Abstract
Depletion of yeast/fly Ataxin-2 rescues TDP-43 overexpression toxicity. In mouse models of Amyotrophic Lateral Sclerosis via TDP-43 overexpression, depletion of its ortholog ATXN2 mitigated motor neuron degeneration and extended lifespan from 25 days to >300 days. There is another ortholog in mammals, named ATXN2L (Ataxin-2-like), which is almost uncharacterized but also functions in RNA surveillance at stress granules. We generated mice with Crispr/Cas9-mediated deletion of Atxn2l exons 5-8, studying homozygotes prenatally and heterozygotes during aging. Our novel findings indicate that ATXN2L absence triggers mid-gestational embryonic lethality, affecting female animals more strongly. Weight and development stages of homozygous mutants were reduced. Placenta phenotypes were not apparent, but brain histology showed lamination defects and apoptosis. Aged heterozygotes showed no locomotor deficits or weight loss over 12 months. Null mutants in vivo displayed compensatory efforts to maximize Atxn2l expression, which were prevented upon nutrient abundance in vitro. Mouse embryonal fibroblast cells revealed more multinucleated giant cells upon ATXN2L deficiency. In addition, in human neural cells, transcript levels of ATXN2L were induced upon starvation and glucose and amino acids exposure, but this induction was partially prevented by serum or low cholesterol administration. Neither ATXN2L depletion triggered dysregulation of ATXN2, nor a converse effect was observed. Overall, this essential role of ATXN2L for embryogenesis raises questions about its role in neurodegenerative diseases and neuroprotective therapies.
1. Introduction
In all eukaryotic organisms, at least one copy of the Ataxin-2 gene (gene symbol ATXN2 in humans) is phylogenetically conserved and serves roles during nutrient stress for RNA surveillance [1]. A conserved Lsm and LsmAD motif enables direct interaction with RNAs, and a PAM2 motif mediates association with the poly(A)-binding protein PABPC1 [2,3]. Thus, most Ataxin-2 protein localizes with mRNAs at the rough endoplasmic reticulum with marker Ribosomal Protein S6 (RPS6 aka S6R) during cell growth periods [4], where its absence leads to expression adaptations of the associated ribosomal translation machinery [5] and modulates the phosphorylation control of translation [6]. During cell stress, e.g., from nutrient deprivation, Ataxin-2 is transcriptionally induced [6] and relocates with the small ribosomal subunit and PABPC1 to stress granules (SG) [7]. The RNA surveillance function of Ataxin-2 seems to be relevant to protect against the translation of viral RNAs, given that poliovirus is optimized to cleave Ataxin-2 [8]. In mammals, all these protein structure domains are also found in its paralog Ataxin-2-like (gene symbol ATXN2L in humans). Both ATXN2 and ATXN2L mRNAs also conserved an alternatively spliced exon, which encodes a proline-rich domain (PRD) that mediates its direct association with SH3 motifs in components of the growth factor receptor endocytosis apparatus [9,10,11]. Abnormal ATXN2 splicing and alternative polyadenylation were documented in diseases with RNA toxicity, such as amyotrophic lateral sclerosis (ALS) [12]. The common ancestor of both proteins in yeast and Caenorhabditis elegans was observed to suppress growth signaling via mTORC1, modulating cell size, and lipid stores [13,14,15]. This reprogramming of nutrient metabolism is accompanied by an important influence on the mitochondrial breakdown of fatty acids and amino acids, as well as glucose utilization [16,17,18,19], probably mediated by the direct protein interaction of ATXN2 with the cytosolic enzyme BCAT1 [20] as the rate-limiting factor in the breakdown of leucine, isoleucine, and valine.
The role of ATXN2 in neurodegenerative diseases has triggered intense research over the past 25 years. Exclusively in human ATXN2, an N-terminal domain with 22 consecutive glutamines (polyQ) exists, which can undergo expansion mutations across generations. Large expansions beyond the size of 32Q trigger the multi-system nervous tissue atrophy Spinocerebellar ataxia type 2 (SCA2), while intermediate expansions of sizes 27Q-32Q increase the risk to be affected by motor neuron diseases such as ALS, fronto-temporal lobar dementia (FTLD) [21,22,23,24,25] or by other tauopathies and Parkinson’s disease variants like progressive supranuclear palsy (PSP) [26,27]. Conversely, the depletion of ATXN2 by knock-out or by injection of antisense-oligonucleotides has a massive neuroprotective effect in yeast/fly/mouse models of ALS and FTLD, as well as in SCA2 and SCA1 fly models [24,27,28,29]. In addition, in yeast, depletion of the ATXN2/ATXN2L ortholog PBP1 rescues the lethal effect of poly(A)-binding protein deletions [30]. The constitutive knock-out of Atxn2 in mice leads to progressive weight gain with excessive storage of lipid droplets and glycogen in the liver, elevated cholesterol and other lipids in the blood, beta-cell hyperplasia in the pancreas with hyperinsulinemia and insulin resistance, increased ganglioside and sulfatide lipids in the brain myelin, locomotor hyperactivity, and mild infertility with gender-dependent impairment of embryogenesis [31,32].
In view of the importance of ATXN2 orthologs for stress response, redundancy occurred in land plants and in vertebrates (except birds) by the co-existence of two homologous genes, named CID3-CID4 in Arabidopsis thaliana weed and ATXN2-ATXN2L in humans and rodents [33]. ATXN2L protein dimerizes with ATXN2 in yeast-two-hybrid tests, and is also a regulator of SGs and mRNA processing during starvation periods, but shows more co-localization with the nuclear splice apparatus than ATXN2 due to an arginine-dimethylation [34,35]. Similar to ATXN2, ATXN2L associates with plasma membrane receptors in dependence on their phosphorylation status, is involved in epidermal-growth-factor (EGF)-receptor signaling, and exists in several isoforms [36,37,38]. Little more is known at present about ATXN2L. Database mining at the STRING web platform for Protein-Protein Interaction Networks and Functional Enrichment Analysis, available online at: https://string-db.org/ [39] confirms that human and mouse ATXN2L show direct protein-protein-interaction with the poly(A)-binding-proteins PABPC1/2, PABPC1L/PABPC3/PABPC4/PABPC6, with further stress-granule-components such as G3BP2, DDX6, and LSM12 [40,41], as well as nuclear RNA-binding protein NUFIP2 and nuclear transcription modulator NFATC2IP. The RNA helicase activity of DDX6 enhances the expression of growth factor receptors such as HER2 and FGFR2 [42]. Possibly by this mechanism, ATXN2 was found to modulate surface receptors [43]. A recent global protein interactome study via FLAG-tag affinity purification and mass spectrometry in HEK293 cells also showed ATXN2L to associate with DDX6 and the proline-rich-domain-containing RNA-binding Ras-GTPase G3BP2, but in addition, demonstrated ATXN2L binding to KLHL20 [20], which is responsible for the degradation of the autophagy/growth modulators DAPK1 and ULK1, as well as of the Rho Guanine nucleotide exchange factor and glutamate-transport-/neurite-growth-modulator ARHGEF11. STRING also lists a potential interaction (based on association in lower species) with SLC9A3R2/NHERF2 that is known to act as PSD-95 scaffold to control albumin endocytosis, the glutamate transporter GLAST, and the metabotropic glutamate receptor mGluR5, in addition to activating Src phosphorylation and modulating the high density lipoprotein receptor SR-B1 that is crucial for cholesterol metabolism [44,45,46,47,48].
Further data mining at the Allen brain atlas, available online at: https://mouse.brain-map.org/ [49] retrieves evidence that Atxn2l mRNA has a particularly strong expression in cerebellar Purkinje neurons. Protein data in different cell types from the Human Protein Atlas, available online at: https://www.proteinatlas.org/ [50] demonstrate its prominent abundance in testis. Its moderate abundance in postmitotic neurons from the cortex, cerebellum, hippocampus, and caudate nucleus, as well as glia and muscle cells were detected by the Atlas antibody HPA041506 targeting ATXN2L at aa 456-547 (in the Uniprot Q8WWM7-1 sequence), which exhibits a single immunoblot band of predicted size and has siRNA-controlled specificity. However, it was not detected by Atlas antibody HPA043391, which targets human ATXN2L aa 572-644, but does not exhibit immunoblot signals of correct size and was not tested by siRNA, as can be seen within the Human Protein Atlas online at: https://www.proteinatlas.org/ENSG00000168488-ATXN2L/tissue/primary+data [50]. Subcellular analyses with both antibodies showed a diffuse cytosolic signal. Among cell lines, a particularly strong expression is detected under lymphoid differentiation, in good agreement with the importance of RNA surveillance for the innate and adaptive immune system. The upregulation of ATXN2L in cancer tissue is an unfavorable marker, in agreement with its published induction by EGF growth signaling [38]. A co-expression survey at the Coexpedia database, online available at: www.coexpedia.org [51] shows that mouse Atxn2l mRNA is regulated together with pathways of thin myocardium, open neural tube, improved glucose tolerance, immune defense, and cell cycle. The RNA binding spliceosome component Srrm2 and the proline-rich inflammatory splicing factor Prrc2a show the highest co-expression score.
In a recently generated mouse with the aggregation of ATXN2 due to a polyglutamine (Q100) expansion knock-in [52], we observed a significant accumulation of ATXN2L peptides in cerebellar global proteome profiles from old animals. It was difficult to know if the impact of this anomaly has pathogenic or compensatory roles, and it was impossible to study it further in view of the unavailability of antibodies shown to be specific, of tagged recombinant constructs and mutant cells. We decided to start an exploratory project into the physiological functions of ATXN2L. Working at organism level, we generated novel Atxn2l-ablated mice to assess their phenotypes in growth, histology, and behavior, in comparison to our previous study of Atxn2-ablated mice [31]. Working at cell culture level, we examined the cellular phenotype of Atxn2l−/− mouse embryonal fibroblasts (MEF), and whether the regulation of human ATXN2L and ATXN2 mRNA levels is similar in response to stressors for neural cells. Overall, the data indicate that ATXN2L is much more important than ATXN2 for growth at early embryonal age, particularly in females. Atxn2l exon 5-8 deletion triggered a growth deficit phenotype rather than the weight excess observed in mice with Atxn2 exon 1 deletion. Apparently, ATXN2L has a similar function as ATXN2, which is not only transcriptionally induced upon deprivation from nutrients and especially lipids, but also upon exposure to excess glucose or amino acids. Hence, we believe that elucidating the native role of ATXN2L throughout development and in response to stress provides important insights into the complementary spatio-temporal roles of ATXN2-ATXN2L and will further our understanding of neurodegenerative pathomechanisms.
2. Results
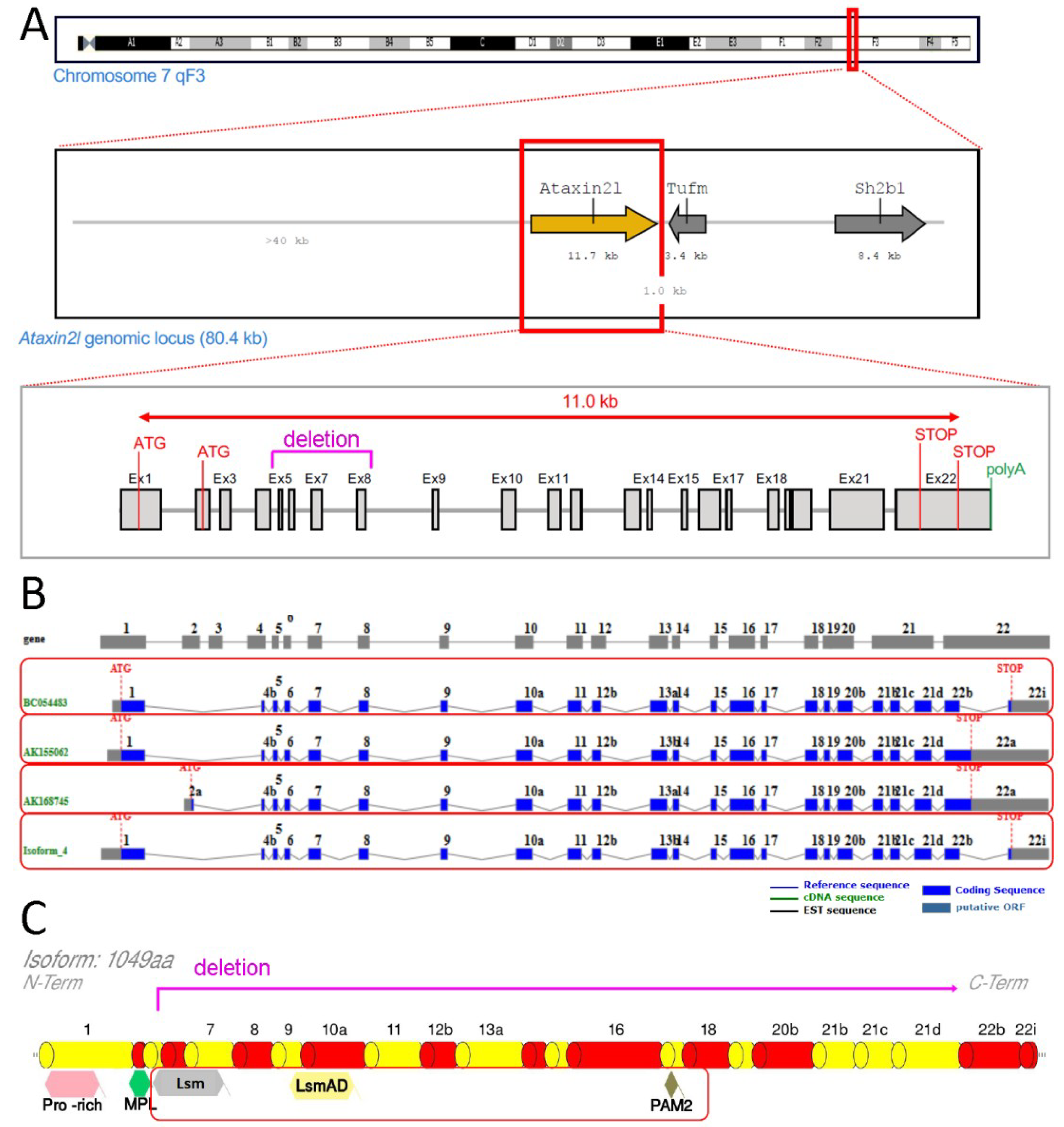
We obtained a null allele of murine Atxn2l via Crispr/Cas9-mediated deletion between two sgRNA sites flanking the exons 5-8 of the gene (outsourced work by TIGM, College Station, TX, USA) (see Figure 1).
