Discrimination of CpG Methylation Status and Nucleotide Differences in Tissue Specimen DNA by Oligoribonucleotide Interference-PCR
Abstract
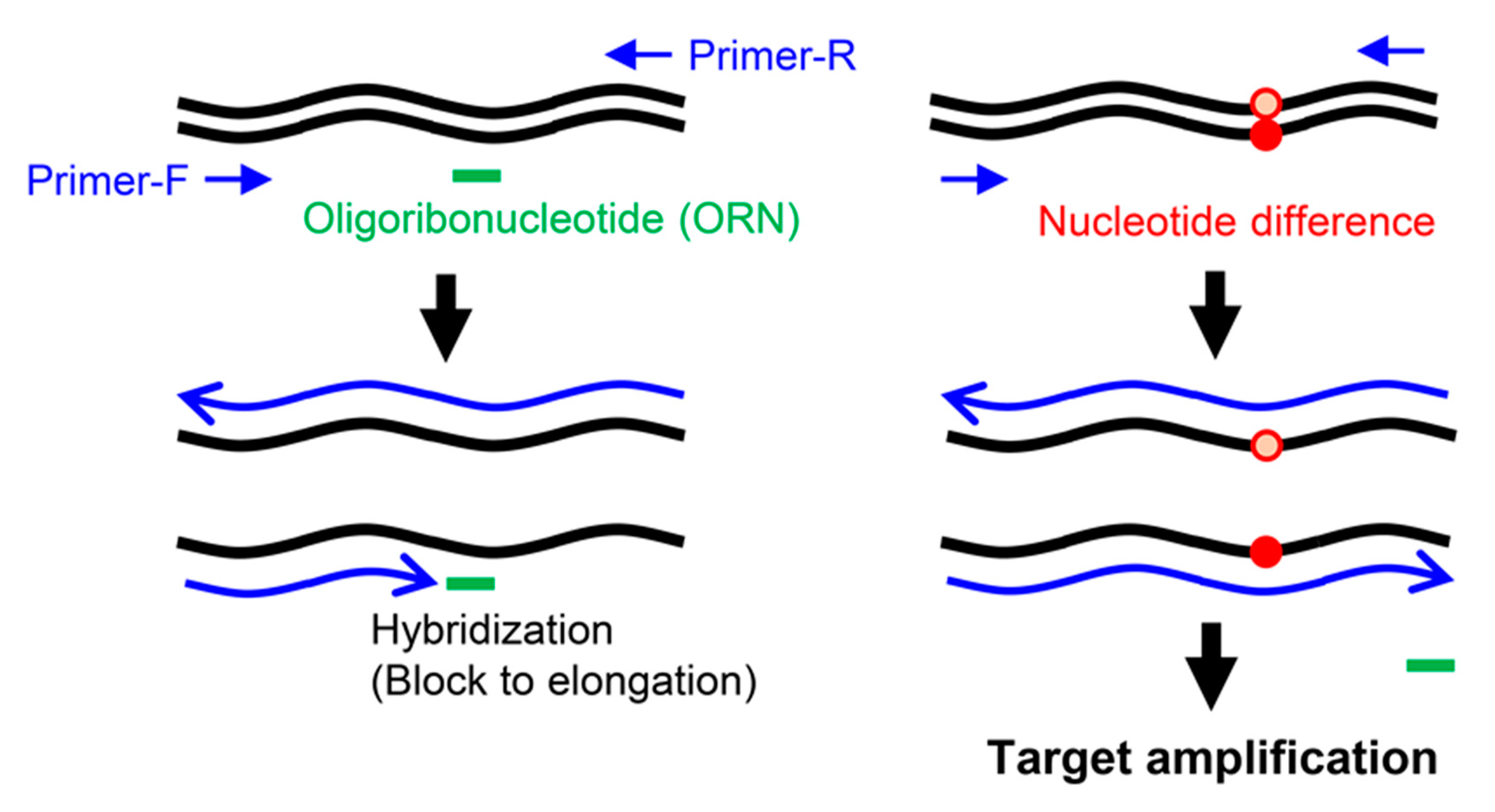
1. Introduction
2. Results
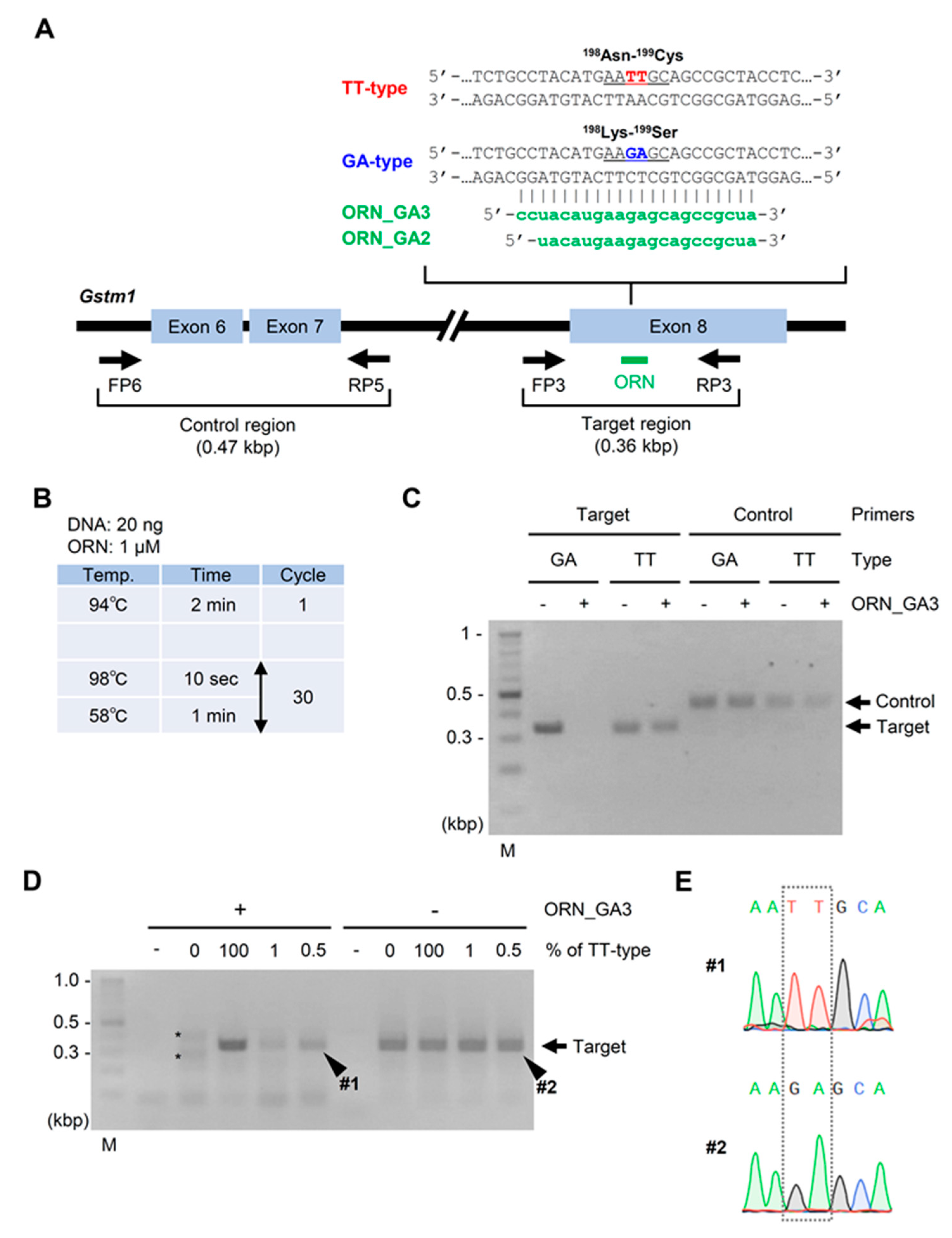
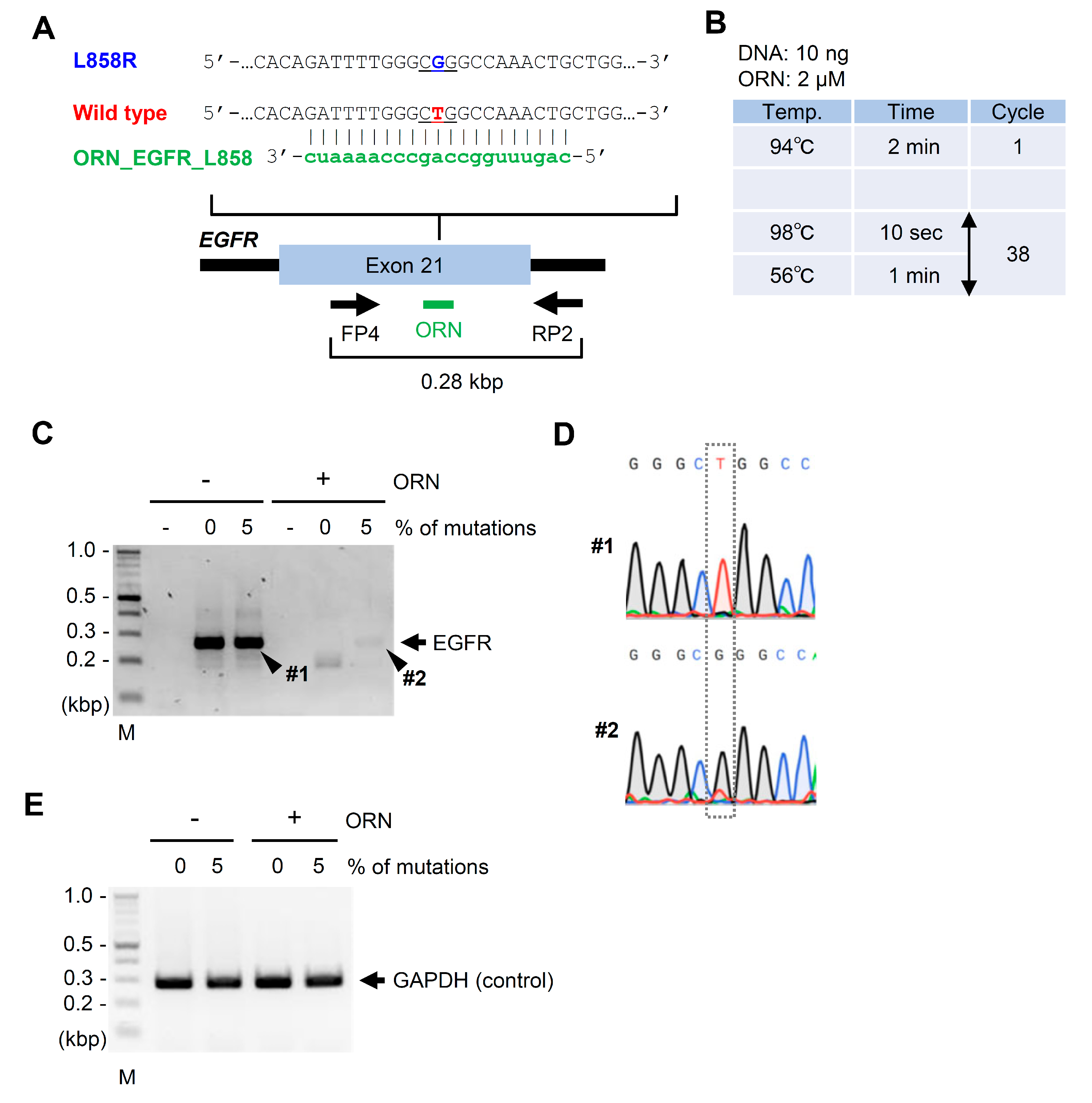
2.1. ORNi-PCR Using AFPE or FFPE Specimen DNA
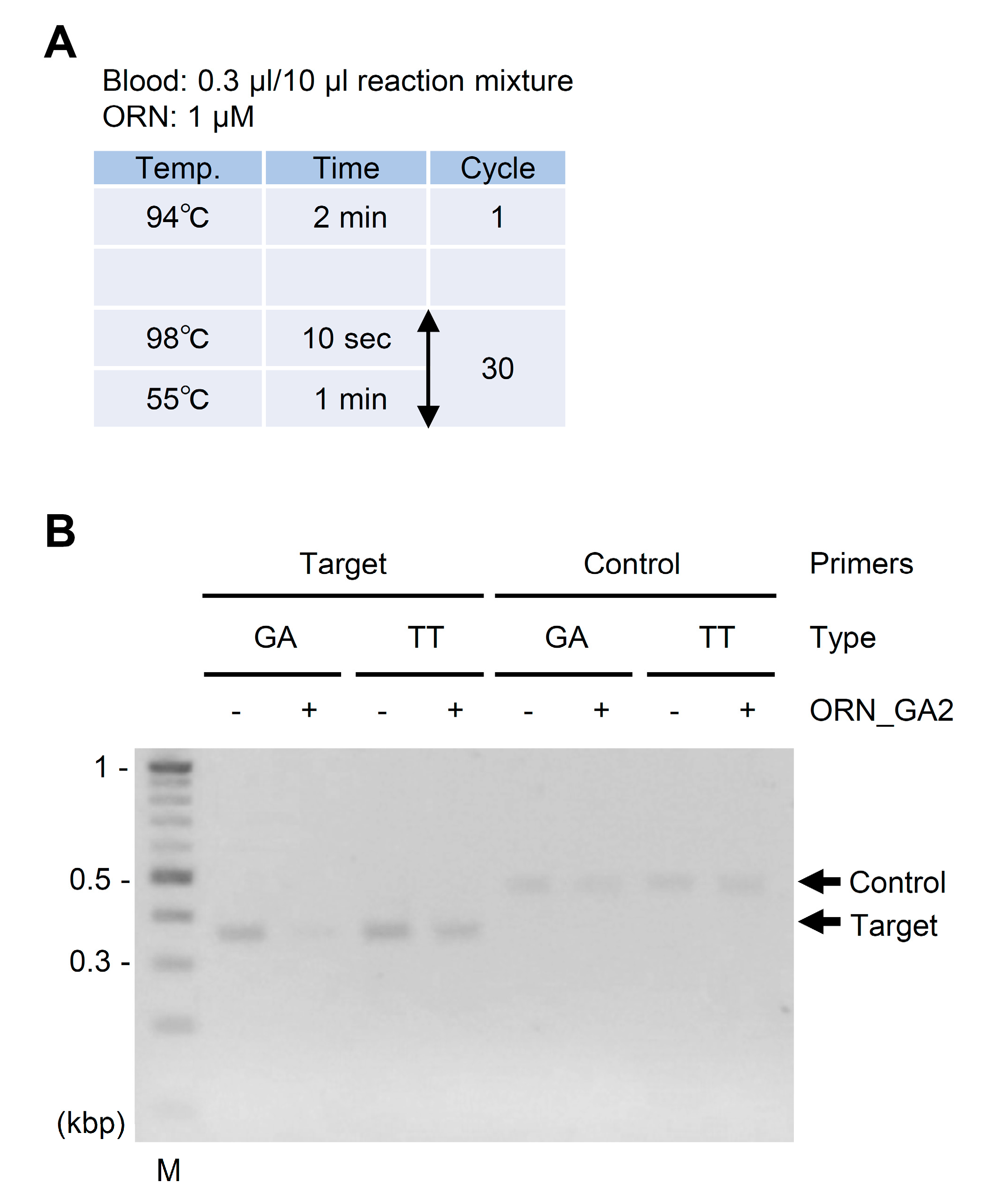
2.2. ORNi-PCR Using Whole Blood Specimens
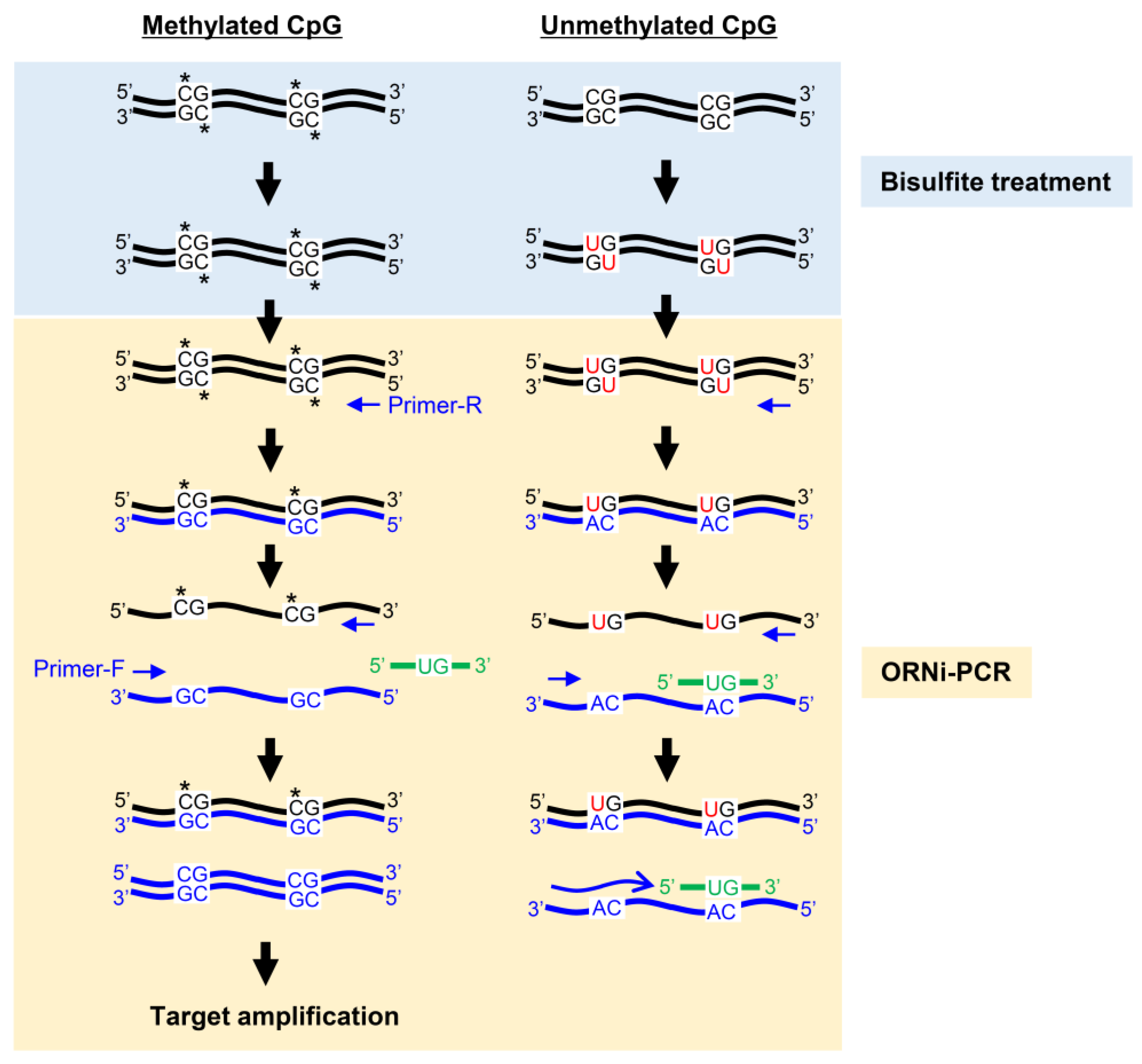
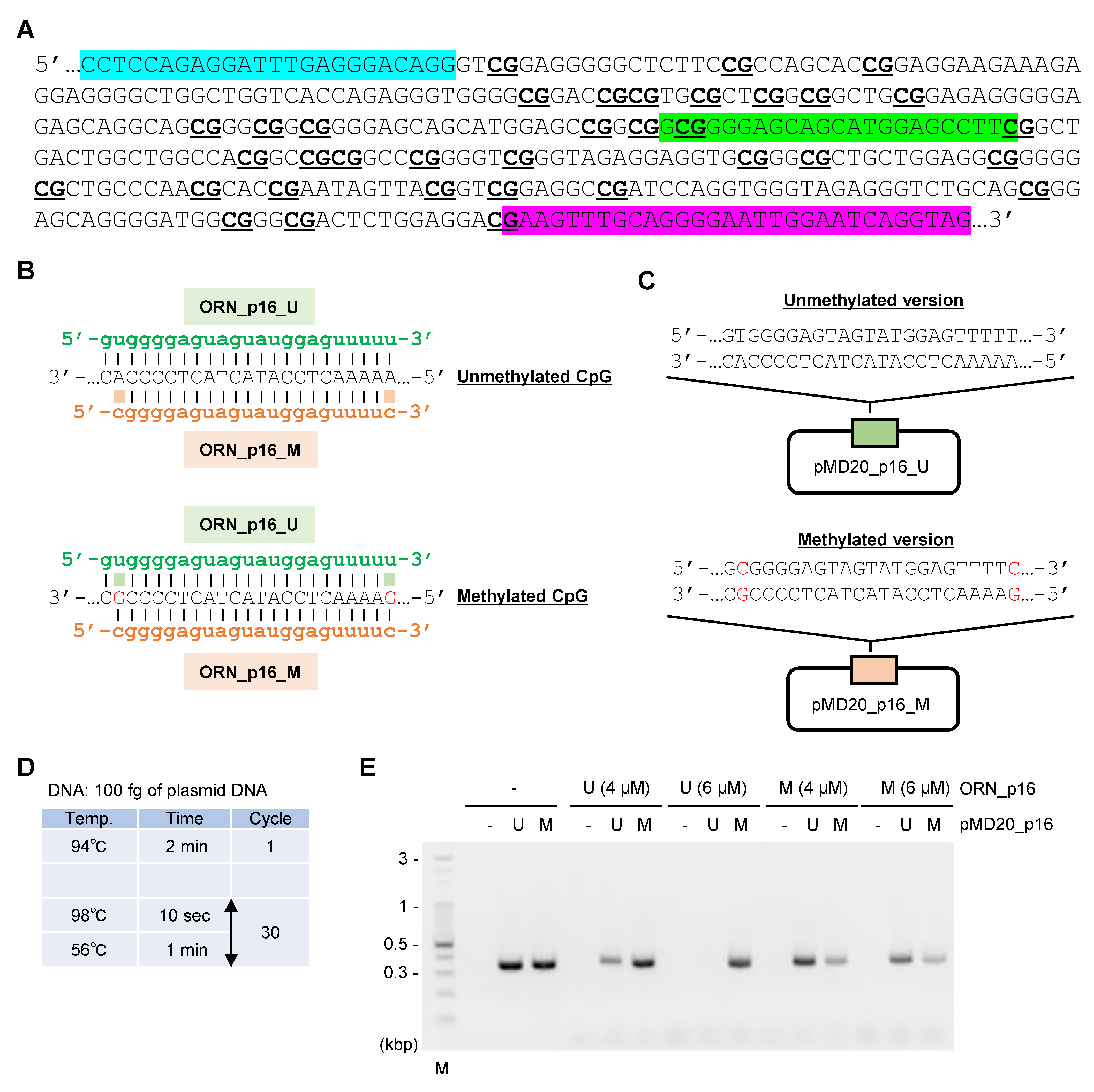
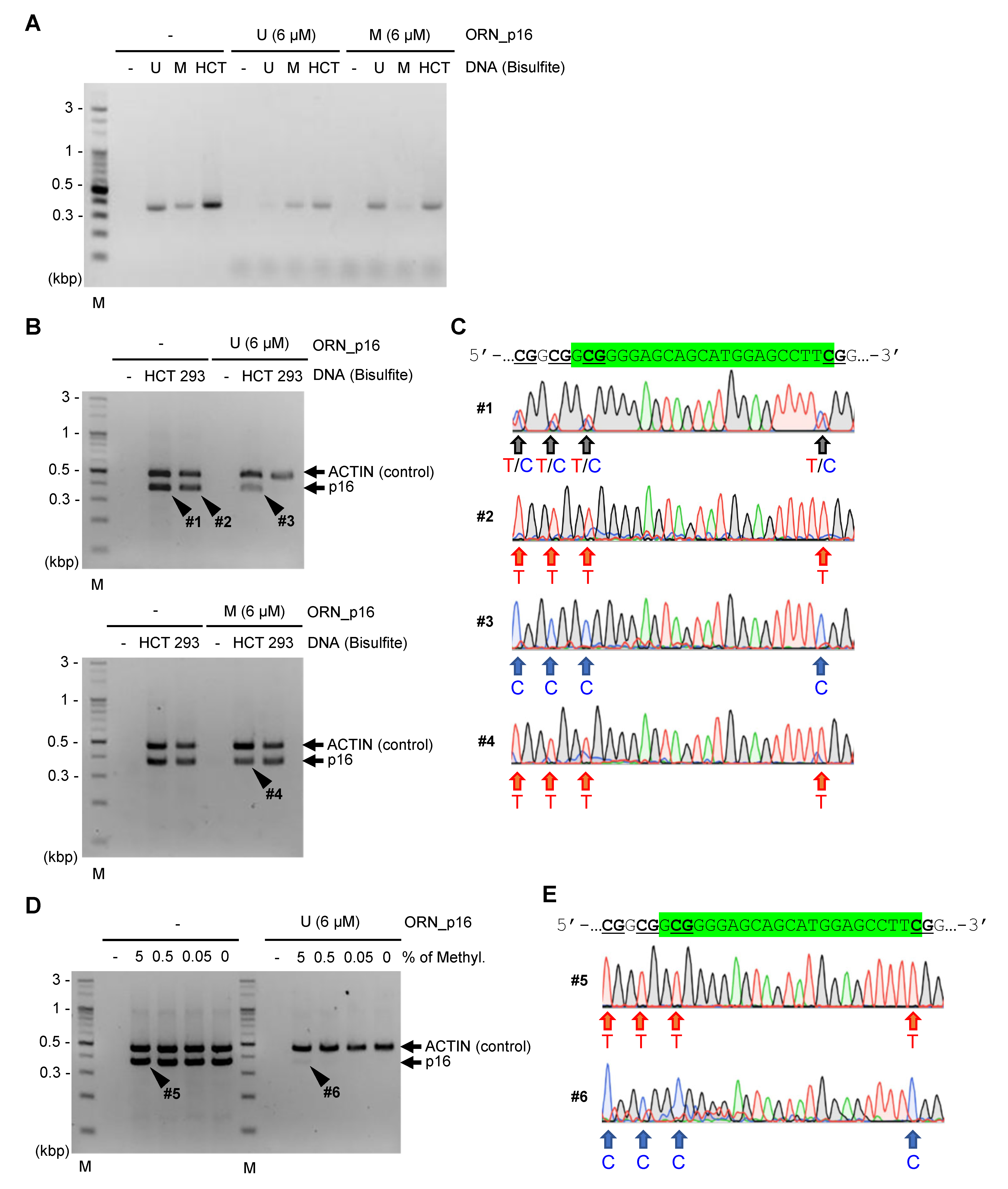
2.3. ORNi-PCR Using Bisulfite-Treated DNA
3. Discussion
4. Materials and Methods
4.1. Oligonucleotides
4.2. Extraction of DNA From Rat Liver Specimens
4.3. Extraction of DNA From FFPE Control Specimens
4.4. ORNi-PCR With AFPE or FFPE Specimen DNA
4.5. ORNi-PCR With Whole Blood Specimens
4.6. Plasmid Construction
4.7. ORNi-PCR With Plasmid DNA or Bisulfite-Treated DNA
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Powledge, T.M. The polymerase chain reaction. Adv. Physiol. Educ. 2004, 28, 44–50. [Google Scholar] [CrossRef]
