Epigenetic Regulation of Verticillium dahliae Virulence: Does DNA Methylation Level Play A Role?
Abstract
1. Introduction
2. Results
2.1. Verticillium Dahliae D Isolates Show A ‘Continuum of Virulence’ in Olive
2.2. Assembly and Annotation of the V. dahliae D Genomes
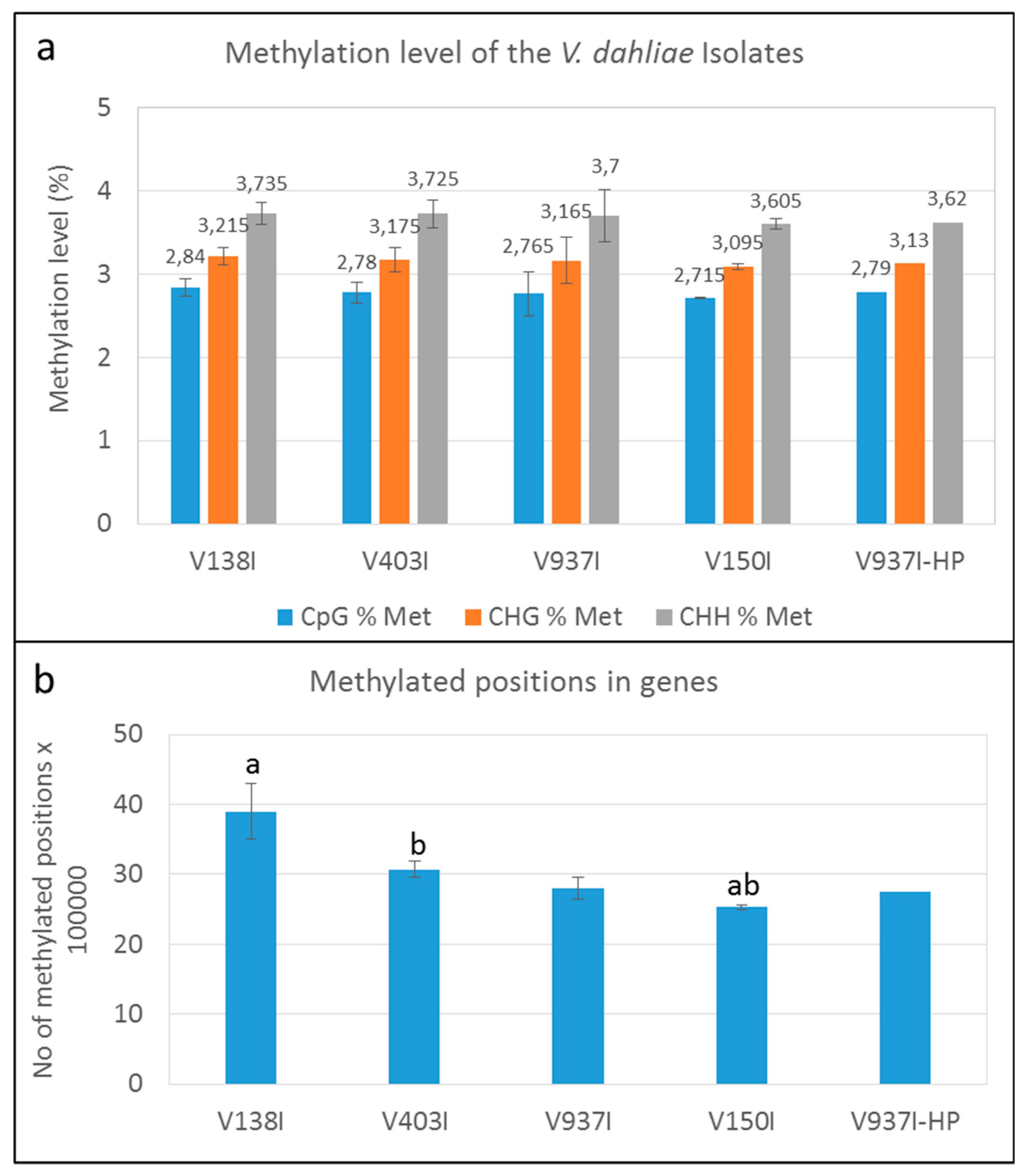
2.3. Type and Level of DNA Methylation in the V. dahliae D Genomes
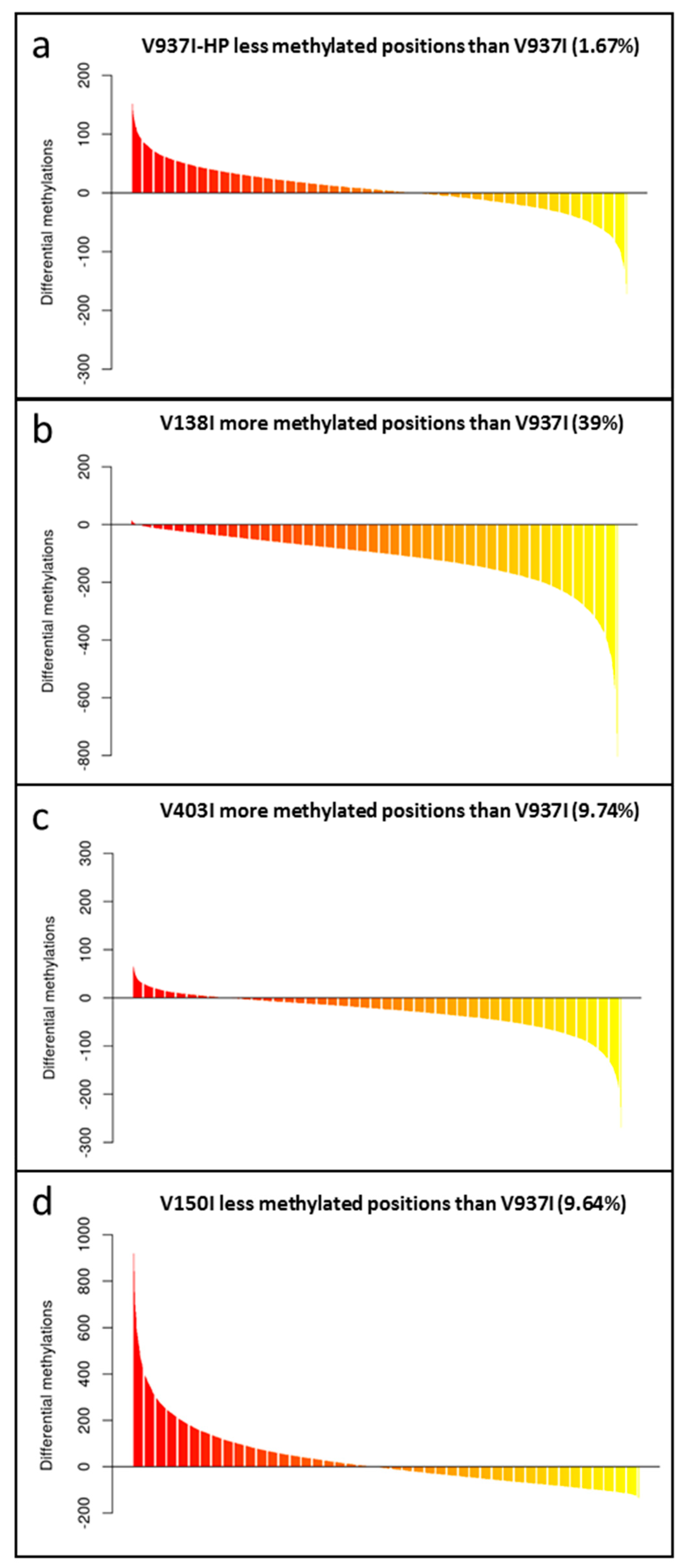
2.4. The Genes of the Highly Virulent V937I-HP Isolate are, on Average, Less Methylated than in Parental Isolate V937I
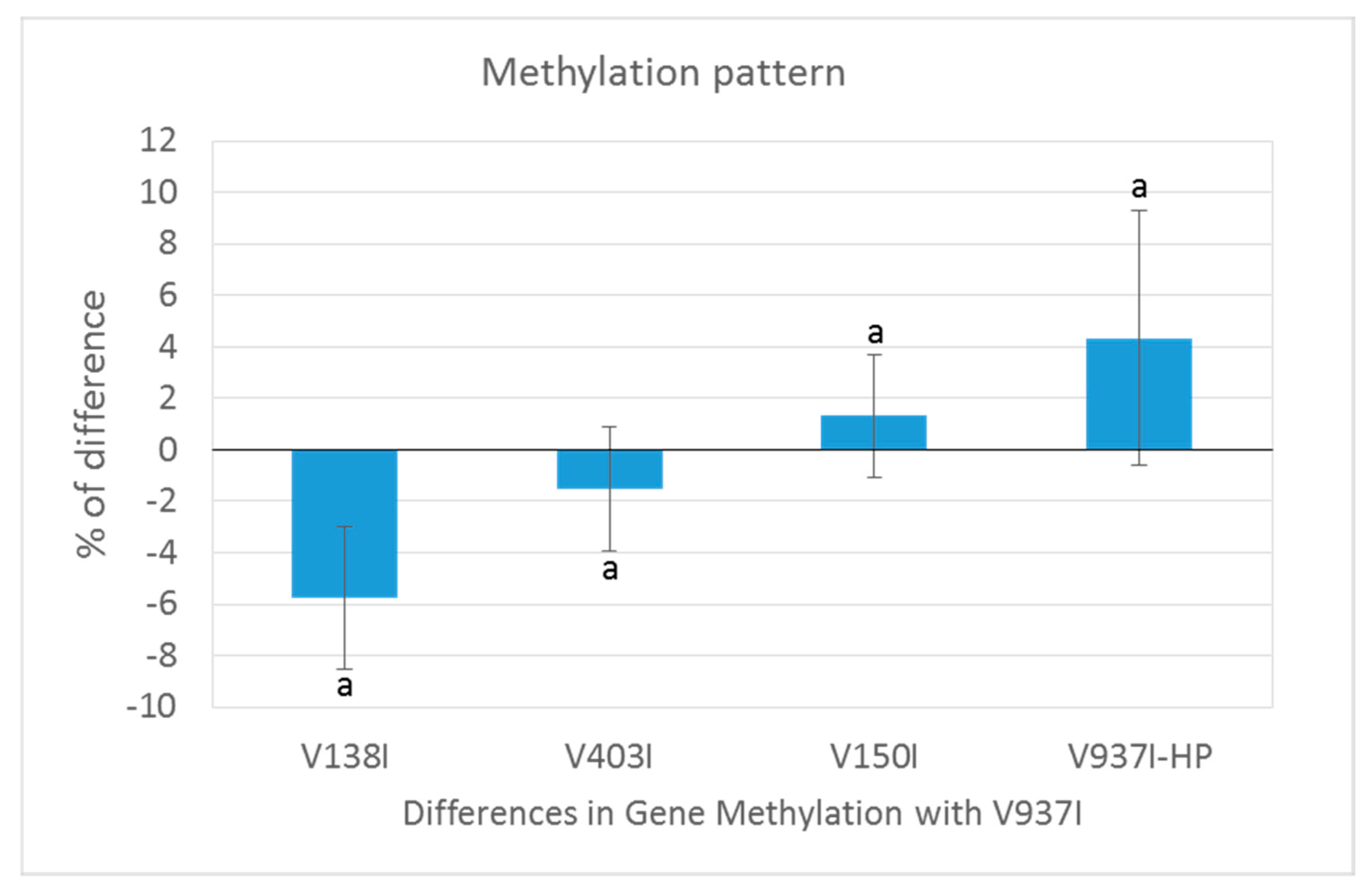
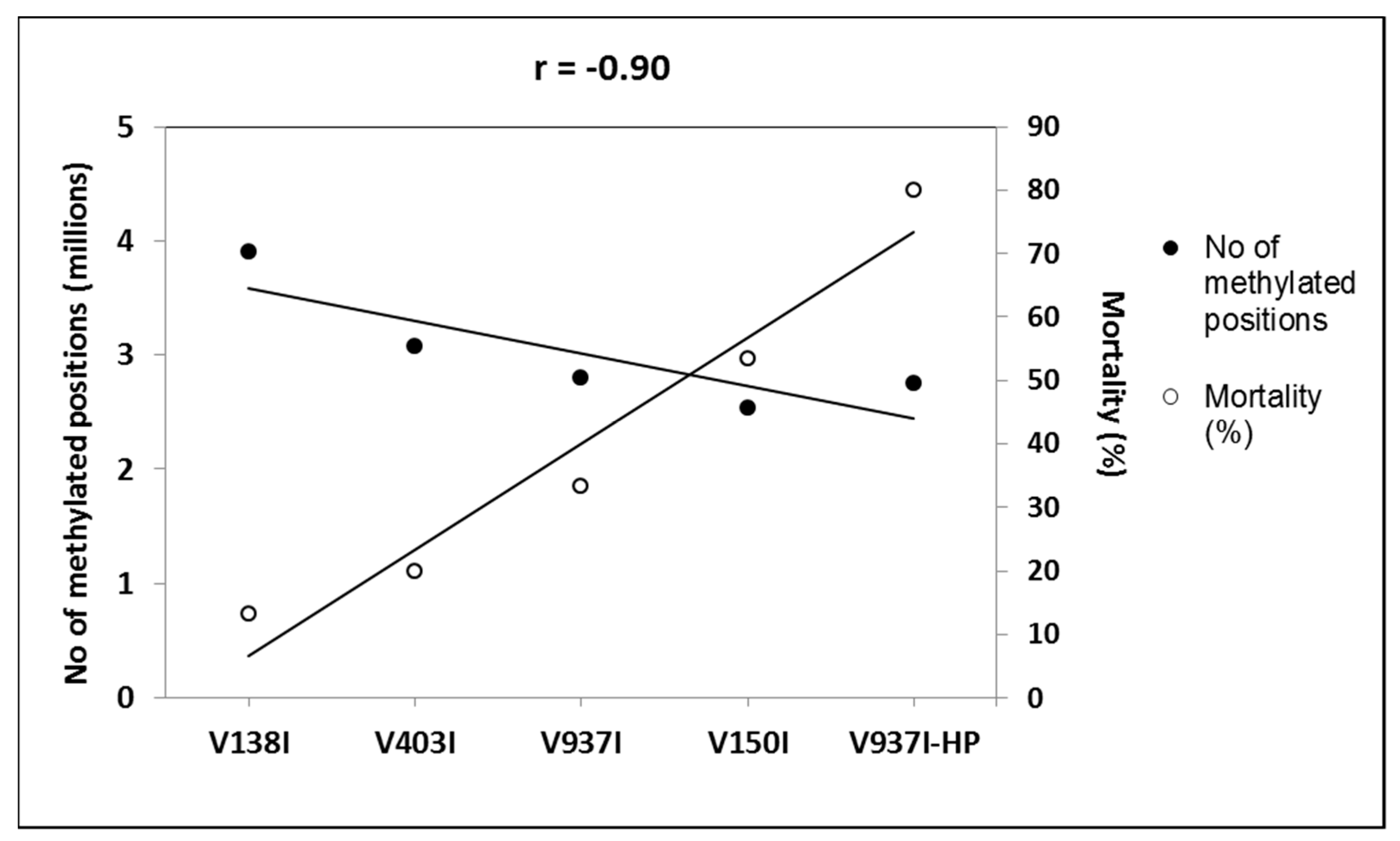
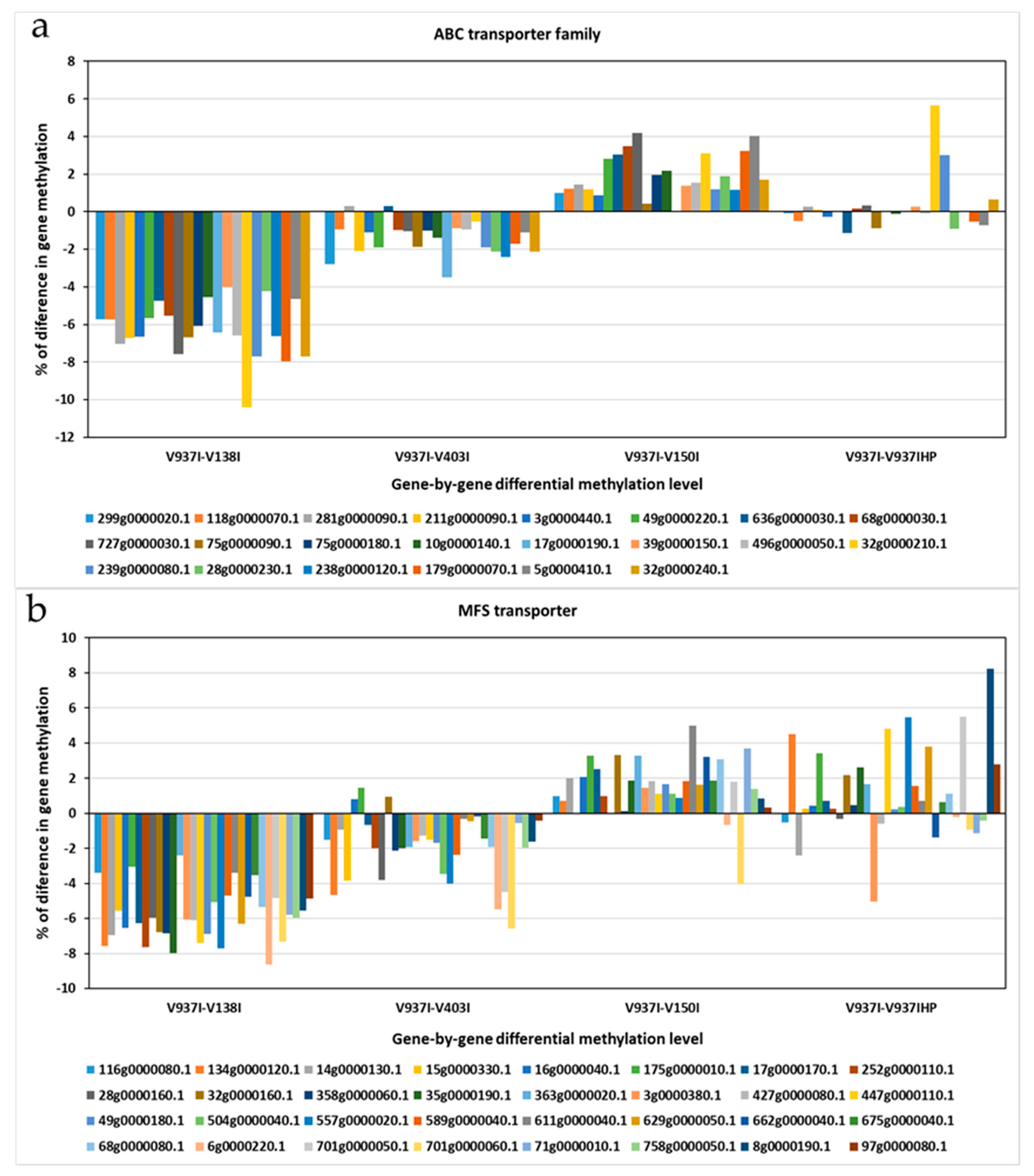
2.5. The Methylation Level of Many Genes Inversely Correlates with D Isolate Virulence
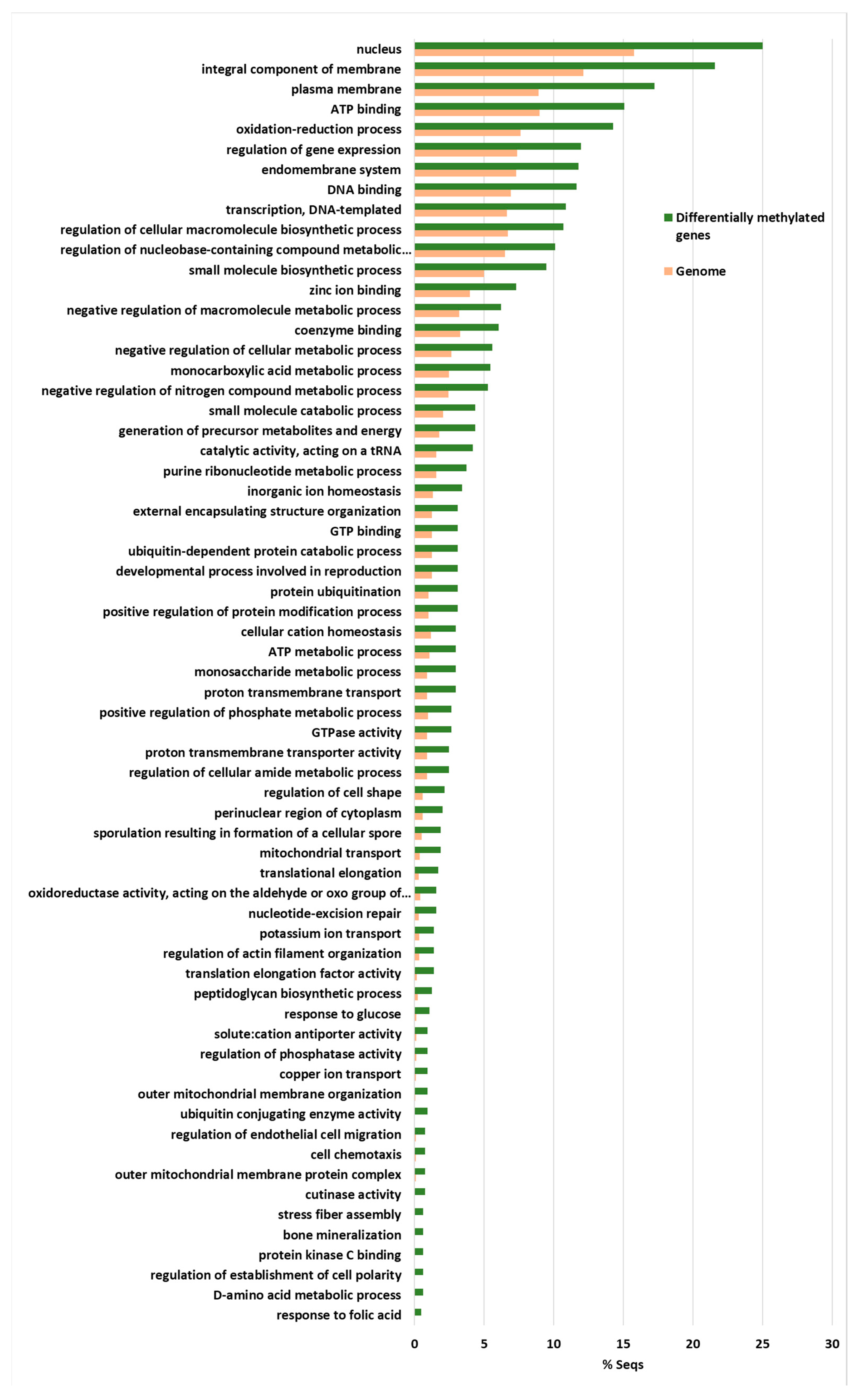
2.6. Analysis of Less Methylated Genes in the D Isolates Showing Higher Virulence in Olive
3. Discussion
4. Methods
4.1. Verticillium Dahliae Isolates
4.2. Verticillium Dahliae-Olive Bioassay
4.3. DNA Extraction
4.4. Genome Sequencing, Assembly and DNA Methylation Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Availability of Data and Material
References
- Fradin, E.F.; Thomma, B.P.H.J. Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol. Plant Pathol. 2006, 7, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Daayf, F. Verticillium wilts in crop plants: Pathogen invasion and host defense responses. Can. J. Plant Pathol. 2015, 37, 8–20. [Google Scholar] [CrossRef]
- Pegg, G.F.; Brady, B.L. Verticillium Wilts; CAB International: Wallingford, UK, 2002. [Google Scholar]
- Jiménez-Ruiz, J.; Leyva-Pérez, M.D.L.O.; Gómez-Lama Cabanás, C.; Barroso, J.B.; Luque, F.; Mercado-Blanco, J. The transcriptome of Verticillium dahliae responds differentially depending on the disease susceptibility level of the olive (Olea europaea L.) cultivar. Genes 2019, 10, 251. [Google Scholar] [CrossRef]
- López-Escudero, F.J.; Mercado-Blanco, J. Verticillium wilt of olive: A case study to implement an integrated strategy to control a soil-borne pathogen. Plant Soil 2011, 344, 1–50. [Google Scholar]