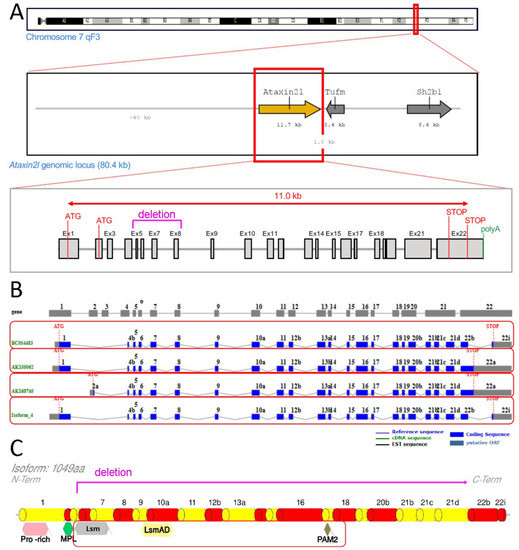
Figure 1.
Scheme of genetic ablation within Atxn2l, with its splicing and translation effects. Ataxin-2-like (A) at the DNA level with neighbor genes Tufm and Sh2b1, indicating the deletion region in letters and lines of magenta color (also showing alternative translation start codons (ATG), structure from exon 1 to exon 22 with introns, alternative translation STOP codons, start of poly(A) tail); (B) at the mRNA level with alternatively spliced isoforms in separate red boxes; (C) at the protein level with relevant known sequence motifs (pro-rich refers to proline-rich-domain, MPL motif was reported by Meunier et al. 2002), highlighting the phylogenetically conserved Lsm, LsmAD and PAM2 motifs in a red box (deletion and frameshift are limiting translation to an N-terminal fragment until magenta line).
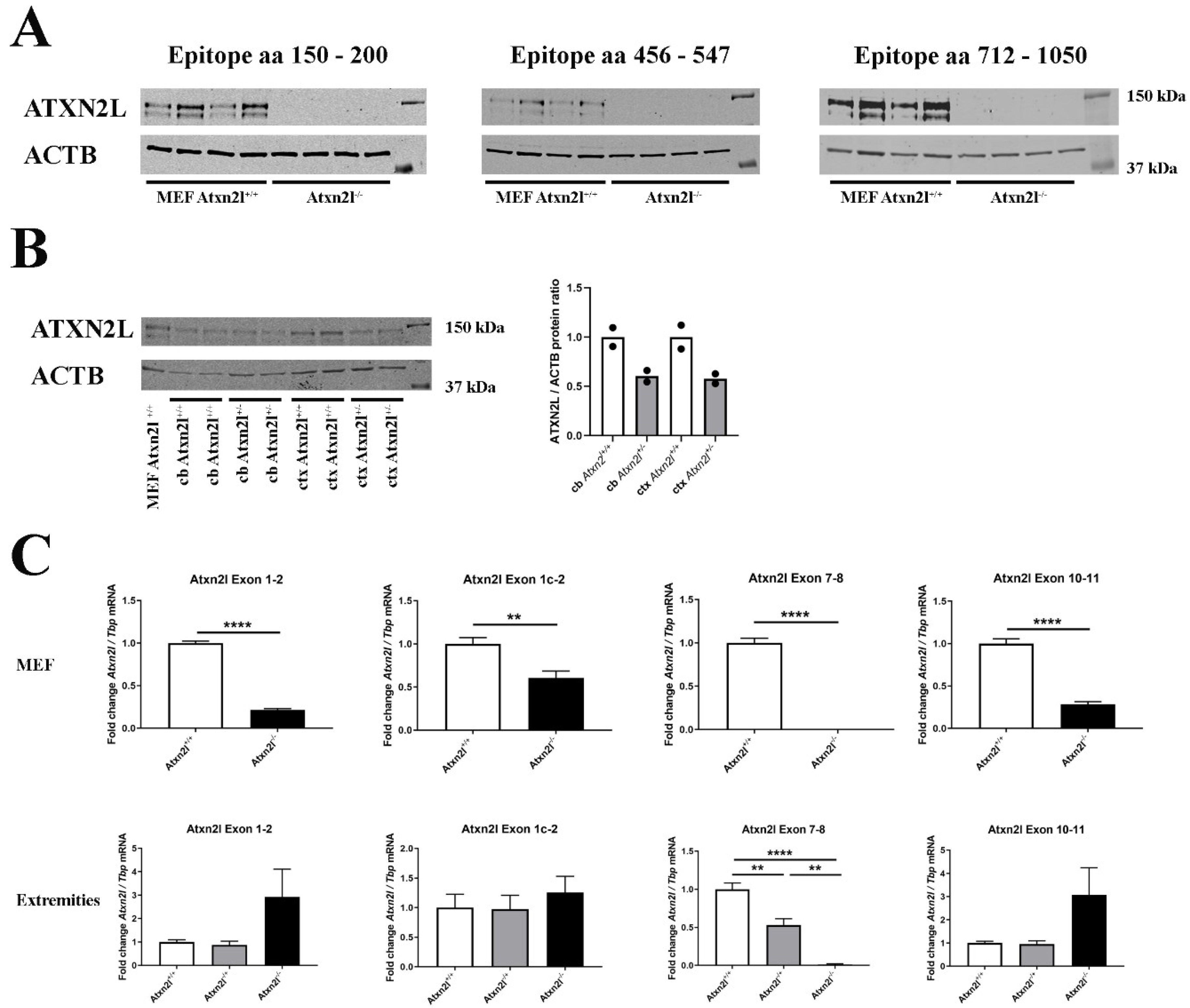
The absence of ATXN2L protein epitopes from residue 150 until residue 1050 was confirmed in Atxn2l−/− mouse embryonal fibroblast (MEF) (Figure 2A). ATXN2L abundance was higher in cerebral cortex than in cerebellum. While MEFs showed two specific ATXN2L bands in immunoblots, in brain tissue the larger band dominated (Figure 2B). The separate quantification of several Atxn2l mRNA exon junctions, namely 7-8 to represent the deleted region, 10-11 further downstream, and 1-2 upstream versus the alternatively spliced 1c-2 suggested the Atxn2l−/− limb cells to sense the loss-of-function and either trigger the Atxn2l promoter or minimize the Atxn2l mRNA decay, thus maximizing the levels of the main Atxn2l mRNA variant. This feedback did not occur in MEF lines that are maintained in nutrient abundance (Figure 2C), suggesting that deprivation of overall energy or specific nutrients modulates Atxn2l transcription.
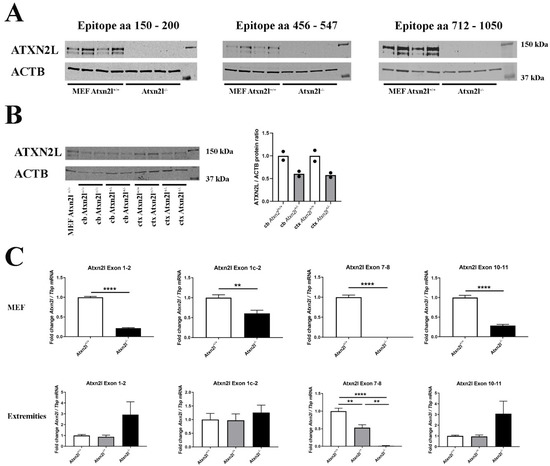
Figure 2.
(A) Protein abundance of 3 different epitopes spanning ATXN2L in control Atxn2l+/+ and homozygous Atxn2l−/− MEF (n = 4vs4). The protein was completely absent in three assessed epitopes from residue 150 until the most C-terminal region (aa: amino acid). (B) Protein abundance of ATXN2L in heterozygous Atxn2l+/− cerebellum (cb) compared to wildtype littermates, or in heterozygous Atxn2l+/− cortex (ctx), in comparison to one wildtype Atxn2l+/+ MEF line. MEFs exhibit two specific bands of ATXN2L, in contrast to brain tissue where the larger band dominates. The overall ATXN2L levels were decreased to approx. 50% in heterozygous tissues (n = 2). Detection was done for the epitope aa 712-1050. (C) In Atxn2l−/− MEF (upper row, n = 4vs4), RT-qPCR showed that mRNA expression of Atxn2l was absent at the boundary between exons 7-8 that was genetically deleted, and significantly reduced at the boundaries of exons 1-2 and 10-11, possibly due to nonsense-mediated RNA decay. In tissue from fore-/hind-limbs [lower row, 3 wildtype (Atxn2l+/+, white bars), 4 heterozygous (Atxn2l+/−, grey bars), 2 Atxn2l-null embryos (Atxn2−/−, black bars)], the Atxn2l mRNA expression measured at the boundary of exons 7-8 was again absent in Atxn2l−/− and reduced by 50% in Atxn2l−/+ mice, whereas 5′-upstream and 3′-downstream from the deletion the increased Atxn2l expression level of null mutants suggested transcript upregulation efforts either via the Atxn2l promoter or via altered mRNA stabilization/degradation to compensate the loss of protein function. The exon 1c-2 assay detects an alternatively spliced isoform. Asterisks represent significance (p < 0.01 **, p < 0.0001 ****).
While the heterozygous mice bred normally, a lethality at the embryonic/prenatal stage was observed for the homozygous mutants. There was also reduced viability of heterozygous mice, particularly for females at the prenatal stage (see Table 1 and Table 2).

Table 1.
Postnatal genotype distribution for offspring from Atxn2l+/− intercrosses.

Table 2.
Embryonic day E11-20 genotype distribution for offspring from Atxn2l+/− intercrosses. Most lethality occurred before E11, with females dying before males.
The dissection at different stages of pregnancy showed the death of homozygous mutants to have mostly occurred by embryonic day E14-16. Female Atxn2l−/− embryos were more severely affected, most of them did not reach E11 (Table 2). These observations were in good agreement with reports in C. elegans worms that embryonal patterning is arrested, stem cell proliferation is reduced, and germline abnormally masculinized upon depletion of the ortholog ATX-2 [53,54]. Interestingly, at the 2-4 cell stage of bovine embryos, a 2.4-fold expression difference of Atxn2l was recently observed to correlate with fertility and that the knock-down of Atxn2l enhanced the development of blastocysts [55].
In the male homozygotes surviving beyond E11, there was a weight loss of 25% on average (Table 3).

Table 3.
Weight of embryos in mg directly after dissection at three different embryonal ages. n: Atxn2l+/+ = 1-2, Atxn2l+/− = 4-6, Atxn2l−/− = 1, E = embryonal day.
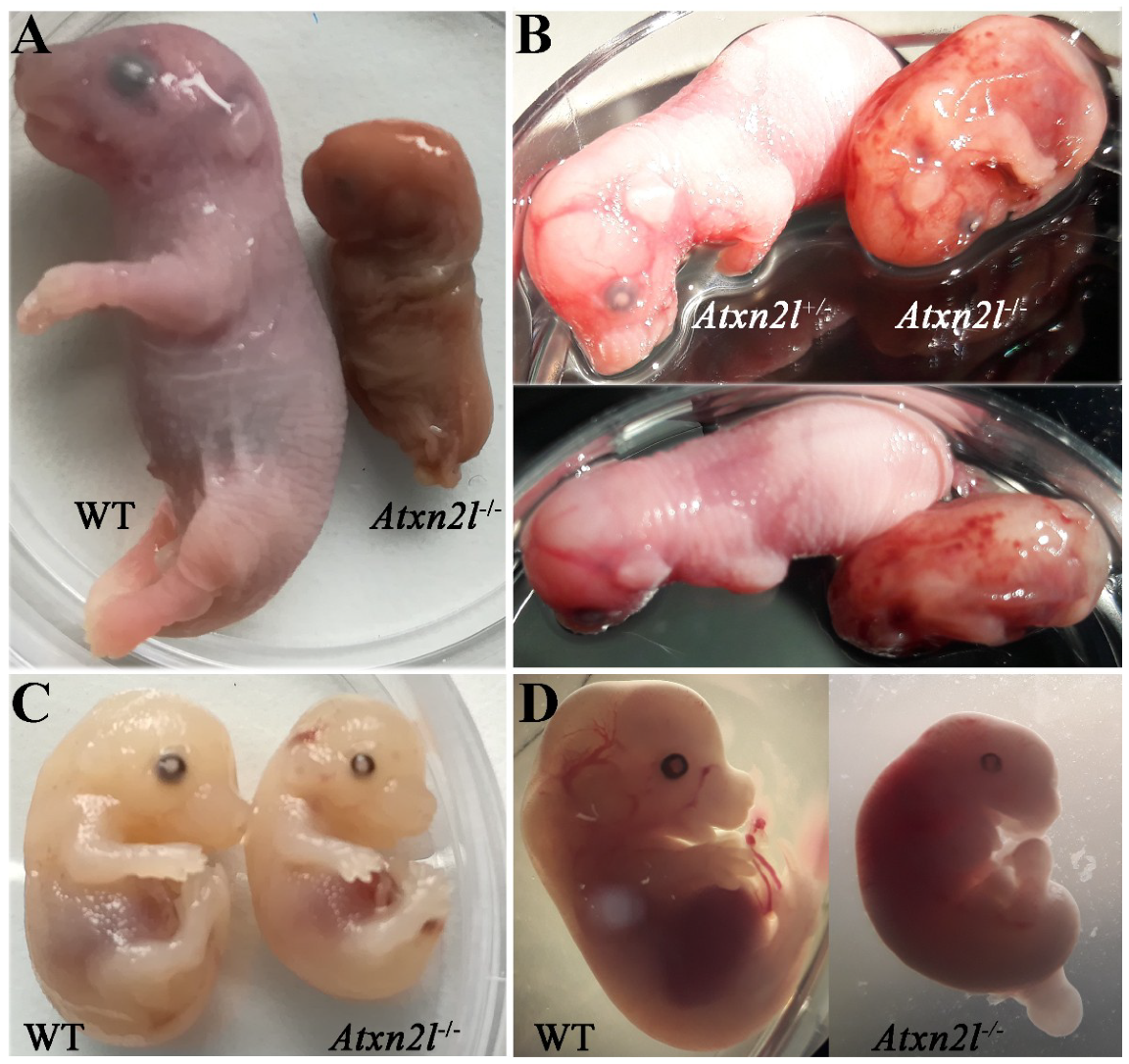
Only one dead male Atxn2l−/− embryo was identified at E20 (Figure 3A, right side), exhibiting disintegrating tissue surrounded by dense liquid as expected during resorption processes [56]. An Atxn2l−/− embryo at E16 was observed to display retarded growth (Figure 3B, right side). The Atxn2l−/− embryos were not pale, had livers of usual size, and showed normal blood-filling of liver tissue (Figure 3C, right side), so there was no evidence for the cardio-vascular or blood circulation deficits that are frequent causes of early or mid-gestational lethality [57]. Strong developmental differences between Atxn2l−/− versus WT littermates (Figure 3D) sometimes amounted to several Theiler stages [58,59,60]. No selective organ affection in gross morphology was noticed during embryogenesis (Figure 3A–D), so an evaluation by high-resolution episcopic microscopy [61] was deemed unnecessary. In general, many fetal sacs in utero were found to contain only placenta rests and a fluid-filled cavity.
