Unusual Occurrence of Two Bona-Fide CCA-Adding Enzymes in Dictyostelium discoideum
Abstract
1. Introduction
2. Results
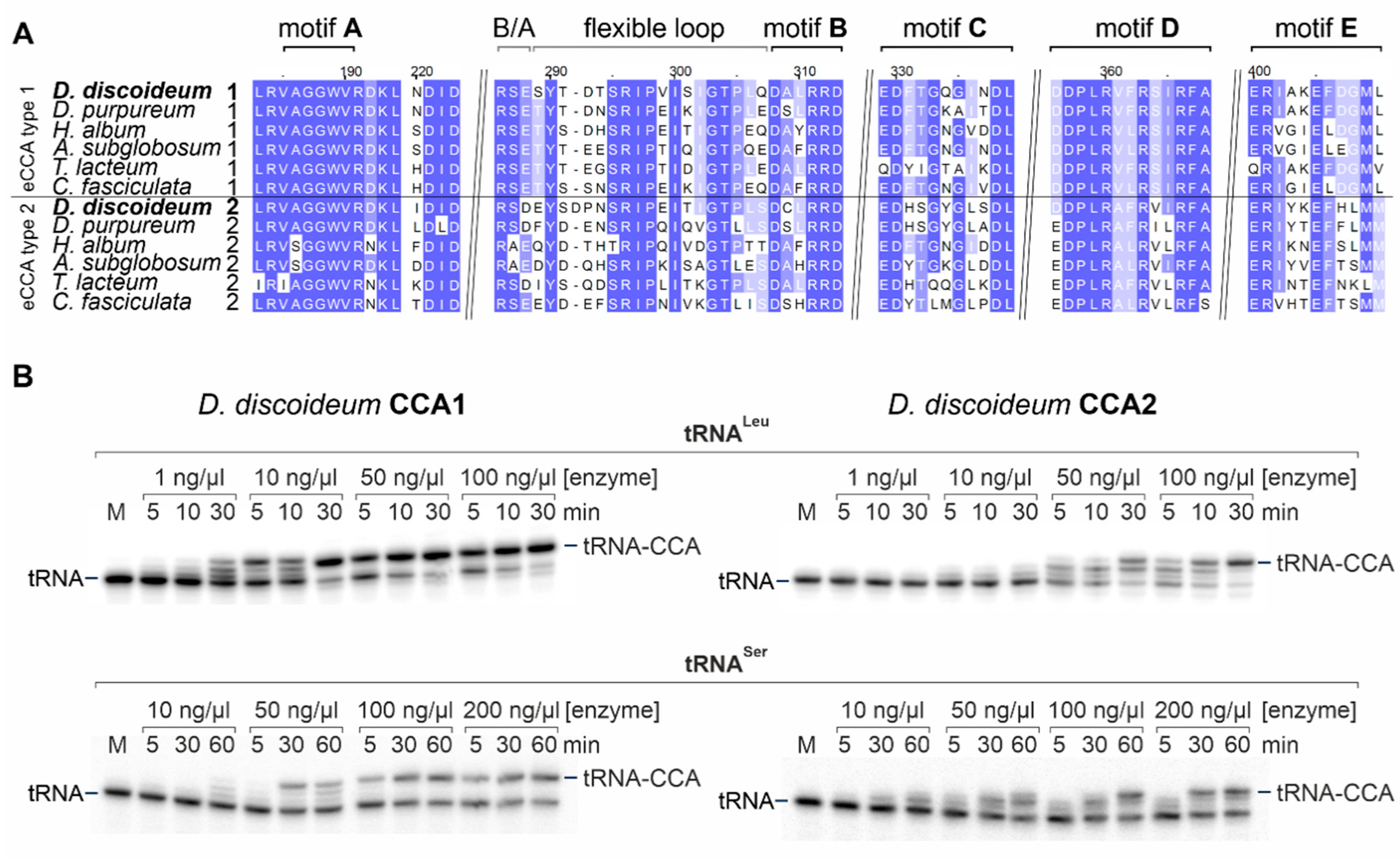
2.1. Dictyostelia Possess Two Genes for CCA-Adding Enzymes
2.2. Both Enzymes of D. discoideum Are Fully Active CCA-Adding Enzymes
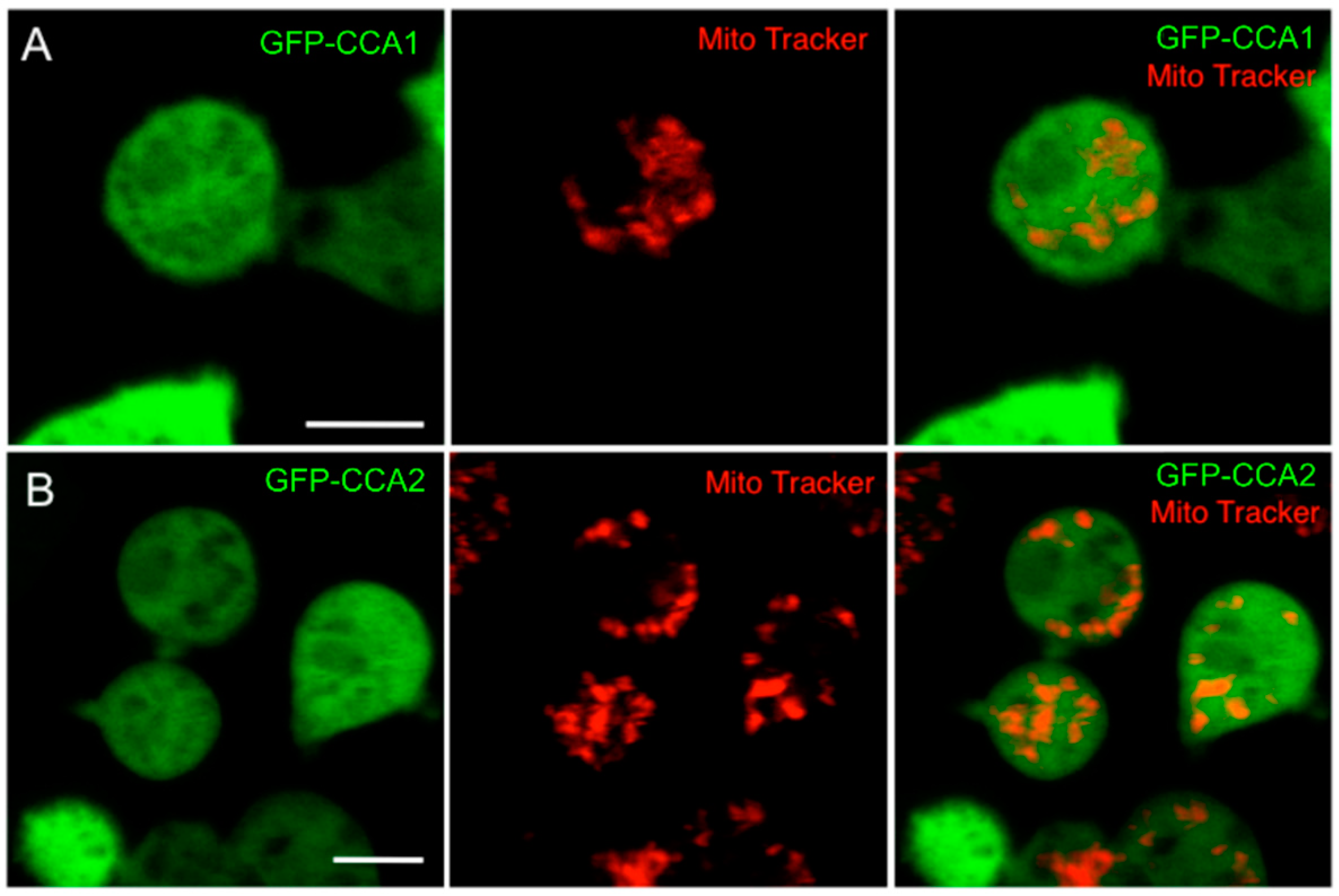
2.3. A Knock-Out of CCA1 and CCA2 Was not Successful, and Both Gene Products Show Similar Localization Patterns
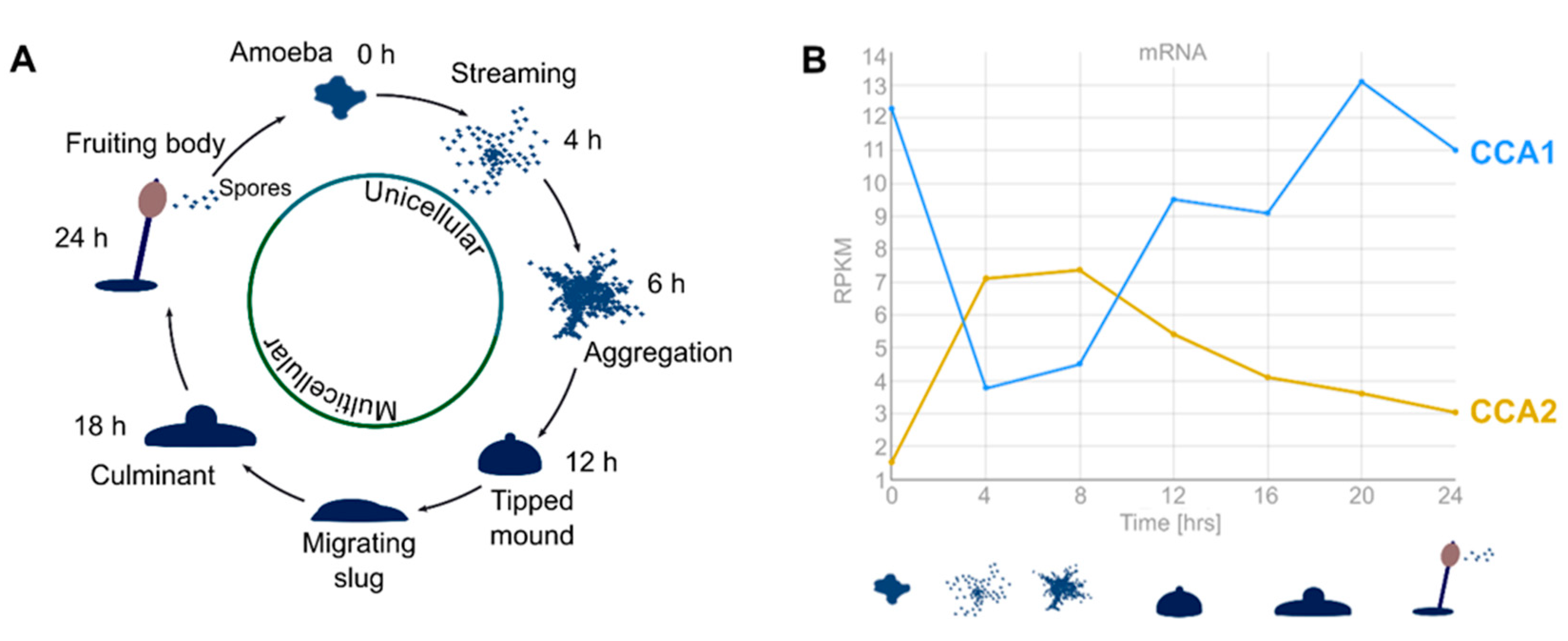
2.4. Expression of CCA1 and CCA2 is Cell-Cycle-Dependent and Inversely Regulated
2.5. CCA1 and CCA2 Have Identical Substrate Specificities
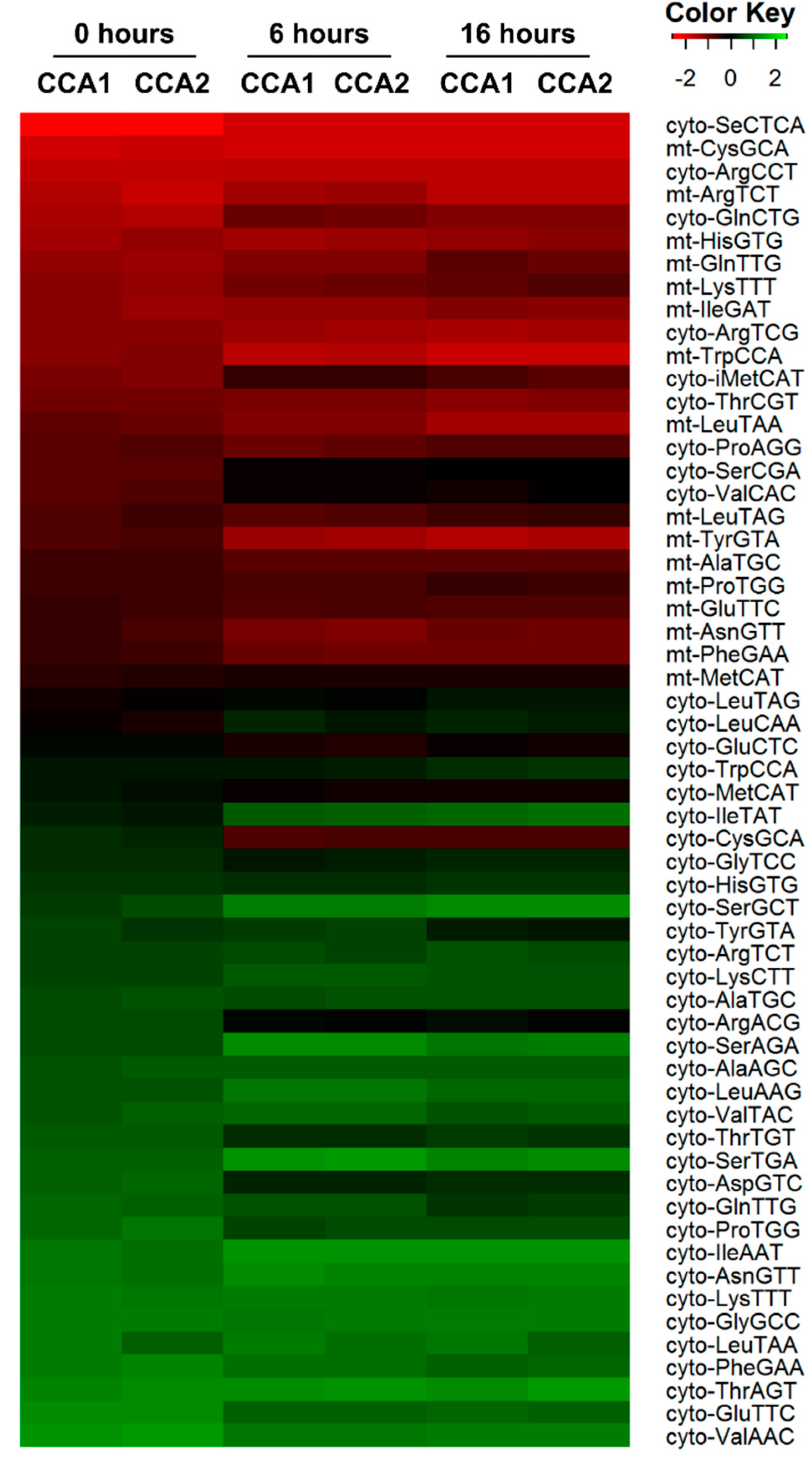
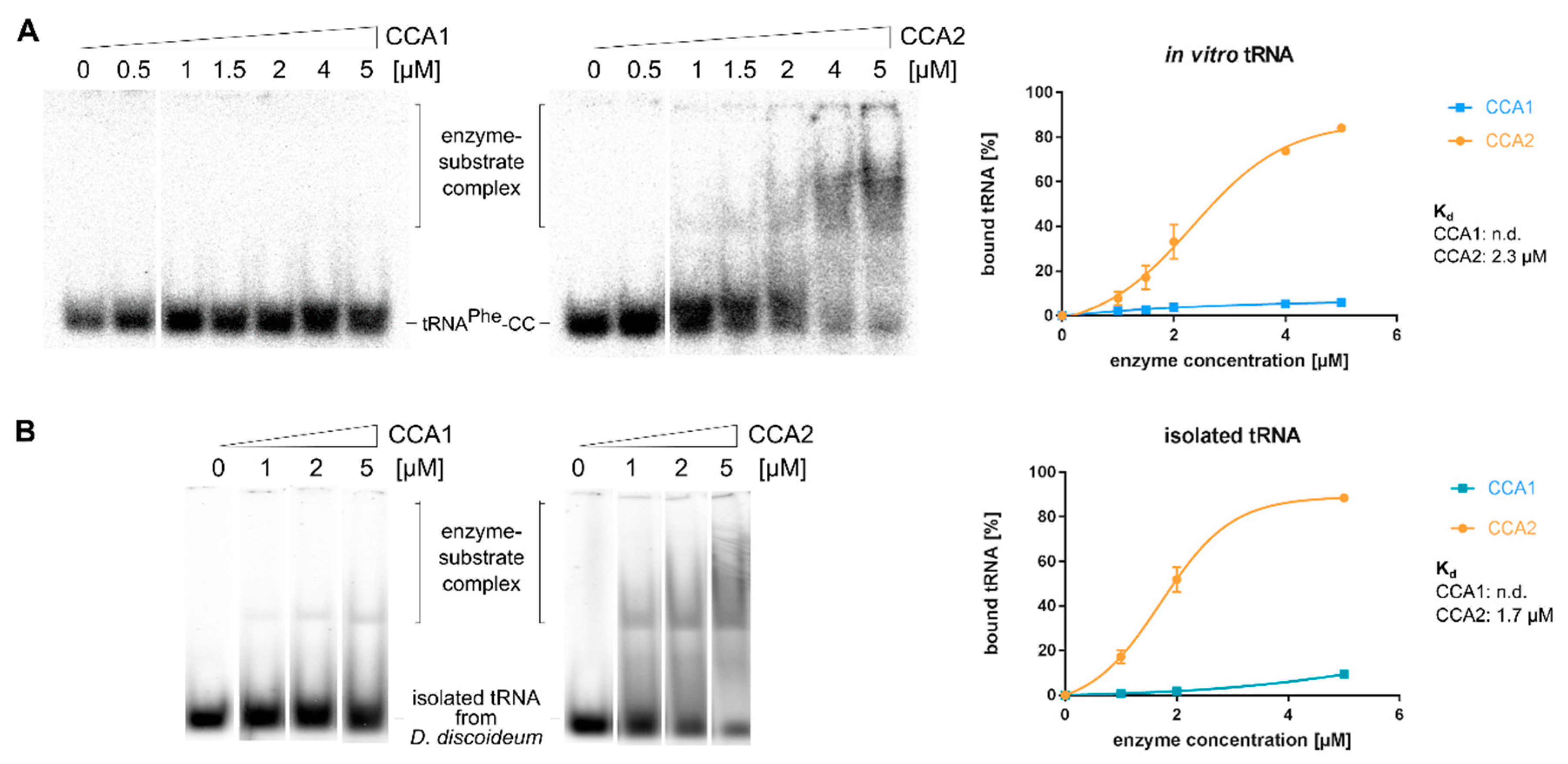
2.6. CCA1 and CCA2 Differ in Their Affinities for tRNA Substrates
3. Discussion
4. Material and Methods
4.1. Isolation of D. discoideum Total RNA and tRNA
4.2. Degradation of the 3′-CCA-end of tRNAs with Snake Venom Phosphodiesterase I
4.3. Expression and Purification of D. discoideum tRNA Nucleotidyltransferases
4.4. In Vitro Activity Test of tRNA Nucleotidyltransferases
4.5. High-Throughput Sequencing of tRNAs with 3′-CCA End
4.6. Electrophoretic Mobility Shift Assay (EMSA)
4.7. Localization of the CCA-Adding Enzymes in Amoebozoal D. discoideum by GFP-Labeling
4.8. Gene Deletion by Homologous Recombination
4.9. Phylogenetic Analysis by Splitstree Network
4.10. Bioinformatic Analysis of the High-Throughput Sequencing Data
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| cca1 | gene encoding enzyme CCA1 |
| cca2 | gene encoding enzyme CCA2 |
| GFP | green fluorescent protein |
| HGT | horizontal gene transfer |
| ORF | open reading frame |
References
- Holm, L.; Sander, C. DNA polymerase beta belongs to an ancient nucleotidyltransferase superfamily. Trends Biochem. Sci. 1995, 20, 345–347. [Google Scholar] [CrossRef]
- Aravind, L.; Koonin, E.V. DNA polymerase beta-like nucleotidyltransferase superfamily: Identification of three new families, classification and evolutionary history. Nucleic Acids Res. 1999, 27, 1609–1618. [Google Scholar] [CrossRef] [PubMed]
- Betat, H.; Rammelt, C.; Mörl, M. tRNA nucleotidyltransferases: Ancient catalysts with an unusual mechanism of polymerization. Cell. Mol. Life Sci. 2010, 67, 1447–1463. [Google Scholar] [CrossRef]
- Xiong, Y.; Steitz, T.A. A story with a good ending: TRNA 3′-end maturation by CCA-adding enzymes. Curr. Opin. Struct. Biol. 2006, 16, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Sprinzl, M.; Cramer, F. The-C-C-A end of tRNA and its role in protein biosynthesis. Prog. Nucleic Acid Res. Mol. Biol. 1979, 22, 1–69. [Google Scholar]
- Liu, J.C.; Liu, M.; Horowitz, J. Recognition of the universally conserved 3′-CCA end of tRNA by elongation factor EF-Tu. RNA 1998, 4, 639–646. [Google Scholar] [CrossRef][Green Version]
