Abstract
Aptamers have gained an increasing role as the molecular recognition element (MRE) in diagnostic assay development, since their first conception thirty years ago. The process to screen for nucleic acid-based binding elements (aptamers) was first described in 1990 by the Gold Laboratory. In the last three decades, many aptamers have been identified for a wide array of targets. In particular, the number of reports on investigating single-stranded DNA (ssDNA) aptamer applications in biosensing and diagnostic platforms have increased significantly in recent years. This review article summarizes the recent (2015 to 2020) progress of ssDNA aptamer research on bacteria, proteins, and lipids of bacterial origins that have implications for human infections. The basic process of aptamer selection, the principles of aptamer-based biosensors, and future perspectives will also be discussed.
1. Introduction
Bacterial infections in humans continue to be a significant challenge in both the community and hospital. In relation to bacterial infection is the increasing emergence of antibiotic resistance. The U.S. Centers for Disease Control and Prevention (CDC) reported more than 2.8 million cases of antibiotic-resistant infections were observed in the U.S. each year, and more than 35,000 cases had led to death in their latest antibiotic resistance threat report [1]. One of the solutions to decrease antibiotic resistance is through the timely and sensitive diagnosis of bacterial infections. This can help to guide antibiotics selection in infected patients, and thus reduce antibiotic resistance. Currently, the bacterial cultural test is a standard method to obtain a confirmative result of the bacterial species [2,3]. However, some bacteria genera, such as mycobacteria, are difficult to culture. In these cases, molecular diagnostics techniques, such as polymerase chain reaction (PCR), or enzyme-linked immunosorbent assay (ELISA), or other antibody-based assays, are used to facilitate the bacterial diagnosis [4,5].
In the last three decades, molecular recognition elements (MRE) that are alternatives to antibodies have been heavily investigated in the area of biosensing and diagnostics. One of the most promising MREs is nucleic acid-based binding elements due to high affinity and specificity. The first reported method of screening an RNA binding element was reported by the Gold Laboratory [6]. Shortly after, Szostak and Ellington reported a different RNA binding element and termed it aptamer [7]. Aptamers can bind to a variety of bacterial targets, including whole cells, polysaccharides, proteins, toxins, and even spores that are generated by bacterium [8]. Due to the versatile nature of aptamers, there has been an exponential increase in aptamer therapeutics and diagnostics research [9].
Because DNA molecules are generally more resistant to nuclease degradation than RNA molecules in vitro, single-stranded DNA (ssDNA) aptamers have received a higher amount of attention in their in vitro diagnostic and biosensing applications [10,11]. The process for identifying aptamers has been extensively reviewed previously [8,12,13,14]. In brief, aptamers are screened through the in vitro selection process, termed, systematic evolution of ligand by exponential enrichment, or commonly abbreviated as SELEX. It is an iterative process whereby a population of 1013 to 1015 different library molecules are subjected to repeated steps of incubation with the target molecule, separation, and amplification of bound library molecules. In the selection process of ssDNA aptamers, library molecules contain a random region flanked by two constant regions for PCR amplifications. The number of bases in the random region may vary, but usually, there is between 20–80 bases. The SELEX process has been modified to contain both positive (target), and negative (counter-targets) rounds, to direct the specificity of the library during the screening (Figure 1).
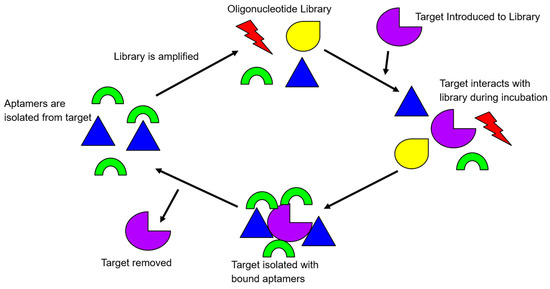
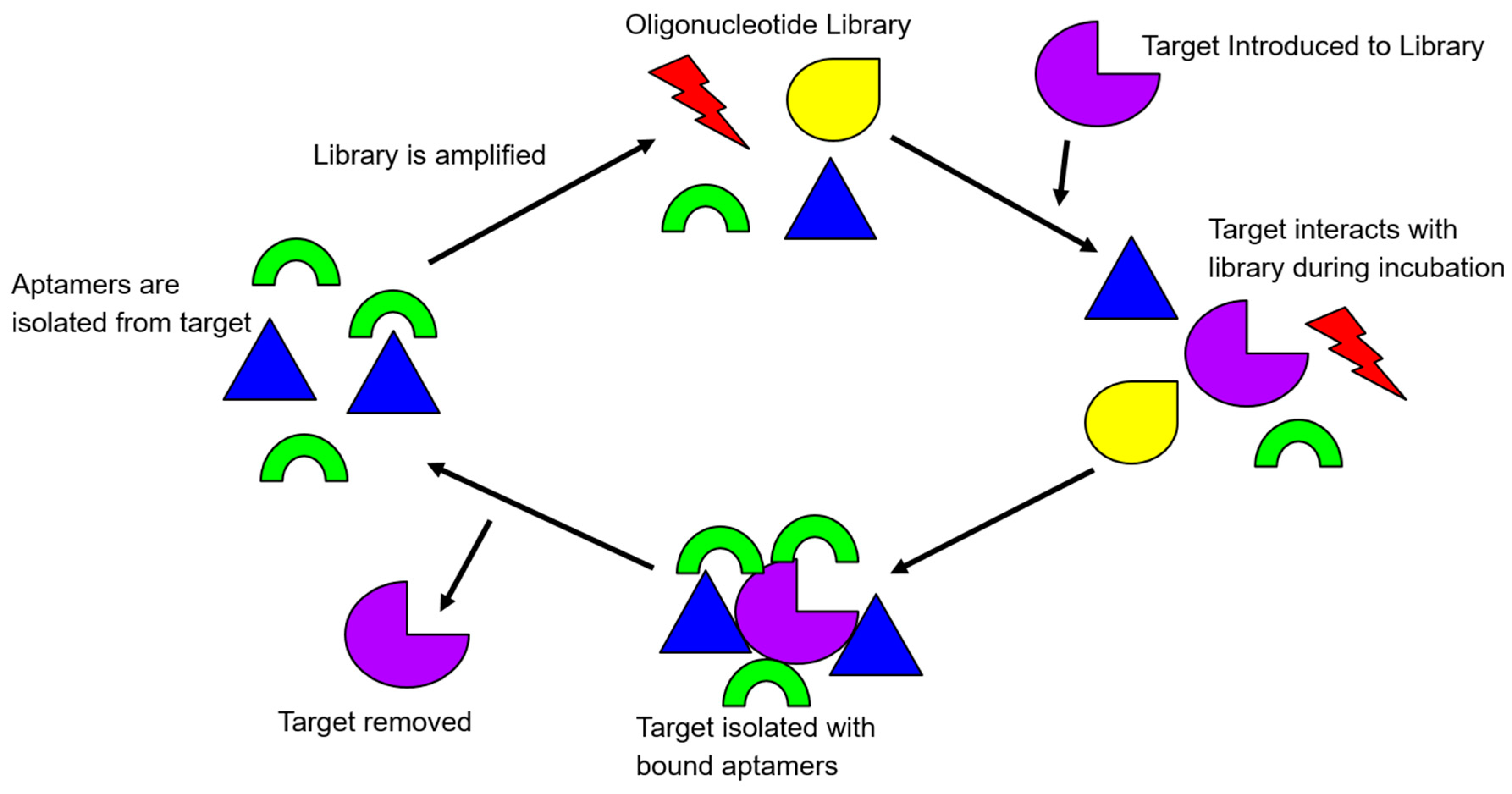
Figure 1.
An illustration of the rudimentary principle of the Systematic Evolution of Ligands by Exponential Enrichment (SELEX) process. Target-bound library molecules are subjected to repeated cycles of incubation, partitioning, and amplification, to increase the library affinity for the target.