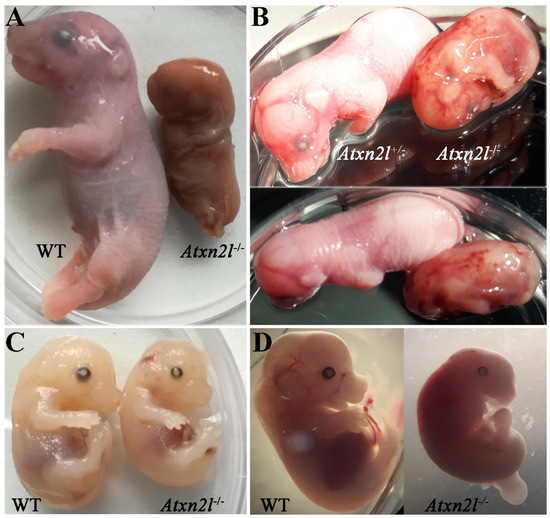
Figure 3.
Comparison of Atxn2l−/− embryos with their littermates, at different gestational stages. Embryonal days reflect approximate values. (A) The only Atxn2l−/− mouse (male) identified at E20 in comparison to a male WT brother. (B) An Atxn2l−/− mouse (male) reaching E16, exhibiting retarded development, and decreased weight, in comparison to the heterozygous Atxn2l+/− female sibling. The petechial bleeds were not seen in other null mutants. (C) An Atxn2l−/− mouse (male) at E15-16 with liver size and blood filling similar to male WT brother. (D) An Atxn2l−/− female embryo at E13 with retarded growth and development and its male WT sibling.
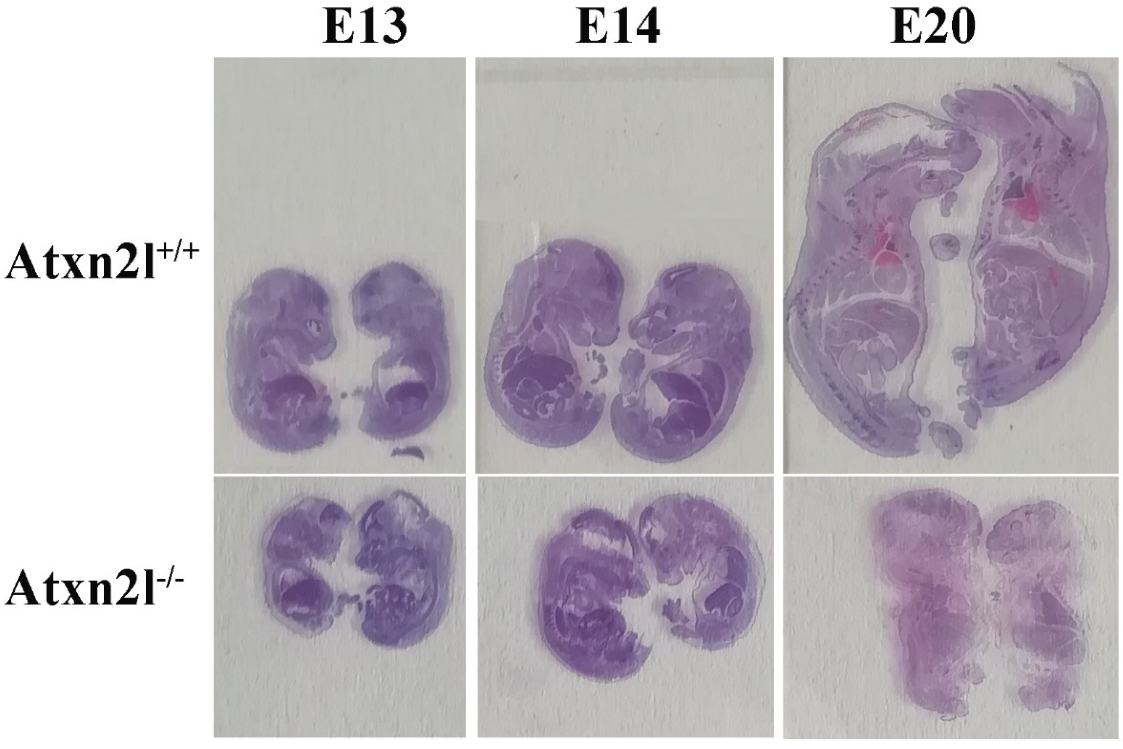
Histological analyses at different embryonic stages from approximately E13 via E14 to E20 (Figure 4) confirmed that the growth deficit is generalized throughout the embryos.
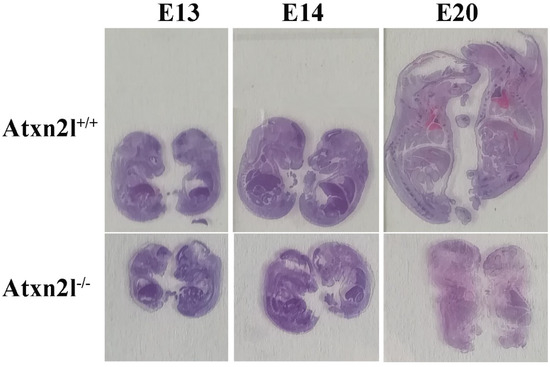
Figure 4.
Evident growth/weight phenotype: Atxn2l+/+ embryos with Atxn2l−/− littermates (WT above, homozygote mutants below, E13 on left side, E14 in center, E20 on right side).
The most frequent causes of early/mid-gestation embryonic lethality before E14 include deficient placentation, followed by blood vessel/heart, skeleton/joint and nervous system malformations [62]. In Atxn2l−/− embryos, no obvious phenotypes apart from developmental delay were detected upon hematoxylin/eosin (H&E) staining of placenta (Supplementary Materials Figure S1), heart, and bones. Interestingly, mice with genetic ablation of the EGF receptor, which was observed in cell lines to influence ATXN2L expression, also exhibit a mid-gestational lethality. However, they display reduced placenta size [63,64] in contrast to the normal Atxn2l−/− placentas. Mid-gestation embryonic lethality was recently also documented for factors of endocytic uptake, such as DENND1A and VPS54 [65,66].
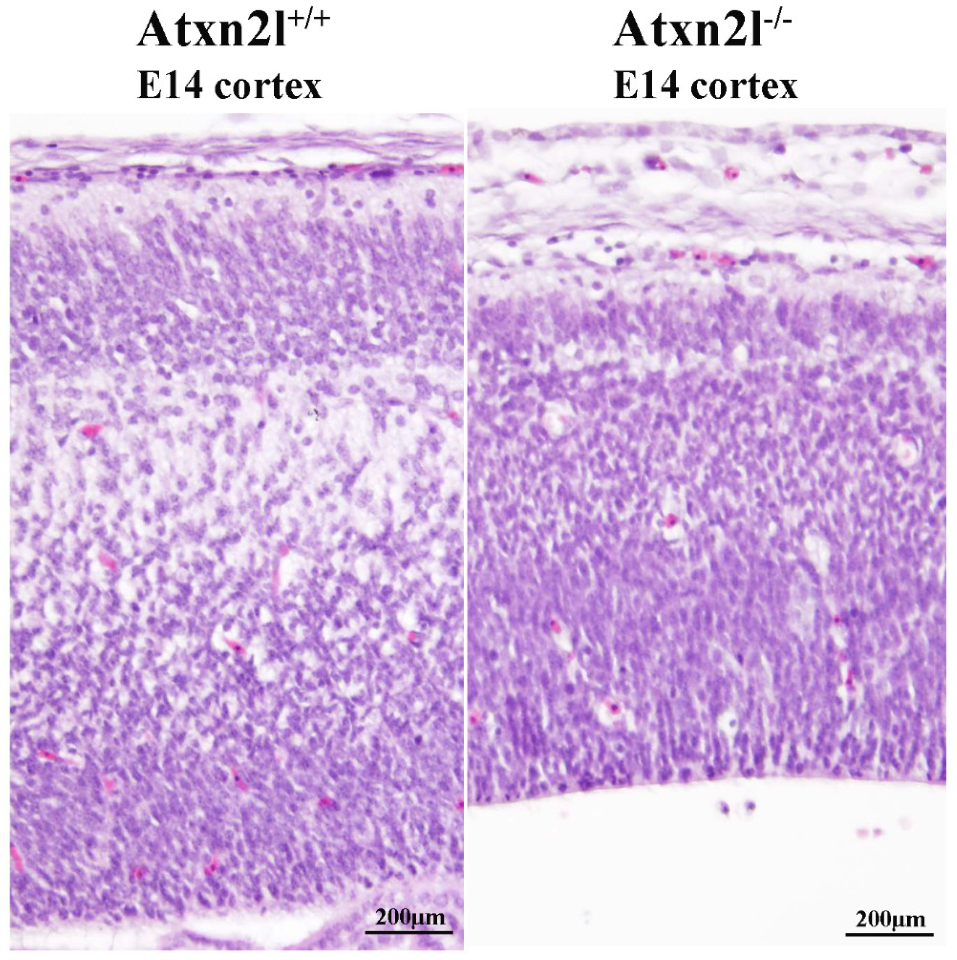
In Atxn2l−/− embryos, preferentially the nervous tissue showed frequent apoptosis, together with reduced cortical layer formation (Figure 5). Similar mid-gestation lethality with abnormal development of the brain cortex was observed, e.g., upon deficient methylation of eukaryotic mRNAs [67]. Neurons more than other cells have a need to modify the 3′-untranslated regions of mRNAs via alternative polyadenylation and microRNA repression [68,69] in a pathway that involves the ATXN2/ATXN2L indirect interactor TDP-43 and stress granules.
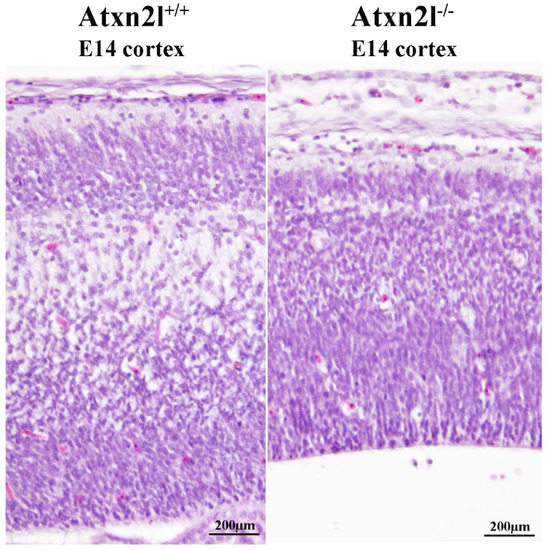
Figure 5.
H&E histology at E14 showed good separation of neuron layers in the cortex of Atxn2l+/+ brain (left); in littermate Atxn2l−/− mice (right), less lamina definition and many neurons were observed with nuclear condensation, which that is indicative of apoptosis; the diameter of brain cortex was also thinner.
In view of the postnatal survival of Atxn2l−/+ mice, a cohort of 12 WT animals and 12 heterozygous littermates was aged until 12 months and assessed every 3 months regarding survival, growth, and locomotor behavior. This preliminary survey found no increased mortality or weight loss, and detected no prominent deficits in spontaneous movements in the open-field paradigm or in muscle coordination under stress on an accelerating rotarod (Supplementary Materials Figure S2).
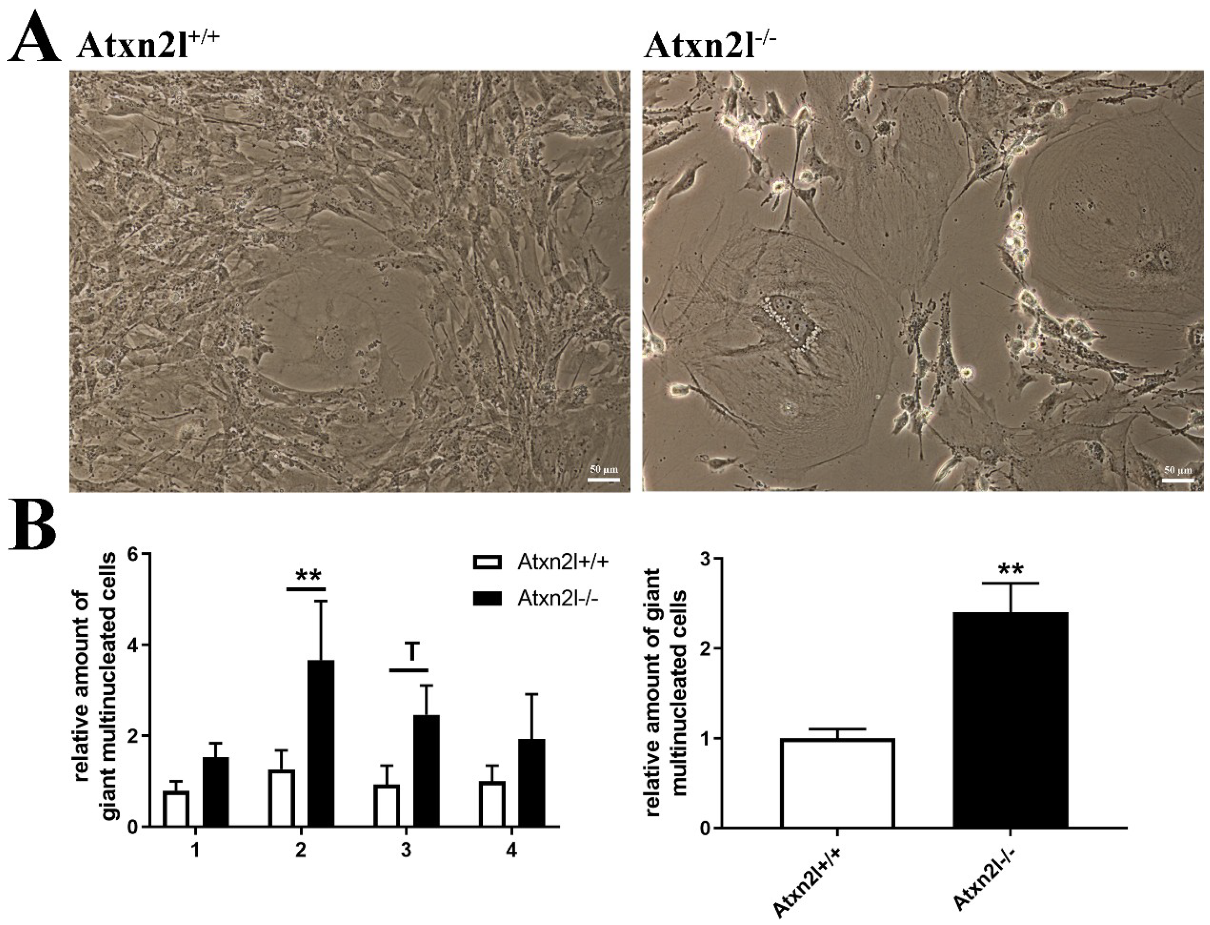
Analyzing cell growth in four ATXN2L-null MEF lines versus four sex-matched WT littermates that were derived from dissected embryos, we noticed the presence many multinucleated giant cells. This cell type occurs in primary cell cultures when nuclear division is not followed by cytokinesis [70]. In ATXN2L-null MEF lines, their frequency was consistently increased, with a >2-fold effect size across all lines (Figure 6A,B). Cholesterol is necessary for cytokinesis, its deficiency induces polyploid cell formation, so it is noteworthy that cholesterol anomalies are well documented in patients and mice with ATXN2 mutations [31,71,72,73]. In addition, the furrow progression during cytokinesis depends on receptor tyrosine kinase activation of SRC and on vesicle endocytosis, where ATXN2 is known to play a modulating role [9,10,74,75,76].