- Dupasquier, M.; Kim, S.; Halkidis, K.; Gamper, H.; Hou, Y.-M. tRNA integrity is a prerequisite for rapid CCA addition: Implication for quality control. J. Mol. Biol. 2008, 379, 579–588. [Google Scholar] [CrossRef]
- Wilusz, J.E.; Whipple, J.M.; Phizicky, E.M.; Sharp, P.A. tRNAs marked with CCACCA are targeted for degradation. Science 2011, 334, 817–821. [Google Scholar] [CrossRef]
- Wellner, K.; Czech, A.; Ignatova, Z.; Betat, H.; Mörl, M. Examining tRNA 3′-ends in Escherichia coli: Teamwork between CCA-adding enzyme, RNase T, and RNase, R. RNA 2018, 24, 361–370. [Google Scholar] [CrossRef]
- Deutscher, M.P. Ribonucleases, tRNA nucleotidyltransferase, and the 3′ processing of tRNA. Prog. Nucleic Acid Res. Mol. Biol. 1990, 39, 209–240. [Google Scholar]
- Li, F.; Xiong, Y.; Wang, J.; Cho, H.D.; Tomita, K.; Weiner, A.M.; Steitz, T.A. Crystal Structures of the Bacillus stearothermophilus CCA-Adding Enzyme and Its Complexes with ATP or CTP. Cell 2002, 111, 815–824. [Google Scholar] [CrossRef]
- Aebi, M.; Kirchner, G.; Chen, J.Y.; Vijayraghavan, U.; Jacobson, A.; Martin, N.C.; Abelson, J. Isolation of a temperature-sensitive mutant with an altered tRNA nucleotidyltransferase and cloning of the gene encoding tRNA nucleotidyltransferase in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1990, 265, 16216–16220. [Google Scholar] [PubMed]
- Jühling, F.; Mörl, M.; Hartmann, R.K.; Sprinzl, M.; Stadler, P.F.; Pütz, J. tRNAdb 2009: Compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009, 37, D159–D162. [Google Scholar] [CrossRef] [PubMed]
- Chan, P.P.; Lowe, T.M. GtRNAdb: A database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009, 37, D93–D97. [Google Scholar] [CrossRef]
- Yue, D.; Maizels, N.; Weiner, A.M. CCA-adding enzymes and poly(A) polymerases are all members of the same nucleotidyltransferase superfamily: Characterization of the CCA-adding enzyme from the archaeal hyperthermophile Sulfolobus shibatae. RNA 1996, 2, 895–908. [Google Scholar] [PubMed]
- Yue, D.; Weiner, A.M.; Maizels, N. The CCA-adding enzyme has a single active site. J. Biol. Chem. 1998, 273, 29693–29700. [Google Scholar] [CrossRef]
- Martin, G.; Keller, W. RNA-specific ribonucleotidyl transferases. RNA 2007, 13, 1834–1849. [Google Scholar] [CrossRef]
- Brautigam, C.A.; Steitz, T.A. Structural and functional insights provided by crystal structures of DNA polymerases and their substrate complexes. Curr. Opin. Struct. Biol. 1998, 8, 54–63. [Google Scholar] [CrossRef]
- Steitz, T.A. A mechanism for all polymerases. Nature 1998, 391, 231–232. [Google Scholar] [CrossRef]
- Toh, Y.; Takeshita, D.; Numata, T.; Fukai, S.; Nureki, O.; Tomita, K. Mechanism for the definition of elongation and termination by the class II CCA-adding enzyme. EMBO J. 2009, 28, 3353–3365. [Google Scholar] [CrossRef]