To initiate a positive round of SELEX, the library of oligonucleotides is exposed to the target in question. After allowing the library to interact with the target, the target-bound complex is then washed of all non-binding library molecules. The library molecules that have effectively bound to the positive target are amplified via PCR. Subsequent rounds of SELEX start after the retrieval of the amplified library single-stranded DNA. Negative rounds of SELEX are usually integrated with the positive rounds of SELEX after several rounds of positive SELEX have been completed. This is to ensure the evolved library is robust enough to be screened by negative targets. A negative target is often similar to the target, “fooling” the less selective library oligonucleotides that have progressed through SELEX until that current point. The oligonucleotides that are less specific and bind to the negative target are, therefore, removed from the library. After completing approximately twelve to fifteen rounds of both positive and negative SELEX, a library can be deemed selective and specific, or whenever the number of target-bound library molecules plateaus [8].
Single-stranded DNA aptamers have many advantages over similar competitors. Firstly, they are more stable than RNA aptamers and still contain unique stem-loop structural variations [15]. Although antibodies have similar high affinity and specificity binding profiles when compared to single-stranded DNA aptamers, they are less thermally stable. Antibodies and other peptide-based binding elements are also irreversible after target recognition in diagnostics and biosensing applications. In addition, the process for selecting a single-stranded DNA aptamer can be performed independently of a living system, as opposed to antibodies. Lastly, single-stranded DNA aptamers can be chemically synthesized cost-effectively with minimal variation, adding yet another advantage to their usage [8].
Many variations can be made to single-stranded DNA aptamers to ease their biosensing and diagnostic applications, for example, the 3′ or 5′ ends of the aptamers may be modified to carry tags. These modifications allow the DNA aptamers to fluoresce or generate redox reactions upon target bindings [16]. This article focuses on reviewing recently (mid-2015 to early 2020) reported new single-stranded DNA aptamers and their usage within bacterial biosensing and diagnostics, that aim to facilitate the detection of bacterial infections in humans.
2. In Vitro Selection of ssDNA Aptamers
2.1. Overview of SELEX Methodology for Aptamer Specific for Bacterial Related Targets
The SELEX methodology has evolved over the last three decades from its original form, depending on the target for selection. Protein targets secreted from bacteria are commonly immobilized on magnetic beads or other solid platforms, such as 96-well plates for bound library molecule partitions. The libraries are often tagged with fluorescence dyes for monitoring library enrichment, such as the FluMag-SELEX [12]. Target-immobilization free techniques, such as the graphene oxide-based SELEX (GO-SELEX) are also useful in selection against small molecules, proteins, and viral particles [17,18,19]. However, when the targets are whole-cell bacteria, the Cell-SELEX, or Whole Cell-SELEX is the most commonly used technique. Frequently, the bacterial species is incubated with the library molecules, and followed by centrifugation partitioning of the bacterial pallet with the bound library [20]. Table 1 summarizes the recent aptamer selection reports on bacteria and bacterial related targets (Table 1). Studies discussing aptamer selection and their diagnostic applications will be summarized in a separate table.

Table 1.
Summary table of new single-stranded DNA (ssDNA) aptamers selected against bacteria and bacterial related targets.
2.2. Highlights on Recent ssDNA Aptamers Specific to Bacterial Origins
2.2.1. Staphylococcus Aureus and Its Related Proteins
Staphylococcus aureus is a common skin-dwelling bacteria. It acidifies the surface of the human body and can cause major infections when it enters into the internal human body. It is the cause of many infections, even including sepsis in severe cases. It is commonly seen as skin infections that cause swelling, warmth, and redness on the outer layers of skin. It has many components that make it toxic to humans in large doses, including secreted enterotoxins as well as cell surface proteins. Sepsis is often a very severe and life-threatening infection, and levels of bacteria need to be tested via blood samples. Several bacteria can cause this disease, but a very common culprit is S. aureus.
A study performed by Graziani et al. tested aptamers against multiple different bacteria that are known to cause sepsis [31]. The Kd estimated for the Staphylococcus aureus specific aptamer was 170.1 nM. The value was determined via SYBR Green real-time quantitative PCR. Another study identified aptamers specific to whole cells of Staphylococcus aureus after performing ten rounds of positive selection and three rounds with a negative target. The aptamers that were generated had a Kd between 34 and 128 nM [45].
Protein A is a common surface protein of S. aureus. Stoltenberg et al. used FluMag SELEX to generate a protein A binding aptamer. The reported Kd of the full length and truncated aptamers determined by surface plasmon resonance were in the micromolar range [43]. The group later utilized enzyme-linked oligonucleotide assays (ELONA) to obtain Kd values in the nanomolar range [46]. Noticeably, different binding experiment setups yield a wide range of affinity data. The same research group further studied this SELEX experiment, by analyzing the selected candidate pool with next-generation sequencing. They stated the originally obtained aptamer remained to be the best candidate for protein A [47].
S. aureus can also produce a group of enterotoxins that are responsible for staphylococcal food poisoning in many areas of the world. Several new aptamer selection studies were published for enterotoxin A, B, and C1.
A novel method of SELEX termed the Staggered Target SELEX (ST-SELEX) was used to identify an aptamer for enterotoxin A (SEA) [48]. The reported dissociation constant value was 7.44 ± 0.6 nM after ten rounds of selection. ST-SELEX involved separate steps of classical SELEX, and a second SELEX aimed to reduce the library binding to enterotoxins that were homologous to enterotoxin A. The same research group also performed a different classical SELEX experiment to identify a different aptamer specific to SEA, with a reported Kd of 8.5 ± 0.91 nM [49].
Enterotoxin B, secreted by staphylococcal cells, generates a large reaction from T cells within the body, and can, therefore, cause Staphylococcal toxic shock syndrome. Wong et al. identified an aptamer with a Kd of 64 nM and tested its antagonistic effect in diminishing the toxin effect [44].
Enterotoxin C1 is another secretory enterotoxin of S. aureus. Huang et al. identified an aptamer with a Kd value of 65.14 ± 11.64 nM after twelve rounds of SELEX, using magnetic separation techniques [50]. The group also developed a graphene oxide-based assay to quantify enterotoxin C1 in food samples.
2.2.2. Pseudomonas aeruginosa
Pseudomonas aeruginosa is also a bacteria that is commonly causing human infection in a variety of organs. This bacterium can infect the digestive tract, the urinary tract, the lungs, and other sites of elderly or immunocompromised individuals. Soundy et al. discovered an aptamer that bound to live P. aeruginosa cells, and with multiple reported dissociation constants in the low nanomolar range [36]. The candidates also demonstrated high specificity and were able to bind to both biofilm and planktonic growth of P. aeruginosa. The group reported the candidates lacked bacteriostatic and bactericidal activity, though it would be useful in diagnostic and biosensing applications.
2.2.3. Mycobacterium Species and Related Proteins
Mycobacterium tuberculosis is the causative bacteria for tuberculosis. While it is uncommon in developed countries, it remains a problem in many other areas of the world. Multiple aptamers have been developed that target strains of this bacterium or virulence factors for its havoc in humans. Mozioglu et al. isolated the H37Ra strain as the positive target in their SELEX study [33]. Both ultrafiltration and centrifugation methods were used to partition the bound and unbound library molecules. At the conclusion of the study, an aptamer with a Kd of 5.09 ± 1.43 nM was obtained.
Zhang et al. selected an aptamer specific for the H37Rv strain M. tuberculosis with a Kd of 37 ± 4 nM [51]. The research group also developed a multichannel series piezoelectric quartz crystal sensor using the selected aptamer, single-walled carbon nanotube, and gold electrode, to achieve a detection limit of 100 CFU/mL in seventy minutes.
Aimaiti et al. identified an aptamer for the H37Rv strain, with a Kd of 12.02 nM after ten rounds of selection [52]. The group also developed a sandwich enzyme-linked immunosorbent assay using a combination of five aptamers for capturing and detecting the specific strain.
In addition to selecting aptamers specific for the whole cell M. tuberculosis, there has been an aptamer developed for a particular component of the bacterium, the mannose-capped lipoarabinomannan (ManLAM) on the outer surface. This lipoglycan serves a vital role in cell to cell communication. Tang et al. identified an aptamer specific for T. tuberculosis ManLam lipoglycan, with a dissociation constant of 668 ± 159 nM. The author also developed an enzyme-linked oligonucleotide assay (ELONA) to detect the ManLAM antigen in patient serum and sputum samples with active TB infections.
Sun et al. selected an aptamer that bound to ManLAM molecules of Mycobacterium bovis, or commonly termed bacillus Calmette–Guerin (BCG). The reported Kd of this aptamer was 8.59 ± 1.23 nM. The author stated the potential use of the aptamer as an immune enhancer for BCG.
