. Drug Dev Res. 2020. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Liu, Y.; Yang, Y.; Zhang, P.; Zhong, W.; Wang, Y.; Wang, Q.; Xu, Y.; Li, M.; Li, X.; et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B 2020, 10, 766–788. [Google Scholar] [CrossRef] [PubMed]
- Vaduganathan, M.; Vardeny, O.; Michel, T.; McMurray, J.J.V.; Pfeffer, M.A.; Solomon, S.D. Renin-Angiotensin-Aldosterone System Inhibitors in Patients with Covid-19. N. Engl. J. Med. 2020, 382, 1653–1659. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, H.R.; Adhikari, S.; Pulgarin, C.; Troxel, A.B.; Iturrate, E.; Johnson, S.B.; Hausvater, A.; Newman, J.D.; Berger, J.S.; Bangalore, S.; et al. Renin-Angiotensin-Aldosterone System Inhibitors and Risk of Covid-19. N. Engl. J. Med. 2020, 382, 2441–2448. [Google Scholar] [CrossRef]
- Mancia, G.; Rea, F.; Ludergnani, M.; Apolone, G.; Corrao, G. Renin-Angiotensin-Aldosterone System Blockers and the Risk of Covid-19. N. Engl. J. Med. 2020, 382, 2431–2440. [Google Scholar] [CrossRef]
- Sidaway, J.E.; Davidson, R.G.; McTaggart, F.; Orton, T.C.; Scott, R.C.; Smith, G.J.; Brunskill, N.J. Inhibitors of 3-hydroxy-3-methylglutaryl-CoA reductase reduce receptor-mediated endocytosis in opossum kidney cells. J. Am. Soc. Nephrol. 2004, 15, 2258–2265. [Google Scholar] [CrossRef]
- Verhulst, A.; D’Haese, P.C.; De Broe, M.E. Inhibitors of HMG-CoA reductase reduce receptor-mediated endocytosis in human kidney proximal tubular cells. J. Am. Soc. Nephrol. 2004, 15, 2249–2257. [Google Scholar] [CrossRef]
- Reiner, Ž.; Hatamipour, M.; Banach, M.; Pirro, M.; Al-Rasadi, K.; Jamialahmadi, T.; Radenkovic, D.; Montecucco, F.; Sahebkar, A. Statins and the COVID-19 main protease: In silico evidence on direct interaction. Arch Med. Sci. 2020, 16, 490–496. [Google Scholar] [CrossRef]
- Wösten-van Asperen, R.M.; Bos, A.P.; Bem, R.A.; Dierdorp, B.S.; Dekker, T.; van Goor, H.; Kamilic, J.; van der Loos, C.M.; van den Berg, E.; Bruijn, M.; et al. Imbalance between pulmonary angiotensin-converting enzyme and angiotensin-converting enzyme 2 activity in acute respiratory distress syndrome. Pediatric Crit. Care Med. 2013, 14, e438–e441. [Google Scholar] [CrossRef]
- Fedson, D.S.; Opal, S.M.; Rordam, O.M. Hiding in Plain Sight: An Approach to Treating Patients with Severe COVID-19 Infection. mBio 2020, 11, e00398-20. [Google Scholar] [CrossRef]
- Yuan, S. Statins May Decrease the Fatality Rate of Middle East Respiratory Syndrome Infection. mBio 2015, 6, e01120. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava-Ranjan, P.; Flint, M.; Bergeron, É.; McElroy, A.K.; Chatterjee, P.; Albariño, C.G.; Nichol, S.T.; Spiropoulou, C.F. Statins Suppress Ebola Virus Infectivity by Interfering with Glycoprotein Processing. mBio 2018, 9, e00660-18. [Google Scholar] [CrossRef] [PubMed]
- Españo, E.; Nam, J.H.; Song, E.J.; Song, D.; Lee, C.K.; Kim, J.K. Lipophilic statins inhibit Zika virus production in Vero cells. Sci. Rep. 2019, 9, 11461. [Google Scholar] [CrossRef] [PubMed]
- Fedson, D.S. Pandemic influenza: A potential role for statins in treatment and prophylaxis. Clin. Infect. Dis. 2006, 43, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Bryan-Marrugo, O.L.; Arellanos-Soto, D.; Rojas-Martinez, A.; Barrera-Saldaña, H.; Ramos-Jimenez, J.; Vidaltamayo, R.; Rivas-Estilla, A.M. The anti-dengue virus properties of statins may be associated with alterations in the cellular antiviral profile expression. Mol. Med. Rep. 2016, 14, 2155–2163. [Google Scholar] [CrossRef]
- Phadke, M.; Saunik, S. COVID-19 treatment by repurposing drugs until the vaccine is in sight. Drug Dev. Res. 2020. [Google Scholar] [CrossRef]
- De Spiegeleer, A.; Bronselaer, A.; Teo, J.T.; Byttebier, G.; De Tré, G.; Belmans, L.; Dobson, R.; Wynendaele, E.; Van De Wiele, C.; Vandaele, F.; et al. The Effects of ARBs, ACEis, and Statins on Clinical Outcomes of COVID-19 Infection Among Nursing Home Residents. J. Am. Med. Dir. Assoc. 2020, 21, 909–914.e2. [Google Scholar] [CrossRef]
- Zanasi, A.; Mazzolini, M.; Kantar, A. A reappraisal of the mucoactive activity and clinical efficacy of bromhexine. Multidiscip. Respir. Med. 2017, 12, 7. [Google Scholar] [CrossRef]
- Lucas, J.M.; Heinlein, C.; Kim, T.; Hernandez, S.A.; Malik, M.S.; True, L.D.; Morrissey, C.; Corey, E.; Montgomery, B.; Mostaghel, E.; et al. The androgen-regulated protease TMPRSS2 activates a proteolytic cascade involving components of the tumor microenvironment and promotes prostate cancer metastasis. Cancer Discov. 2014, 4, 1310–1325. [Google Scholar] [CrossRef]
- Iwata-Yoshikawa, N.; Okamura, T.; Shimizu, Y.; Hasegawa, H.; Takeda, M.; Nagata, N. TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. J. Virol. 2019, 93, e01815-18. [Google Scholar] [CrossRef]
- Shen, L.W.; Mao, H.J.; Wu, Y.L.; Tanaka, Y.; Zhang, W. TMPRSS2: A potential target for treatment of influenza virus and coronavirus infections. Biochimie 2017, 142, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Maggio, R.; Corsini, G.U. Repurposing the mucolytic cough suppressant and TMPRSS2 protease inhibitor bromhexine for the prevention and management of SARS-CoV-2 infection. Pharmacol. Res. 2020, 157, 104837. [Google Scholar] [CrossRef] [PubMed]
- Markus, D.; Gottfried, L.; Markus, M.; Marina, R.; Dario, B.; Danielle de, V. A SARS-CoV-2 prophylactic and treatment; A counter argument against the sole use of chloroquine. Am J Biomed Sci. 2020, 248–351. [Google Scholar]
- Seifart, C.; Clostermann, U.; Seifart, U.; Müller, B.; Vogelmeier, C.; von Wichert, P.; Fehrenbach, H. Cell-specific modulation of surfactant proteins by ambroxol treatment. Toxicol. Appl. Pharmacol. 2005, 203, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, M.L.; Nuttall, J.; Hart, P.A.; TACTIC Investigative Team. A phase 1/2 trial to evaluate the pharmacokinetics, safety, and efficacy of NI-03 in patients with chronic pancreatitis: Study protocol for a randomized controlled trial on the assessment of camostat treatment in chronic pancreatitis (TACTIC). Trials 2019, 20, 501. [Google Scholar] [CrossRef]
- Maekawa, A.; Kakizoe, Y.; Miyoshi, T.; Wakida, N.; Ko, T.; Shiraishi, N.; Adachi, M.; Tomita, K.; Kitamura, K. Camostat mesilate inhibits prostasin activity and reduces blood pressure and renal injury in salt-sensitive hypertension. J. Hypertens. 2009, 27, 181–189. [Google Scholar] [CrossRef]
- Okuno, M.; Kojima, S.; Akita, K.; Matsushima-Nishiwaki, R.; Adachi, S.; Sano, T.; Takano, Y.; Takai, K.; Obora, A.; Yasuda, I.; et al. Retinoids in liver fibrosis and cancer. Front. Biosci. 2002, 7, d204-18. [Google Scholar] [CrossRef]
- Hsieh, H.P.; Hsu, J.T. Strategies of development of antiviral agents directed against influenza virus replication. Curr. Pharm. Des. 2007, 13, 3531–3542. [Google Scholar] [CrossRef]
- Kawase, M.; Shirato, K.; van der Hoek, L.; Taguchi, F.; Matsuyama, S. Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. J. Virol. 2012, 86, 6537–6545. [Google Scholar] [CrossRef]
- Uno, Y. Camostat mesilate therapy for COVID-19. Intern. Emerg. Med. 2020, 1–2. [Google Scholar] [CrossRef]
- Coote, K.; Atherton-Watson, H.C.; Sugar, R.; Young, A.; MacKenzie-Beevor, A.; Gosling, M.; Bhalay, G.; Bloomfield, G.; Dunstan, A.; Bridges, R.J.; et al. Camostat attenuates airway epithelial sodium channel function in vivo through the inhibition of a channel-activating protease. J. Pharmacol. Exp. Ther. 2009, 329, 764–774. [Google Scholar] [CrossRef]
- Hirota, M.; Shimosegawa, T.; Kitamura, K.; Takeda, K.; Takeyama, Y.; Mayumi, T. Continuous regional arterial infusion versus intravenous administration of the protease inhibitor nafamostat mesilate for predicted severe acute pancreatitis: A multicenter, randomized, open-label, phase 2 trial. J. Gastroenterol. 2020, 55, 342–352. [Google Scholar] [CrossRef]