- Ernst, F.G.M.; Rickert, C.; Bluschke, A.; Betat, H.; Steinhoff, H.-J.; Mörl, M. Domain movements during CCA-addition: A new function for motif C in the catalytic core of the human tRNA nucleotidyltransferases. RNA Biol. 2015, 12, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Betat, H.; Mede, T.; Tretbar, S.; Steiner, L.; Stadler, P.F.; Mörl, M.; Prohaska, S.J. The ancestor of modern Holozoa acquired the CCA-adding enzyme from Alphaproteobacteria by horizontal gene transfer. Nucleic Acids Res. 2015, 43, 6739–6746. [Google Scholar] [CrossRef]
- Erber, L.; Franz, P.; Betat, H.; Prohaska, S.J.; Mörl, M. Divergent Evolution of Eukaryotic CC- and A-Adding Enzymes. IJMS 2020, 21, 462. [Google Scholar] [CrossRef] [PubMed]
- Preston, M.A.; Porter, D.F.; Chen, F.; Buter, N.; Lapointe, C.P.; Keles, S.; Kimble, J.; Wickens, M. Unbiased screen of RNA tailing activities reveals a poly(UG) polymerase. Nat. Methods 2019, 16, 437–445. [Google Scholar] [CrossRef] [PubMed]
- Reid, N.E.; Ngou, J.S.; Joyce, P.B.M. Schizosaccharomyces pombe contains separate CC- and A-adding tRNA nucleotidyltransferases. Biochem. Biophys. Res. Commun. 2019, 508, 785–790. [Google Scholar] [CrossRef] [PubMed]
- Tomita, K.; Weiner, A.M. Collaboration between CC- and A-adding enzymes to build and repair the 3’-terminal CCA of tRNA in Aquifex aeolicus. Science 2001, 294, 1334–1336. [Google Scholar] [CrossRef] [PubMed]
- Bralley, P.; Chang, S.A.; Jones, G.H. A phylogeny of bacterial RNA nucleotidyltransferases: Bacillus halodurans contains two tRNA nucleotidyltransferases. J. Bacteriol. 2005, 187, 5927–5936. [Google Scholar] [CrossRef]
- Neuenfeldt, A.; Just, A.; Betat, H.; Mörl, M. Evolution of tRNA nucleotidyltransferases: A small deletion generated CC-adding enzymes. Proc. Natl. Acad. Sci. USA 2008, 105, 7953–7958. [Google Scholar] [CrossRef]
- Tretbar, S.; Neuenfeldt, A.; Betat, H.; Mörl, M. An inhibitory C-terminal region dictates the specificity of A-adding enzymes. Proc. Natl. Acad. Sci. USA 2011, 108, 21040–21045. [Google Scholar] [CrossRef]
- Kessin, R.H. Dictyostelium: Evolution, Cell Biology, and the Development of Multicellularity; Cambridge University Press: Cambridge, UK, 2001. [Google Scholar]
- Cosson, P.; Soldati, T. Eat, kill or die: When amoeba meets bacteria. Curr. Opin. Microbiol. 2008, 11, 271–276. [Google Scholar] [CrossRef]
- van Driessche, N.; Shaw, C.; Katoh, M.; Morio, T.; Sucgang, R.; Ibarra, M.; Kuwayama, H.; Saito, T.; Urushihara, H.; Maeda, M.; et al. A transcriptional profile of multicellular development in Dictyostelium discoideum. Development 2002, 129, 1543–1552. [Google Scholar]
- Chisholm, R.L.; Firtel, R.A. Insights into morphogenesis from a simple developmental system. Nat. Rev. Mol. Cell Biol. 2004, 5, 531–541. [Google Scholar] [CrossRef] [PubMed]
- Parikh, A.; Miranda, E.R.; Katoh-Kurasawa, M.; Fuller, D.; Rot, G.; Zagar, L.; Curk, T.; Sucgang, R.; Chen, R.; Zupan, B.; et al. Conserved developmental transcriptomes in evolutionarily divergent species. Genome Biol. 2010, 11, R35. [Google Scholar] [CrossRef] [PubMed]