M. tuberculosis is also capable of secreting proteins to further the course of human infection, such as the MPT64 secretory protein. Following ten cycles of SELEX, an aptamer with a Kd of 8.92 nM was identified [32]. The identified aptamer was also incorporated into an ELONA assay for the specific and sensitive detection of MPT64 in clinical sputum samples.
Another study was conducted to detect the Mycobacterium Ag85A secretory complex. An aptamer was selected using magnetic beads [53]. The reported Kd was 63 nM. The author also developed a rapid fluorescent assay with the aptamer and graphene oxide. The reported limit of detection was 1.5 nM.
2.2.4. Escherichia coli
Pathogenic strains of Escherichia coli continue to generate interest in many aptamer research groups.
Marton et al. identified four aptamers specific to a laboratory strain of E. coli [26]. The reported Kd values ranged from 11.97 nM to 161 nM. The author stated that one of the twelve aptamer candidates was highly specific to E. coli and was able to bind to meningitis/sepsis associated E. coli (MNEC). They indicated the potential for the candidate aptamer to be used in diagnosing MNEC associated infections.
Masoum et al. isolated an aptamer specific for E. coli O157:H7, a pathogenic strain that causes foodborne illnesses in humans [24]. Following nine rounds of positive selection and a final tenth round of negative selection, the author reported a highly specific aptamer with a Kd of 107.6 ± 67.8 pM. The author stated the potential for using it in diagnostic and biosensing of bacteria in food.
Zou et al. further examined the possibility of identifying an aptamer specific for different stages of the O157:H7 strain [25]. The reported aptamer had a Kd of 9.04 ± 2.80 nM. The author concluded lipopolysaccharide (LPS) could potentially be the molecular target of the aptamer.
Yu et al. again selected an aptamer for the O157:H7 strain E. coli [54]. After nineteen total rounds of SELEX with six rounds of negative selection, the reported aptamer had a dissociation constant of 10.3 nM. The author also developed a quartz crystal microbalance aptasensor to detect the strain. The limit of detection was 1.46 × 103 CFU/mL.
A similar strain, O78:K80:H11, was also chosen for the aptamer selection experiment. Kaur et al. utilized a total of twelve rounds of SELEX, with rounds one through five consisting of positive selections and rounds six through eleven consisting of negative selections [55]. The reported dissociation constant was approximately 14 nM. The aptamer was also incorporated into a label-free impedimetric aptasensor for the sensitive detection of the strain in liquid samples. The reported limit of detection was about 10 CFU/mL.
Another E. coli strain, DH5α, was examined as a target by Renders et al. [27]. The author obtained a sensitive aptamer, with a Kd of 27.4 ± 18.7 nM, after performing twelve rounds of whole cell SELEX.
Song et al. took a different approach to identify aptamers that were specific to not only E. coli, but to other disease-causing gram-positive and gram-negative bacteria [38]. This includes Enterobacter aerogenes, Klebsiella pneumoniae, Citrobacter freundii, Bacillus subtilis, and Staphylococcus epidermidis. This was performed via a novel SELEX, termed the sequential toggle cell SELEX (STC-SELEX). In this SELEX methodology, one of the six bacterial groups was incubated with the library at once. After the library was exposed to one bacterial group, it was allowed to be exposed to the next, and so on. The isolated aptamers had reported dissociation constants in the low nanomolar range. They were also only specific to the bacteria species that were included in the selection.
2.2.5. Streptococcus Species
Streptococcus pyogenes can overgrow in the throat, particularly on the tonsils. These symptoms are accompanied by soreness in the throat, as well as a fever, and is commonly called strep throat. Hamula et al. subjected two sets of libraries to look for aptamers specific for S. pyogenes. Several aptamers were selected, with dissociation constants ranging from 7 nM to 71 nM [28].
Streptococcus mutans is an oral infective agent in humans. A specific aptamer with a Kd of 69.45 ± 38.53 nM was identified after nine rounds of whole cell SELEX. This aptamer was highly specific to cariogenic clinical S. mutans (Streptococcus mutans) strains [30].
Group A streptococcus expresses serotype M3 on its surface. This surface protein generates a more lethal infection for human hosts, and therefore this is a more severe pathogenic strain of the species. Using this specific strain of streptococcus, Alfavian et al. identified an aptamer with a Kd value of 7.47 ± 1.72 pM [29].
Streptococcus pneumoniae can cause fever and chest congestion, as well as airway irritation, swelling, and cough. The associated infection can be fatal in elderly and immunocompromised patients. Bayrac et al. reported several aptamers capable of binding specifically to this species of streptococcus [56]. After ten rounds of whole cell SELEX, three aptamers were selected with the following Kd values, Lyd−1 = 844.7 + 123.6 nM Lyd−2 = 1984.8 + 347.5 nM Lyd−3 = 661.8 + 111.3 nM. A graphene oxide based fluorescent assay was developed using the Lyd−3 aptamer. The reported limit of detection was 15 CFU/mL.
2.2.6. Bacillus Anthracis Virulence Factors
Anthrax, although rare, can have dire consequences of infection. Biondi et al. reported using an artificially expanded genetic information system (AEGIS) to perform a SELEX experiment with two additional artificial nucleotides [40]. The author identified an aptamer with high affinity and specificity toward the protective antigen (PA63) of Bacillus anthracis. After the completion of fourteen rounds of selection, the author reported a specific aptamer with a dissociation constant of 35 nM.
Lahousse et al. identified an aptamer against a different virulence factor, the lethal factor (LF) of B. anthracis (Bacillus anthracis) [39]. The reported Kd was 11 ± 2.7 nM. The aptamer selected also had a potent inhibition on B. anthracis. The author expressed the potential of using the aptamer as a gateway to effective treatments in the future.
2.2.7. Vibrio Species
Vibrio species bacteria reside mostly in oceans and can cause dangerous food borne illnesses in humans. Vibrio parahemolyticus is a common foodborne pathogen. Song et al. utilized a modified graphene oxide SELEX, coupled with isothermal rolling circle amplification to identify an aptamer specific for V. parahemolyticus (Vibrio parahemolyticus) [22]. The best aptamer had a reported Kd of 10.3 ± 2.5 nM. Specificity of this aptamer was tested via flow cytometry. Both negative and positive rounds were utilized to identify the aptamers, with only six rounds of selection being performed total.
Vibrio alginolyticus is capable of infecting both marine lives as well as humans. Yu et al. identified two aptamers specific for the species [21]. The reported dissociation constants were 14.31 ± 4.26 nM and 90 ± 13.51 nM. The author noted that the aptamers did not have cytotoxic effects both in vitro and in vivo and could be used as a probe for sensitive detection of V. alginolyticus.
Yan et al. selected an aptamer specific for Vibrio vulnificus [23]. After ten positive and three negative rounds of selection, an aptamer was identified. The reported Kd was 26.8 + 5.3 nM. The author stated the aptamer could specifically detect the pathogen at a range between 8 to 2.0 × 108 CFU/mL.
2.2.8. Helicobacter pylori
Another bacterial species that normally resides within humans is Helicobacter pylori. It usually resides within the stomach of humans. It can cause infection and symptoms in humans when there is extreme growth. Yan et al. selected a specific aptamer against the recombinant surface protein of H. pylori [42]. The reported dissociation constant was 26.48 ± 5.72 nM. Although the aptamer was selected against a protein target, it showed specific binding to H. pylori cells.
3. Diagnostic and Biosensing Applications of ssDNA Aptamer for Bacterial Infection
3.1. Overview of Common Detection Principles
In general, the majority of the investigational ssDNA aptamer-based diagnostic and biosensing platforms can be divided into two broad categories, (1) optical and (2) electrical/electrochemical. The former category includes fluorescence, chemiluminescence, and colorimetric and surface plasmon resonance detections. The latter category most commonly includes amperometric and impedimetric sensors.