- Milgroom, M.G.; Jiménez-Gasco, M.M.; Olivares-García, C.; Jiménez-Díaz, R.M. Clonal expansion and migration of a highly virulent, defoliating lineage of Verticillium dahliae. Phytopathology 2016, 106, 1036–1046. [Google Scholar] [CrossRef]
- Dervis, S.; Mercado-Blanco, J.; Erten, L.; Valverde-Corredor, A.; Pérez-Artés, E. Verticillium wilt of olive in Turkey: A survey on disease importance, pathogen diversity and susceptibility of relevant olive cultivars. Eur. J. Plant Pathol. 2010, 127, 287–301. [Google Scholar] [CrossRef]
- López-Escudero, F.J.; Mercado-Blanco, J.; Roca, J.M.; Valverde-Corredor, A.; Blanco-López, M.A. Verticillium wilt of olive in the Guadalquivir Valley (Southern Spain): Relations with some agronomical factors and spread of Verticillium dahliae. Phytopathol. Mediterr. 2010, 49, 370–380. [Google Scholar]
- Triki, M.A.; Krid, S.; Hsairi, H.; Hammemi, I.; Ioos, R.; Gdoura, R.; Rhouma, A. Occurrence of Verticillium dahliae defoliating pathotypes on olive trees in Tunisia. Phytopathol. Mediterr. 2011, 50, 267–272. [Google Scholar]
- Schnathorst, W.C.; Sibbet, G.S. The relation of strains of Verticillium albo-atrum to severity of verticillium wilt in Gossypium hirsutum and Olea europaea in California. Plant Dis. Rep. 1971, 55, 780–782. [Google Scholar]
- Rodríguez-Jurado, D.; Blanco-López, M.A.; Rapoport, H.F.; Jiménez-Díaz, R.M. Present status of verticillium wilt of olive in Andalucía (southern Spain). EPPO Bull. 1993, 23, 513–516. [Google Scholar] [CrossRef]
- Maldonado-González, M.M.; Bakker, P.A.H.M.; Prieto, P.; Mercado-Blanco, J. Arabidopsis thaliana as a tool to identify traits involved in Verticillium dahliae biocontrol by the olive root endophyte Pseudomonas fluorescens PICF7. Front. Microbiol. 2015, 6, 266. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Xie, G.; Lo, C.; Starkenburg, S.R.; Chain, P.S.G. Pathogen comparative genomics in the next-generation sequencing era: Genome alignments, pangenomics and metagenomics. Brief. Funct. Genom. 2011, 10, 322–333. [Google Scholar] [CrossRef]