- Hiraishi, M.; Yamazaki, Z.; Ichikawa, K.; Kanai, F.; Idezuki, Y.; Onishi, K. Plasma collection using nafamostat mesilate and dipyridamole as an anticoagulant. Int. J. Artif. Organs 1988, 11, 212–216. [Google Scholar] [CrossRef]
- Maruyama, Y.; Yoshida, H.; Uchino, S.; Yokoyama, K.; Yamamoto, H.; Takinami, M.; Hosoya, T. Nafamostat mesilate as an anticoagulant during continuous veno-venous hemodialysis: A three-year retrospective cohort study. Int. J. Artif. Organs 2011, 34, 571–576. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Matsuyama, S.; Li, X.; Takeda, M.; Kawaguchi, Y.; Inoue, J.I. Identification of nafamostat as a potent inhibitor of middle east respiratory syndrome coronavirus S protein-mediated membrane fusion using the split-protein-based cell-cell fusion assay. Antimicrob. Agents Chemother. 2016, 60, 6532–6539. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Schroeder, S.; Kleine-Weber, H.; Müller, M.A.; Drosten, C.; Pöhlmann, S. Nafamostat mesylate blocks activation of SARS-CoV-2: New treatment option for COVID-19. Antimicrob. Agents Chemother. 2020, e00754-20. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Lee, K.E.; Oh, J.H.; Jung, C.S.; Choi, D.; Kim, Y.; Jeon, J.S.; Han, D.C.; Noh, H. Cardiac arrest caused by nafamostat mesilate. Kidney Res. Clin Pract. 2016, 35, 187–189. [Google Scholar] [CrossRef]
- Nakatsuka, M.; Asagiri, K.; Noguchi, S.; Habara, T.; Kudo, T. Nafamostat mesilate, a serine protease inhibitor, suppresses lipopolysaccharide-induced nitric oxide synthesis and apoptosis in cultured human trophoblasts. Life Sci. 2000, 67, 1243–1250. [Google Scholar] [CrossRef]
- Kang, M.W.; Song, H.J.; Kang, S.K.; Kim, Y.; Jung, S.B.; Jee, S.; Moon, J.Y.; Suh, K.S.; Lee, S.D.; Jeon, B.H.; et al. Nafamostat mesilate inhibits TNF-alpha-induced vascular endothelial cell dysfunction by inhibiting reactive oxygen species production. Korean J. Physiol. Pharmacol. 2015, 19, 229–234. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.; Rhee, J.Y. Three cases of treatment with Nafamostat in elderly patients with COVID-19 pneumonia who need oxygen therapy. Int. J. Infect. Dis. 2020, 96, 500–502. [Google Scholar] [CrossRef] [PubMed]
- Sonawane, K.; Barale, S.S.; Dhanavade, M.J.; Waghmare, S.R.; Nadaf, N.H.; Kamble, S.A.; Mohammed, A.A.; Makandar, A.M.; Fandilolu, P.M.; Dound, A.S.; et al. Homology modeling and docking studies of TMPRSS2 with experimentally known inhibitors camostat mesylate, nafamostat and bromhexine hydrochloride to control SARS-Coronavirus-2. ChemRxiv 2020. [Google Scholar] [CrossRef]
- Spížek, J.; Řezanka, T. Lincomycin, clindamycin and their applications. Appl. Microbiol. Biotechnol. 2004, 64, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Brook, I.; Lewis, M.A.; Sándor, G.K.; Jeffcoat, M.; Samaranayake, L.P.; Vera Rojas, J. Clindamycin in dentistry: More than just effective prophylaxis for endocarditis? Oral. Surg. Oral. Med. Oral. Pathol. Oral. Radiol. Endod. 2005, 100, 550–558. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, S.; Madke, B.; Kabra, P.; Singh, A.L. Anti-inflammatory and immunomodulatory effects of antibiotics and their use in dermatology. Indian J. Dermatol. 2016, 61, 469–481. [Google Scholar]
- Blaising, J.; Polyak, S.J.; Pecheur, E.I. Arbidol as a broad-spectrum antiviral: An update. Antivir. Res. 2014, 107, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Haviernik, J.; Stefanik, M.; Fojtikova, M.; Kali, S.; Tordo, N.; Rudolf, I.; Hubalek, Z.; Eyer, L.; Ruzek, D. Arbidol (Umifenovir): A broad-spectrum antiviral drug that inhibits medically important arthropod-borne Flaviviruses. Viruses 2018, 10, 184. [Google Scholar] [CrossRef]
- Hulseberg, C.E.; Fénéant, L.; Szymańska-de Wijs, K.M.; Kessler, N.P.; Nelson, E.A.; Shoemaker, C.J.; Schmaljohn, C.S.; Polyak, S.J.; White, J.M. Arbidol and other low-molecular-weight drugs that inhibit Lassa and Ebola viruses. J. Virol. 2019, 93, e02185-18. [Google Scholar] [CrossRef]
- Fink, S.L.; Vojtech, L.; Wagoner, J.; Slivinski, N.S.J.; Jackson, K.J.; Wang, R.; Khadka, S.; Luthra, P.; Basler, C.F.; Polyak, S.J. The antiviral drug arbidol inhibits Zika virus. Sci. Rep. 2018, 8, 8989. [Google Scholar] [CrossRef]
- Pecheur, E.I.; Borisevich, V.; Halfmann, P.; Morrey, J.D.; Smee, D.F.; Prichard, M.; Mire, C.E.; Kawaoka, Y.; Geisbert, T.W.; Polyak, S.J. The synthetic antiviral drug arbidol inhibits globally prevalent pathogenic viruses. J. Virol. 2016, 90, 3086–3092. [Google Scholar] [CrossRef]
- Li, M.K.; Liu, Y.Y.; Wei, F.; Shen, M.X.; Zhong, Y.; Li, S.; Chen, L.J.; Ma, N.; Liu, B.Y.; Mao, Y.D.; et al. Antiviral activity of arbidol hydrochloride against herpes simplex virus I in vitro and in vivo. Int. J. Antimicrob. Agents 2018, 51, 98–106. [Google Scholar] [CrossRef]
- Herod, M.R.; Adeyemi, O.O.; Ward, J.; Bentley, K.; Harris, M.; Stonehouse, N.J.; Polyak, S.J. The broad-spectrum antiviral drug arbidol inhibits foot-and-mouth disease virus genome replication. J. Gen. Virol. 2019, 100, 1293–1302. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Hu, S.; Gao, J. Discovering drugs to treat coronavirus disease 2019 (COVID-19). Drug Discov. Ther. 2020, 14, 58–60. [Google Scholar] [CrossRef] [PubMed]
- Kadam, R.U.; Wilson, I.A. Structural basis of influenza virus fusion inhibition by the antiviral drug Arbidol. Proc. Natl. Acad. Sci. USA 2017, 114, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.Y.; Yang, J.; Liu, S. Investigational hemagglutinin-targeted influenza virus inhibitors. Expert Opin. Investig. Drugs. 2017, 26, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Blaising, J.; Levy, P.L.; Polyak, S.J.; Stanifer, M.; Boulant, S.; Pecheur, E.I. Arbidol inhibits viral entry by interfering with clathrin-dependent trafficking. Antivir. Res. 2013, 100, 215–219. [Google Scholar] [CrossRef]
- Teissier, E.; Zandomeneghi, G.; Loquet, A.; Lavillette, D.; Lavergne, J.P.; Montserret, R.; Cosset, F.L.; Bockmann, A.; Meier, B.H.; Penin, F.; et al. Mechanism of inhibition of enveloped virus membrane fusion by the antiviral drug arbidol. PLoS ONE 2011, 6, e15874. [Google Scholar] [CrossRef]
- Vankadari, N. Arbidol: A potential antiviral drug for the treatment of SARS-CoV-2 by blocking the trimerization of viral spike glycoprotein? Int. J. Antimicrob. Agents 2020, 105998. [Google Scholar] [CrossRef]
- Zhu, Z.; Lu, Z.; Xu, T.; Chen, C.; Yang, G.; Zha, T.; Lu, J.; Xue, Y. Arbidol monotherapy is superior to lopinavir/ritonavir in treating COVID-19. J. Infect. 2020, 81, e21–e23. [Google Scholar] [CrossRef]
- Deng, L.; Li, C.; Zeng, Q.; Liu, X.; Li, X.; Zhang, H.; Hong, Z.; Xia, J. Arbidol combined with LPV/r versus LPV/r alone against Corona Virus Disease 2019: A retrospective cohort study. J. Infect. 2020, 81, e1–e5. [Google Scholar] [CrossRef]
- Lian, N.; Xie, H.; Lin, S.; Huang, J.; Zhao, J.; Lin, Q. Umifenovir treatment is not associated with improved outcomes in patients with coronavirus disease 2019: A retrospective study. Clin. Microbiol. Infect. 2020, 26, 917–921. [Google Scholar] [CrossRef]
- Chen, C.; Zhang, Y.; Huang, J.; Yin, P.; Cheng, Z.; Wu, J.; Chen, S.; Zhang, Y.; Bo Chen, B.; Lu, M.; et al. Favipiravir versus arbidol for COVID-19: A randomized clinical trial. MedRxiv 2020. [Google Scholar] [CrossRef]
- Li, Y.; Xie, Z.; Lin, W.; Cai, W.; Wen, C.; Guan, Y.; Mo, X.; Wang, J.; Wang, Y.; Peng, P.; et al. Efficacy and safety of lopinavir/ritonavir or arbidol in adult patients with mild/moderate COVID-19: An exploratory randomized controlled trial. Med. J. 2020. [Google Scholar] [CrossRef]
- Fohner, A.E.; Sparreboom, A.; Altman, R.B.; Klein, T.E. PharmGKB summary: Macrolide antibiotic pathway, pharmacokinetics/pharmacodynamics. Pharm. Genom. 2017, 27, 164–167. [Google Scholar] [CrossRef] [PubMed]