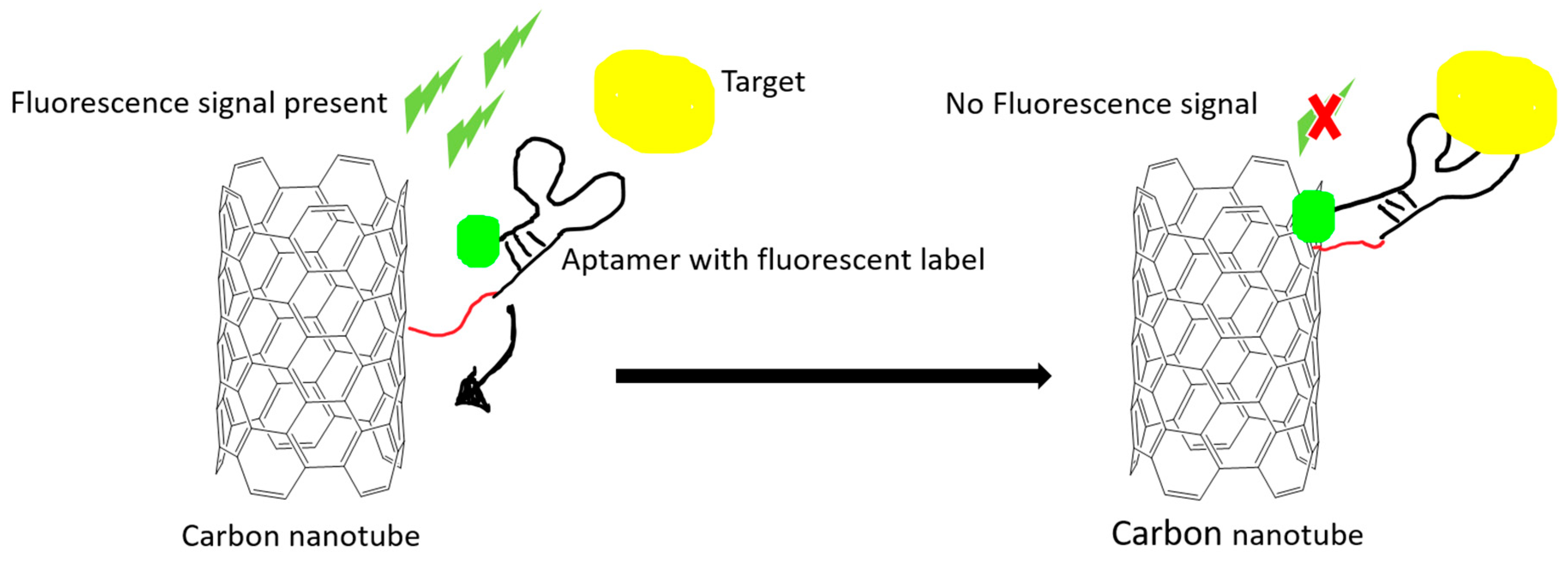
Optical proof-of-principle detections are relatively easy to develop and operate. The aptamer is often conjugated with various fluorescence molecules, such as 6-carboxy-fluorescein (FAM), cyanine, and quantum dots. Using quantum dots has the advantage of overcoming photobleaching effects that are common in fluorescent dyes. Förster or fluorescence resonance energy transfer (FRET) is a special phenomenon that relies on energy transfer between two light sensitive molecules in a very short distance. Carbon nanomaterials have been utilized as a signal quencher in FRET-based aptamer sensors (Figure 2).
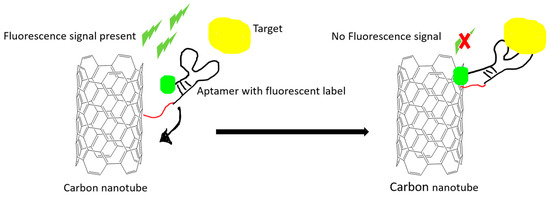
Figure 2.
An example illustration of how carbon nanomaterials are used as a fluorescence quencher to turn off the signal. The fluorescence signal is quenched after the target-induced conformational change in the aptamer. The quenching effect is due to the Förster resonance energy transfer (FRET) effect.
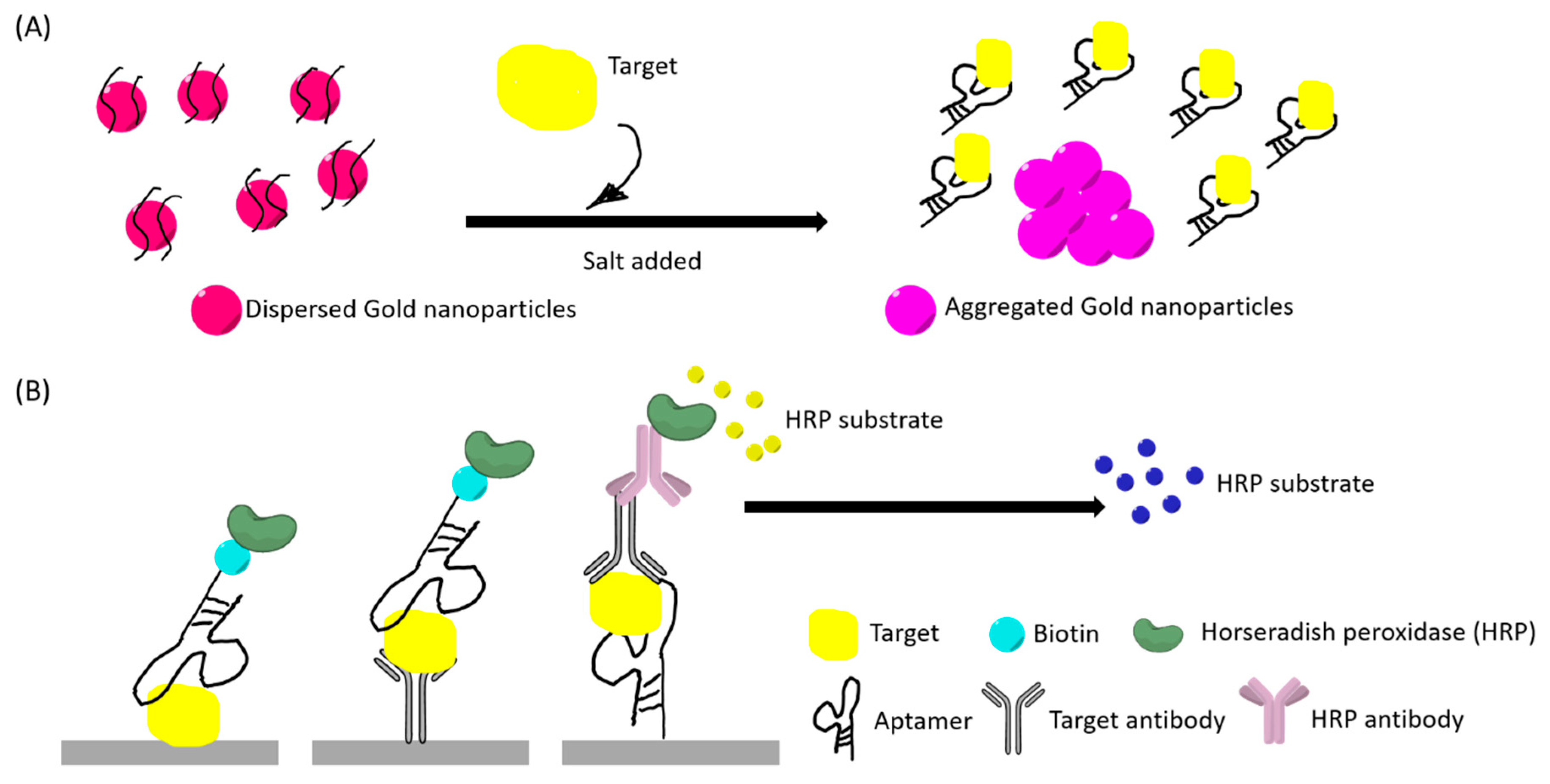
The colorimetric detection method often utilizes gold nanoparticles (AuNP) for rapid detection. This method is generally low-cost and user-friendly. Salt addition can induce changes in the aggregated size of the nanoparticles and lead to a red to purple color shift that is visible to the naked eye (Figure 3A). These factors have led to many studies developing aptamer-based colorimetric assays. Aptamers can also be incorporated into peroxidase enzyme-linked assays, capable of generating color changes that are readable as absorbance values (Figure 3B).