- Loomis, W.F. Genetic control of morphogenesis in Dictyostelium. Dev. Biol. 2015, 402, 146–161. [Google Scholar] [CrossRef] [PubMed]
- González-Velasco, Ó.; de Las Rivas, J.; Lacal, J. Proteomic and Transcriptomic Profiling Identifies Early Developmentally Regulated Proteins in Dictyostelium Discoideum. Cells 2019, 8. [Google Scholar] [CrossRef]
- Loomis, W.F. Dictyostelium Discoideum. In A Developmental System; Academic Press: New York, NY, USA, 1975. [Google Scholar]
- Dimond, R.L.; Mayer, M.; Loomis, W.F. Characterization and developmental regulation of β-galactosidase isozymes in Dictyostelium discoideum. Dev. Biol. 1976, 52, 74–82. [Google Scholar] [CrossRef]
- Bennett, V.D.; Dimond, R.L. Biosynthesis of two developmentally distinct acid phosphatase isozymes in Dictyostelium discoideum. J. Biol. Chem. 1986, 261, 5355–5362. [Google Scholar]
- Bürki, E.; Anjard, C.; Scholder, J.-C.; Reymond, C.D. Isolation of two genes encoding putative protein kinases regulated during Dictyostelium discoideum development. Gene 1991, 102, 57–65. [Google Scholar] [CrossRef]
- Uchiyama, S.; Isobe, K.; Nagai, S. Isozymes of ribonuclease and the changes in their relative levels during development in the cellular slime mould Dictyostelium discoideum. Biochem. Cell Biol. 1991, 69, 84–87. [Google Scholar] [CrossRef]
- Liaw, S.H.; Kuo, I.; Eisenberg, D. Discovery of the ammonium substrate site on glutamine synthetase, a third cation binding site. Protein Sci. 1995, 4, 2358–2365. [Google Scholar] [CrossRef]
- Gamper, M.; Howard, P.K.; Hunter, T.; Firtel, R.A. Multiple roles of the novel protein tyrosine phosphatase PTP3 during Dictyostelium growth and development. Mol. Cell. Biol. 1996, 16, 2431–2444. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Leibovitch, M.; Bublak, D.; Hanic-Joyce, P.J.; Tillmann, B.; Flinner, N.; Amsel, D.; Scharf, K.-D.; Mirus, O.; Joyce, P.B.M.; Schleiff, E. The folding capacity of the mature domain of the dual-targeted plant tRNA nucleotidyltransferase influences organelle selection. Biochem. J. 2013, 453, 401–412. [Google Scholar] [CrossRef] [PubMed]
- Malinovska, L.; Palm, S.; Gibson, K.; Verbavatz, J.-M.; Alberti, S. Dictyostelium discoideum has a highly Q/N-rich proteome and shows an unusual resilience to protein aggregation. Proc. Natl. Acad. Sci. USA 2015, 112, E2620–E2629. [Google Scholar] [CrossRef] [PubMed]
- Wende, S.; Bonin, S.; Götze, O.; Betat, H.; Mörl, M. The identity of the discriminator base has an impact on CCA addition. Nucleic Acids Res. 2015, 43, 5617–5629. [Google Scholar] [CrossRef] [PubMed]
- Kelly, K.O.; Deutscher, M.P. The presence of only one of five exoribonucleases is sufficient to support the growth of Escherichia coli. J. Bacteriol. 1992, 174, 6682–6684. [Google Scholar] [CrossRef] [PubMed]
- Bechhofer, D.H.; Deutscher, M.P. Bacterial ribonucleases and their roles in RNA metabolism. Crit. Rev. Biochem. Mol. Biol. 2019, 54, 242–300. [Google Scholar] [CrossRef]