- Dinos, G.P. The macrolide antibiotic renaissance. Br. J. Pharmacol. 2017, 174, 2967–2983. [Google Scholar] [CrossRef] [PubMed]
- McMullan, B.J.; Mostaghim, M. Prescribing azithromycin. Aust. Prescr. 2015, 38, 87–89. [Google Scholar] [CrossRef] [PubMed]
- Andreani, J.; Le Bideau, M.; Duflot, I.; Jardot, P.; Rolland, C.; Boxberger, M.; Wurtz, N.; Rolain, J.M.; Colson, P.; La Scola, B.; et al. In vitro testing of Hydroxychloroquine and Azithromycin on SARS-CoV-2 shows 1 synergistic effect 2. Lung 2020, 21, 22. [Google Scholar]
- Touret, F.; Gilles, M.; Barral, K.; Nougairède, A.; Decroly, E.; de Lamballerie, X.; Coutard, B. In vitro screening of a FDA approved chemical library reveals potential inhibitors of SARS-CoV-2 replication. bioRxiv 2020. [Google Scholar] [CrossRef]
- Poschet, J.; Perkett, E.; Timmins, G.; Deretic, V. Azithromycin and ciprofloxacin have a chloroquine-like effect on respiratory epithelial cells. bioRxiv 2020. [Google Scholar] [CrossRef]
- Bosseboeuf, E.; Aubry, M.; Nhan, T.; de Pina, J.J.; Rolain, J.M.; Raoult, D.; Musso, D. Azithromycin inhibits the replication of Zika virus. J. Antivir. Antiretrovir. 2018, 10, 6–11. [Google Scholar] [CrossRef]
- Retallack, H.; Di Lullo, E.; Arias, C.; Knopp, K.A.; Laurie, M.T.; Sandoval-Espinosa, C.; Mancia Leon, W.R.; Krencik, R.; Ullian, E.M.; Spatazza, J.; et al. Zika virus cell tropism in the developing human brain and inhibition by azithromycin. Proc. Natl. Acad. Sci. USA 2016, 113, 14408–14413. [Google Scholar] [CrossRef]
- Li, C.; Zu, S.; Deng, Y.Q.; Li, D.; Parvatiyar, K.; Quanquin, N.; Shang, J.; Sun, N.; Su, J.; Liu, Z.; et al. Azithromycin protects against Zika virus infection by upregulating virus-induced type I and III interferon responses. Antimicrob. Agents Chemother. 2019, 63, e00394-19. [Google Scholar] [CrossRef] [PubMed]
- Madrid, P.B.; Panchal, R.G.; Warren, T.K.; Shurtleff, A.C.; Endsley, A.N.; Green, C.E.; Kolokoltsov, A.; Davey, R.; Manger, I.D.; Gilfillan, L.; et al. Evaluation of Ebola Virus inhibitors for drug repurposing. ACS Infect. Dis. 2015, 1, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Tran, D.H.; Sugamata, R.; Hirose, T.; Suzuki, S.; Noguchi, Y.; Sugawara, A.; Ito, F.; Yamamoto, T.; Kawachi, S.; Akagawa, K.S.; et al. Azithromycin, a 15-membered macrolide antibiotic, inhibits influenza A (H1N1)pdm09 virus infection by interfering with virus internalization process. J. Antibiot. (Tokyo) 2019, 72, 759–768. [Google Scholar] [CrossRef] [PubMed]
- Bermejo-Martin, J.F.; Kelvin, D.J.; Eiros, J.M.; Castrodeza, J.; Ortiz de Lejarazu, R. Macrolides for the treatment of severe respiratory illness caused by novel H1N1 swine influenza viral strains. J. Infect. Dev. Ctries. 2009, 3, 159–161. [Google Scholar] [CrossRef] [PubMed]
- Altenburg, J.; de Graaff, C.S.; Stienstra, Y.; Sloos, J.H.; van Haren, E.H.; Koppers, R.J.; van der Werf, T.S.; Boersma, W.G. Effect of azithromycin maintenance treatment on infectious exacerbations among patients with non-cystic fibrosis bronchiectasis: The BAT randomized controlled trial. JAMA 2013, 309, 1251–1259. [Google Scholar] [CrossRef]
- Feola, D.J.; Garvy, B.A.; Cory, T.J.; Birket, S.E.; Hoy, H.; Hayes DJr Murphy, B.S. Azithromycin alters macrophage phenotype and pulmonary compartmentalization during lung infection with Pseudomonas. Antimicrob. Agents Chemother. 2010, 54, 2437–2447. [Google Scholar] [CrossRef]
- Min, J.Y.; Jang, Y.J. Macrolide therapy in respiratory viral infections. Mediat. Inflamm. 2012, 2012, 649570. [Google Scholar] [CrossRef]
- Li, H.; Liu, D.H.; Chen, L.L.; Zhao, Q.; Yu, Y.Z.; Ding, J.J.; Miao, L.Y.; Xiao, Y.L.; Cai, H.R.; Zhang, D.P.; et al. Meta-analysis of the adverse effects of long-term azithromycin use in patients with chronic lung diseases. Antimicrob. Agents Chemother. 2014, 58, 511–517. [Google Scholar] [CrossRef]
- Gielen, V.; Johnston, S.L.; Edwards, M.R. Azithromycin induces anti-viral responses in bronchial epithelial cells. Eur. Respir. J. 2010, 36, 646–654. [Google Scholar] [CrossRef]
- Krempaska, K.; Barnowski, S.; Gavini, J.; Hobi, N.; Ebener, S.; Simillion, C.; Stokes, A.; Schliep, R.; Knudsen, L.; Geiser, T.K.; et al. Azithromycin has enhanced effects on lung fibroblasts from idiopathic pulmonary fibrosis (IPF) patients compared to controls. Respir. Res. 2020, 21, 25. [Google Scholar] [CrossRef]
- Kawamura, K.; Ichikado, K.; Yasuda, Y.; Anan, K.; Suga, M. Azithromycin for idiopathic acute exacerbation of idiopathic pulmonary fibrosis: A retrospective single-center study. BMC Pulm. Med. 2017, 17, 94. [Google Scholar] [CrossRef] [PubMed]
- Hiratsuka, T.; Mukae, H.; Iiboshi, H.; Ashitani, J.; Nabeshima, K.; Minematsu, T.; Chino, N.; Ihi, T.; Kohno, S.; Nakazato, M. Increased concentrations of human beta-defensins in plasma and bronchoalveolar lavage fluid of patients with diffuse panbronchiolitis. Thorax 2003, 58, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, K.; Ichikado, K.; Takaki, M.; Eguchi, Y.; Anan, K.; Suga, M. Adjunctive therapy with azithromycin for moderate and severe acute respiratory distress syndrome: A retrospective, propensity score-matching analysis of prospectively collected data at a single center. Int. J. Antimicrob. Agents 2018, 51, 918–924. [Google Scholar] [CrossRef] [PubMed]
- Gautret, P.; Lagier, J.C.; Parola, P.; Hoang, V.T.; Meddeb, L.; Mailhe, M.; Doudier, B.; Courjon, J.; Giordanengo, V.; Vieira, V.E.; et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: Results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents 2020, 105949. [Google Scholar] [CrossRef]
- Millán-Oñate, J.; Millan, W.; Mendoza, L.A.; Sánchez, C.G.; Fernandez-Suarez, H.; Bonilla-Aldana, D.K.; Rodríguez-Morales, A.J. Successful recovery of COVID-19 pneumonia in a patient from Colombia after receiving chloroquine and clarithromycin. Ann. Clin. Microbiol. Antimicrob. 2020, 19, 16. [Google Scholar] [CrossRef]
- Jiang, E.; Jiang, X.; Jiang, X.; XIA, M. Use of Carrimycin and Pharmaceutically Acceptable Salt Thereof for Manufacture of Medicament for Treatment and/or Prevention of Tumors. U.S. Patent 16/500,967, 30 January 2020. [Google Scholar]
- Wang, Y.; Jiang, Y.; Zhao, X.; He, W. Use of Carrimycin in Mycobacterium Tuberculosis Infection Resistance. U.S. Patent 16/067,327, 3 January 2019. [Google Scholar]
- Crump, A. Ivermectin: Enigmatic multifaceted ‘wonder’ drug continues to surprise and exceed expectations. J. Antibiot (Tokyo) 2017, 70, 495–505. [Google Scholar] [CrossRef]
- McCavera, S.; Rogers, A.T.; Yates, D.M.; Woods, D.J.; Wolstenholme, A.J. An ivermectin-sensitive glutamate-gated chloride channel from the parasitic nematode Haemonchus contortus. Mol. Pharmacol. 2009, 75, 1347–1355. [Google Scholar] [CrossRef]
- Wagstaff, K.M.; Sivakumaran, H.; Heaton, S.M.; Harrich, D.; Jans, D.A. Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem. J. 2012, 443, 851–856. [Google Scholar] [CrossRef]
- Yang, S.N.Y.; Atkinson, S.C.; Wang, C.; Lee, A.; Bogoyevitch, M.A.; Borg, N.A.; Jans, D.A. The broad spectrum antiviral ivermectin targets the host nuclear transport importin α/β1 heterodimer. Antivir. Res. 2020, 177, 104760. [Google Scholar] [CrossRef]
- Wulan, W.N.; Heydet, D.; Walker, E.J.; Gahan, M.E.; Ghildyal, R. Nucleocytoplasmic transport of nucleocapsid proteins of enveloped RNA viruses. Front. Microbiol. 2015, 6, 553. [Google Scholar] [CrossRef] [PubMed]
- Rowland, R.R.; Chauhan, V.; Fang, Y.; Pekosz, A.; Kerrigan, M.; Burton, M.D. Intracellular localization of the severe acute respiratory syndrome coronavirus nucleocapsid protein: Absence of nucleolar accumulation during infection and after expression as a recombinant protein in vero cells. J. Virol. 2005, 79, 11507–11512. [Google Scholar] [CrossRef] [PubMed]