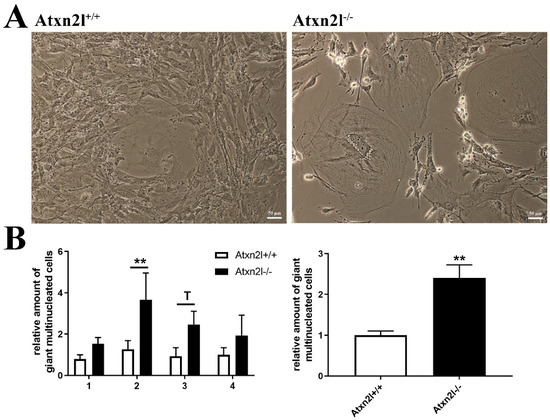
Figure 6.
(A) Cell culture images of Atxn2l+/+ and Atxn2l−/− MEF illustrating the increased number of giant multinucleated cells in the absence of Atxn2l. (B) Quantification of giant cells for 4 different age- matched littermate lines with 3 technical replicates each. The diagram on the left side shows the increase with consistency for the individual MEF lines 1-4, while the diagram on the right side represents the overall increase across all lines. Statistical testing was done by two-way ANOVA and t-test with Welch’s correction, respectively, significance levels were illustrated by ** for p < 0.01, by T for 0.05 < p <0.1.
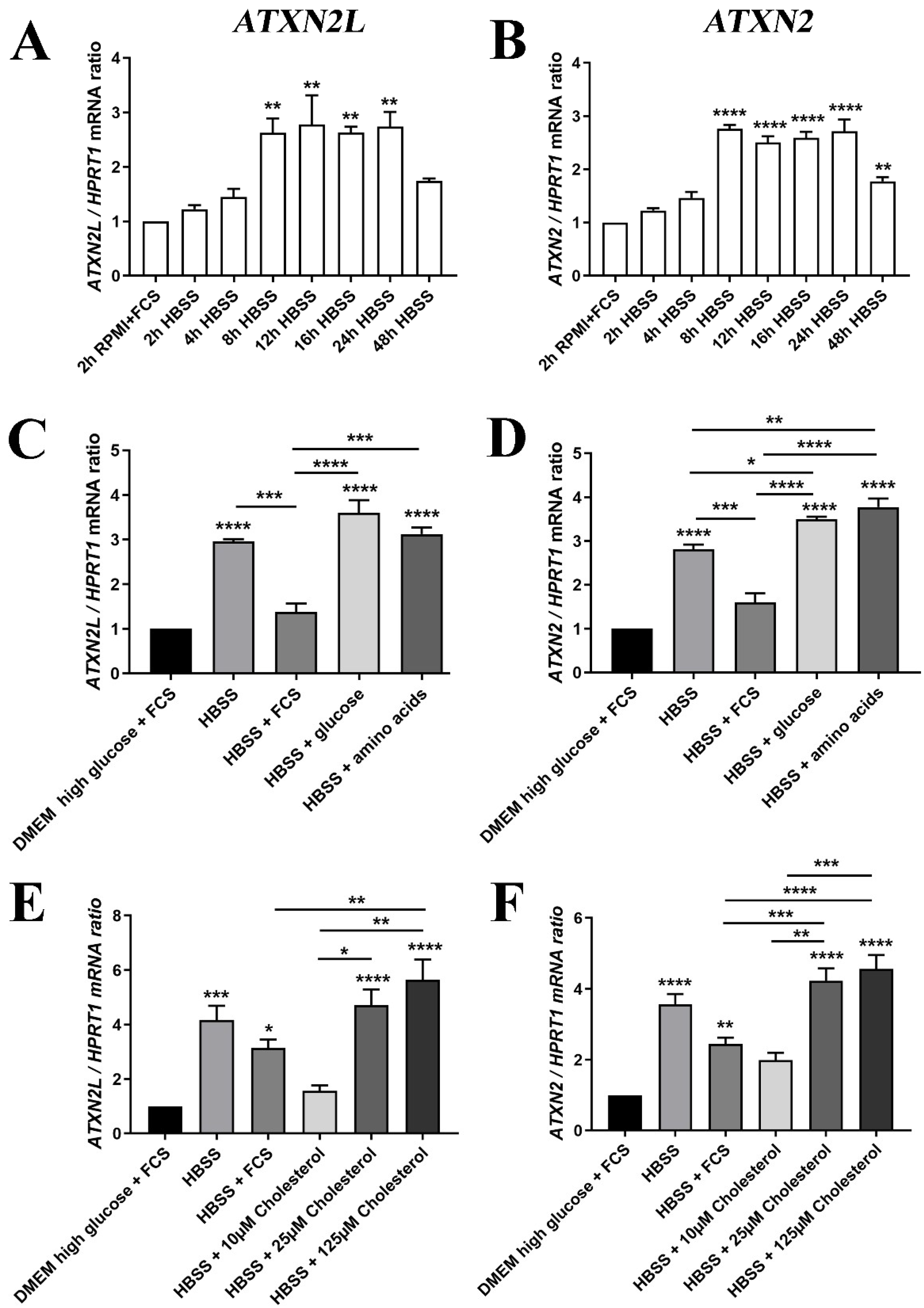
Then, we assessed whether the deprivation of nutrients, such as glucose and lipids, indeed alter the expression of Atxn2l, as was previously observed for its homolog Atxn2 [31]. The human neuronal cell line SH-SY5Y, first cultured in high-glucose DMEM medium with fetal calf serum (FCS) supplement, was switched to the starvation medium HBSS without FCS for a time course of 48 h. The sudden exposure to conditions of low glucose, no amino acids, no FCS (containing growth factors/apolipoproteins with lipids/transferrin with iron, etcetera) triggered a phasic induction that peaked at 8–24 h with a 3-fold Atxn2l mRNA increase, similar to the induction of Atxn2 (Figure 7A,B). Preliminary testing in MEF cells suggest that RNA levels of Atxn2 levels decreased slightly upon to stress by a toxic RNA-analogon, Poly(I:C), while Atxn2l failed to respond (Supplementary Materials Figure S3E–H). These MEF analyses provided no evidence that the depletion of ATXN2 triggers compensatory dysregulation of its homolog Ataxin-2-like at mRNA or protein level, or that conversely the depletion of ATXN2L triggers expression or abundance changes of Ataxin-2 (Supplementary Materials Figure S3E–J).
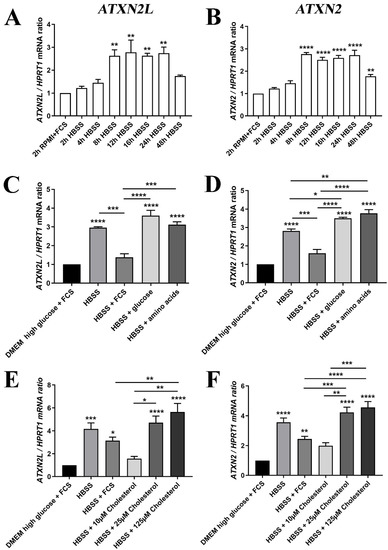
Figure 7.
(A) The mRNA expression of ATXN2L in human SH-SY5Y neuroblastoma cells was induced by nutrient deprivation in HBSS medium over 48 h with low glucose/no amino acids, in the absence of lipids normally supplied via FCS, similar to the expression of (B) ATXN2 mRNA. (C) For the induction peak at 24 h, the expression upregulation of ATXN2L was partially prevented upon supplementation of 10% FCS, while glucose or amino acids alone had no rescue effect; (D) FCS addition also diminished ATXN2 expression, while glucose and amino acid administration enhanced the expression further; (E) The HBSS induction of ATXN2L was reduced at least as much by low cholesterol concentrations as by FCS, while higher dosage of cholesterol elicited even stronger upregulations. (F) This effect was similarly observed for ATXN2. A-D: n = 3; E/F: n = 8. Statistical testing was done by one-way ANOVA, significance levels were illustrated by asterisks: * p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001. Comparison was always in respect to nutrient abundant control condition, if not stated otherwise.
Further analysis of individual nutrients demonstrated that the transcriptional induction of ATXN2L in HBSS starvation medium could be diminished by the supplementation with FCS (which contains components such as insulin, transferrin, and apolipoproteins), and by low level cholesterol administration, two effects that were similarly observed for ATXN2. This was in contrast to supplementation with excess glucose, amino acids, or excess cholesterol that were unable to downregulate ATXN2L or further enhanced its levels (Figure 7C,E). The increased transcription of ATXN2 upon glucose and amino acid addition was significant, as well as the induction by cholesterol excess (Figure 7D,F).
Further analyses about components of FCS that could be responsible for the rescue effect in the induction of ATXN2L are shown in Supplementary Materials Figure S3. Different cell culture media were used as basal media (DMEM having the highest abundance of nutrients, MEM containing fewer nutrients such as vitamins or amino acids, and HBSS containing no nutrients apart from low glucose amounts). The supplements included SPITE (a mixture of sodium selenite, sodium pyruvate, bovine insulin, human transferrin, and ethanolamine), ITS (a mixture of bovine insulin, human transferrin, and sodium selenite), and SITE+3 (containing analogous components as SPITE, but 5 mg/mL BSA, linoleic acid and oleic acid as a lipid energy source instead of pyruvate). However, none of these supplements was sufficient for a significant rescue of the starvation-triggered ATXN2L induction, to a degree that approached FCS. The comparison of SPITE versus SITE+3 supplementation suggested that fatty acid administration is more helpful than pyruvate for the reduction of ATXN2L and ATXN2 mRNA levels (Supplementary Materials Figure S3C,D). Overall, these in vitro observations confirm the notion derived from limb tissues in vivo and from MEF cells, that either the promoter activity of the Ataxin-2-like gene is upregulated or the Atxn2l/Atxn2 mRNA decay is downregulated during periods of growth deficits and then this regulation can be reversed upon availability of specific nutrients. In this context, it is noteworthy that the RegRNA2.0 webtool predicts both transcripts to contain AU-rich elements in their 3′-untranslated region, which are known to modulate the stability of selected mRNA according to growth needs. It is also relevant to mention that the promoters/enhancers of human Ataxin-2/Ataxin-2-like share 476 transcription factor binding sites, according to predictions found at the GeneCards website, including stress-response regulator families that are strongly implicated in neurodegeneration, such as ATF1/2/3/4/7, E2F1/5/6/8/ELF1/3/4/ETS1/ETV1/4/5/6, HNRNP-H1/K/L/LL/UL1, or IRF1/2/3/4.
A dysregulation of ATXN2L levels upon depletion of its ortholog ATXN2 was not observed in MEF cells, nor a converse dysregulation of ATXN2 upon depletion of ATXN2L, neither at protein nor at mRNA level (Supplementary Materials Figure S3E–J). As mentioned in the Introduction however, unpublished global proteome mass spectrometry data in our lab documented a significant increase of ATXN2L in the brain of Atxn2-CAG100-knockin mice. This finding may therefore be specific to the CAG-repeat expansion mutation and be caused by the polyQ aggregation process in stress granules, where ATXN2 and ATXN2L have common protein interactors such as DDX6 and may co-precipitate.
3. Discussion
ATXN2L was described as direct RNA-binding protein with sequence homology to nuclear spliceosome factors [37,77]. Its alternatively spliced isoforms may interact with tyrosine kinase receptor signaling at the plasma membrane [36]. The constitutive isoform of ATXN2L was mainly characterized for its role as surveillance factor during transient periods of cell damage, which promotes the formation of stress granules in the cytosol [34]. Now, our ATXN2L-deficient mouse data demonstrate that homozygous loss of its RNA-interaction domains is incompatible with embryonic development from early stages. Their absence delays the cytokinesis stage of cell division and impairs the layer formation and survival of brain neurons.
Human data agree with selective roles of ATXN2L for the growth process, the nervous system, and immunity. Genome-wide association studies (GWAS) of single nucleotide polymorphism (SNP) alleles versus phenotypes in healthy populations (summarized at the NHGRI-EBI Catalog of human genome-wide association studies, online available at: https://www.ebi.ac.uk/gwas/ [78] demonstrated significant effects in several independent studies of the ATXN2L gene locus. Its impact on growth was reflected by associations with body mass index (in diverse reports for different polymorphisms p = 4e−29; 2e−15; 9e−15; 2e−13; 3e−8; 2e−7) [79,80,81], body weight (p = 2e−8) [81], and waist circumference (p = 4e−8) [81]. Its impact on the nervous system was reflected by associations with cognitive function (p = 8e−35) [82], grip strength (p = 1e−25) [83], intelligence (p = 1e−19; 2e−9; 1e−8; 2e−8; 4e−8) [84,85,86,87], and educational attainment (p = 4e−14; 7e−11; 1e−7; 1e−6) [85,88,89]. Its impact on immunity was reflected by associations with albumin/globulin ratio (p = 6e−10) [90], inflammatory bowel disease (p = 2e−9) [91], and pediatric autoimmune diseases (p = 6e−7) [92].