- Wiegand, S.; Kruse, J.; Gronemann, S.; Hammann, C. Efficient generation of gene knockout plasmids for Dictyostelium discoideum using one-step cloning. Genomics 2011, 97, 321–325. [Google Scholar] [CrossRef]
- Sekine, R.; Kawata, T.; Muramoto, T. CRISPR/Cas9 mediated targeting of multiple genes in Dictyostelium. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef]
- Gaudet, P.; Pilcher, K.E.; Fey, P.; Chisholm, R.L. Transformation of Dictyostelium discoideum with plasmid DNA. Nat. Protoc. 2007, 2, 1317–1324. [Google Scholar] [CrossRef]
- Rot, G.; Parikh, A.; Curk, T.; Kuspa, A.; Shaulsky, G.; Zupan, B. dictyExpress: A Dictyostelium discoideum gene expression database with an explorative data analysis web-based interface. BMC Bioinform. 2009, 10, 265. [Google Scholar] [CrossRef]
- Deutscher, M.P. Reactions at the 3′ Terminus of Transfer Ribonucleic Acid: III. Catalytic Properties of two Purified Rabbit Liver Transfer Ribonucleic Acid Nucleotidyltransferases. J. Biol. Chem. 1972, 247, 459–468. [Google Scholar] [PubMed]
- Erber, L.; Hoffmann, A.; Fallmann, J.; Betat, H.; Stadler, P.F.; Mörl, M. LOTTE-seq (Long hairpin oligonucleotide based tRNA high-throughput sequencing): Specific selection of tRNAs with 3’-CCA end for high-throughput sequencing. RNA Biol. 2019, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Tomita, K.; Weiner, A.M. Closely related CC- and A-adding enzymes collaborate to construct and repair the 3′-terminal CCA of tRNA in Synechocystis sp. and Deinococcus radiodurans. J. Biol. Chem. 2002, 277, 48192–48198. [Google Scholar] [CrossRef] [PubMed]
- Bralley, P.; Cozad, M.; Jones, G.H. Geobacter sulfurreducens contains separate C- and A-adding tRNA nucleotidyltransferases and a poly(A) polymerase. J. Bacteriol. 2009, 191, 109–114. [Google Scholar] [CrossRef]
- Schaap, P.; Winckler, T.; Nelson, M.; Alvarez-Curto, E.; Elgie, B.; Hagiwara, H.; Cavender, J.; Milano-Curto, A.; Rozen, D.E.; Dingermann, T.; et al. Molecular phylogeny and evolution of morphology in the social amoebas. Science 2006, 314, 661–663. [Google Scholar] [CrossRef]
- Heidel, A.J.; Lawal, H.M.; Felder, M.; Schilde, C.; Helps, N.R.; Tunggal, B.; Rivero, F.; John, U.; Schleicher, M.; Eichinger, L.; et al. Phylogeny-wide analysis of social amoeba genomes highlights ancient origins for complex intercellular communication. Genome Res. 2011, 21, 1882–1891. [Google Scholar] [CrossRef]
- Malinovska, L.; Alberti, S. Protein misfolding in Dictyostelium: Using a freak of nature to gain insight into a universal problem. Prion 2015, 9, 339–346. [Google Scholar] [CrossRef]
- Santarriaga, S.; Petersen, A.; Ndukwe, K.; Brandt, A.; Gerges, N.; Bruns Scaglione, J.; Scaglione, K.M. The Social Amoeba Dictyostelium discoideum Is Highly Resistant to Polyglutamine Aggregation. J. Biol. Chem. 2015, 290, 25571–25578. [Google Scholar] [CrossRef]