- Coleman, W.B.; Tsongalis, G.J. The polymerase chain reaction. In Molecular Diagnostics for the Clinical Laboratorian, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2006; pp. 47–55. [Google Scholar]
- Frayling, I.M.; Monk, E.; Butler, R. PCR-Based Methods for Mutation Detection. Mol. Diagn. 2006, 65–74. [Google Scholar] [CrossRef]
- Vestheim, H.; Deagle, B.E.; Jarman, S.N. Application of Blocking Oligonucleotides to Improve Signal-to-Noise Ratio in a PCR. Methods Mol. Biol. 2011, 687, 265–274. [Google Scholar] [CrossRef]
- Tanigawa, N.; Fujita, T.; Fujii, H. Oligoribonucleotide (ORN) Interference-PCR (ORNi-PCR): A Simple Method for Suppressing PCR Amplification of Specific DNA Sequences Using ORNs. PLoS ONE 2014, 9, e113345. [Google Scholar] [CrossRef] [PubMed]
- Fujita, T.; Yuno, M.; Kitaura, F.; Fujii, H. Detection of genome-edited cells by oligoribonucleotide interference-PCR. DNA Res. 2018, 25, 395–407. [Google Scholar] [CrossRef] [PubMed]
- Fujita, T.; Yuno, M.; Kitaura, F.; Fujii, H. A refined two-step oligoribonucleotide interference-PCR method for precise discrimination of nucleotide differences. Sci. Rep. 2018, 8, 17195. [Google Scholar] [CrossRef] [PubMed]
- Fujita, T.; Motooka, D.; Fujii, H. Target enrichment from a DNA mixture by oligoribonucleotide interference-PCR (ORNi-PCR). Boil. Methods Protoc. 2019, 4, bpz009. [Google Scholar] [CrossRef]
- Baba, K.; Fujita, T.; Tasaka, S.; Fujii, H. Simultaneous Detection of the T790M and L858R Mutations in the EGFR Gene by Oligoribonucleotide Interference-PCR. Int. J. Mol. Sci. 2019, 20, 4020. [Google Scholar] [CrossRef]
- Ben-Ezra, J.; Johnson, D.A.; Rossi, J.; Cook, N.; Wu, A. Effect of fixation on the amplification of nucleic acids from paraffin-embedded material by the polymerase chain reaction. J. Histochem. Cytochem. 1991, 39, 351–354. [Google Scholar] [CrossRef]
- Gillespie, J.W.; Best, C.J.M.; Bichsel, V.E.; Cole, K.A.; Greenhut, S.F.; Hewitt, S.M.; Ahram, M.; Gathright, Y.B.; Merino, M.J.; Strausberg, R.L.; et al. Evaluation of Non-Formalin Tissue Fixation for Molecular Profiling Studies. Am. J. Pathol. 2002, 160, 449–457. [Google Scholar] [CrossRef]