- Timani, K.A.; Liao, Q.; Ye, L.; Zeng, Y.; Liu, J.; Zheng, Y.; Ye, L.; Yang, X.; Lingbao, K.; Gao, J.; et al. Nuclear/ nucleolar localization properties of C-terminal nucleocapsid protein of SARS coronavirus. Virus Res. 2005, 114, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Caly, L.; Druce, J.D.; Catton, M.G.; Jans, D.A.; Wagstaff, K.M. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir. Res. 2020, 178, 104787. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Mook, R.A., Jr.; Premont, R.T.; Wang, J. Niclosamide: Beyond an antihelminthic drug. Cell Signal. 2018, 41, 89–96. [Google Scholar] [CrossRef]
- Pan, J.X.; Ding, K.; Wang, C.Y. Niclosamide, an old antihelminthic agent, demonstrates antitumor activity by blocking multiple signaling pathways of cancer stem cells. Chin. J. Cancer 2012, 31, 178–184. [Google Scholar] [CrossRef]
- Pizzorno, A.; Terrier, O.; Nicolas de Lamballerie, C.; Julien, T.; Padey, B.; Traversier, A.; Roche, M.; Hamelin, M.E.; Rhéaume, C.; Croze, S.; et al. Repurposing of Drugs as Novel Influenza Inhibitors from Clinical Gene Expression Infection Signatures. Front. Immunol. 2019, 10, 60. [Google Scholar] [CrossRef]
- Wu, C.J.; Jan, J.T.; Chen, C.M.; Hsieh, H.P.; Hwang, D.R.; Liu, H.W.; Liu, C.Y.; Huang, H.W.; Chen, S.C.; Hong, C.F.; et al. Inhibition of severe acute respiratory syndrome coronavirus replication by niclosamide. Antimicrob. Agents Chemother. 2004, 48, 2693–2696. [Google Scholar] [CrossRef]
- Xu, J.; Shi, P.Y.; Li, H.; Zhou, J. Broad Spectrum Antiviral Agent Niclosamide and Its Therapeutic Potential. ACS Infect. Dis. 2020, 6, 909–915. [Google Scholar] [CrossRef]
- Jeon, S.; Ko, M.; Lee, J.; Choi, I.; Byun, S.Y.; Park, S.; Shum, D.; Kim, S. Identification of Antiviral Drug Candidates Against SARS-CoV-2 From FDA-approved Drugs. Antimicrob. Agents Chemother. 2020, 6, 909–915. [Google Scholar] [CrossRef]
- Pindiprolu, S.K.S.S.; Pindiprolu, S.H. Plausible mechanisms of Niclosamide as an antiviral agent against COVID-19. Med. Hypotheses 2020, 140, 109765. [Google Scholar] [CrossRef]
- Griffin, M.O.; Fricovsky, E.; Ceballos, G.; Villarreal, F. Tetracyclines: A pleitropic family of compounds with promising therapeutic properties. Review of the literature. Am. J. Physiol. Cell Physiol. 2010, 299, C539–C548. [Google Scholar] [CrossRef] [PubMed]
- Zakeri, B.; Wright, G.D. Chemical biology of tetracycline antibiotics. Biochem. Cell Biol. 2008, 86, 124–136. [Google Scholar] [CrossRef] [PubMed]
- Humar, A.; McGilvray, I.; Phillips, M.J.; Levy, G.A. Severe acute respiratory syndrome and the liver. Hepatology 2004, 39, 291–294. [Google Scholar] [CrossRef]
- Phillips, J.M.; Gallagher, T.; Weiss, S.R. Neurovirulent murine coronavirus JHM.SD uses cellular zinc metalloproteases for virus entry and cell-cell fusion. J. Virol. 2017, 91, e01564-16. [Google Scholar] [CrossRef] [PubMed]
- Rothan, H.A.; Mohamed, Z.; Paydar, M.; Rahman, N.A.; Yusof, R. Inhibitory effect of doxycycline against dengue virus replication in vitro. Arch. Virol. 2014, 159, 711–718. [Google Scholar] [CrossRef]
- Henehan, M.; Montuno, M.; De Benedetto, A. Doxycycline as an anti-inflammatory agent: Updates in dermatology. J. Eur. Acad. Dermatol. Venereol. 2017, 31, 1800–1808. [Google Scholar] [CrossRef]
- Kritas, S.K.; Ronconi, G.; Caraffa, A.; Gallenga, C.E.; Ross, R.; Conti, P. Mast cells contribute to coronavirus-induced inflammation: New anti-inflammatory strategy. J. Biol. Regul. Homeost. Agents 2020, 34. [Google Scholar] [CrossRef]
- Sandler, C.; Nurmi, K.; Lindstedt, K.A.; Sorsa, T.; Golub, L.M.; Kovanen, P.T.; Eklund, K.K. Chemically modified tetracyclines induce apoptosis in cultured mast cells. Int. Immunopharmacol. 2005, 5, 1611–1621. [Google Scholar] [CrossRef]
- Sandler, C.; Ekokoski, E.; Lindstedt, K.A.; Vainio, P.J.; Finel, M.; Sorsa, T.; Kovanen, P.T.; Golub, L.M.; Eklund, K.K. Chemically modified tetracycline (CMT)-3 inhibits histamine release and cytokine production in mast cells: Possible involvement of protein kinase C. Inflamm. Res. 2005, 54, 304–312. [Google Scholar] [CrossRef]
- Lee, V.S.; Chong, W.L.; Sukumaran, S.D.; Nimmanpipug, P.; Letchumanan, V.; Goh, B.-H.; Lee, L.H.; Zain, S.M.; Abd Rahman, N. Computational screening and identifying binding interaction of anti-viral and anti-malarial drugs: Toward the potential cure for SARS-CoV-2. Prog. Drug Discov. Biomed. Sci. 2020, 3, a0000065. [Google Scholar] [CrossRef]
- Boyd-Kimball, D.; Gonczy, K.; Lewis, B.; Mason, T.; Siliko, N.; Wolfe, J. Classics in Chemical Neuroscience: Chlorpromazine. ACS Chem. Neurosci. 2019, 10, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Greenblatt, H.K.; Greenblatt, D.J. Use of antipsychotics for the treatment of behavioral symptoms of dementia. J. Clin. Pharmacol. 2016, 56, 1048–1057. [Google Scholar] [CrossRef] [PubMed]
- Pu, Y.; Zhang, X. Mouse hepatitis virus type 2 enters cells through a clathrin-mediated endocytic pathway independent of Eps15. J. Virol. 2008, 82, 8112–8123. [Google Scholar] [CrossRef] [PubMed]
- Chu, V.C.; McElroy, L.J.; Chu, V.; Bauman, B.E.; Whittaker, G.R. The avian coronavirus infectious bronchitis virus undergoes direct low-pH-dependent fusion activation during entry into host cells. J. Virol. 2006, 80, 3180–3188. [Google Scholar] [CrossRef] [PubMed]
- Plaze, M.; Attali, D.; Petit, A.C.; Blatzer, M.; Simon-Loriere, E.; Vinckier, F.; Cachia, A.; Chrétien, F.; Gaillard, R. Repurposing of chlorpromazine in COVID-19 treatment: The reCoVery study. Encephale 2020. [Google Scholar] [CrossRef] [PubMed]
- Weston, S.; Christopher, M.; Coleman, C.M.; Haupt, R.; Logue, J.; Matthews, K.; Frieman, M.B. Broad anti-coronaviral activity of FDA approved drugs against SARS-CoV-2 in vitro and SARS-CoV in vivo. bioRxiv 2020. [Google Scholar] [CrossRef]
- Hamilton Sr, D.; Nandkeolyar, S.; Lan, H.; Desai, P.; Evans, J.; Hauschild, C.; Choksi, D.; Abudayyeh, I.; Contractor, T.; Hilliard, A. Amiodarone: A Comprehensive Guide for Clinicians. Am. J. Cardiovasc. Drugs 2020. [Google Scholar] [CrossRef]
- Godfraind, T. Discovery and development of calcium channel blockers. Front. Pharmacol. 2017, 8, 286. [Google Scholar] [CrossRef]
- Gehring, G.; Rohrmann, K.; Atenchong, N.; Mittler, E.; Becker, S.; Dahlmann, F.; Pöhlmann, S.; Vondran, F.W.; David, S.; Manns, M.P.; et al. The clinically approved drugs amiodarone, dronedarone and verapamil inhibit filovirus cell entry. J. Antimicrob. Chemother. 2014, 69, 2123–2131. [Google Scholar] [CrossRef]
- Salata, C.; Baritussio, A.; Munegato, D.; Calistri, A.; Ha, H.R.; Bigler, L.; Fabris, F.; Parolin, C.; Palù, G.; Mirazimi, A. Amiodarone and metabolite MDEA inhibit Ebola virus infection by interfering with the viral entry process. Pathog. Dis. 2015, 73, ftv032. [Google Scholar] [CrossRef]
- Stadler, K.; Ha, H.R.; Ciminale, V.; Spirli, C.; Saletti, G.; Schiavon, M.; Bruttomesso, D.; Bigler, L.; Follath, F.; Pettenazzo, A.; et al. Amiodarone alters late endosomes and inhibits SARS coronavirus infection at a post-endosomal level. Am. J. Respir. Cell Mol. Biol. 2008, 39, 142–149. [Google Scholar] [CrossRef]
- Cheng, Y.L.; Lan, K.H.; Lee, W.P.; Tseng, S.H.; Hung, L.R.; Lin, H.C.; Lee, F.Y.; Lee, S.D.; Lan, K.H. Amiodarone inhibits the entry and assembly steps of hepatitis C virus life cycle. Clin. Sci. 2013, 125, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Aimo, A.; Baritussio, A.; Emdin, M.; Tascini, C. Amiodarone as a possible therapy for coronavirus infection. Eur. J. Prev. Cardiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Al-Horani, R.A.; Desai, U.R. Recent advances on plasmin inhibitors for the treatment of fibrinolysis-related disorders. Med Res Rev. 2014, 34, 1168–1216. [Google Scholar] [CrossRef]
- Leading Biosciences. Available online: https://leadingbiosciences.com/lb1148/ (accessed on 28 June 2020).