Our studies of the expression regulation of Atxn2l and Atxn2 by diverse nutrients indicate a strong similarity in their role, with a transcriptional induction being triggered by nutrient deprivation as well as by an excess of cholesterol and glucose (both of which have a role for plasma membrane flexibility/stickiness), while the transcription was reduced upon the supplementation of FCS. Given that the depletion of the ATXN2L ortholog in flies is neuroprotective, there may be therapeutic value in the knowledge that its expression can be downregulated in mammalian cells by the abundance of specific nutrients. The observations here may be relevant for the treatment of ALS, where hypercaloric feeding was shown to be beneficial, and evaluations on the relative contribution from carbohydrates versus lipids are ongoing [93,94]. An experimental artifact was observed upon nutrient deprivation and upon Poly(I:C) stress in vitro: Although the induction strength of Atxn2 and Atxn2l showed limited variance during repeated experiments by a student with a SH-SY5Y aliquot and a specific FCS batch, it could however show quite different fold-changes months later in different hands using a separate SY-SY5Y stock with another FCS batch. It remained unclear to what degree passage number, clonal mutation drift or handling variance played a role here (compare Figure 7C vs. E, and Figure S2 F vs. H).
Publicly available expression data on Atxn2l and Atxn2 by RNA sequencing and further processing at the Broad Institute within the Genotype-Tissue-Expression project database, online available at: https://gtexportal.org/home/ [95] with normalization to transcripts per million reads (TPM) confirm that Atxn2 and Atxn2l are both preferentially expressed in cerebellar hemispheres and total cerebellum (median TPM value = 30/33 vs. 184/229, respectively), in comparison with other regions of the nervous system. Interestingly, our observations on sex differences in the phenotype of null mutants are also reflected by these public expression data: they document that Atxn2 expression is stronger in most female organs (36/31/30/29/28/24 TPM in ovary/uterus/endocervix/FallopianTube/ectocervix/vagina, respectively) than in cerebellum. In contrast, Atxn2l shows the highest expression in testis tissue (TPM 296), as opposed to the average expression in female organs (e.g., ovary TPM 124). Overall, Atxn2l in cerebellum shows a 6-fold higher expression than Atxn2, and the preferential expression of Atxn2l in male testis contrasts with the pronounced expression of Atxn2 in several female sexual organs. Given that the biosynthesis of all sex hormones starts from cholesterol, the effects of ATXN2L and ATXN2 on the cholesterol homeostasis may account for the sex difference of Atxn2l-null phenotypes, the increased formation of giant multinucleated MEF cells, and the expression regulation by cholesterol. It is important to note that Atxn2-mutations were already observed to affect cholesterol homeostasis and trigger gender-dependent or hormonal effects [31,73,96,97]. Giant multinucleated cells can be due to increased mTOR activity [98,99,100]. Given that ATXN2 was reported to inhibit mTOR in yeast, nematodes and mice; also the absence of ATXN2L might lead to enhanced mTOR signaling and, in this way, increase the appearance of multinucleated giant cells in MEF cultures. These phenotypes may be downstream effects of the reported protein interaction between ATXN2L and NHERF2 as a known modulator of cholesterol metabolism and Src-dependent phosphorylation signals for growth. While a gene duplication event in mammals led to the coexistence of two homologous genes, in plants a coexistence of four homologous genes from the ataxin-2 family raises the possibility that mTOR-dependent growth can be regulated by each of them in response to different stressors or to different nutrients, via an RNA-binding mechanism.
Clearly, the regulation of Atxn2l and Atxn2 mRNA levels by nutrients is almost parallel in our cell culture analyses, so it seems paradoxical that our null mutant of ATXN2L has a growth deficit phenotype, while the null mutant of ATXN2 was found by two independent teams to show a converse nutrient excess phenotype [31,32]. An important difference in the recombinant design of these mutants should be noted here. The KO of Atxn2 was targeting exon 1 and removed all of the protein domains, while the ablation in Atxn2l targeted exons 5-8, with a resulting shift of reading frame downstream. In the mutant ATXN2L, the N-terminal Pro-rich domain across residues 4-61 and the MPL interaction domain across residues 96-119 would still be synthesized as a fragment, the available commercial antibodies cannot detect this N-terminal region. In view of the increase of Atxn2l non-targeted exons in the limb tissue of homozygous mutants (Figure 2B), this hypothetical fragment could exist in overdose. The excess amounts of such an N-terminal fragment would interact with plasma membrane receptors and with the endocytosis machinery. In analogy to the published observations for ATXN2 overexpression, it is expected to impair trophic uptake mechanisms [9]. At the same time, the absent middle and C-terminal part of ATXN2L would fail to interact with RNAs in the appropriate manner. Overall, our mouse mutant might represent not only a loss of the C-terminal functions of ATXN2L; it is conceivable that it also involves a toxic gain of N-terminal functions of ATXN2L. Perhaps this speculation provides a clue why our mouse mutant has this unexpected phenotype, which is converse to Atxn2-KO mice.
In the future, antibodies against the N-terminal portion of ATXN2L should be generated to obtain evidence regarding this doubt, and a new ATXN2L mutant that targets exons 1-4 would also help to clarify this issue. It will be crucial to generate conditional mutants where the postnatal role of ATXN2L for tissues such as the aging brain can be studied selectively. Moreover, it is interesting to note that commercial antibodies such as HPA041506/PA5-59601 in U2OS cells showed only the band of predicted size, and our immunoblots in brain tissue also detected this putative full-length band, while we observed two ATXN2L bands with three different antibodies in MEF cells, and also the A301-369A and 24822-1-AP antibodies detected a smaller band in HeLa/HEK293T and Jurkat cells, according to manufacturer datasheets. Thus, further studies will be necessary to understand the usage of the two alternative start codons and of the alternative splice variants in various tissues/cell populations and after different stressors. Clearly, our data indicated that the depletion of ATXN2 or ATXN2L in MEF cells fail to trigger a compensatory upregulation of the other homolog, so the unpublished accumulation of ATXN2L in Atxn2-CAG100-KIN brain appears to be an expansion-driven effect and ATXN2L immunoreactivity should be assessed within the Q100-ATXN2 aggregates. Since TDP-43 is essential for embryonic development in mouse and its aggregation drives the decisive toxicity for motor neuron degeneration, it is conceivable that ATXN2L—a factor that is also essential for embryonic development—could co-aggregate with ATXN2 and drive toxicity in cerebellar Purkinje neurons where it has selective abundance.
4. Materials and Methods
4.1. Generation, Breeding, Maintenance, and Dissection of Atxn2l−/− Mice
To specifically target sites flanking exons 5-8 of the Atxn2l gene, this pair of sgRNAs was used: upstreamGRNA085CCT (atgtggcagggtcagtatcag), downstreamGRNA02CCC (gttacctggataccaagct). To distinguish the long wildtype from the short mutant allele, primers Atxn2l-F (gagagtgtgtgttgggtgac), Atxn2l-F2 (gaagcacagtgctgttcagc), and Atxn2l-R (gtaatttgcagcaacagtagagc) were employed. Heterozygous breeders were shipped, crossed at the ZFE animal facility of Goethe University Medical Faculty in Frankfurt as described [101] and health-monitored with sentinel animals at quarterly intervals. Congenic heterozygous mice were then crossed among them. The studies were ethically approved by Regierungspräsidium Darmstadt, code V54-19c20/15-FK/1083, on 11 March 2019.
4.2. Genotyping
DNA was extracted from adult ear punches or embryo tail biopsies for genotyping by incubating at 95 °C in 100 µL of 25 mM NaOH/0.2 mM EDTA for 30 min before neutralizing with 100 µL 40 mM Tris–HCl, pH 5.0. For the subsequent genotyping reactions, 1 µL of lysed tissue sample was used per reaction. The Atxn2l alleles were amplified by PCR with the primers Atxn2l-F (GAGAGTGTGTGTTGGGTGAC), Atxn2l-F2 (GAAGCACAGTGCTGTTCAGC) and Atxn2l-R (GCTCTACTGTTGCTGCAAATTAC), the PCR products 217 bp and 309 bp corresponding to Atxn2l wt and Atxn2l mutant respectively were separated in 1.5% agarose gel.
For the determination of the sex, primers Rmb31F (CACCTTAAGAAGCCAATACA) and Rbm31R (GGCTTGTCCTGAAAACATTTGG) were used to yield a 269 bp product from X chromosome and a 353 bp product from the Y chromosome [102].
4.3. Behavioral Tests
Behavioral tests were done as described in [103]. Twelve Atxn2l+/+ and twelve Atxn2l+/− male mice were aged and assessed at ages between 3 months and 12 months.
4.4. Generation and Cell Culture of MEF Cells
MEF were derived from Atxn2l−/− embryos at day 14-16 and their littermate Atxn2l+/+ controls. As previously described [104], the skin of the embryos was dissected, homogenized, and trypsinized for 10 min at 37 °C. The cells (passage 2-3) were cultured in DMEM, containing 4.5 g/L D-glucose, 1% L-glutamine, 1% Penicillin/Streptomycin and 10% FCS. Atxn2−/− MEF cells were described before [9].
4.5. Poly(I:C) Treatment
Atxn2+/+ and Atxn2−/− MEF (3 vs. 3 cell lines), as well as Atxn2l+/+ and Atxn2l−/− MEF (4 vs. 4 cell lines) were treated with 1 µg/mL Poly(I:C) (Invivogen) for 16 h with subsequent RNA extraction and RT-qPCR [105].
4.6. RNA Extraction and Expression Analysis
RNA extraction from limbs and MEF was performed with TRIzol Reagent (Sigma Aldrich) according to the user manual. RNA from cell culture experiments with SH-SY5Y cells was performed with Extractme Total RNA Kit (7Bioscience EM09.1-250). Synthesis of cDNA from 1 µg of total RNA template was performed with the SuperScript IV VILO kit (ThermoFisher) according to the manufacturer’s instructions. cDNA from 25 ng total RNA was used for each RT-PCR reaction with 0.5 µL TaqMan® Assay, 5 µL FastStart Universal Probe Master 2x (Rox) Mix and ddH2O up to 10 µL of total volume. The mouse-specific TaqMan® Assays utilized for this study: Atxn2l (Mm00805548_m1, Mm00805539_m1, Mm01165459_m1, Mm01276208_m1), Atxn2 (Mm01199894_m1), and Tbp (Mm00446973_m1).
4.7. Immunoblotting
Protein was isolated with RIPA buffer as described before [106]. Samples with 20 µg protein were separated on 8% SDS gels and blotted on nitrocellulose membranes. Membranes were incubated overnight at 4 °C with the following antibodies: ATXN2L (Bethyl A301-369), ATXN2L (Invitrogen PA5-59601), ATXN2L (Proteintech 24822-1-AP), ATXN2 (Proteintech 21776-1-AP) or beta-Actin (Sigma A5441), and for 1 h with the respective secondary antibodies (Li-Cor). Fluorescence was detected with the Li-Cor Odyssey Classic Instrument and bands were analyzed with Image Studio Lite, Version 5.2.
4.8. Sections and Staining
All tissue specimens were fixed in 4% paraformaldehyde (PFA) for at least 48 h. All specimens were embedded in paraffin and aligned as displayed on the respective images (see Figure 4 and Figure 5 and Figure S1). The 4-µm thick sections were made using a microtome. Slides were stained with hematoxylin and eosin according to standard protocols. Images were taken using an Olympus BX50 microscope and an Olympus DP72 camera.
4.9. Microscopy and Cell Counting of MEF
175.000 MEF cells were seeded in 6-well plates and grown overnight (n = 4 Atxn2l+/+ vs. 4 Atxn2l−/− lines in 3 technical replicates each). For each well, 5 random areas were microscopically visualized (Leica) and all giant cells were counted. Means for each cell line was calculated and statistically evaluated. Images were taken with a Nikon microscope after changing the cell culture medium to PBS.
4.10. Cell Culture Experiments
SH-SY5Y cells were purchased from ECACC, and cultured as described [6] in Dulbecco’s Modified Eagle Medium (DMEM) containing 4.5 g/L D-glucose, 2 mM L-glutamine with 10% FCS. The cells were starved of trophic factors by incubating them in Hanks’ balanced salt solution (HBSS) for a range of different periods (time course) to determine when the cells show the strongest deprivation effect. In different experiments, HBSS was supplied with 10% FCS, glucose 4.5%, MEM non-essential amino acids solution 1×, or cholesterol (SIGMA L4646) at 10 µM, 25 µM, cholesterol 125 µM (Figure 7). For the experiments shown in Supplementary Figure S3, cells were incubated with the indicated basal media (DMEM, MEF, or HBSS), supplemented with SPITE medium supplement 1x (Sigma S5666), ITS liquid media supplement 1x (Sigma I3146), or SITE+3 liquid media supplement 1x (Sigma S5295). After 24 h, cells were collected and RNA was extracted with EXTRACTME (Blirt, Gdańsk, Poland). Synthesis of cDNA and RT-qPCR was performed as previously described with human-specific TaqMan® Assay ATXN2L (Hs00944485_g1), ATXN2 (Hs00268077_m1) and HPRT1 (Hs99999909_m1). The numbers of cell lines used is stated in respective figure legends.
4.11. Statistical Analysis
All statistical tests were performed as unpaired Student’s t-test with Welch’s correction or one-way or two-way ANOVA using GraphPad Prism software version 7 after establishing that each population was normally distributed (one-sided Kolmogorov–Smirnov test). Figures display mean values and standard error of the mean (s.e.m.). Values of p < 0.05 were considered significant and marked with asterisks p < 0.05 *, p < 0.01 **, p < 0.001 ***, p < 0.0001 ****.
5. Conclusions
Overall, this first analysis of ATXN2L-depletion in mouse demonstrates its essential role for embryonic development, due to growth delays with prominent vulnerability of brain lamination, especially in females. A preferential affection of female embryos and an increased occurrence of multinucleated giant cells in MEF culture were observed, which may reflect a role of ATXN2L in cholesterol and hormone homeostasis. This mouse mutant will provide an important tool to dissect the role of ATXN2L domains, such as the impact of LSM and PAM2 motifs on RNA surveillance, which were ablated here, versus the effects of PRD and MPL interaction motifs on trophic endocytosis, during stress periods and aging. Since the data show ATXN2L to be more important than ATXN2 for cell growth, it will be highly interesting to study its role in neurodegenerative disorders due to RNA surveillance problems, such as ALS, where the depletion of mammalian ATXN2 and of the yeast/fly ortholog also of ATXN2L was already shown to be neuroprotective.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/14/5124/s1.