- Van Essen, H.F.; Verdaasdonk, M.A.M.; Elshof, S.M.; De Weger, R.A.; Van Diest, P.J. Alcohol based tissue fixation as an alternative for formaldehyde: Influence on immunohistochemistry. J. Clin. Pathol. 2010, 63, 1090–1094. [Google Scholar] [CrossRef] [PubMed]
- Guyard, A.; Boyez, A.; Pujals, A.; Robe, C.; Van Nhieu, J.T.; Allory, Y.; Moroch, J.; Georges, O.; Fournet, J.-C.; Zafrani, E.-S.; et al. DNA degrades during storage in formalin-fixed and paraffin-embedded tissue blocks. Virchows Arch. 2017, 471, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Raizis, A.M.; Schmitt, F.; Jost, J.P. A Bisulfite Method of 5-Methylcytosine Mapping That Minimizes Template Degradation. Anal. Biochem. 1995, 226, 161–166. [Google Scholar] [CrossRef]
- Grunau, C.; Clark, S.J.; Rosenthal, A. Bisulfite genomic sequencing: Systematic investigation of critical experimental parameters. Nucleic Acids Res. 2001, 29, e65. [Google Scholar] [CrossRef] [PubMed]
- Dempo, K.; Kottel, R.H.; Fishman, W.H. Demonstration of species difference of placental alkaline phosphatase isozymes in acetone-fixed paraffin-embedded tissues. J. Histochem. Cytochem. 1980, 28, 282–284. [Google Scholar] [CrossRef]
- Hølund, B.; Clausen, P.P.; Clemmensen, I. The influence of fixation and tissue preparation on the immunohistochemical demonstration of fibronectin in human tissue. Histochemistry 1981, 72, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Fan, Y.; Yamana, D.; Miura, T.; Nanashima, N.; Yamada, T.; Tsuchida, S. Glutathione S-transferase A4 is a positive marker for rat hepatic foci induced by clofibrate and genotoxic carcinogens. Cancer Sci. 2010, 101, 1093–1098. [Google Scholar] [CrossRef] [PubMed]
- Kumano, T.; Kimura, J.; Hayakari, M.; Yamazaki, T.; Sawamura, D.; Tsuchida, S. Polymorphism of the glutathione transferase subunit 3 in Sprague–Dawley rats involves a reactive cysteine residue. Biochem. J. 2000, 350, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Miura, M.; Tanigawa, C.; Fujii, Y.; Kaneko, S. Comparison of six commercially-available DNA polymerases for direct PCR. Rev. Inst. Med. Trop. São Paulo 2013, 55, 401–406. [Google Scholar] [CrossRef]