- Sica, D.A. Pharmacokinetics and pharmacodynamics of mineralocorticoid blocking agents and their effects on potassium homeostasis. Heart Fail. Rev. 2005, 10, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Millington, K.; Liu, E.; Chan, Y.M. The Utility of Potassium Monitoring in Gender-Diverse Adolescents Taking Spironolactone. J. Endocr. Soc. 2019, 3, 1031–1038. [Google Scholar] [CrossRef]
- Wambier, C.G.; Goren, A. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection is likely to be androgen mediated. J. Am. Acad. Dermatol. 2020, 83, 308–309. [Google Scholar] [CrossRef]
- Cadegiani, F.A. Can spironolactone be used to prevent COVID-19-induced acute respiratory distress syndrome in patients with hypertension? Am. J. Physiol. Endocrinol. Metab. 2020, 318, E587–E588. [Google Scholar] [CrossRef]
- Gheblawi, M.; Wang, K.; Viveiros, A.; Nguyen, Q.; Zhong, J.C.; Turner, A.J.; Raizada, M.K.; Grant, M.B.; Oudit, G.Y. Angiotensin-Converting Enzyme 2: SARS-CoV-2 Receptor and Regulator of the Renin-Angiotensin System: Celebrating the 20th Anniversary of the Discovery of ACE2. Circ. Res. 2020, 126, 1456–1474. [Google Scholar] [CrossRef]
- Nelson, A.M.; Gilliland, K.L.; Cong, Z.; Thiboutot, D.M. 13-cis Retinoic acid induces apoptosis and cell cycle arrest in human SEB-1 sebocytes. J. Investig. Dermatol. 2006, 126, 2178–2189. [Google Scholar] [CrossRef]
- Layton, A. The use of isotretinoin in acne. Dermatoendocrinology 2009, 1, 162–169. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.C.K.; Schäffer, A.; Aldape, K.; Schiff, E.; Ruppin, E. Systematic cell line-based identification of drugs modifying ACE2 expression. Preprints 2020. [Google Scholar] [CrossRef]
- Norris, D.A.; Osborn, R.; Robinson, W.; Tonnesen, M.G. Isotretinoin produces significant inhibition of monocyte and neutrophil chemotaxis in vivo in patients with cystic acne. J. Investig. Dermatol. 1987, 89, 38–43. [Google Scholar] [CrossRef] [PubMed]
- McClellan, K.; Perry, C.M. Oseltamivir: A review of its use in influenza. Drugs 2001, 61, 263–283. [Google Scholar] [CrossRef] [PubMed]
- Whitley, R.J.; Hayden, F.G.; Reisinger, K.S.; Young, N.; Dutkowski, R.; Ipe, D.; Mills, R.G.; Ward, P. Oral oseltamivir treatment of influenza in children. Pediatric Infect. Dis. J. 2001, 20, 127–133. [Google Scholar] [CrossRef]
- McKimm-Breschkin, J.L. Influenza neuraminidase inhibitors: Antiviral action and mechanisms of resistance. Influenza Other Respir. Viruses 2013, 7 (Suppl. 1), 25–36. [Google Scholar] [CrossRef]
- Hay, A.J.; Hayden, F.G. Oseltamivir resistance during treatment of H7N9 infection. Lancet 2013, 381, 2230–2232. [Google Scholar] [CrossRef]
- Tan, E.L.; Ooi, E.E.; Lin, C.Y.; Tan, H.C.; Ling, A.E.; Lim, B.; Stanton, L.W. Inhibition of SARS coronavirus infection in vitro with clinically approved antiviral drugs. Emerg. Infect. Dis. 2004, 10, 581–586. [Google Scholar] [CrossRef]
- Scott, L.J. Linagliptin: In type 2 diabetes mellitus. Drugs 2011, 71, 611–624. [Google Scholar] [CrossRef]
- Wang, N.; Shi, X.; Jiang, L.; Zhang, S.; Wang, D.; Tong, P.; Guo, D.; Fu, L.; Cui, Y.; Liu, X.; et al. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 2013, 23, 986–993. [Google Scholar] [CrossRef]
- Solerte, S.B.; Di Sabatino, A.; Galli, M.; Fiorina, P. Dipeptidyl peptidase-4 (DPP4) inhibition in COVID-19. Acta Diabetol. 2020, 57, 779–783. [Google Scholar] [CrossRef] [PubMed]
- Assunção-Miranda, I.; Cruz-Oliveira, C.; Neris, R.L.; Figueiredo, C.M.; Pereira, L.P.; Rodrigues, D.; Araujo, D.F.; Da Poian, A.T.; Bozza, M.T. Inactivation of Dengue and Yellow Fever Viruses by Heme, Cobalt-Protoporphyrin IX and Tin-Protoporphyrin IX. J. Appl. Microbiol. 2016, 120, 790–804. [Google Scholar] [CrossRef] [PubMed]
- Neris, R.L.S.; Figueiredo, C.M.; Higa, L.M.; Araujo, D.F.; Carvalho, C.A.M.; Verçoza, B.R.F.; Silva, M.O.L.; Carneiro, F.A.; Tanuri, A.; Gomes, A.M.O.; et al. Co-protoporphyrin IX and Sn-protoporphyrin IX inactivate Zika, Chikungunya and other arboviruses by targeting the viral envelope. Sci. Rep. 2018, 8, 9805. [Google Scholar] [CrossRef] [PubMed]
- Ibáñez, L.; Alcaraz, M.J.; Maicas, N.; Guede, D.; Caeiro, J.R.; Koenders, M.I.; van den Berg, W.B.; Ferrándiz, M.L. Up-regulation of the Inflammatory Response by Ovariectomy in Collagen-Induced Arthritis. Effects of Tin Protoporphyrin IX. Inflammation 2011, 34, 585–596. [Google Scholar] [CrossRef] [PubMed]
- Kaizu, T.; Tamaki, T.; Tanaka, M.; Uchida, Y.; Tsuchihashi, S.; Kawamura, A.; Kakita, A. Preconditioning with tin-protoporphyrin IX attenuates ischemia/reperfusion injury in the rat kidney. Kidney Int. 2003, 63, 1393–1403. [Google Scholar] [CrossRef] [PubMed]
- Braza-Boïls, A.; Alcaraz, M.J.; Ferrándiz, M.L. Regulation of the inflammatory response by tin protoporphyrin IX in the rat anterior cruciate ligament transection model of osteoarthritis. J. Orthop. Res. 2011, 29, 1375–1382. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Geng, Y.; Duan, W.; Wang, D.; Fu, M.; Wang, X. Ionic liquid-based ultrasound-assisted extraction of fangchinoline and tetrandrine from Stephaniae tetrandrae. J. Sep. Sci. 2009, 32, 3550–3554. [Google Scholar] [CrossRef]
- Bhagya, N.; Chandrashekar, K.R. Tetrandrine-A molecule of wide bioactivity. Phytochemistry 2016, 125, 5–13. [Google Scholar] [CrossRef]
- Hu, S.; Dutt, J.; Zhao, T.; Foster, C.S. Tetrandrine potently inhibits herpes simplex virus type-1-induced keratitis in BALB/c mice. Ocul. Immunol. Inflamm. 1997, 5, 173–180. [Google Scholar] [CrossRef]