- De Jonge, R.; van Esse, H.P.; Maruthachalam, K.; Bolton, M.D.; Santhanam, P.; Saber, M.K.; Zhang, Z.; Usami, T.; Lievens, B.; Subbarao, K.V.; et al. Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc. Natl. Acad. Sci. USA 2012, 109, 5110–5115. [Google Scholar] [CrossRef]
- De Jonge, R.; Bolton, M.D.; Kombrink, A.; van den Berg, G.C.; Yadeta, K.A.; Thomma, B.P. Extensive chromosomal reshuffling drives evolution of virulence in an asexual pathogen. Genome Res. 2013, 23, 1271–1282. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-Y.; Liu, C.; Gui, Y.J.; Si, K.W.; Zhang, D.D.; Wang, J.; Short, D.P.G.; Huang, J.Q.; Li, N.Y.; Liang, Y.; et al. Comparative genomics reveals cotton-specific virulence factors in flexible genomic regions in Verticillium dahliae and evidence of horizontal gene transfer from Fusarium. New Phytol. 2018, 217, 756–770. [Google Scholar] [CrossRef]
- Shi-Kunne, X.; van Kooten, M.; Depotter, J.; Thomma, B.P.H.J.; Seidl, M.F. The genome of the fungal pathogen Verticillium dahliae reveals extensive bacterial to fungal gene transfer. Genome Biol. Evol. 2019, 11, 855–868. [Google Scholar] [CrossRef] [PubMed]
- Keykhasaber, M. Unravelling Aspects of Spatial and Temporal Distribution of Verticillium dahliae in Olive, Maple and Ash Trees and Improvement of Detection Methods. Ph.D. Thesis, University of Wageningen, Wageningen, The Netherlands, 2017. [Google Scholar] [CrossRef][Green Version]
- Zhang, D.-D.; Wang, J.; Wang, D.; Kong, Z.Q.; Zhou, L.; Zhang, G.Y.; Gui, Y.-J.; Li, J.-J.; Huang, J.-Q.; Wang, B.-L.; et al. Population genomics demystifies the defoliation phenotype in the plant pathogen Verticillium dahliae. New Phytol. 2019, 222, 1012–1029. [Google Scholar] [CrossRef]
- Seidl, M.F.; Thomma, B.P.H.J. Sex or no sex: Evolutionary adaptation occurs regardless. BioEssays 2014, 36, 335–345. [Google Scholar] [CrossRef]
- Faino, L.; Seidl, M.F.; Datema, E.; van den Berg, G.C.; Janssen, A.; Wittenberg, A.H.; Thomma, B.P. Single-molecule real-time sequencing combined with optical mapping yields completely finished fungal genome. mBio 2015, 6, e00936-15. [Google Scholar] [CrossRef]