- Herman, J.G.; Graff, J.R.; Myöhänen, S.; Nelkin, B.D.; Baylin, S.B. Methylation-specific PCR: A novel PCR assay for methylation status of CpG islands. Proc. Natl. Acad. Sci. USA 1996, 93, 9821–9826. [Google Scholar] [CrossRef]
- Dobrovic, A. Methods for Analysis of DNA Methylation. Mol. Diagn. 2006, 149–160. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Hansen, L.L. PCR-Based Methods for Detecting Single-Locus DNA Methylation Biomarkers in Cancer Diagnostics, Prognostics, and Response to Treatment. Clin. Chem. 2009, 55, 1471–1483. [Google Scholar] [CrossRef] [PubMed]
- Myöhänen, S.K.; Baylin, S.B.; Herman, J.G. Hypermethylation can selectively silence individual p16ink4A alleles in neoplasia. Cancer Res. 1998, 58, 591–593. [Google Scholar] [PubMed]
- Cottrell, S.E.; Distler, J.; Goodman, N.S.; Mooney, S.H.; Kluth, A.; Olek, A.; Schwope, I.; Tetzner, R.; Ziebarth, H.; Berlin, K. A real-time PCR assay for DNA-methylation using methylation-specific blockers. Nucleic Acids Res. 2004, 32, e10. [Google Scholar] [CrossRef] [PubMed]