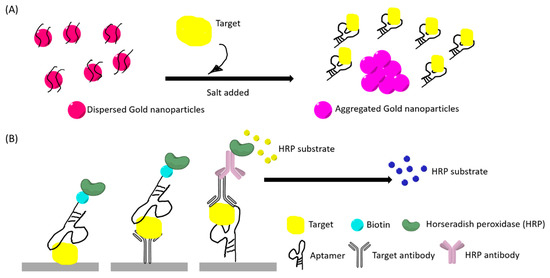
Figure 3.
(A) An example illustration of a “red to purple” colorimetric sensor using gold nanoparticles (AuNPs). The left had side illustrates the initial stage where AuNPs are dispersed by aptamer-coating, thus having a red appearance. AuNPs aggregated after target bind to the aptamer and salt addition. (B) An example illustration of aptamer enzyme-linked sorbent assays. Primary antibody for the target and secondary antibody can be used in different sandwich assays. The color changes in both (A) and (B) can be observed with naked eyes and quantified by optical instruments.
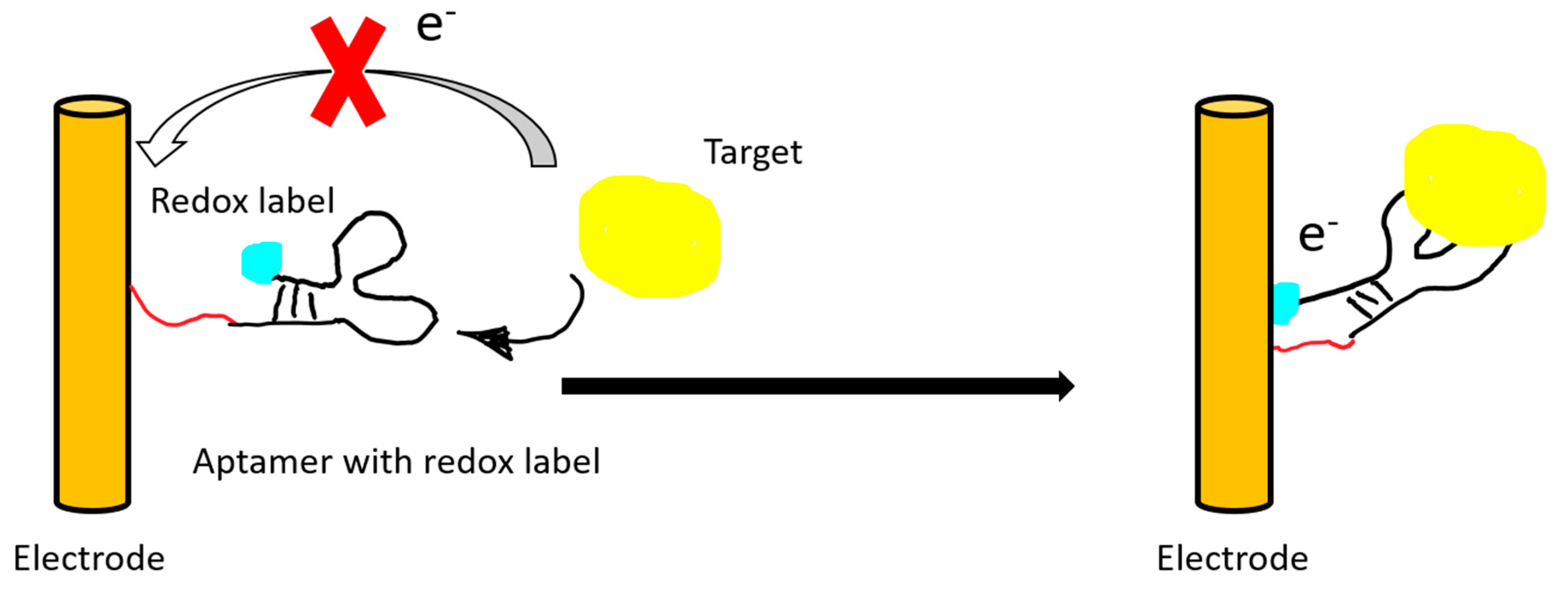
Electric or electrochemical aptasensors can be label-free or labeled. In a labeled aptasensor, a redox tag on the aptamer is needed for transferring the electron upon target binding. The electron transfer can then be translated into an amperometric or an impedimetric reading (Figure 4).
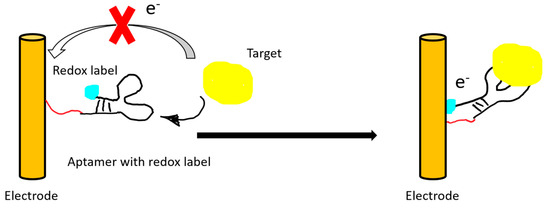
Figure 4.
An illustration of a “signal on” electrochemical sensor. The electrical signal of the redox label is recorded after target binding and subsequent conformational change of the aptamer.
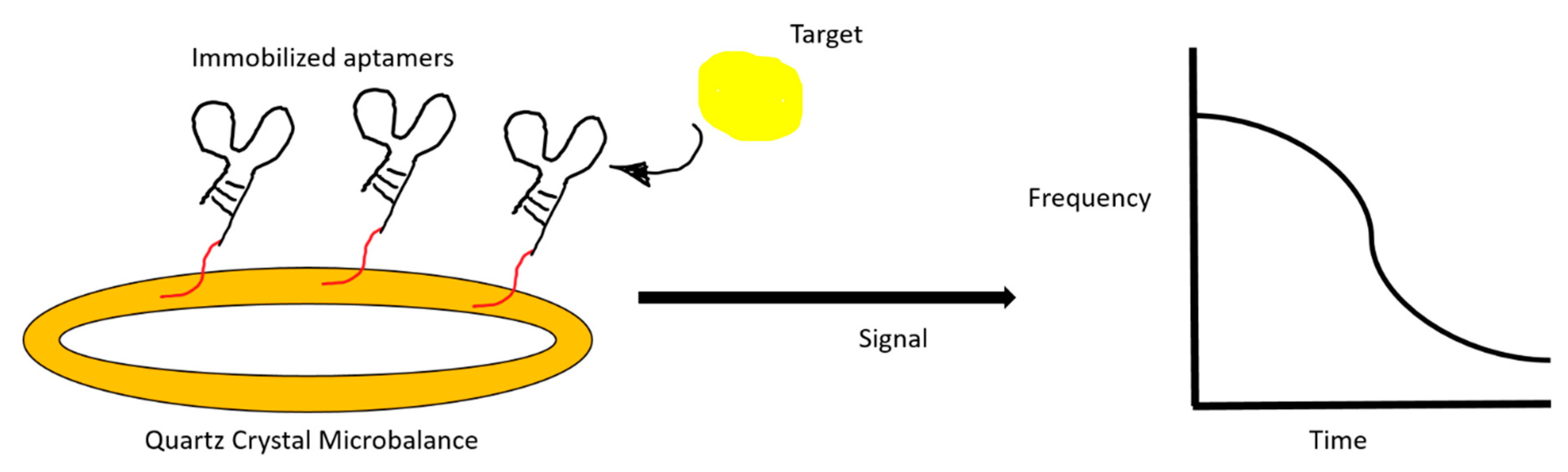
A label-free aptasensor can utilize mass-sensitive techniques, such as quartz crystal microbalance (QCM) and micromechanical cantilever. These piezoelectric biosensors can generate a resonance frequency change after target binding (Figure 5).
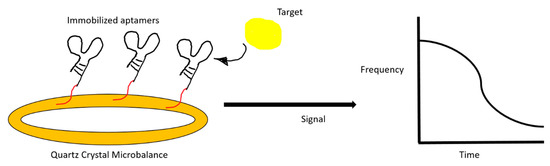
Figure 5.
An illustration of a quartz crystal microbalance aptasensor. The change in resonance frequency after target binding is registered on a monitor.