- Dubey, A.; Jeon, J. Epigenetic regulation of development and pathogenesis in fungal plant pathogens. Mol. Plant Pathol. 2017, 18, 887–898. [Google Scholar] [CrossRef]
- Allshire, R.C.; Ekwall, K. Epigenetic regulation of chromatin states in Schizosaccharomyces pombe. CSH Perspect. Biol. 2015, 7, a018770. [Google Scholar]
- Aramayo, R.; Selker, C.U. Neurospora crassa, a model system for epigenetics research. CSH Perspect. Biol. 2013, 5, a017921. [Google Scholar] [CrossRef] [PubMed]
- Grunstein, M.; Gasser, S.M. Epigenetics in Saccharomyces Cerevisiae. CSH Perspect. Biol. 2013, 5, a017491. [Google Scholar] [CrossRef] [PubMed]
- Gijzen, M.; Ishmael, S.; Shrestha, S.D. Epigenetic control of effectors in plant pathogens. Front. Plant Sci. 2014, 5, 638. [Google Scholar] [CrossRef]
- Jin, Y.; Zhao, J.H.; Zhao, P.; Zhang, T.; Wang, S.; Guo, H.S. A fungal milRNA mediates epigenetic repression of a virulence gene in Verticillium dahliae. Phil. Trans. R. Soc. B 2019, 374, 20180309. [Google Scholar] [CrossRef] [PubMed]
- Hohl, H.R.; Balsiger, S. A Model System for the Study of Fungus—Host Surface Interactions: Adhesion of Phytophthora Megasperma to Protoplasts and Mesophyll Cells of Soybean. In Recognition in Microbe-Plant Symbiotic and Pathogenic Interactions; Lugtenberg, B., Ed.; NATO ASI Series (Series H: Cell Biology); Springer: Berlin/Heidelberg, Germany, 1986; Volume 4, pp. 259–272. [Google Scholar]
- Krueger, F.; Andrews, S.R. Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 1571, 2011, 27–1572. [Google Scholar] [CrossRef]
- Christodoulidou, A.; Briza, P.; Ellinger, A.; Bouriotis, V. Yeast ascospore wall assembly requires two chitin deacetylase isozymes. FEBS Lett. 1999, 460, 275–279. [Google Scholar] [CrossRef]
- Yamada, M.; Kurano, M.; Inatomi, S.; Taguchi, G.; Okazaki, M.; Shimosaka, M. Isolation and characterization of a gene coding for chitin deacetylase specifically expressed during fruiting body development in the basidiomycete Flammulina velutipes and its expression in the yeast Pichia pastoris. FEMS Microbiol. Lett. 2008, 298, 130–137. [Google Scholar] [CrossRef]