- Mauger, F.; Kernaleguen, M.; Lallemand, C.; Kristensen, V.N.; Deleuze, J.-F.; Tost, J. Enrichment of methylated molecules using enhanced-ice-co-amplification at lower denaturation temperature-PCR (E-ice-COLD-PCR) for the sensitive detection of disease-related hypermethylation. Epigenomics 2018, 10, 525–537. [Google Scholar] [CrossRef] [PubMed]
- Fiala, C.; Diamandis, E.P. New approaches for detecting cancer with circulating cell-free DNA. BMC Med. 2019, 17, 159. [Google Scholar] [CrossRef]
- Rand, K.; Qu, W.; Ho, T.; Clark, S.J.; Molloy, P. Conversion-specific detection of DNA methylation using real-time polymerase chain reaction (ConLight-MSP) to avoid false positives. Methods 2002, 27, 114–120. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shimizu, T.; Fujita, T.; Fukushi, S.; Horino, Y.; Fujii, H. Discrimination of CpG Methylation Status and Nucleotide Differences in Tissue Specimen DNA by Oligoribonucleotide Interference-PCR. Int. J. Mol. Sci. 2020, 21, 5119. https://doi.org/10.3390/ijms21145119
Shimizu T, Fujita T, Fukushi S, Horino Y, Fujii H. Discrimination of CpG Methylation Status and Nucleotide Differences in Tissue Specimen DNA by Oligoribonucleotide Interference-PCR. International Journal of Molecular Sciences. 2020; 21(14):5119. https://doi.org/10.3390/ijms21145119
Chicago/Turabian StyleShimizu, Takeshi, Toshitsugu Fujita, Sakie Fukushi, Yuri Horino, and Hodaka Fujii. 2020. "Discrimination of CpG Methylation Status and Nucleotide Differences in Tissue Specimen DNA by Oligoribonucleotide Interference-PCR" International Journal of Molecular Sciences 21, no. 14: 5119. https://doi.org/10.3390/ijms21145119
APA StyleShimizu, T., Fujita, T., Fukushi, S., Horino, Y., & Fujii, H. (2020). Discrimination of CpG Methylation Status and Nucleotide Differences in Tissue Specimen DNA by Oligoribonucleotide Interference-PCR. International Journal of Molecular Sciences, 21(14), 5119. https://doi.org/10.3390/ijms21145119