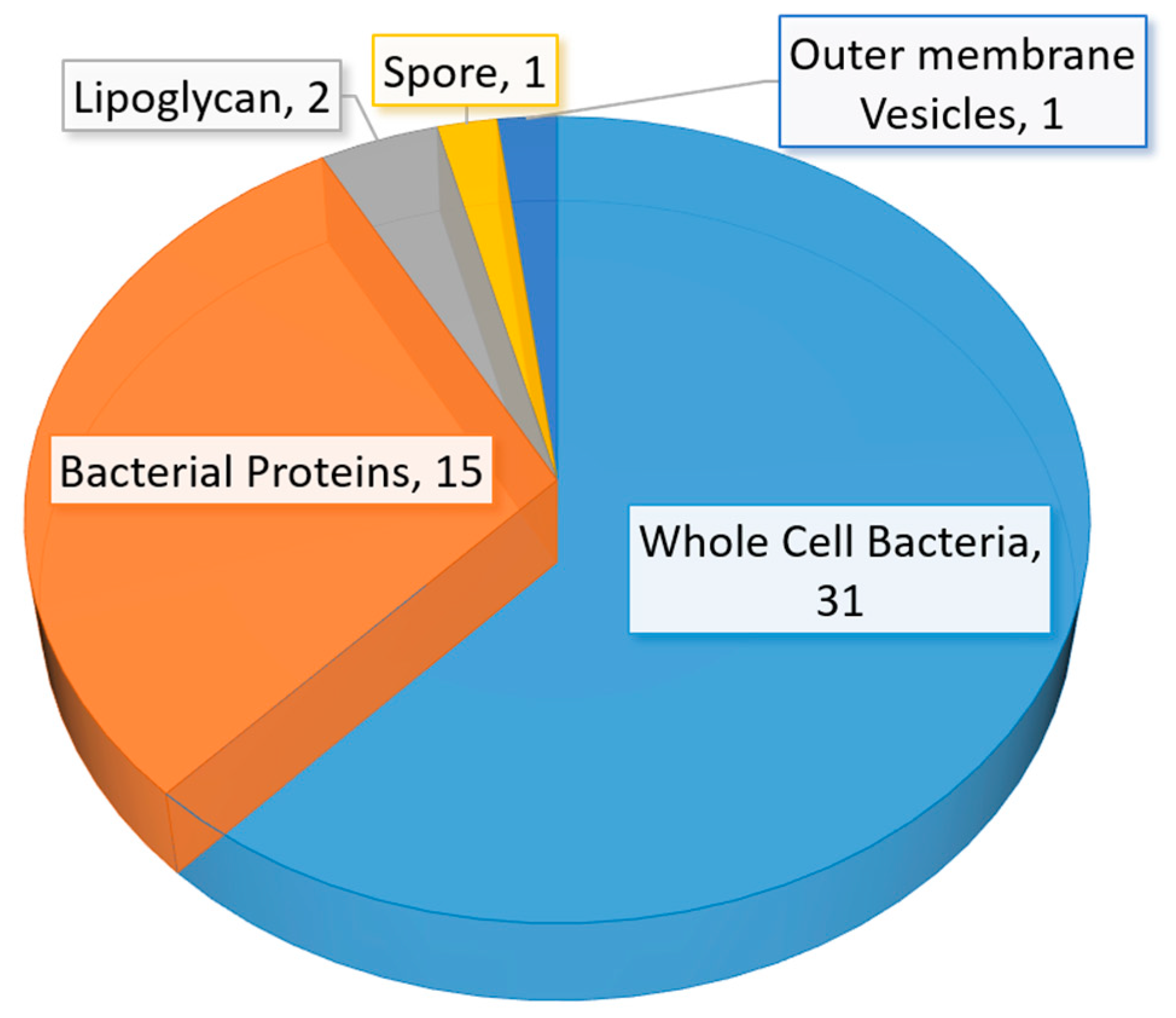
A significant amount of research related to aptamer selections and diagnostic applications were published between mid-2015 to early 2020. The number of studies in aptamer-based diagnostic and biosensor development is also beyond the capability of a full discussion in this review. Thus, we have elected to discuss selected studies based on the area or location of the infection in humans. The summary of recent (mid-2015 to early 2020) bacterial-related aptamer selection and diagnostic application studies are listed in Table 2 (Table 2). The total number of newly identified ssDNA aptamers for bacteria and bacterial components are illustrated in a pie chart (Figure 6). Likewise, bacterial diagnostic sensor studies in this period are summarized in Table 3. Aptasensors designed solely for pathogen detection in food and drink samples are excluded in this table (Table 3). The types of aptasensor and the categories of the detected target are illustrated in Figure 7.

Table 2.
Summary table of new ssDNA aptamers selected against bacteria and bacterial related targets and their diagnostic and biosensing applications.
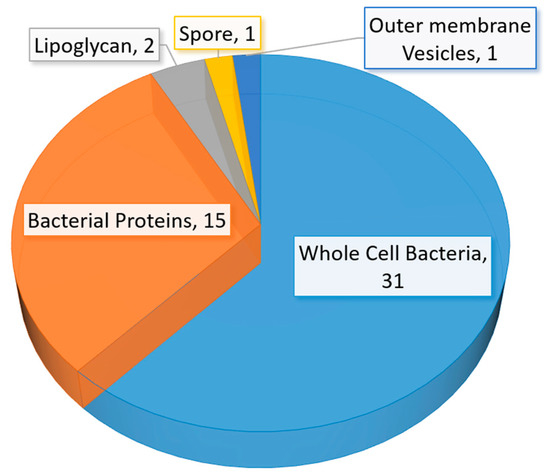

Table 3.
Summary table of new ssDNA aptamer-based biosensors intended for the diagnostic of bacterial infection in humans.
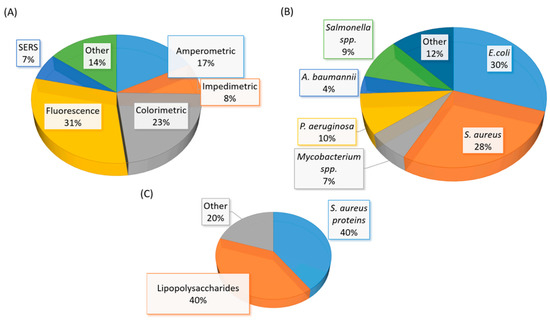
Figure 7.
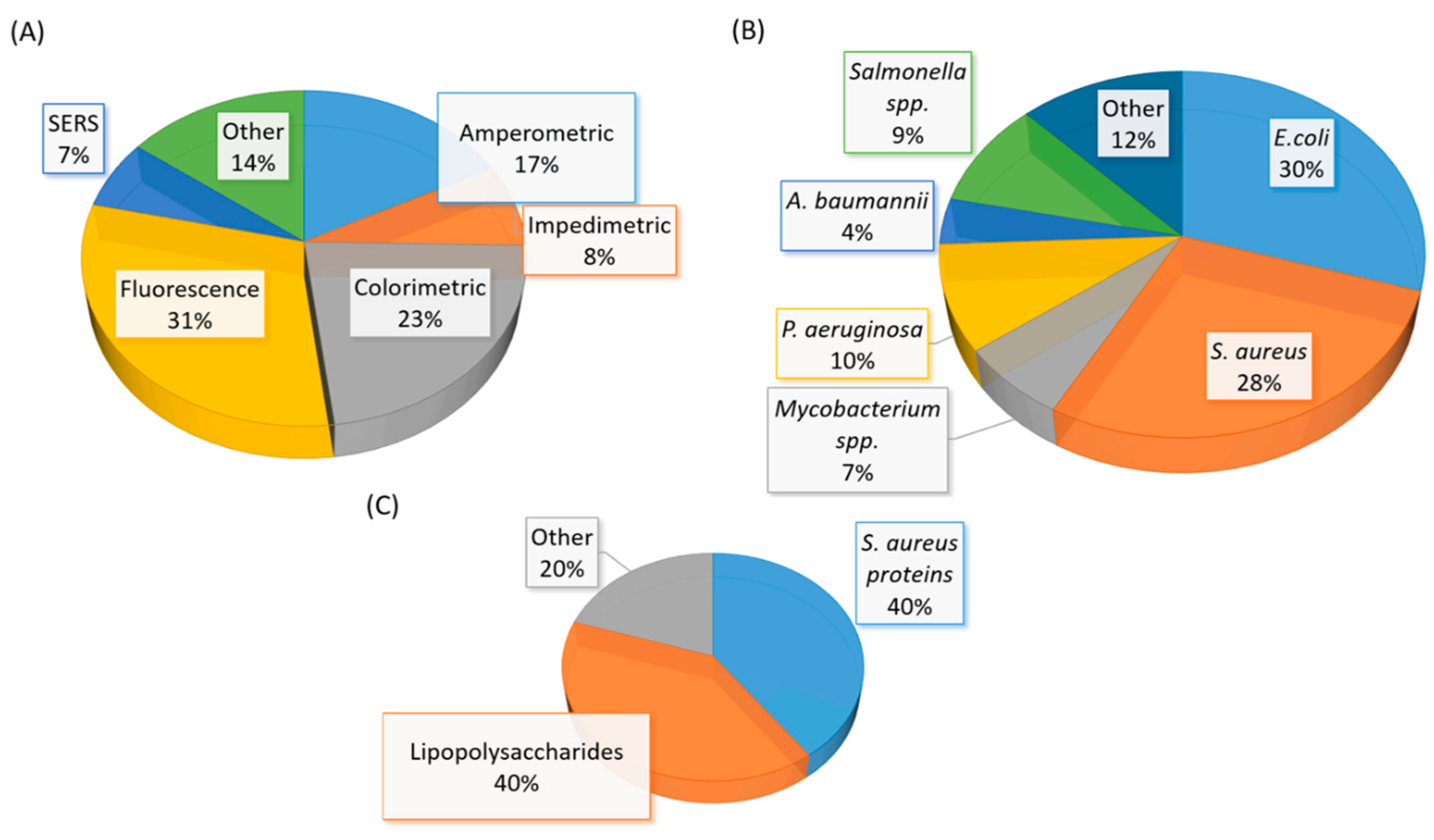
(A) A graphical representation of the type of aptasensor summarized in Table 3. (B) A graphical representation of the whole-cell bacterial species detected in Table 3. (C) A graphical representation of the types of bacterial component detected in Table 3. The total number of articles summarized in Table 3 is fifty-eight. Note: Eight studies reported the detection of more than one bacterial species (spp.)