Author Contributions
Conceptualization, G.A. and S.G.; methodology, J.K., N.-E.S., P.N.H. and S.G.; software, J.K., S.G. and G.A.; validation, J.K., E.G. and S.G.; formal analysis, J.K. and S.G.; investigation, J.K., G.A. and S.G.; resources, G.A. and S.G.; data curation, G.A.; writing—original draft preparation, G.A.; writing—review and editing, J.K., N.-E.S., G.A. and S.G.; visualization, J.K., G.A. and S.G.; supervision, P.N.H., E.G., G.A. and S.G.; project administration, G.A.; funding acquisition, G.A. All authors have read and agreed to the published version of the manuscript.
Funding
The study was financed by the team budget at Goethe University Frankfurt/M, Germany.
Acknowledgments
We are grateful to Gabriele Köpf, Aleksandar Arsovic, and Tatjana Starzetz for technical assistance, and to the staff at the animal facility ZFE at Frankfurt university school of medicine. The assistance of Dr. Kader Thiam and his team at Genoway (Lyon, France) in the design and evaluation of targeted mutations within Atxn2l (Figure 1) is highly appreciated.
Conflicts of Interest
The authors declare no conflict of interest. G.A. advises RochePharma and TakedaPharma regarding ATXN2 research, receiving honoraria from them. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Abbreviations
| °C | Degrees Celsius (temperature) |
| aa | amino acid |
| ACTB | beta-actin |
| Adnp | Activity-Dependent Neuroprotector Homeobox Protein |
| ALS | Amyotrophic Lateral Sclerosis |
| ANOVA | Analysis of variance |
| ARHGEF11 | Rho Guanine Nucleotide Exchange Factor (GEF) 11 |
| ATF1 | Activating Transcription Factor 1 |
| ATG | Start codon Adenine-Thymidine/Uracil-Guanine |
| ATX-2 | the single nematode ortholog of mammalian Ataxin-2 and Ataxin-2-like |
| ATXN2 | Ataxin-2 |
| ATXN2L | Ataxin-2-like, aka: Ataxin-2-Related Protein, A2LP, A2RP or A2D |
| BDNF | Brain-Derived Neurotrophic Factor |
| bp | Base pair |
| BSA | Bovine serum albumin |
| Carm1 | Coactivator Associated Arginine Methyltransferase 1 |
| Cart1 | Cartilage paired class homeoprotein 1 (aka Alx1) |
| Cb | Cerebellum |
| Cbp | cAMP-response element binding protein (Creb) binding protein (aka Crebbp) |
| cDNA | complementary Deoxyribo-Nucleic Acid |
| CID3 | CTC-interacting domain 3 protein in Arabidopsis thaliana |
| CID4 | CTC-interacting domain 4 protein in Arabidopsis thaliana |
| Cited2 | Cbp/P300 Interacting Transactivator With Glu/Asp Rich Carboxy-Terminal Domain 2 |
| Crispr/Cas9 | Clustered Regularly Interspaced Short Palindromic Repeats/Crispr-associated protein 9 |
| C-term | C-terminal end of the protein |
| Ctx | Cortex |
| DAPK1 | Death-Associated Protein Kinase 1 |
| DDX6 | DEAD/H (Asp-Glu-Ala-Asp/His) Box Polypeptide 6 (RNA Helicase, 54kD) |
| DENND1A | DENN/MADD Domain Containing 1A (RAB35-activating guanine nucleotide exchange factor) |
| DMEM | Dulbecco’s modified essential medium |
| DNA | Desoxyribo-Nucleic-Acid chain |
| E | Embryonic development day |
| E2F1 | E2F Transcription Factor 1 |
| e.g., | Exemplo gratia (for example, in Latin language) |
| EGF | Epidermal growth factor |
| ELF1 | E74 Like ETS Transcription Factor 1 |
| Ep300 | E1A Binding Protein, Histone Acetyltransferase P300 |
| EST | Expressed sequence tag |
| Ex1 | Exon 1 |
| FCS | Fetal calf serum |
| FTLD | Fronto-temporal lobar dementia |
| Foxj2 | Forkhead Box Protein J2, a transcriptional activator |
| g | Gram |
| G3BP2 | Ras-GTPase Activating Protein SH3 Domain-Binding Protein 2 |
| GLAST GWAS | Sodium-Dependent Glutamate/Aspartate Transporter 1 (in astrocytes) Genome wide association studies |
| h | hour |
| H&E | Hematoxylin and Eosin staining |
| HBSS | Hank’s buffered salt solution |
| Het | Heterozygote |
| HNRNP | Heterogeneous Nuclear Ribonucleoprotein |
| Kb | kilo-base |
| IRF1 | Interferon Regulatory Factor 1 |
| ITS | Insulin-Transferrin-Selenium |
| KD | Knock-Down |
| KO | Knock-Out |
| L | liter |
| lncRNA | long non-coding RNA |
| Lsm | Like “Smith antigen” protein domain |
| LSM12 | Homolog of yeast cytosolic LSM12 protein |
| LsmAD | Lsm-associated domain |
| mg | milliGram |
| mm | milliMeter |
| mM | milliMolar |
| µg | microGram |
| µL | microLiter |
| µM | microMolar |
| MEF | Mouse embryonal fibroblasts |
| MEM | Minimal Essential Medium |
| MGI mGluR5 | Mouse Genome Informatics metabotropic Glutamate Receptor 5 |
| MPL | Interactor domain with Myelo-Proliferative Leukemia gene, encodes Thrombopoietin receptor |
| mRNA | Messenger ribonucleic acid |
| mTORC1 | Mechanistic “target of rapamycin” complex 1 |
| Neat1 | Nuclear Paraspeckle Assembly Transcript 1 |
| NFATC2IP | Nuclear Factor of Activated T-Cells, Cytoplasmic, Calcineurin-Dependent 2 Interacting Protein |
| ng | nanoGram |
| NHGRI-EBI N-term | American National Human Genetics Research Institute & European Bioinformatics Institute N-terminal end of the protein |
| NUFIP2 | Nuclear Fragile X Mental Retardation Protein 1 Interacting Protein 2 |
| ORF | open reading frame |
| p | probability value for the occurrence of a given observation by chance |
| PABPC1 | Poly(A)-binding-protein cytosolic 1 |
| PAM2 | Poly(A)-binding mediator domain 2 |
| PBS | Phosphate buffered saline |
| PFA | Paraformaldehyde |
| poly(A) tail | adenosine-repeat at the 3′ end of mRNA |
| Poly(I:C) | Poly-Inosinic: Poly-Cytidylic acid chain |
| Pro | Proline |
| PRD | Proline-rich domain |
| PSD-95 PSP | Postsynaptic Density Protein 95 (aka DLK4, Disks Large Homolog 4) progressive supranuclear palsy (aka Parkinson plus) |
| Q, 22Q | glutamine, repeat of 22 consecutive glutamines |
| RA | Retinoic acid |
| RT-qPCR | Reverse-Transcriptase quantitative Polymerase Chain Reaction |
| S6R | Small ribosomal subunit protein 6 |
| SCA1 | Spinocerebellar Ataxia type 1 |
| SCA2 | Spinocerebellar Ataxia type 2 |
| SDS | Sodium-Dodecyl-Sulfate detergent |
| s.e.m. | Standard error of the mean |
| Setd5 | Histone-Lysine N-Methyltransferase, SET domain containing 5 |
| SG | Stress granule |
| sgRNA | Single guide Ribo-Nucleic-Acid chain |
| Sh2b1 | Pro-Rich, PH And SH2 Domain-Containing Signaling Mediator gene |
| SH3 | Src-homology 3 domain |
| SH-SY5Y | Cell line subcloned from SK-N-SH line from 4-year-old female neuroblastoma patient |
| SITE+3 | Selenite, Insulin, Transferrin, Ethanolamine +3 |
| SLC9A3R2 SNP | Solute Carrier Family 9 (Sodium/Hydrogen Exchange) Member 3 Regulator 2 (aka NHERF2) Single nucleotide polymorphism |
| SPITE | Selenite, Pyruvate, Insulin, Transferrin, Ethanolamine +3 |
| SR-B1 Src | Scavenger Receptor Class B Member 1 (aka CD36, Thrombospondin Receptor-Like 1) V-Src Avian Sarcoma (Schmidt-Ruppin A-2) Viral Oncogene |
| Tbp | TATA-binding factor of transcription |
| TDP-43 | TAR (transcription active response element) DNA-Binding Protein 43 (encoded by TARDBP) |
| Tfap2a | Transcription Factor Activator-Protein-2 alpha |
| TIGM, TX | Texas Institute of Genomic Medicine, Texas, United States of America |
| TPM | Transcripts per million (expression unit as normalization method in RNA-sequencing) |
| Tufm | Tu Translation Elongation Factor, Mitochondrial gene |
| ULK1 | Unc-51 Like Autophagy Activating Kinase 1 |
| VPS54 | Vacuolar Protein Sorting-Associated Protein 54 (endosomal factor for retrograde transport) |
| WT | Wild-Type |
References
- Auburger, G.; Sen, N.E.; Meierhofer, D.; Basak, A.N.; Gitler, A.D. Efficient Prevention of Neurodegenerative Diseases by Depletion of Starvation Response Factor Ataxin-2. Trends Neurosci. 2017, 40, 507–516. [Google Scholar] [CrossRef] [PubMed]
- Satterfield, T.F.; Pallanck, L.J. Ataxin-2 and its Drosophila homolog, ATX2, physically assemble with polyribosomes. Hum. Mol. Genet. 2006, 15, 2523–2532. [Google Scholar] [CrossRef] [PubMed]
- Yokoshi, M.; Li, Q.; Yamamoto, M.; Okada, H.; Suzuki, Y.; Kawahara, Y. Direct binding of Ataxin-2 to distinct elements in 3’ UTRs promotes mRNA stability and protein expression. Mol. Cell 2014, 55, 186–198. [Google Scholar] [CrossRef] [PubMed]
- van de Loo, S.; Eich, F.; Nonis, D.; Auburger, G.; Nowock, J. Ataxin-2 associates with rough endoplasmic reticulum. Exp. Neurol. 2009, 215, 110–118. [Google Scholar] [CrossRef]
- Fittschen, M.; Lastres-Becker, I.; Halbach, M.V.; Damrath, E.; Gispert, S.; Azizov, M.; Walter, M.; Muller, S.; Auburger, G. Genetic ablation of ataxin-2 increases several global translation factors in their transcript abundance but decreases translation rate. Neurogenetics 2015, 16, 181–192. [Google Scholar] [CrossRef]
- Lastres-Becker, I.; Nonis, D.; Eich, F.; Klinkenberg, M.; Gorospe, M.; Kotter, P.; Klein, F.A.; Kedersha, N.; Auburger, G. Mammalian ataxin-2 modulates translation control at the pre-initiation complex via PI3K/mTOR and is induced by starvation. Biochim. Biophys. Acta 2016, 1862, 1558–1569. [Google Scholar] [CrossRef]
- Nonhoff, U.; Ralser, M.; Welzel, F.; Piccini, I.; Balzereit, D.; Yaspo, M.L.; Lehrach, H.; Krobitsch, S. Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol. Biol. Cell 2007, 18, 1385–1396. [Google Scholar] [CrossRef]
- Jagdeo, J.M.; Dufour, A.; Klein, T.; Solis, N.; Kleifeld, O.; Kizhakkedathu, J.; Luo, H.; Overall, C.M.; Jan, E. N-Terminomics TAILS Identifies Host Cell Substrates of Poliovirus and Coxsackievirus B3 3C Proteinases That Modulate Virus Infection. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Nonis, D.; Schmidt, M.H.H.; van de Loo, S.; Eich, F.; Dikic, I.; Nowock, J.; Auburger, G. Ataxin-2 associates with the endocytosis complex and affects EGF receptor trafficking. Cell. Signal. 2008, 20, 1725–1739. [Google Scholar] [CrossRef]
- Drost, J.; Nonis, D.; Eich, F.; Leske, O.; Damrath, E.; Brunt, E.R.; Lastres-Becker, I.; Heumann, R.; Nowock, J.; Auburger, G. Ataxin-2 modulates the levels of Grb2 and SRC but not ras signaling. J. Mol. Neurosci. 2013, 51, 68–81. [Google Scholar] [CrossRef]