- Kim, D.E.; Min, J.S.; Jang, M.S.; Lee, J.Y.; Shin, Y.S.; Song, J.H.; Kim, H.R.; Kim, S.; Jin, Y.H.; Kwon, S. Natural bis-benzylisoquinoline alkaloids-Tetrandrine, fangchinoline, and cepharanthine, inhibit human coronavirus OC43 infection of MRC-5 human lung cells. Biomolecules 2019, 9, 696. [Google Scholar] [CrossRef]
- Sakurai, Y.; Kolokoltsov, A.A.; Chen, C.-C.; Tidwell, M.W.; Bauta, W.E.; Klugbauer, N.; Grimm, C.; Wahl-Schott, C.; Biel, M.; Davey, R.A. Two-pore channels control Ebola virus host cell entry and are drug targets for disease treatment. Science 2015, 347, 995–998. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.M.; He, T.Z.; Chen, X. Tetrandrine inhibits differentiation of proinflammatory subsets of T helper cells but spares de novo differentiation of iTreg cells. Int. Immunopharmacol. 2019, 69, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Ceribelli, A.; Motta, F.; De Santis, M.; Ansari, A.A.; Ridgway, W.M.; Gershwin, M.E.; Selmi, C. Recommendations for coronavirus infection in rheumatic diseases treated with biologic therapy. J. Autoimmun. 2020, 109, 102442. [Google Scholar] [CrossRef] [PubMed]
- Richardson, P.; Griffin, I.; Tucker, C.; Smith, D.; Oechsle, O.; Phelan, A.; Stebbing, J. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet 2020, 395, e30–e31. [Google Scholar] [CrossRef]
- Zhang, W.; Zhao, Y.; Zhang, F.; Wang, Q.; Li, T.; Liu, Z.; Wang, J.; Qin, Y.; Zhang, X.; Yan, X.; et al. The use of anti-inflammatory drugs in the treatment of people with severe coronavirus disease 2019 (COVID-19): The perspectives of clinical immunologists from China. Clin. Immunol. 2020, 214, 108393. [Google Scholar] [CrossRef]
- Harrison, C.; Vannucchi, A.M. Ruxolitinib: A potent and selective Janus kinase 1 and 2 inhibitor in patients with myelofibrosis. An update for clinicians. Ther. Adv. Hematol. 2012, 3, 341–354. [Google Scholar] [CrossRef]
- Stark, G.R.; Darnell, J.E., Jr. The JAK-STAT pathway at twenty. Immunity 2012, 36, 503–514. [Google Scholar] [CrossRef]
- Elli, E.M.; Baratè, C.; Mendicino, F.; Palandri, F.; Palumbo, G.A. Mechanisms underlying the anti-inflammatory and immunosuppressive activity of ruxolitinib. Front. Oncol. 2019, 9, 1186. [Google Scholar] [CrossRef]
- Cantini, F.; Niccoli, L.; Matarrese, D.; Nicastri, E.; Stobbione, P.; Goletti, D. Baricitinib therapy in COVID-19: A pilot study on safety and clinical impact. J. Infect. 2020. [Google Scholar] [CrossRef]
- Mesa, R.A.; Yasothan, U.; Kirkpatrick, P. Ruxolitinib. Nat. Rev. Drug Discov. 2012, 11, 103–104. [Google Scholar] [CrossRef]
- Harrison, C.; Mesa, R.; Ross, D.; Mead, A.; Keohane, C.; Gotlib, J.; Verstovsek, S. Practical management of patients with myelofibrosis receiving ruxolitinib. Expert Rev. Hematol. 2013, 6, 511–523. [Google Scholar] [CrossRef] [PubMed]
- Vannucchi, A.M.; Kiladjian, J.J.; Griesshammer, M.; Masszi, T.; Durrant, S.; Passamonti, F.; Harrison, C.N.; Pane, F.; Zachee, P.; Mesa, R.; et al. Ruxolitinib versus standard therapy for the treatment of polycythemia vera. N. Engl. J. Med. 2015, 372, 426–435. [Google Scholar] [CrossRef] [PubMed]
- Wollenhaupt, J.; Silverfield, J.; Lee, E.B.; Curtis, J.R.; Wood, S.P.; Soma, K.; Nduaka, C.I.; Benda, B.; Gruben, D.; Nakamura, H.; et al. Safety and efficacy of tofacitinib, an oral janus kinase inhibitor, for the treatment of rheumatoid arthritis in open-label, long term extension studies. J. Rheumatol. 2014, 41, 837–852. [Google Scholar] [CrossRef] [PubMed]
- Onishi, A.; St Ange, K.; Dordick, J.S.; Linhardt, R.J. Heparin and anticoagulation. Front. Biosci. (Landmark Ed) 2016, 21, 1372–1392. [Google Scholar] [PubMed]
- Liu, J.; Thorp, S.C. Cell surface heparan sulfate and its roles in assisting viral infections. Med. Res. Rev. 2002, 22, 1–25. [Google Scholar] [CrossRef]
- Milewska, A.; Zarebski, M.; Nowak, P.; Stozek, K.; Potempa, J.; Pyrc, K. Human coronavirus NL63 utilizes heparan sulfate proteoglycans for attachment to target cells. J. Virol. 2014, 88, 13221–13230. [Google Scholar] [CrossRef]
- Lang, J.; Yang, N.; Deng, J.; Liu, K.; Yang, P.; Zhang, G.; Jiang, C. Inhibition of SARS pseudovirus cell entry by lactoferrin binding to heparan sulfate proteoglycans. PLoS ONE. 2011, 6, e23710. [Google Scholar] [CrossRef]
- Mycroft-West, C.J.; Su, D.; Pagani, I.; Rudd, T.R.; Elli, S.; Guimond, S.E.; Gavin Miller, G.; Meneghetti, M.C.Z.; Nader, H.B.; Li, Y.; et al. Heparin inhibits cellular invasion by SARS-CoV-2: Structural dependence of the interaction of the surface protein (spike) S1 receptor binding domain with heparin. bioRxiv 2020. [Google Scholar] [CrossRef]
- Mycroft-West, C.J.; Su, D.; Elli, S.; Guimond, S.; Miller, G.; Turnbull, J.; Yates, E.; Guerrini, M.; Fernig, D.; Lima, M.; et al. The 2019 coronavirus (SARS-CoV-2) surface protein (Spike) S1 Receptor Binding Domain undergoes conformational change upon heparin binding. bioRxiv 2020. [Google Scholar] [CrossRef]
- Li, T.; Lu, H.; Zhang, W. Clinical observation and management of COVID-19 patients. Emerg. Microbes Infect. 2020, 9, 687–690. [Google Scholar] [CrossRef]
- Barrett, C.D.; Moore, H.B.; Yaffe, M.B.; Moore, E.E. ISTH interim guidance on recognition and management of coagulopathy in COVID-19: A comment. J. Thromb. Haemost. 2020. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Liu, W.; Liu, K.; Fang, Y.Y.; Shang, J.; Zhou, L.; Wang, K.; Leng, F.; Wei, S.; Chen, L.; et al. Clinical characteristics of fatal and recovered cases of coronavirus disease 2019 in Wuhan, China: A retrospective study. Chin. Med. J. (Engl) 2020, 133, 1261–1267. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Chen, X.; Cai, Y.; Xia, J.; Zhou, X.; Xu, S.; Huang, H.; Zhang, L.; Zhou, X.; Du, C.; et al. Risk Factors Associated with Acute Respiratory Distress Syndrome and Death in Patients with Coronavirus Disease 2019 Pneumonia in Wuhan, China. JAMA Intern. Med. 2020, 180, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ranucci, M.; Ballotta, A.; Di Dedda, U.; Bayshnikova, E.; Dei Poli, M.; Resta, M.; Falco, M.; Albano, G.; Menicanti, L. The procoagulant pattern of patients with COVID-19 acute respiratory distress syndrome. J. Thromb. Haemost. 2020. [Google Scholar] [CrossRef] [PubMed]