- Kreppel, L.; Fey, P.; Gaudet, P.; Just, E.; Kibbe, W.A.; Chisholm, R.L.; Kimmel, A.R. dictyBase: A new Dictyostelium discoideum genome database. Nucleic Acids Res. 2004, 32, D332–D333. [Google Scholar] [CrossRef]
- Ogawa, S.; Yoshino, R.; Angata, K.; Iwamoto, M.; Pi, M.; Kuroe, K.; Matsuo, K.; Morio, T.; Urushihara, H.; Yanagisawa, K.; et al. The mitochondrial DNA of Dictyostelium discoideum: Complete sequence, gene content and genome organization. Mol. Gen. Genet. 2000, 263, 514–519. [Google Scholar] [CrossRef]
- Wojtkowska, M.; Buczek, D.; Suzuki, Y.; Shabardina, V.; Makałowski, W.; Kmita, H. The emerging picture of the mitochondrial protein import complexes of Amoebozoa supergroup. BMC Genom. 2017, 18, 997. [Google Scholar] [CrossRef] [PubMed]
- Shaw, D.R.; Richter, H.; Giorda, R.; Ohmachi, T.; Ennis, H.L. Nucleotide sequences of Dictyostelium discoideum developmentally regulated cDNAs rich in (AAC) imply proteins that contain clusters of asparagine, glutamine, or threonine. Mol. Gen. Genet. 1989, 218, 453–459. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, M.H.; Wanker, E.E.; Andrade-Navarro, M.A. Evolution and function of CAG/polyglutamine repeats in protein-protein interaction networks. Nucleic Acids Res. 2012, 40, 4273–4287. [Google Scholar] [CrossRef] [PubMed]
- Zurawel, A.A.; Kabeche, R.; DiGregorio, S.E.; Deng, L.; Menon, K.M.; Opalko, H.; Duennwald, M.L.; Moseley, J.B.; Supattapone, S. CAG Expansions Are Genetically Stable and Form Nontoxic Aggregates in Cells Lacking Endogenous Polyglutamine Proteins. mBio 2016, 7. [Google Scholar] [CrossRef]
- Scala, C.; Tian, X.; Mehdiabadi, N.J.; Smith, M.H.; Saxer, G.; Stephens, K.; Buzombo, P.; Strassmann, J.E.; Queller, D.C. Amino acid repeats cause extraordinary coding sequence variation in the social amoeba Dictyostelium discoideum. PLoS ONE 2012, 7, e46150. [Google Scholar] [CrossRef] [PubMed]
- de Hostos, E.L.; McCaffrey, G.; Sucgang, R.; Pierce, D.W.; Vale, R.D. A developmentally regulated kinesin-related motor protein from Dictyostelium discoideum. Mol. Biol. Cell 1998, 9, 2093–2106. [Google Scholar] [CrossRef][Green Version]
- Müller, S.; Windhof, I.M.; Maximov, V.; Jurkowski, T.; Jeltsch, A.; Förstner, K.U.; Sharma, C.M.; Gräf, R.; Nellen, W. Target recognition, RNA methylation activity and transcriptional regulation of the Dictyostelium discoideum Dnmt2-homologue (DnmA). Nucleic Acids Res. 2013, 41, 8615–8627. [Google Scholar] [CrossRef]
- Lee, S.K.; Yu, S.L.; Garcia, M.X.; Alexander, H.; Alexander, S. Differential developmental expression of the rep B and rep D xeroderma pigmentosum related DNA helicase genes from Dictyostelium discoideum. Nucleic Acids Res. 1997, 25, 2365–2374. [Google Scholar] [CrossRef]
- Platt, J.L.; Rogers, B.J.; Rogers, K.C.; Harwood, A.J.; Kimmel, A.R. Different CHD chromatin remodelers are required for expression of distinct gene sets and specific stages during development of Dictyostelium discoideum. Development 2013, 140, 4926–4936. [Google Scholar] [CrossRef]