3.2. Highlights of ssDNA Aptamer-Based Diagnostics for Bacterial Skin Infections
Bacterial skin infections in the hospital are extremely prevalent and can be easily transmissible from patient to patient, or even patients to hospital staff. A common culprit of skin infections is Staphylococcus aureus. This bacteria is the most common cause of skin infections, as it is normally present on the skin, but if it overgrows, it can cause infections that can even cause sepsis [146]. There was a reported aptasensor that could detect S. aureus very sensitively at 1.5 CFU/mL [79]. This aptasensor was based upon the surface-enhanced Raman scattering principle or SERS. The aptamer changed its configuration to optimally bind to the S. aureus bacteria, and then this complex acted as a template for gold nanoparticles to bind to. An infrared light was utilized to beam through the sample. A unique shift in vibrational energy was generated in the presence of gold-bound aptamer and bacteria complex, therefore confirming the presence of the bacteria.
S. aureus infection can be so detrimental and difficult to treat due to a commonly secreted enterotoxin [146]. An aptasensor specific for staphylococcus enterotoxin B was created utilizing the multicolor time-resolved fluorescence method [139]. It utilized graphene oxide as a resonance acceptor. The presence of the target bacteria caused a signal quench, thus turning off the sensor. The reported toxin concentration detection limit was at 1 ng/mL.
Methicillin-resistant Staphylococcus aureus (MRSA) infections in humans are often resistant to several antibiotic treatments and can require a longer course of medication [147]. A study reported detecting this bacterium in a microfluidic system and achieved a limit of detection at 450 CFU/mL [117]. In order to recognize this bacteria target, a nitrocellulose membrane attached to a glass slide was coated with the aptamer, and the bacteria were then incubated on the membrane. Unbound materials were washed away, and the captured bacteria that remained on the membrane received another round of aptamer exposure and additional rounds of washing for unbound materials. The secondary aptamer was labeled with biotin, and after washing, the tetramethylbenzidine substrate reacted with the horseradish peroxidase. The change in color was observed, and the absorbance level was measured to approximate the bacterial concentration [117].
3.3. Highlights of ssDNA Aptamer-Based Diagnostics for Bacterial Respiratory System Infections
It is generally more difficult to infect the respiratory tract, because of an array of defense mechanisms present along the tract. However, bacteria such as Mycobacterium tuberculosis can cause lung infection in humans. The thick sputum can harbor the bacteria and related virulence factors [148]. An ultrasensitive electrochemical aptasensor was developed recently [73]. It utilized gold nanoparticles decorated fullerene-doped polyaniline as the redox label to generate an amplified electrochemical signal upon binding to the M. tuberculosis antigen, MPT64. The reported limit of detection was at 20 fg/mL. It also demonstrated specificity and sensitivity for MPT64 in clinical serum samples [73].
Acinetobacter baumanii is another prevalent bacterium that continues to cause respiratory tract infections. It can develop and frequently be transmitted between hospitalized patients. This bacterium can originate in the skin, soil, water, or food, and can rapidly be transferred to generate multidrug-resistant strains [149]. An enzyme-linked aptamer sorbent assay (ELASA) was utilized to measure the fluorescence signal upon aptamer binding to the bacteria [62]. The assay showed good specificity over other commonly observed bacteria in healthcare settings, such as P. aeruginosa, and E. coli. The sandwich ELASA assay demonstrated a sensitivity of 95.47% on clinical isolates [62].
Perhaps one of the most commonly seen respiratory infection is caused by Streptococcus pneumoniae. It mainly infects people with compromised lung function or the elderly. It has proven to have a high rate of mortality [150]. A fluorescent assay was developed using graphene oxide. The aptamers showed selectivity against E. coli, S. pyogenes, and S. aureus. The limit of detection for this apparatus was as low as 15 CFU/mL [56].
3.4. Highlights of ssDNA Aptamer-Based Diagnostics for Other Infections
In addition to the infection types listed above, bacteria continue to cause a plethora of symptoms in human hosts. Not simply limited to one organ system, these bacteria can cause a unique array of symptoms, and this can, therefore, make them even more deadly.
For example, Pseudomonas aeruginosa is a bacteria that can be found in soil, food, and water, and can cause a wide array of human symptoms like urinary tract infections, pneumonia, lung disease, and cystic fibrosis [151]. Because of its broad scope of symptoms, this bacterium is particularly troublesome for their human hosts and often results in more severe infections. An aptasensor was created for this bacterium that was unique because it utilized two different strategies, Surface-enhanced Raman spectroscopy (SERS), and colorimetry [113]. The aptamer was bound to gold nanoparticles, with the larger gold nanoparticles and aptamer complexes acting for colorimetry, and the smaller complexes acting as signalers for SERS. When the bacteria were not present, the two complexes bound together, and no signal was transmitted. This aptasensor could detect as low as 20 CFU/mL of the bacteria in solution [113].
Another non-traditional bacteria that can infect humans is Bacillus anthracis. Its spores spread its ability to transmit infection, and therefore its spores were the target of sensitive detection. This bacterium is known for its ability to be a biological weapon and can cause general signs of infection, such as fever, shortness of breath, sweating, nausea, and vomiting [152]. These spores were known as B. cereus spores, and the limit of detection of a reported impedimetric aptasensor was at 3 × 103 CFU/mL [135]. The aptamer was first immobilized on gold electrodes, with a redox label, Fe(CN6), immersed in the environment. When the target was introduced into thesystem, the Fe(CN6) was displaced from the aptamer, and the decrease of resistance was measured [135].
An additional bacteria species that can uniquely cause disease in humans is Vibrio alginolyticus. It is resistant to salt and can thrive in oceans. It can cause infections in open wounds and ears in humans [153]. Zhao et al. described a method of sensing this bacteria using magnetic beads [141]. With a “capture DNA” fixed on the surface of the magnetic beads, the aptamer combined with the complex and bound to the target when it was present. The electrode sensed the aptamer detached from the beads as a result of target-binding. It measured the electrostatic interactions with a limit of detection of 10 CFU/mL [141].
4. Conclusions and Future Perspective
Pertinent and emerging new diseases will require precise and rapid diagnosis to facilitate proper treatment. Since the first description of the aptamer technology three decades ago, aptamers have shown to be very adaptive in binding to a wide range of targets. This feature has allowed them to be investigated as the recognition and binding element in diagnostic and biosensing applications. A major advantage of aptamer technologies resides within the selection process. It can identify new binding elements for previously unknown sites or motifs on the bacterium. Newly discovered aptamers can help to map novel binding sites that could have therapeutic potentials, in addition to diagnostic implications. Aptamers specific for bacterial virulence factors have shown to demonstrate neutralization abilities [44,49]. These aptamers could have the potential to be applied as a combined therapeutic and diagnostic agent, or a theranostic agent.