- Abe, H.; Shimoda, C. Autoregulated Expression of Schizosaccharomyces pombe meiosis-specific transcription factor Mei4 and a genome-wide search for its target genes. Genetics 2000, 154, 1497–1508. [Google Scholar]
- Gómez-Lama Cabanás, C.; Schilirò, E.; Valverde-Corredor, A.; Mercado-Blanco, J. Systemic responses in a tolerant olive (Olea europaea L.) cultivar upon root colonization by the vascular pathogen Verticillium dahliae. Front. Microbiol. 2015, 6, 928. [Google Scholar] [CrossRef]
- Gowher, H.; Ehrlich, K.C.; Jeltsch, A. DNA from Aspergillus flavus contains 5-methylcytosine. FEMS Microbiol. Lett. 2001, 205, 151–155. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.L.; Zhuang, Z.H.; Zhang, F.; Song, F.Q.; Zhong, H.; Ran, F.L.; Yu, S.; Xu, G.P.; Lan, F.X.; Wang, S.H. Inhibition of aflatoxin metabolism and growth of Aspergillus flavus in liquid culture by a DNA methylation inhibitor. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 2015, 32, 554–563. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.Y.; Lin, J.Q.; Wu, H.L.; Wang, C.C.; Huang, S.J.; Luo, Y.F.; Sun, J.H.; Zhou, J.X.; Yan, S.J.; He, J.G.; et al. Bisulfite sequencing reveals that Aspergillus flavus holds a hollow in DNA methylation. PLoS ONE 2012, 7, e30349. [Google Scholar] [CrossRef] [PubMed]
- Kostlánová, N.; Mitchell, E.P.; Lortat-Jacob, H.; Oscarson, S.; Lahmann, M.; Gilboa-Garber, N.; Chambat, G.; Wimmerová, M.; Lmberty, A. The fucose-binding lectin from Ralstonia solanacearum. A new type of beta-propeller architecture formed by oligomerization and interacting with fucoside, fucosyllactose, and plant xyloglucan. J. Biol. Chem. 2005, 2809, 2783–27849. [Google Scholar]
- Houser, J.; Komarek, J.; Kostlanova, N.; Cioci, G.; Varrot, A.; Kerr, S.C.; Lahmann, M.; Balloy, V.; Fahy, J.V.; Chignard, M.; et al. A soluble fucose-specific lectin from Aspergillus fumigatus conidia-structure, specificity and possible role in fungal pathogenicity. PLoS ONE 2013, 8, e83077. [Google Scholar] [CrossRef]