- Lastres-Becker, I.; Nonis, D.; Nowock, J.; Auburger, G. New alternative splicing variants of the ATXN2 transcript. Neurol. Res. Pract. 2019, 1, 22. [Google Scholar] [CrossRef]
- Prudencio, M.; Belzil, V.V.; Batra, R.; Ross, C.A.; Gendron, T.F.; Pregent, L.J.; Murray, M.E.; Overstreet, K.K.; Piazza-Johnston, A.E.; Desaro, P.; et al. Distinct brain transcriptome profiles in C9orf72-associated and sporadic ALS. Nat. Neurosci. 2015, 18, 1175–1182. [Google Scholar] [CrossRef] [PubMed]
- DeMille, D.; Badal, B.D.; Evans, J.B.; Mathis, A.D.; Anderson, J.F.; Grose, J.H. PAS kinase is activated by direct SNF1-dependent phosphorylation and mediates inhibition of TORC1 through the phosphorylation and activation of Pbp1. Mol. Biol. Cell 2015, 26, 569–582. [Google Scholar] [CrossRef] [PubMed]
- Bar, D.Z.; Charar, C.; Dorfman, J.; Yadid, T.; Tafforeau, L.; Lafontaine, D.L.; Gruenbaum, Y. Cell size and fat content of dietary-restricted Caenorhabditis elegans are regulated by ATX-2, an mTOR repressor. Proc. Natl. Acad. Sci. USA 2016, 113, E4620–E4629. [Google Scholar] [CrossRef]
- Takahara, T.; Maeda, T. Transient sequestration of TORC1 into stress granules during heat stress. Mol. Cell 2012, 47, 242–252. [Google Scholar] [CrossRef]
- Meierhofer, D.; Halbach, M.; Sen, N.E.; Gispert, S.; Auburger, G. Ataxin-2 (Atxn2)-Knock-Out Mice Show Branched Chain Amino Acids and Fatty Acids Pathway Alterations. Mol. Cell. Proteom 2016, 15, 1728–1739. [Google Scholar] [CrossRef] [PubMed]
- Sen, N.E.; Drost, J.; Gispert, S.; Torres-Odio, S.; Damrath, E.; Klinkenberg, M.; Hamzeiy, H.; Akdal, G.; Gulluoglu, H.; Basak, A.N.; et al. Search for SCA2 blood RNA biomarkers highlights Ataxin-2 as strong modifier of the mitochondrial factor PINK1 levels. Neurobiol. Dis. 2016, 96, 115–126. [Google Scholar] [CrossRef]
- Seidel, G.; Meierhofer, D.; Sen, N.E.; Guenther, A.; Krobitsch, S.; Auburger, G. Quantitative Global Proteomics of Yeast PBP1 Deletion Mutants and Their Stress Responses Identifies Glucose Metabolism, Mitochondrial, and Stress Granule Changes. J. Proteome Res. 2017, 16, 504–515. [Google Scholar] [CrossRef]
- Wang, X.; Chen, X.J. A cytosolic network suppressing mitochondria-mediated proteostatic stress and cell death. Nature 2015, 524, 481–484. [Google Scholar] [CrossRef] [PubMed]
- Huttlin, E.L.; Ting, L.; Bruckner, R.J.; Gebreab, F.; Gygi, M.P.; Szpyt, J.; Tam, S.; Zarraga, G.; Colby, G.; Baltier, K.; et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell 2015, 162, 425–440. [Google Scholar] [CrossRef] [PubMed]
- Elden, A.C.; Kim, H.J.; Hart, M.P.; Chen-Plotkin, A.S.; Johnson, B.S.; Fang, X.; Armakola, M.; Geser, F.; Greene, R.; Lu, M.M.; et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 2010, 466, 1069–1075. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.; Li, Y.R.; Ingre, C.; Weber, M.; Grehl, T.; Gredal, O.; de Carvalho, M.; Meyer, T.; Tysnes, O.B.; Auburger, G.; et al. Ataxin-2 intermediate-length polyglutamine expansions in European ALS patients. Hum. Mol. Genet. 2011, 20, 1697–1700. [Google Scholar] [CrossRef] [PubMed]
- Gispert, S.; Kurz, A.; Waibel, S.; Bauer, P.; Liepelt, I.; Geisen, C.; Gitler, A.D.; Becker, T.; Weber, M.; Berg, D.; et al. The modulation of Amyotrophic Lateral Sclerosis risk by ataxin-2 intermediate polyglutamine expansions is a specific effect. Neurobiol. Dis. 2012, 45, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Becker, L.A.; Huang, B.; Bieri, G.; Ma, R.; Knowles, D.A.; Jafar-Nejad, P.; Messing, J.; Kim, H.J.; Soriano, A.; Auburger, G.; et al. Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice. Nature 2017, 544, 367–371. [Google Scholar] [CrossRef] [PubMed]
- Lahut, S.; Omur, O.; Uyan, O.; Agim, Z.S.; Ozoguz, A.; Parman, Y.; Deymeer, F.; Oflazer, P.; Koc, F.; Ozcelik, H.; et al. ATXN2 and its neighbouring gene SH2B3 are associated with increased ALS risk in the Turkish population. PLoS ONE 2012, 7, e42956. [Google Scholar] [CrossRef] [PubMed]
- Ross, O.A.; Rutherford, N.J.; Baker, M.; Soto-Ortolaza, A.I.; Carrasquillo, M.M.; DeJesus-Hernandez, M.; Adamson, J.; Li, M.; Volkening, K.; Finger, E.; et al. Ataxin-2 repeat-length variation and neurodegeneration. Hum. Mol. Genet. 2011, 20, 3207–3212. [Google Scholar] [CrossRef]
- Shulman, J.M.; Feany, M.B. Genetic modifiers of tauopathy in Drosophila. Genetics 2003, 165, 1233–1242. [Google Scholar]
- Scoles, D.R.; Meera, P.; Schneider, M.D.; Paul, S.; Dansithong, W.; Figueroa, K.P.; Hung, G.; Rigo, F.; Bennett, C.F.; Otis, T.S.; et al. Antisense oligonucleotide therapy for spinocerebellar ataxia type 2. Nature 2017, 544, 362–366. [Google Scholar] [CrossRef]
- Al-Ramahi, I.; Perez, A.M.; Lim, J.; Zhang, M.; Sorensen, R.; de Haro, M.; Branco, J.; Pulst, S.M.; Zoghbi, H.Y.; Botas, J. dAtaxin-2 mediates expanded Ataxin-1-induced neurodegeneration in a Drosophila model of SCA1. PLoS Genet. 2007, 3, e234. [Google Scholar] [CrossRef]
- Mangus, D.A.; Amrani, N.; Jacobson, A. Pbp1p, a factor interacting with Saccharomyces cerevisiae poly(A)-binding protein, regulates polyadenylation. Mol. Cell. Biol. 1998, 18, 7383–7396. [Google Scholar] [CrossRef]
- Lastres-Becker, I.; Brodesser, S.; Lutjohann, D.; Azizov, M.; Buchmann, J.; Hintermann, E.; Sandhoff, K.; Schurmann, A.; Nowock, J.; Auburger, G. Insulin receptor and lipid metabolism pathology in ataxin-2 knock-out mice. Hum. Mol. Genet. 2008, 17, 1465–1481. [Google Scholar] [CrossRef] [PubMed]
- Kiehl, T.R.; Nechiporuk, A.; Figueroa, K.P.; Keating, M.T.; Huynh, D.P.; Pulst, S.M. Generation and characterization of Sca2 (ataxin-2) knockout mice. Biochem. Biophys. Res. Commun. 2006, 339, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Lopez, D.; Guzman, P. Insights into the evolution and domain structure of Ataxin-2 proteins across eukaryotes. BMC Res. Notes 2014, 7, 453. [Google Scholar] [CrossRef]
- Kaehler, C.; Isensee, J.; Nonhoff, U.; Terrey, M.; Hucho, T.; Lehrach, H.; Krobitsch, S. Ataxin-2-like is a regulator of stress granules and processing bodies. PLoS ONE 2012, 7, e50134. [Google Scholar] [CrossRef] [PubMed]
- Kaehler, C.; Guenther, A.; Uhlich, A.; Krobitsch, S. PRMT1-mediated arginine methylation controls ATXN2L localization. Exp. Cell Res. 2015, 334, 114–125. [Google Scholar] [CrossRef]
- Meunier, C.; Bordereaux, D.; Porteu, F.; Gisselbrecht, S.; Chretien, S.; Courtois, G. Cloning and characterization of a family of proteins associated with Mpl. J. Biol. Chem. 2002, 277, 9139–9147. [Google Scholar] [CrossRef]
- Figueroa, K.P.; Pulst, S.M. Identification and expression of the gene for human ataxin-2-related protein on chromosome 16. Exp. Neurol. 2003, 184, 669–678. [Google Scholar] [CrossRef]
- Lin, L.; Li, X.; Pan, C.; Lin, W.; Shao, R.; Liu, Y.; Zhang, J.; Luo, Y.; Qian, K.; Shi, M.; et al. ATXN2L upregulated by epidermal growth factor promotes gastric cancer cell invasiveness and oxaliplatin resistance. Cell Death Dis. 2019, 10, 173. [Google Scholar] [CrossRef]
- STRING Web Platform. Available online: https://string-db.org/ (accessed on 17 July 2020).
- Lee, J.; Yoo, E.; Lee, H.; Park, K.; Hur, J.H.; Lim, C. LSM12 and ME31B/DDX6 Define Distinct Modes of Posttranscriptional Regulation by ATAXIN-2 Protein Complex in Drosophila Circadian Pacemaker Neurons. Mol. Cell 2017, 66, 129–140.e127. [Google Scholar] [CrossRef]
- Dougherty, J.D.; Tsai, W.C.; Lloyd, R.E. Multiple Poliovirus Proteins Repress Cytoplasmic RNA Granules. Viruses 2015, 7, 6127–6140. [Google Scholar] [CrossRef]
- Tajirika, T.; Tokumaru, Y.; Taniguchi, K.; Sugito, N.; Matsuhashi, N.; Futamura, M.; Yanagihara, K.; Akao, Y.; Yoshida, K. DEAD-Box Protein RNA-Helicase DDX6 Regulates the Expression of HER2 and FGFR2 at the Post-Transcriptional Step in Gastric Cancer Cells. Int. J. Mol. Sci. 2018, 19, 2005. [Google Scholar] [CrossRef] [PubMed]
- Hansen, M.; Zeddies, S.; Meinders, M.; di Summa, F.; van Alphen, F.P.J.; Hoogendijk, A.J.; Moore, K.S.; Halbach, M.; Gutierrez, L.; van den Biggelaar, M.; et al. The RNA-Binding Protein ATXN2 is Expressed during Megakaryopoiesis and May Control Timing of Gene Expression. Int. J. Mol. Sci. 2020, 21, 967. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; He, L.; Zhou, Q.; Wang, M.; Shen, W.-J.; Azhar, S.; Pan, F.; Guo, Z.; Hu, Z. Nherf1 and nherf2 regulation of sr-b1 stability via ubiquitination and proteasome degradation. Biochem. Biophys. Res. Commun. 2017, 490, 1168–1175. [Google Scholar] [CrossRef] [PubMed]
- Hryciw, D.H.; Jenkin, K.A.; Simcocks, A.C.; Grinfeld, E.; McAinch, A.J.; Poronnik, P. The interaction between megalin and clc-5 is scaffolded by the na+–h+ exchanger regulatory factor 2 (nherf2) in proximal tubule cells. Int. J. Biochem. Cell Biol. 2012, 44, 815–823. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.; Rayfield, A.; Hryciw, D.H.; Ma, T.A.; Wang, D.; Pow, D.; Broer, S.; Yun, C.; Poronnik, P. Na+–h+ exchanger regulatory factor 1 is a pdz scaffold for the astroglial glutamate transporter glast. Glia 2007, 55, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Filippov, A.K.; Simon, J.; Barnard, E.A.; Brown, D.A. The scaffold protein nherf2 determines the coupling of p2y1 nucleotide and mglur5 glutamate receptor to different ion channels in neurons. J. Neurosci. 2010, 30, 11068–11072. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.J.; Jeon, E.S.; Lee, H.J.; Oh, Y.-S.; Suh, P.-G.; Jung, J.S.; Donowitz, M.; Kim, J.H. Nherf2 increases platelet-derived growth factor-induced proliferation through pi-3-kinase/akt-, erk-, and src family kinase-dependent pathway. Cell. Signal. 2004, 16, 791–800. [Google Scholar] [CrossRef]
- Allen Brain Atlas. Available online: https://mouse.brain-map.org/ (accessed on 17 July 2020).
- Human Protein Atlas. Available online: https://www.proteinatlas.org/ (accessed on 17 July 2020).
- Coexpedia Database. Available online: www.coexpedia.org (accessed on 17 July 2020).