- Levi, M.; Thachil, J.; Iba, T.; Levy, J.H. Coagulation abnormalities and thrombosis in patients with COVID-19. Lancet Haematol. 2020, 7, e438–e440. [Google Scholar] [CrossRef]
- Liu, J.; Li, J.; Arnold, K.; Pawlinski, R.; Key, N.S. Using heparin molecules to manage COVID-2019. Res. Pract. Thromb. Haemost. 2020, 4, 518–523. [Google Scholar] [CrossRef]
- Young, E. The anti-inflammatory effects of heparin and related compounds. Thromb. Res. 2008, 122, 743–752. [Google Scholar] [CrossRef]
- Shi, C.; Wang, C.; Wang, H.; Yang, C.; Cai, F.; Zeng, F.; Cheng, F.; Liu, Y.; Zhou, T.; Bin Deng, B.; et al. The potential of low molecular weight heparin to mitigate cytokine storm in severe COVID-19 patients: A retrospective clinical study. medRxiv 2020. [Google Scholar] [CrossRef]
- Marjuki, H.; Mishin, V.P.; Chesnokov, A.P.; De La Cruz, J.A.; Fry, A.M.; Villanueva, J.; Gubareva, L.V. An investigational antiviral drug, DAS181, effectively inhibits replication of zoonotic influenza A virus subtype H7N9 and protects mice from lethality. J. Infect. Dis. 2014, 210, 435–440. [Google Scholar] [CrossRef]
- Chan, R.W.; Chan, M.C.; Wong, A.C.; Karamanska, R.; Dell, A.; Haslam, S.M.; Sihoe, A.D.; Chui, W.H.; Triana-Baltzer, G.; Li, Q.; et al. DAS181 inhibits H5N1 influenza virus infection of human lung tissues. Antimicrob. Agents Chemother. 2009, 53, 3935–3941. [Google Scholar] [CrossRef]
- Triana-Baltzer, G.B.; Babizki, M.; Chan, M.C.; Wong, A.C.; Aschenbrenner, L.M.; Campbell, E.R.; Li, Q.X.; Chan, R.W.; Peiris, J.S.; Nicholls, J.M.; et al. DAS181, a sialidase fusion protein, protects human airway epithelium against influenza virus infection: An in vitro pharmacodynamic analysis. J. Antimicrob. Chemother. 2010, 65, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Malakhov, M.P.; Aschenbrenner, L.M.; Smee, D.F.; Wandersee, M.K.; Sidwell, R.W.; Gubareva, L.V.; Mishin, V.P.; Hayden, F.G.; Kim, D.H.; Ing, A.; et al. Sialidase fusion protein as a novel broad-spectrum inhibitor of influenza virus infection. Antimicrob. Agents Chemother. 2006, 50, 1470–1479. [Google Scholar] [CrossRef] [PubMed]
- Moscona, A.; Porotto, M.; Palmer, S.; Tai, C.; Aschenbrenner, L.; Triana-Baltzer, G.; Li, Q.X.; Wurtman, D.; Niewiesk, S.; Fang, F. A recombinant sialidase fusion protein effectively inhibits human parainfluenza viral infection in vitro and in vivo. J. Infect. Dis. 2010, 202, 234–241. [Google Scholar] [CrossRef] [PubMed]
- Triana-Baltzer, G.B.; Gubareva, L.V.; Klimov, A.I.; Wurtman, D.F.; Moss, R.B.; Hedlund, M.; Larson, J.L.; Belshe, R.B.; Fang, F. Inhibition of neuraminidase inhibitor-resistant influenza virus by DAS181, a novel sialidase fusion protein. PLoS ONE. 2009, 4, e7838. [Google Scholar] [CrossRef] [PubMed]
- Monteil, V.; Kwon, H.; Prado, P.; Hagelkrüys, A.; Wimmer, R.A.; Stahl, M.; Leopoldi, A.; Garreta, E.; Hurtado Del Pozo, C.; Romero, J.P.; et al. Inhibition of SARS-CoV-2 Infections in Engineered Human Tissues Using Clinical-Grade Soluble Human ACE2. Cell 2020, 181, 905–913.e7. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Benthin, C.; Zeno, B.; Albertson, T.E.; Boyd, J.; Christie, J.D.; Hall, R.; Poirier, G.; Ronco, J.J.; Tidswell, M.; et al. A pilot clinical trial of recombinant human angiotensin-converting enzyme 2 in acute respiratory distress syndrome. Crit. Care. 2017, 21, 234. [Google Scholar] [CrossRef]
- Hansen, J.; Baum, A.; Pascal, K.E.; Russo, V.; Giordano, S.; Wloga, E.; Fulton, B.O.; Yan, Y.; Koon, K.; Patel, K.; et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science 2020, eabd0827. [Google Scholar] [CrossRef]
- Bloch, E.M.; Shoham, S.; Casadevall, A.; Sachais, B.S.; Shaz, B.; Winters, J.L.; van Buskirk, C.; Grossman, B.J.; Joyner, M.; Henderson, J.P.; et al. Deployment of convalescent plasma for the prevention and treatment of COVID-19. J. Clin. Investig. 2020, 130, 2757–2765. [Google Scholar] [CrossRef]
- Roback, J.D.; Guarner, J. Convalescent Plasma to Treat COVID-19: Possibilities and Challenges. JAMA 2020. [Google Scholar] [CrossRef]
- Casadevall, A.; Pirofski, L.A. The convalescent sera option for containing COVID-19. J. Clin. Investig. 2020, 130, 1545–1548. [Google Scholar] [CrossRef]
- Cunningham, A.C.; Goh, H.P.; Koh, D. Treatment of COVID-19: Old tricks for new challenges. Crit. Care. 2020, 24, 91. [Google Scholar] [CrossRef] [PubMed]
- Rojas, M.; Rodríguez, Y.; Monsalve, D.M.; Acosta-Ampudia, Y.; Camacho, B.; Gallo, J.E.; Rojas-Villarraga, A.; Ramírez-Santana, C.; Díaz-Coronado, J.C.; Manrique, R.; et al. Convalescent plasma in Covid-19: Possible mechanisms of action. Autoimmun. Rev. 2020, 19, 102554. [Google Scholar] [CrossRef] [PubMed]
- Wong, V.W.; Dai, D.; Wu, A.K.; Sung, J.J. Treatment of severe acute respiratory syndrome with convalescent plasma. Hong Kong Med. J. 2003, 9, 199–201. [Google Scholar] [PubMed]
- Cheng, Y.; Wong, R.; Soo, Y.O.; Wong, W.S.; Lee, C.K.; Ng, M.H.; Chan, P.; Wong, K.C.; Leung, C.B.; Cheng, G. Use of convalescent plasma therapy in SARS patients in Hong Kong. Eur. J. Clin. Microbiol. Infect. Dis. 2005, 24, 44–46. [Google Scholar] [CrossRef] [PubMed]