- Kuboi, S.; Ishimaru, T.; Tamada, S.; Bernard, E.M.; Perlin, D.S.; Armstrong, D. Molecular characterization of AfuFleA, an L-fucose-specific lectin from Aspergillus fumigatus. J. Infect. Chemother. 2013, 19, 1021–1028. [Google Scholar] [CrossRef]
- Tsigos, I.; Bouriotis, V. Purification and characterization of chitin deacetylase from Colletotrichum lindemuthianum. J. Biol. Chem. 1995, 2706, 26286–26291. [Google Scholar] [CrossRef]
- Kolattukudy, P.E.; Podila, G.K.; Mohan, R. Molecular basis of the early events in plant–fungus interaction. Genome 1989, 31, 342–349. [Google Scholar] [CrossRef]
- Fan, C.-Y.; Köller, W. Diversity of cutinases from plant pathogenic fungi: Differential and sequential expression of cutinlytic esterases by Alternaria brassicicola. FEMS Microbiol. Lett. 1998, 158, 33–38. [Google Scholar] [CrossRef]
- St Leger, R.J.; Joshi, L.; Roberts, D.W. Adaptation of proteases and carbohydrases of saprophytic, phytopathogenic and entomopathogenic fungi to the requirements of their ecological niches. Microbiology 1997, 143, 1983–1992. [Google Scholar] [CrossRef]
- Collins, A.; Mercado-Blanco, J.; Jiménez-Díaz, R.M.; Olivares, C.; Clewes, E.; Barbara, D.J. Correlation of molecular markers and biological properties in Verticillium dahliae and the possible origins of some isolates. Plant Pathol. 2005, 54, 549–557. [Google Scholar] [CrossRef]
- Collado-Romero, M.; Mercado-Blanco, J.; Olivares-Garcíam, C.; Jiménez-Díaz, R.M. Phylogenetic analysis of Verticillium dahliae vegetative compatibility groups. Phytopathology 2008, 98, 1019–1028. [Google Scholar] [CrossRef]
- Jiménez-Díaz, R.M.; Mercado-Blanco, J.; Olivares-García, C.; Collado-Romero, M.; Bejarano-Alcázar, J.; Rodríguez-Jurado, D.; Garca-Jimnez, J.; Armengol, J. Genetic and virulence diversity in Verticillium dahliae populations infecting artichoke in eastern-central Spain. Phytopathology 2006, 96, 288–298. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Collado-Romero, M.; Mercado-Blanco, J.; Olivares-García, C.; Valverde-Corredor, A.; Jiménez-Díaz, R.M. Molecular variability within and among Verticillium dahliae vegetative compatibility groups determined by fluorescent amplified fragment length polymorphism and polymerase chain reaction markers. Phytopathology 2006, 96, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Lama Cabanás, C.; Sesmero, R.; Valverde-Corredor, A.; López-Escudero, F.J.; Mercado-Blanco, J. A split-root system to assess biocontrol effectiveness and defense-related genetic responses in above-ground tissues during the tripartite interaction Verticillium dahliae-olive-Pseudomonas fluorescens PICF7 in roots. Plant Soil 2017, 417, 433–452. [Google Scholar] [CrossRef]
- Maldonado-González, M.M.; Schilirò, E.; Prieto, P.; Mercado-Blanco, J. Endophytic colonization and biocontrol performance of Pseudomonas fluorescens PICF7 in olive (Olea europaea L.) are determined neither by pyoverdine production nor swimming motility. Environ. Microbiol. 2015, 17, 3139–3153. [Google Scholar] [CrossRef]
- Campbell, C.L.; Madden, L.V. Introduction to Plant Disease Epidemiology; John Wiley & Sons: New York, NY, USA, 1990. [Google Scholar]
- Raeder, U.; Broda, P. Rapid preparation of DNA from filamentous fungi. Lett. Appl. Microbiol. 1985, 1, 17–20. [Google Scholar] [CrossRef]
- Nurk, S.; Bankevich, A.; Antipov, D.; Gurevich, A.A.; Korobeynikov, A.; Lapidus, A.; Prjibelski, A.D.; Pyshkin, A.; Sirotkin, A.; Sirotkin, Y.; et al. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J. Comput. Biol. 2013, 20, 714–737. [Google Scholar] [CrossRef]
- Prjibelski, A.D.; Vasilinetc, I.; Bankevich, A.; Gurevich, A.; Krivosheeva, T.; Nurk, S.; Pham, S.; Korobeynikov, A.; Lapidus, A.; Pevzenr, P.A.; et al. ExSPAnder: A universal repeat resolver for DNA fragment assembly. Bioinformatics 2014, 30, i293–i301. [Google Scholar] [CrossRef]
- Vasilinetc, I.; Prjibelski, A.D.; Gurevich, A.; Korobeynikov, A.; Pevzner, P.A. Assembling short reads from jumping libraries with large insert sizes. Bioinformatics 2015, 31, 3262–3268. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]

| Disease Parameters | |||||
|---|---|---|---|---|---|
| Treatments | S | AUDPC | DI (%) | DII | M (%) |
| V-937I | 1.65 bc | 57.9 bc | 53.3 | 0.41 | 33.3 |
| V-937I-HP | 3.32 a | 133 a | 93.3 | 0.83 | 80 |