- Sen, N.E.; Canet-Pons, J.; Halbach, M.V.; Arsovic, A.; Pilatus, U.; Chae, W.H.; Kaya, Z.E.; Seidel, K.; Rollmann, E.; Mittelbronn, M.; et al. Generation of an Atxn2-CAG100 knock-in mouse reveals N-acetylaspartate production deficit due to early Nat8l dysregulation. Neurobiol. Dis. 2019, 132, 104559. [Google Scholar] [CrossRef]
- Kiehl, T.R.; Shibata, H.; Pulst, S.M. The ortholog of human ataxin-2 is essential for early embryonic patterning in C. elegans. J. Mol. Neurosci. 2000, 15, 231–241. [Google Scholar] [CrossRef]
- Ciosk, R.; DePalma, M.; Priess, J.R. ATX-2, the C. elegans ortholog of ataxin 2, functions in translational regulation in the germline. Development 2004, 131, 4831–4841. [Google Scholar] [CrossRef]
- Gross, N.; Strillacci, M.G.; Penagaricano, F.; Khatib, H. Characterization and functional roles of paternal RNAs in 2-4 cell bovine embryos. Sci. Rep. 2019, 9, 20347. [Google Scholar] [CrossRef] [PubMed]
- Flores, L.E.; Hildebrandt, T.B.; Kuhl, A.A.; Drews, B. Early detection and staging of spontaneous embryo resorption by ultrasound biomicroscopy in murine pregnancy. Reprod. Biol. Endocrinol. 2014, 12, 38. [Google Scholar] [CrossRef] [PubMed]
- Papaioannou, V.E.; Behringer, R.R. Early embryonic lethality in genetically engineered mice: Diagnosis and phenotypic analysis. Vet. Pathol. 2012, 49, 64–70. [Google Scholar] [CrossRef]
- Armit, C.; Richardson, L.; Hill, B.; Yang, Y.; Baldock, R.A. eMouseAtlas informatics: Embryo atlas and gene expression database. Mamm. Genome 2015, 26, 431–440. [Google Scholar] [CrossRef]
- Dhenain, M.; Ruffins, S.W.; Jacobs, R.E. Three-dimensional digital mouse atlas using high-resolution MRI. Dev. Biol. 2001, 232, 458–470. [Google Scholar] [CrossRef] [PubMed]
- Bard, J.L.; Kaufman, M.H.; Dubreuil, C.; Brune, R.M.; Burger, A.; Baldock, R.A.; Davidson, D.R. An internet-accessible database of mouse developmental anatomy based on a systematic nomenclature. Mech. Dev. 1998, 74, 111–120. [Google Scholar] [CrossRef]
- Adams, D.; Baldock, R.; Bhattacharya, S.; Copp, A.J.; Dickinson, M.; Greene, N.D.; Henkelman, M.; Justice, M.; Mohun, T.; Murray, S.A.; et al. Bloomsbury report on mouse embryo phenotyping: Recommendations from the IMPC workshop on embryonic lethal screening. Dis. Models Mech. 2013, 6, 571–579. [Google Scholar] [CrossRef]
- Perez-Garcia, V.; Fineberg, E.; Wilson, R.; Murray, A.; Mazzeo, C.I.; Tudor, C.; Sienerth, A.; White, J.K.; Tuck, E.; Ryder, E.J.; et al. Placentation defects are highly prevalent in embryonic lethal mouse mutants. Nature 2018, 555, 463–468. [Google Scholar] [CrossRef]
- Lee, T.C.; Threadgill, D.W. Generation and validation of mice carrying a conditional allele of the epidermal growth factor receptor. Genesis 2009, 47, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Dackor, J.; Caron, K.M.; Threadgill, D.W. Placental and embryonic growth restriction in mice with reduced function epidermal growth factor receptor alleles. Genetics 2009, 183, 207–218. [Google Scholar] [CrossRef]
- Teves, M.E.; Modi, B.P.; Kulkarni, R.; Han, A.X.; Marks, J.S.; Subler, M.A.; Windle, J.; Newall, J.M.; McAllister, J.M.; Strauss, J.F., 3rd. Human DENND1A.V2 Drives Cyp17a1 Expression and Androgen Production in Mouse Ovaries and Adrenals. Int. J. Mol. Sci. 2020, 21, 2545. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, P.; Droce, A.; Moser, J.M.; Cuhlmann, S.; Padilla, C.O.; Heimann, P.; Bartsch, J.W.; Fuchtbauer, A.; Fuchtbauer, E.M.; Schmitt-John, T. Loss of vps54 function leads to vesicle traffic impairment, protein mis-sorting and embryonic lethality. Int. J. Mol. Sci. 2013, 14, 10908–10925. [Google Scholar] [CrossRef]
- Li, M.; Zhao, X.; Wang, W.; Shi, H.; Pan, Q.; Lu, Z.; Perez, S.P.; Suganthan, R.; He, C.; Bjoras, M.; et al. Ythdf2-mediated m(6)A mRNA clearance modulates neural development in mice. Genome Biol. 2018, 19, 69. [Google Scholar] [CrossRef] [PubMed]
- Bae, B.; Miura, P. Emerging Roles for 3’ UTRs in Neurons. Int. J. Mol. Sci. 2020, 21, 3413. [Google Scholar] [CrossRef] [PubMed]
- Pham, J.; Keon, M.; Brennan, S.; Saksena, N. Connecting RNA-Modifying Similarities of TDP-43, FUS, and SOD1 with MicroRNA Dysregulation Amidst A Renewed Network Perspective of Amyotrophic Lateral Sclerosis Proteinopathy. Int. J. Mol. Sci. 2020, 21, 3464. [Google Scholar] [CrossRef] [PubMed]
- Holt, D.J.; Grainger, D.W. Multinucleated giant cells from fibroblast cultures. Biomaterials 2011, 32, 3977–3987. [Google Scholar] [CrossRef]
- Atilla-Gokcumen, G.E.; Muro, E.; Relat-Goberna, J.; Sasse, S.; Bedigian, A.; Coughlin, M.L.; Garcia-Manyes, S.; Eggert, U.S. Dividing cells regulate their lipid composition and localization. Cell 2014, 156, 428–439. [Google Scholar] [CrossRef]
- Fernandez, C.; Lobo Md Mdel, V.; Gomez-Coronado, D.; Lasuncion, M.A. Cholesterol is essential for mitosis progression and its deficiency induces polyploid cell formation. Exp. Cell Res. 2004, 300, 109–120. [Google Scholar] [CrossRef]
- Sen, N.E.; Arsovic, A.; Meierhofer, D.; Brodesser, S.; Oberschmidt, C.; Canet-Pons, J.; Kaya, Z.E.; Halbach, M.V.; Gispert, S.; Sandhoff, K.; et al. In Human and Mouse Spino-Cerebellar Tissue, Ataxin-2 Expansion Affects Ceramide-Sphingomyelin Metabolism. Int. J. Mol. Sci. 2019, 20, 5854. [Google Scholar] [CrossRef]
- Ng, M.M.; Chang, F.; Burgess, D.R. Movement of membrane domains and requirement of membrane signaling molecules for cytokinesis. Dev. Cell 2005, 9, 781–790. [Google Scholar] [CrossRef]
- Kettle, E.; Page, S.L.; Morgan, G.P.; Malladi, C.S.; Wong, C.L.; Boadle, R.A.; Marsh, B.J.; Robinson, P.J.; Chircop, M. A Cholesterol-Dependent Endocytic Mechanism Generates Midbody Tubules During Cytokinesis. Traffic 2015, 16, 1174–1192. [Google Scholar] [CrossRef] [PubMed]
- Feng, B.; Schwarz, H.; Jesuthasan, S. Furrow-specific endocytosis during cytokinesis of zebrafish blastomeres. Exp. Cell Res. 2002, 279, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Neuwald, A.F.; Koonin, E.V. Ataxin-2, global regulators of bacterial gene expression, and spliceosomal snRNP proteins share a conserved domain. J. Mol. Med. 1998, 76, 3–5. [Google Scholar] [CrossRef] [PubMed]
- NHGRI-EBI Catalog of Human Genome-Wide Association Studies. Available online: https://www.ebi.ac.uk/gwas/ (accessed on 22 November 2019).
- Hoffmann, T.J.; Choquet, H.; Yin, J.; Banda, Y.; Kvale, M.N.; Glymour, M.; Schaefer, C.; Risch, N.; Jorgenson, E. A Large Multiethnic Genome-Wide Association Study of Adult Body Mass Index Identifies Novel Loci. Genetics 2018, 210, 499–515. [Google Scholar] [CrossRef]
- Graff, M.; Scott, R.A.; Justice, A.E.; Young, K.L.; Feitosa, M.F.; Barata, L.; Winkler, T.W.; Chu, A.Y.; Mahajan, A.; Hadley, D.; et al. Genome-wide physical activity interactions in adiposity—A meta-analysis of 200,452 adults. PLoS Genet. 2017, 13, e1006528. [Google Scholar] [CrossRef]
- Tachmazidou, I.; Suveges, D.; Min, J.L.; Ritchie, G.R.S.; Steinberg, J.; Walter, K.; Iotchkova, V.; Schwartzentruber, J.; Huang, J.; Memari, Y.; et al. Whole-Genome Sequencing Coupled to Imputation Discovers Genetic Signals for Anthropometric Traits. Am. J. Hum. Genet. 2017, 100, 865–884. [Google Scholar] [CrossRef]
- Lee, J.J.; Wedow, R.; Okbay, A.; Kong, E.; Maghzian, O.; Zacher, M.; Nguyen-Viet, T.A.; Bowers, P.; Sidorenko, J.; Karlsson Linner, R.; et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat. Genet. 2018, 50, 1112–1121. [Google Scholar] [CrossRef]
- Tikkanen, E.; Gustafsson, S.; Amar, D.; Shcherbina, A.; Waggott, D.; Ashley, E.A.; Ingelsson, E. Biological Insights Into Muscular Strength: Genetic Findings in the UK Biobank. Sci. Rep. 2018, 8, 6451. [Google Scholar] [CrossRef]
- Hill, W.D.; Marioni, R.E.; Maghzian, O.; Ritchie, S.J.; Hagenaars, S.P.; McIntosh, A.M.; Gale, C.R.; Davies, G.; Deary, I.J. A combined analysis of genetically correlated traits identifies 187 loci and a role for neurogenesis and myelination in intelligence. Mol. Psychiatry 2019, 24, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Lam, M.; Trampush, J.W.; Yu, J.; Knowles, E.; Davies, G.; Liewald, D.C.; Starr, J.M.; Djurovic, S.; Melle, I.; Sundet, K.; et al. Large-Scale Cognitive GWAS Meta-Analysis Reveals Tissue-Specific Neural Expression and Potential Nootropic Drug Targets. Cell Rep. 2017, 21, 2597–2613. [Google Scholar] [CrossRef]
- Sniekers, S.; Stringer, S.; Watanabe, K.; Jansen, P.R.; Coleman, J.R.I.; Krapohl, E.; Taskesen, E.; Hammerschlag, A.R.; Okbay, A.; Zabaneh, D.; et al. Genome-wide association meta-analysis of 78,308 individuals identifies new loci and genes influencing human intelligence. Nat. Genet. 2017, 49, 1107–1112. [Google Scholar] [CrossRef] [PubMed]
- Coleman, J.R.I.; Bryois, J.; Gaspar, H.A.; Jansen, P.R.; Savage, J.E.; Skene, N.; Plomin, R.; Munoz-Manchado, A.B.; Linnarsson, S.; Crawford, G.; et al. Biological annotation of genetic loci associated with intelligence in a meta-analysis of 87,740 individuals. Mol. Psychiatry 2019, 24, 182–197. [Google Scholar] [CrossRef]
- Okbay, A.; Beauchamp, J.P.; Fontana, M.A.; Lee, J.J.; Pers, T.H.; Rietveld, C.A.; Turley, P.; Chen, G.B.; Emilsson, V.; Meddens, S.F.; et al. Genome-wide association study identifies 74 loci associated with educational attainment. Nature 2016, 533, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Rietveld, C.A.; Medland, S.E.; Derringer, J.; Yang, J.; Esko, T.; Martin, N.W.; Westra, H.J.; Shakhbazov, K.; Abdellaoui, A.; Agrawal, A.; et al. GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science 2013, 340, 1467–1471. [Google Scholar] [CrossRef] [PubMed]
- Kanai, M.; Akiyama, M.; Takahashi, A.; Matoba, N.; Momozawa, Y.; Ikeda, M.; Iwata, N.; Ikegawa, S.; Hirata, M.; Matsuda, K.; et al. Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat. Genet. 2018, 50, 390–400. [Google Scholar] [CrossRef] [PubMed]
- Imielinski, M.; Baldassano, R.N.; Griffiths, A.; Russell, R.K.; Annese, V.; Dubinsky, M.; Kugathasan, S.; Bradfield, J.P.; Walters, T.D.; Sleiman, P.; et al. Common variants at five new loci associated with early-onset inflammatory bowel disease. Nat. Genet. 2009, 41, 1335–1340. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.R.; Li, J.; Zhao, S.D.; Bradfield, J.P.; Mentch, F.D.; Maggadottir, S.M.; Hou, C.; Abrams, D.J.; Chang, D.; Gao, F.; et al. Meta-analysis of shared genetic architecture across ten pediatric autoimmune diseases. Nat. Med. 2015, 21, 1018–1027. [Google Scholar] [CrossRef]
- Wills, A.M.; Hubbard, J.; Macklin, E.A.; Glass, J.; Tandan, R.; Simpson, E.P.; Brooks, B.; Gelinas, D.; Mitsumoto, H.; Mozaffar, T.; et al. Hypercaloric enteral nutrition in patients with amyotrophic lateral sclerosis: A randomised, double-blind, placebo-controlled phase 2 trial. Lancet 2014, 383, 2065–2072. [Google Scholar] [CrossRef]
- Dorst, J.; Cypionka, J.; Ludolph, A.C. High-caloric food supplements in the treatment of amyotrophic lateral sclerosis: A prospective interventional study. Amyotroph. Lateral Scler. Front. Degener. 2013, 14, 533–536. [Google Scholar] [CrossRef]
- Genotype-Tissue-Expression Project Database. Available online: https://gtexportal.org/home/ (accessed on 17 July 2020).
- Almaguer-Mederos, L.E.; Aguilera-Rodriguez, R.; Almaguer-Gotay, D.; Hechavarria-Barzaga, K.; Alvarez-Sosa, A.; Chapman-Rodriguez, Y.; Silva-Ricardo, Y.; Gonzalez-Zaldivar, Y.; Vazquez-Mojena, Y.; Cuello-Almarales, D.; et al. Testosterone Levels Are Decreased and Associated with Disease Duration in Male Spinocerebellar Ataxia Type 2 Patients. Cerebellum 2020. [Google Scholar] [CrossRef]
- Klockgether, T.; Ludtke, R.; Kramer, B.; Abele, M.; Burk, K.; Schols, L.; Riess, O.; Laccone, F.; Boesch, S.; Lopes-Cendes, I.; et al. The natural history of degenerative ataxia: A retrospective study in 466 patients. Brain 1998, 121 Pt 4, 589–600. [Google Scholar] [CrossRef]
- War, A.R.; Dang, K.; Jiang, S.; Xiao, Z.; Miao, Z.; Yang, T.; Li, Y.; Qian, A. Role of cancer stem cells in the development of giant cell tumor of bone. Cancer Cell Int. 2020, 20, 135. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, Y.; Lim, C.X.; Weichhart, T.; Valeyre, D.; Bentaher, A.; Calender, A. Sarcoidosis and the mTOR, Rac1, and Autophagy Triad. Trends Immunol. 2020, 41, 286–299. [Google Scholar] [CrossRef] [PubMed]
- Feliciano, D.M.; Su, T.; Lopez, J.; Platel, J.C.; Bordey, A. Single-cell Tsc1 knockout during corticogenesis generates tuber-like lesions and reduces seizure threshold in mice. J. Clin. Investig. 2011, 121, 1596–1607. [Google Scholar] [CrossRef] [PubMed]
- Key, J.; Mueller, A.K.; Gispert, S.; Matschke, L.; Wittig, I.; Corti, O.; Munch, C.; Decher, N.; Auburger, G. Ubiquitylome profiling of Parkin-null brain reveals dysregulation of calcium homeostasis factors ATP1A2, Hippocalcin and GNA11, reflected by altered firing of noradrenergic neurons. Neurobiol. Dis. 2019, 127, 114–130. [Google Scholar] [CrossRef] [PubMed]
- Tunster, S.J. Genetic sex determination of mice by simplex PCR. Biol. Sex Differ. 2017, 8, 31. [Google Scholar] [CrossRef]
- Damrath, E.; Heck, M.V.; Gispert, S.; Azizov, M.; Nowock, J.; Seifried, C.; Rub, U.; Walter, M.; Auburger, G. ATXN2-CAG42 sequesters PABPC1 into insolubility and induces FBXW8 in cerebellum of old ataxic knock-in mice. PLoS Genet. 2012, 8, e1002920. [Google Scholar] [CrossRef]
- Key, J.; Kohli, A.; Barcena, C.; Lopez-Otin, C.; Heidler, J.; Wittig, I.; Auburger, G. Global Proteome of LonP1(+/-) Mouse Embryonal Fibroblasts Reveals Impact on Respiratory Chain, but No Interdependence between Eral1 and Mitoribosomes. Int. J. Mol. Sci. 2019, 20, 4523. [Google Scholar] [CrossRef]
- Torres-Odio, S.; Key, J.; Hoepken, H.H.; Canet-Pons, J.; Valek, L.; Roller, B.; Walter, M.; Morales-Gordo, B.; Meierhofer, D.; Harter, P.N.; et al. Progression of pathology in PINK1-deficient mouse brain from splicing via ubiquitination, ER stress, and mitophagy changes to neuroinflammation. J. Neuroinflamm. 2017, 14, 154. [Google Scholar] [CrossRef]
- Gispert, S.; Parganlija, D.; Klinkenberg, M.; Drose, S.; Wittig, I.; Mittelbronn, M.; Grzmil, P.; Koob, S.; Hamann, A.; Walter, M.; et al. Loss of mitochondrial peptidase Clpp leads to infertility, hearing loss plus growth retardation via accumulation of CLPX, mtDNA and inflammatory factors. Hum. Mol. Genet. 2013, 22, 4871–4887. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).