- Mair-Jenkins, J.; Saavedra-Campos, M.; Baillie, J.K.; Cleary, P.; Khaw, F.M.; Lim, W.S.; Makki, S.; Rooney, K.D.; Nguyen-Van-Tam, J.S.; Beck, C.R.; et al. The effectiveness of convalescent plasma and hyperimmune immunoglobulin for the treatment of severe acute respiratory infections of viral etiology: A systematic review and exploratory meta-analysis. J. Infect. Dis. 2015, 211, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Soo, Y.O.; Cheng, Y.; Wong, R.; Hui, D.S.; Lee, C.K.; Tsang, K.K.; Ng, M.H.; Chan, P.; Cheng, G.; Sung, J.J. Retrospective comparison of convalescent plasma with continuing high-dose methylprednisolone treatment in SARS patients. Clin. Microbiol. Infect. 2004, 10, 676–678. [Google Scholar] [CrossRef] [PubMed]
- Duan, K.; Liu, B.; Li, C.; Zhang, H.; Yu, T.; Qu, J.; Zhou, M.; Chen, L.; Meng, S.; Hu, Y.; et al. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc. Natl. Acad. Sci. USA 2020, 117, 9490–9496. [Google Scholar] [CrossRef]
- Shen, C.; Wang, Z.; Zhao, F.; Yang, Y.; Li, J.; Yuan, J.; Wang, F.; Li, D.; Yang, M.; Xing, L.; et al. Treatment of 5 Critically Ill Patients with COVID-19 With Convalescent Plasma. JAMA 2020, 323, 1582–1589. [Google Scholar] [CrossRef]
- Salazar, E.; Perez, K.K.; Ashraf, M.; Chen, J.; Castillo, B.; Christensen, P.A.; Eubank, T.; Bernard, D.W.; Eagar, T.N.; Long, S.W.; et al. Treatment of COVID-19 Patients with Convalescent Plasma in Houston, Texas. medRxiv 2020. [Google Scholar] [CrossRef]
- Ye, M.; Fu, D.; Ren, Y.; Wang, F.; Wang, D.; Zhang, F.; Xia, X.; Lv, T. Treatment with convalescent plasma for COVID-19 patients in Wuhan, China. J. Med. Virol. 2020. [Google Scholar] [CrossRef]
- Li, L.; Zhang, W.; Hu, Y.; Tong, X.; Zheng, S.; Yang, J.; Kong, Y.; Ren, L.; Wei, Q.; Mei, H.; et al. Effect of Convalescent Plasma Therapy on Time to Clinical Improvement in Patients with Severe and Life-threatening COVID-19: A Randomized Clinical Trial. JAMA 2020. [Google Scholar] [CrossRef] [PubMed]
- Valk, S.J.; Piechotta, V.; Chai, K.L.; Doree, C.; Monsef, I.; Wood, E.M.; Lamikanra, A.; Kimber, C.; McQuilten, Z.; So-Osman, C.; et al. Convalescent plasma or hyperimmune immunoglobulin for people with COVID-19: A rapid review. Cochrane Database Syst. Rev. 2020, 5, CD013600. [Google Scholar] [PubMed]
- Jawhara, S. Could Intravenous Immunoglobulin Collected from Recovered Coronavirus Patients Protect against COVID-19 and Strengthen the Immune System of New Patients? Int. J. Mol. Sci. 2020, 21, 2272. [Google Scholar] [CrossRef] [PubMed]
- Sanders, J.M.; Monogue, M.L.; Jodlowski, T.Z.; Cutrell, J.B. Pharmacologic Treatments for Coronavirus Disease 2019 (COVID-19): A Review. JAMA 2020. [Google Scholar] [CrossRef]
- Bonam, S.R.; Kaveri, S.V.; Sakuntabhai, A.; Gilardin, L.; Bayry, J. Adjunct Immunotherapies for the Management of Severely Ill COVID-19 Patients. Cell Rep. Med. 2020, 1, 100016. [Google Scholar] [CrossRef]
- Cao, W.; Liu, X.; Bai, T.; Fan, H.; Hong, K.; Song, H.; Han, Y.; Lin, L.; Ruan, L.; Li, T. High-Dose Intravenous Immunoglobulin as a Therapeutic Option for Deteriorating Patients with Coronavirus Disease 2019. Open Forum Infect. Dis. 2020, 7, ofaa102. [Google Scholar] [CrossRef]
- Xie, Y.; Cao, S.; Dong, H.; Li, Q.; Chen, E.; Zhang, W.; Yang, L.; Fu, S.; Wang, R. Effect of regular intravenous immunoglobulin therapy on prognosis of severe pneumonia in patients with COVID-19. J. Infect. 2020. [Google Scholar] [CrossRef]
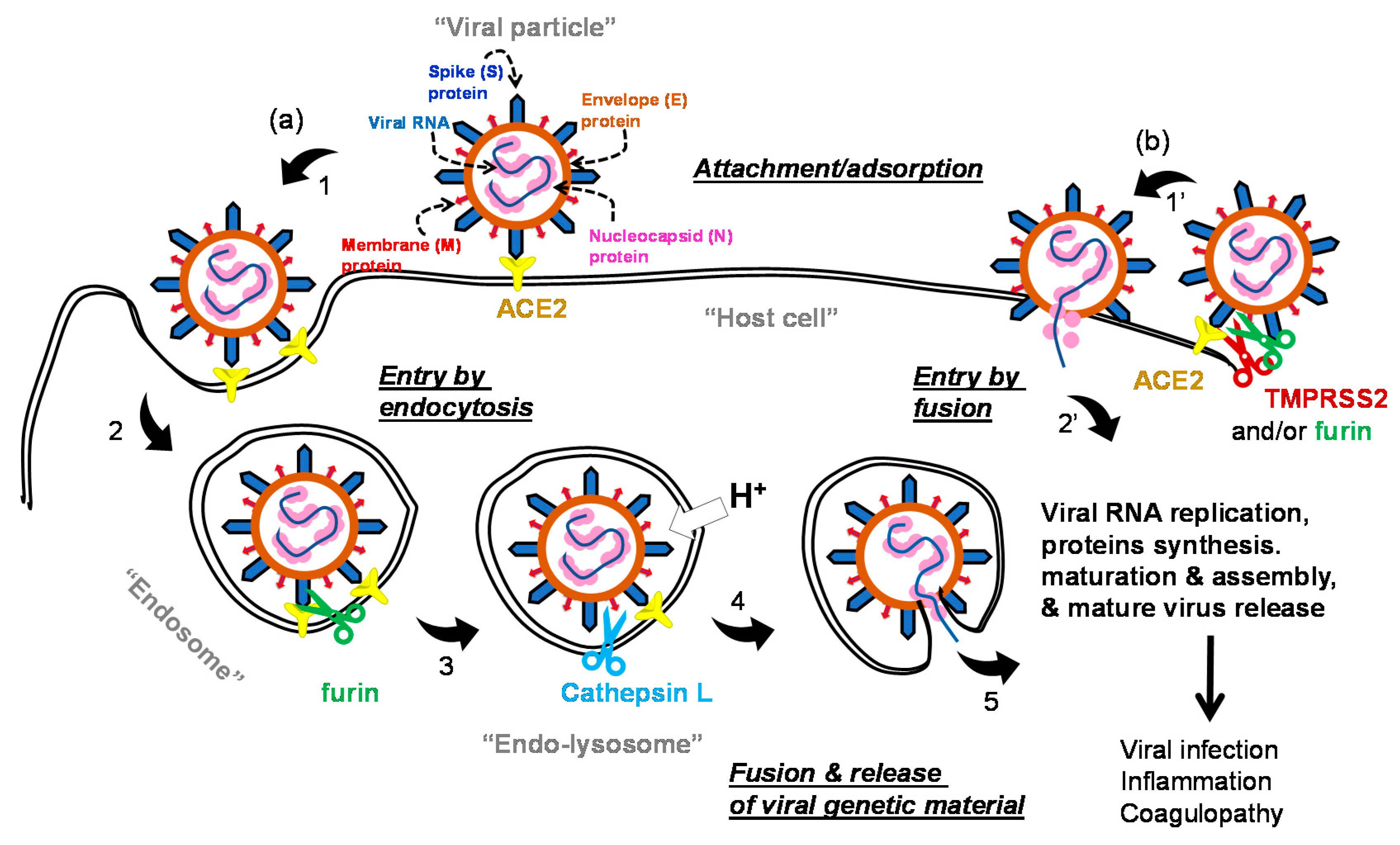
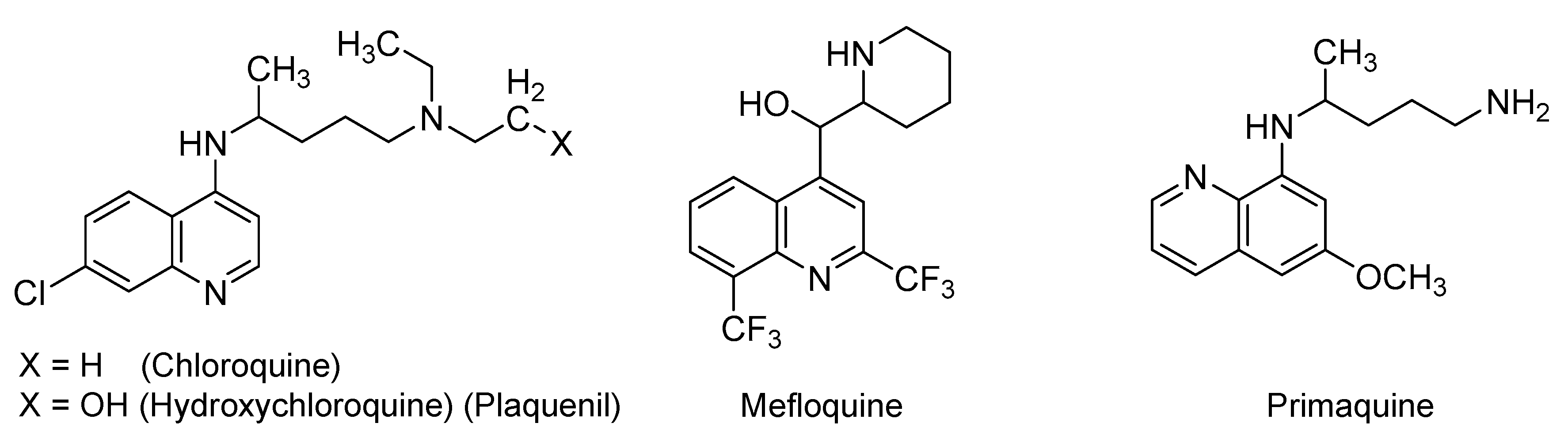
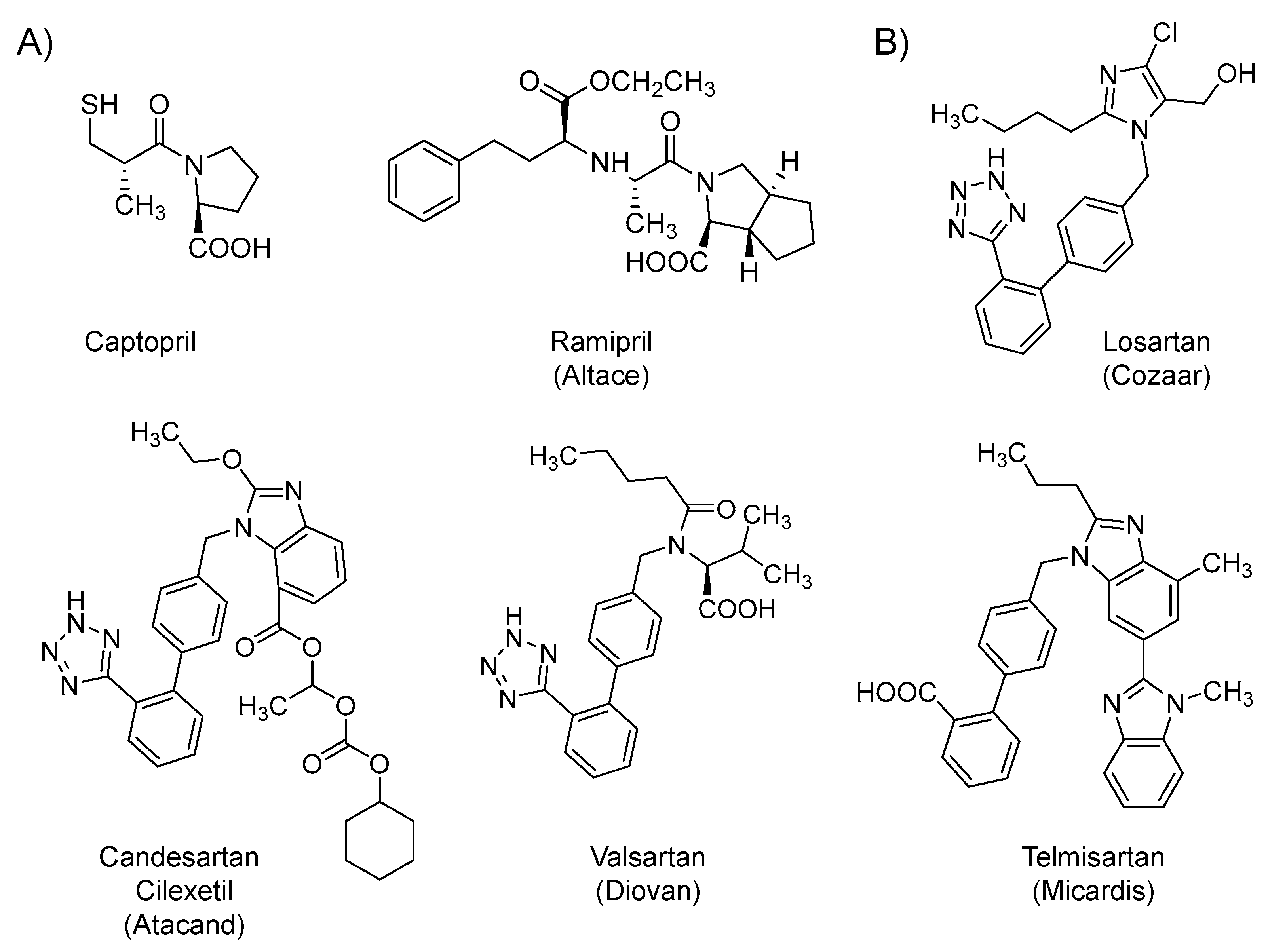
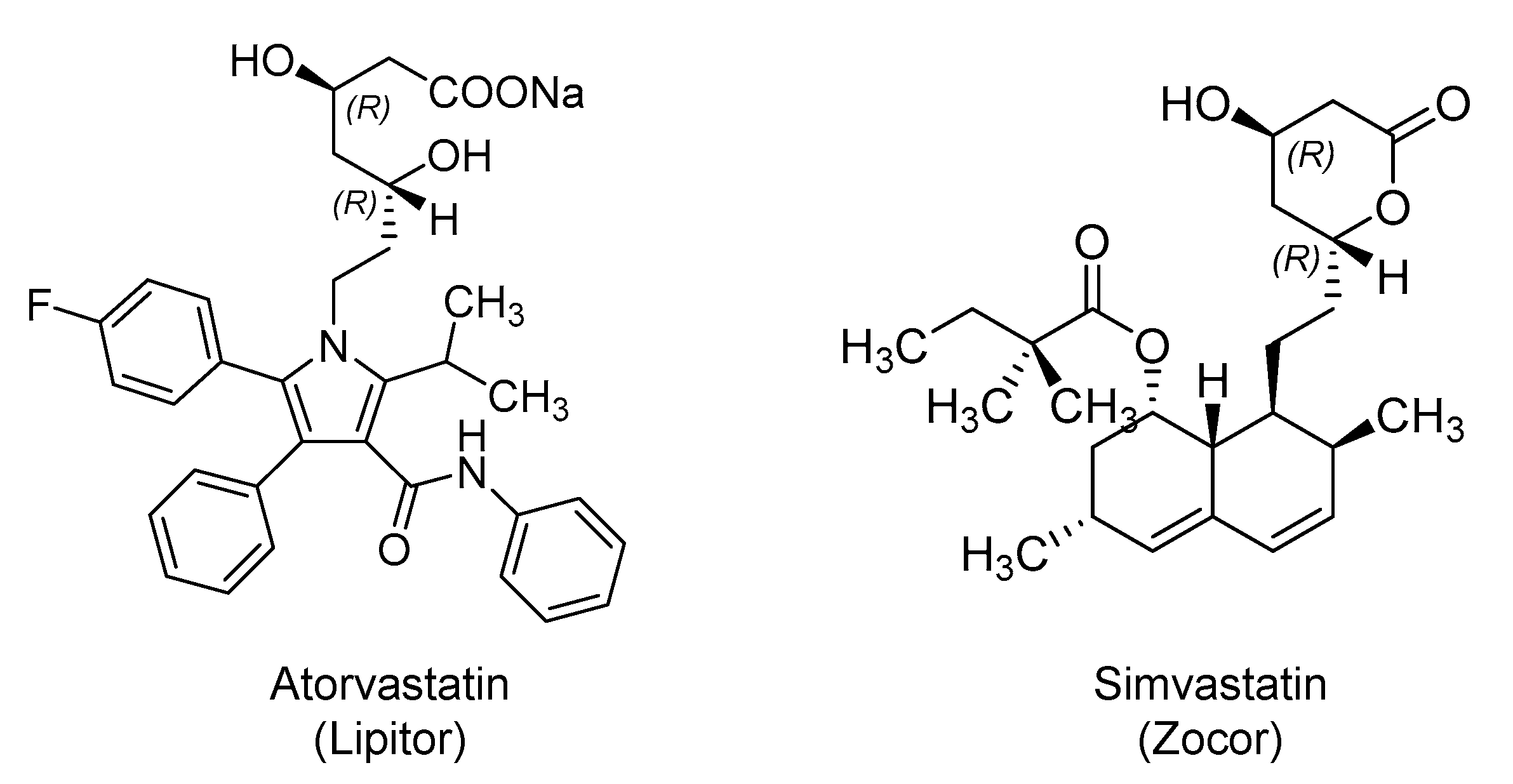
| Small Molecules |
| Quinoline-based drugs: (Hydroxy)chloroquine, mefloquine, & primaquine |
| Renin-angiotensin-aldosterone system (RAAS) modifiers: Captopril, losartan, & others |
| Hydroxy methylglutaryl CoA (HMG CoA) reductase inhibitors: Atorvastatin & simvastatin |
| Transmembrane protease serine 2 (TMPRSS2) inhibitors: Camostat, nafamostat, & others |
| Umifenovir |
| Macrolides: Azithromycin, clarithromycin, & carrimycin |
| Ivermectin |
| Niclosamide |
| Doxycycline |
| Chlorpromazine |
| Amiodarone and verapamil |
| Tranexamic acid |
| Spironolactone |
| Isotretinoin |
| Oseltamivir |
| Dipeptidyl peptidase-4 (DPP-4) inhibitors: Linagliptin and sitagliptin |
| Stannous protoporphyrin |
| Tetrandrine |
| Janus kinases (JAKs) inhibitors: Baricitinib, ruxolitinib, & tofacitinib |
| Macromolecules |
| Unfractionated heparin (UFH) and low molecular weight heparins (LMWHs) |
| DAS181 |
| Recombinant human ACE2 (rhACE2) |
| REGN10933 and REGN10987 |
| COVID-19 convalescent plasma and immunoglobulins |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Horani, R.A.; Kar, S.; Aliter, K.F. Potential Anti-COVID-19 Therapeutics that Block the Early Stage of the Viral Life Cycle: Structures, Mechanisms, and Clinical Trials. Int. J. Mol. Sci. 2020, 21, 5224. https://doi.org/10.3390/ijms21155224
Al-Horani RA, Kar S, Aliter KF. Potential Anti-COVID-19 Therapeutics that Block the Early Stage of the Viral Life Cycle: Structures, Mechanisms, and Clinical Trials. International Journal of Molecular Sciences. 2020; 21(15):5224. https://doi.org/10.3390/ijms21155224
Chicago/Turabian StyleAl-Horani, Rami A., Srabani Kar, and Kholoud F. Aliter. 2020. "Potential Anti-COVID-19 Therapeutics that Block the Early Stage of the Viral Life Cycle: Structures, Mechanisms, and Clinical Trials" International Journal of Molecular Sciences 21, no. 15: 5224. https://doi.org/10.3390/ijms21155224
APA StyleAl-Horani, R. A., Kar, S., & Aliter, K. F. (2020). Potential Anti-COVID-19 Therapeutics that Block the Early Stage of the Viral Life Cycle: Structures, Mechanisms, and Clinical Trials. International Journal of Molecular Sciences, 21(15), 5224. https://doi.org/10.3390/ijms21155224