- Long, Y.; Abad, M.G.; Olson, E.D.; Carrillo, E.Y.; Jackman, J.E. Identification of distinct biological functions for four 3′-5′ RNA polymerases. Nucleic Acids Res. 2016, 44, 8395–8406. [Google Scholar] [CrossRef]
- Finney, R.; Ellis, M.; Langtimm, C.; Rosen, E.; Firtel, R.; Soll, D.R. Gene regulation during dedifferentiation in Dictyostelium discoideum. Dev. Biol. 1987, 120, 561–576. [Google Scholar] [CrossRef]
- Iranfar, N.; Fuller, D.; Loomis, W.F. Genome-wide expression analyses of gene regulation during early development of Dictyostelium discoideum. Eukaryotic Cell 2003, 2, 664–670. [Google Scholar] [CrossRef] [PubMed]
- Shi, P.Y.; Maizels, N.; Weiner, A.M. CCA addition by tRNA nucleotidyltransferase: Polymerization without translocation? EMBO J. 1998, 17, 3197–3206. [Google Scholar] [CrossRef] [PubMed]
- King, O.D.; Gitler, A.D.; Shorter, J. The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res. 2012, 1462, 61–80. [Google Scholar] [CrossRef]
- Alberti, S. Aggregating the message to control the cell cycle. Dev. Cell 2013, 25, 551–552. [Google Scholar] [CrossRef][Green Version]
- Scheibe, M.; Bonin, S.; Hajnsdorf, E.; Betat, H.; Mörl, M. Hfq stimulates the activity of the CCA-adding enzyme. BMC Mol. Biol. 2007, 8, 92. [Google Scholar] [CrossRef]
- Kim, S.; Liu, C.; Halkidis, K.; Gamper, H.B.; Hou, Y.-M. Distinct kinetic determinants for the stepwise CCA addition to tRNA. RNA 2009, 15, 1827–1836. [Google Scholar] [CrossRef]
- Hou, Y.-M. CCA addition to tRNA: Implications for tRNA quality control. IUBMB Life 2010, 62, 251–260. [Google Scholar] [CrossRef]
- Gregg, J.H.; Hackney, A.L.; Krivanek, J.O. Nitrogen Metabolism Of The Slime Mold Dictyostelium Discoideum During Growth And Morphogenesis. Biol. Bull. 1954, 107, 226–235. [Google Scholar] [CrossRef]
- Schindler, J.; Sussman, M. Ammonia determines the choice of morphogenetic pathways in Dictyostelium discoideum. J. Mol. Biol. 1977, 116, 161–169. [Google Scholar] [CrossRef]
- Williams, J.G. Regulation of cellular differentiation during dictyostelium morphogenesis. Curr. Opin. Genet. Dev. 1991, 1, 358–362. [Google Scholar] [CrossRef]
- Davies, L.; Satre, M.; Martin, J.-B.; Gross, J.D. The target of ammonia action in dictyostelium. Cell 1993, 75, 321–327. [Google Scholar] [CrossRef]
- Kumada, Y.; Benson, D.R.; Hillemann, D.; Hosted, T.J.; Rochefort, D.A.; Thompson, C.J.; Wohlleben, W.; Tateno, Y. Evolution of the glutamine synthetase gene, one of the oldest existing and functioning genes. Proc. Natl. Acad. Sci. USA 1993, 90, 3009–3013. [Google Scholar] [CrossRef] [PubMed]
- Dunbar, A.J.; Wheldrake, J.F. Evidence for a developmentally regulated prespore-specific glutamine synthetase in the cellular slime mould Dictyostelium discoideum. Microbiology 1995, 141, 1125–1130. [Google Scholar] [CrossRef]