| V-138I | 1.18 c | 43.7 c | 53.3 | 0.3 | 13.3 |
| V-150I | 2.57 ab | 92.6 ab | 73.3 | 0.64 | 53.3 |
| V-403I | 1.47 bc | 42.3 c | 60 | 0.33 | 20 |
| Isolate DNA | Without Bisulfite Treatment (No Reads) | With Bisulfite Treatment (No Reads in Two Replicates) | |
|---|---|---|---|
| V-937I | 29,352,982 | 18,662,110 | 10,547,437 |
| V-937I-HP | 24,087,863 | 16,318,447 | 11,068,341 |
| V-138I | 22,157,160 | 16,817,943 | 22,237,529 |
| V-150I | 21,766,421 | 12,384,988 | 14,527,876 |
| V-403I | 20,520,224 | 15,113,143 | 13,553,206 |
| Summarized Benchmarks in the BUSCO Notation C: 86%(D:3.4%),F: 6.1%,M:7.5%,n: 3725 | |
|---|---|
| Complete and single-copy BUSCOs | 3085 |
| Complete BUSCOs | 3215 |
| Complete and duplicated BUSCOs | 130 |
| Fragmented BUSCOs | 229 |
| Missing BUSCOs | 281 |
| Total BUSCO groups searched | 3725 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramírez-Tejero, J.A.; Cabanás, C.G.-L.; Valverde-Corredor, A.; Mercado-Blanco, J.; Luque, F. Epigenetic Regulation of Verticillium dahliae Virulence: Does DNA Methylation Level Play A Role? Int. J. Mol. Sci. 2020, 21, 5197. https://doi.org/10.3390/ijms21155197
Ramírez-Tejero JA, Cabanás CG-L, Valverde-Corredor A, Mercado-Blanco J, Luque F. Epigenetic Regulation of Verticillium dahliae Virulence: Does DNA Methylation Level Play A Role? International Journal of Molecular Sciences. 2020; 21(15):5197. https://doi.org/10.3390/ijms21155197
Chicago/Turabian StyleRamírez-Tejero, Jorge A., Carmen Gómez-Lama Cabanás, Antonio Valverde-Corredor, Jesús Mercado-Blanco, and Francisco Luque. 2020. "Epigenetic Regulation of Verticillium dahliae Virulence: Does DNA Methylation Level Play A Role?" International Journal of Molecular Sciences 21, no. 15: 5197. https://doi.org/10.3390/ijms21155197
APA StyleRamírez-Tejero, J. A., Cabanás, C. G.-L., Valverde-Corredor, A., Mercado-Blanco, J., & Luque, F. (2020). Epigenetic Regulation of Verticillium dahliae Virulence: Does DNA Methylation Level Play A Role? International Journal of Molecular Sciences, 21(15), 5197. https://doi.org/10.3390/ijms21155197