- Dunbar, A.J.; Wheldrake, J.F. Analysis of mRNA levels for developmentally regulated prespore specific glutamine synthetase in Dictyostelium discoideum. Dev. Growth Differ. 1997, 39, 617–624. [Google Scholar] [CrossRef] [PubMed]
- Carra, A.; Gambino, G.; Schubert, A. A cetyltrimethylammonium bromide-based method to extract low-molecular-weight RNA from polysaccharide-rich plant tissues. Anal. Biochem. 2007, 360, 318–320. [Google Scholar] [CrossRef] [PubMed]
- Green, M.R.; Sambrook, J. Isolation of DNA Fragments from Polyacrylamide Gels by the Crush and Soak Method. Cold Spring Harb. Protoc. 2019, 2019, pdb.prot100479. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- de Wijn, R.; Hennig, O.; Ernst, F.G.M.; Lorber, B.; Betat, H.; Mörl, M.; Sauter, C. Combining crystallogenesis methods to produce diffraction-quality crystals of a psychrophilic tRNA-maturation enzyme. Acta Crystallogr. F Struct. Biol. Commun. 2018, 74, 747–753. [Google Scholar] [CrossRef]
- Schürer, H.; Lang, K.; Schuster, J.; Mörl, M. A universal method to produce in vitro transcripts with homogeneous 3′ ends. Nucleic Acids Res. 2002, 30, e56. [Google Scholar] [CrossRef]
- Mörl, M.; Hartmann, R.K. Production of RNAs with Homogeneous 5′- and 3′-Ends. In Handbook of RNA Biochemistry, 2nd ed.; Hartmann, R.K., Bindereif, A., Schön, A., Westhof, E., Eds.; Wiley-VCH: Weinheim, Germany, 2014; pp. 29–44. [Google Scholar]
- Veltman, D.M.; Akar, G.; Bosgraaf, L.; van Haastert, P.J.M. A new set of small, extrachromosomal expression vectors for Dictyostelium discoideum. Plasmid 2009, 61, 110–118. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Erber, L.; Hoffmann, A.; Fallmann, J.; Hagedorn, M.; Hammann, C.; Stadler, P.F.; Betat, H.; Prohaska, S.; Mörl, M. Unusual Occurrence of Two Bona-Fide CCA-Adding Enzymes in Dictyostelium discoideum. Int. J. Mol. Sci. 2020, 21, 5210. https://doi.org/10.3390/ijms21155210
Erber L, Hoffmann A, Fallmann J, Hagedorn M, Hammann C, Stadler PF, Betat H, Prohaska S, Mörl M. Unusual Occurrence of Two Bona-Fide CCA-Adding Enzymes in Dictyostelium discoideum. International Journal of Molecular Sciences. 2020; 21(15):5210. https://doi.org/10.3390/ijms21155210
Chicago/Turabian StyleErber, Lieselotte, Anne Hoffmann, Jörg Fallmann, Monica Hagedorn, Christian Hammann, Peter F. Stadler, Heike Betat, Sonja Prohaska, and Mario Mörl. 2020. "Unusual Occurrence of Two Bona-Fide CCA-Adding Enzymes in Dictyostelium discoideum" International Journal of Molecular Sciences 21, no. 15: 5210. https://doi.org/10.3390/ijms21155210
APA StyleErber, L., Hoffmann, A., Fallmann, J., Hagedorn, M., Hammann, C., Stadler, P. F., Betat, H., Prohaska, S., & Mörl, M. (2020). Unusual Occurrence of Two Bona-Fide CCA-Adding Enzymes in Dictyostelium discoideum. International Journal of Molecular Sciences, 21(15), 5210. https://doi.org/10.3390/ijms21155210