Although a vast amount of research has been conducted in the aptamer field in recent years, there is still a lack of commercially available aptasensors for infectious disease diagnosis. One of the challenges that is unique in detecting living organisms lies in the aptamer specificity for the pathogenic strains of the bacterium. Many aptamer selection studies had utilized bacteria from laboratory cultures. These laboratory strains may be different than clinical isolates from patients, and thus could lead to potential false negatives or false positives. In addition to factors related to the aptamer binding profile, sensor setup and detection principles are also crucial to sensitive and specific detection of the target virulence factor or bacterium. Clinical samples such as whole blood, plasma, saliva, and sputum are complex biological matrices. Enzyme degradation, non-specific binding to serum proteins, and non-pathogenic normal flora are all likely to interfere with the binding aptamers. The aptamer selection process must be designed to overcome potential interferences from components in these biological fluid samples. The amount of sample preparation is also a factor that can dictate the type of sensing principle in aptasensors. Whole blood samples are not ideal for colorimetric sensing because of the natural color of hemoglobin. Serum samples containing albumin can also interfere with fluorescence signals. In comparison, electrochemical aptasensors have an advantage in overcoming these optical interferences.
Recently, Wan et al. reported using an aptamer cocktail consisting of multiple aptamers with varying motifs, to detect Mycoplasma in cell culture [154]. This strategy could be useful in the detection of bacteria in clinical samples. The selection process often generates multiple aptamer candidates. Additional motif analysis and experiments could be performed in most bacterial whole-cell SELEX, as there is a high potential for multiple interaction sites between the relatively large bacterial cell and the aptamer library molecules.
It is evident that aptamer-based biosensors or aptasensors require a multidisciplinary collaboration for success. The biologists, chemists, and engineers all play a crucial role in integrating the aptamer technology. As we are moving into the fourth decade of aptamer research, it is reasonable to believe that the limitless future of aptamers remain to be discovered.
Author Contributions
Conceptualization, K.L.H.; writing—original draft preparation, N.E.T.; writing—review and editing, K.L.H.; visualization, N.E.T., K.L.H.; supervision, K.L.H.; project administration, K.L.H.; funding acquisition, K.L.H. Both authors have read and agreed to the published version of the manuscript.
Funding
This research and APC were funded by Wilkes University Research and Scholarship Fund and the Department of Pharmaceutical Sciences at Wilkes University. N.E.T. was in-part funded by Summer Mentoring Grant 2019.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| MRE | Molecular recognition elements |
| LOD | Limit of detection |
| MRSA | Methicillin-resistant Staphylococcus aureus |
| LPS | Lipopolysaccharides |
| SERS | Surface-enhanced Raman scattering |
| SPR | Surface plasmonresonance |
| FRET | Förster resonance energy transfer |
| SELEX | Systematic evolution of ligand by exponential enrichment |
| GO | Graphene oxide |
| AuNP | Gold nanoparticles |
| CFU | Colony forming unit |
| PCR | Polymerase chain reaction |
References
- Antibiotic Resistance Threats in the United States; Centres for Disease Control and Prevention: U.S. Department of Health and Human Services: Washington, DC, USA, 2013.
- Brouwer, M.C.; Tunkel, A.R.; van de Beek, D. Epidemiology, diagnosis, and antimicrobial treatment of acute bacterial meningitis. Clin. Microbiol. Rev. 2010, 23, 467–492. [Google Scholar] [CrossRef] [PubMed]
- Pierre, D.M.; Baron, J.; Yu, V.L.; Stout, J.E. Diagnostic testing for Legionnaires’ disease. Ann. Clin. Microbiol. Antimicrob. 2017, 16, 59. [Google Scholar] [CrossRef]
- Başpınar, E.Ö.; Dayan, S.; Bekçibaşı, M.; Tekin, R.; Ayaz, C.; Deveci, Ö.; Hoşoğlu, S. Comparison of culture and PCR methods in the diagnosis of bacterial meningitis. Braz J. Microbiol. 2017, 48, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Ustinova, V.V.; Smirnova, T.G.; Sochivko, D.G.; Varlamov, D.A.; Larionova, E.E.; Andreevskaya, S.N.; Andrievskaya, I.Y.; Kiseleva, E.A.; Chernousova, L.N.; Ergeshov, A. New assay to diagnose and differentiate between Mycobacterium tuberculosis complex and nontuberculous mycobacteria. Tuberculosis 2019, 114, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Hong, K.L.; Sooter, L.J. Single-Stranded DNA Aptamers against Pathogens and Toxins: Identification and Biosensing Applications. Biomed. Res. Int. 2015, 2015, 419318. [Google Scholar] [CrossRef]
- Ali, M.H.; Elsherbiny, M.E.; Emara, M. Updates on Aptamer Research. Int. J. Mol. Sci. 2019, 20, 2511. [Google Scholar] [CrossRef]
- Li, Z.; Mohamed, M.A.; Vinu Mohan, A.M.; Zhu, Z.; Sharma, V.; Mishra, G.K.; Mishra, R.K. Application of Electrochemical Aptasensors toward Clinical Diagnostics, Food, and Environmental Monitoring: Review. Sensors 2019, 19, 5435. [Google Scholar] [CrossRef]
- Hong, K.L.; Kaur Brar, S.; Hegde, K.; Pachapur, V.L. An overview of DNA/RNA-based monitoring tools and biosensors: Benefits and applications in the environmental toxicology. In Tools, Techniques and Protocols for Monitoring Environmental Contaminants; Elsevier, 2019; pp. 97–124. Available online: https://www.sciencedirect.com/science/article/pii/B9780128146798000054 (accessed on 20 June 2020).
- Pfeiffer, F.; Mayer, G. Selection and Biosensor Application of Aptamers for Small Molecules. Front. Chem. 2016, 4, 25. [Google Scholar] [CrossRef]
- Ruscito, A.; DeRosa, M.C. Small-Molecule Binding Aptamers: Selection Strategies, Characterization, and Applications. Front. Chem. 2016, 4, 14. [Google Scholar] [CrossRef] [PubMed]
- Hori, S.-I.; Herrera, A.; Rossi, J.J.; Zhou, J. Current Advances in Aptamers for Cancer Diagnosis and Therapy. Cancers 2018, 10, 9. [Google Scholar] [CrossRef]
- Potty, A.S.R.; Kourentzi, K.; Fang, H.; Schuck, P.; Willson, R.C. Biophysical characterization of DNA and RNA aptamer interactions with hen egg lysozyme. Int. J. Biol. Macromol. 2011, 48, 392–397. [Google Scholar] [CrossRef] [PubMed]
- Elskens, J.P.; Elskens, J.M.; Madder, A. Chemical Modification of Aptamers for Increased Binding Affinity in Diagnostic Applications: Current Status and Future Prospects. Int. J. Mol. Sci. 2020, 21, 4522. [Google Scholar] [CrossRef] [PubMed]
- Ahmad Raston, N.H.; Nguyen, V.T.; Gu, M.B. A new lateral flow strip assay (LFSA) using a pair of aptamers for the detection of Vaspin. Biosens. Bioelectron. 2017, 93, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Lee, J.; Lee, B.H.; Song, C.S.; Gu, M.B. Specific detection of avian influenza H5N2 whole virus particles on lateral flow strips using a pair of sandwich-type aptamers. Biosens. Bioelectron. 2019, 134, 123–129. [Google Scholar] [CrossRef]
- Nguyen, V.T.; Kwon, Y.S.; Gu, M.B. Aptamer-based environmental biosensors for small molecule contaminants. Curr. Opin. Biotechnol. 2017, 45, 15–23. [Google Scholar] [CrossRef]
- Takahashi, M. Aptamers targeting cell surface proteins. Biochimie 2018, 145, 63–72. [Google Scholar] [CrossRef]
- Yu, Q.; Liu, M.; Su, H.; Xiao, H.; Wu, S.; Qin, X.; Li, S.; Mi, H.; Lu, Z.; Shi, D.; et al. Selection and characterization of ssDNA aptamers specifically recognizing pathogenic Vibrio alginolyticus. J. Fish. Dis. 2019, 42, 851–858. [Google Scholar] [CrossRef]
- Song, S.; Wang, X.; Xu, K.; Li, Q.; Ning, L.; Yang, X. Selection of highly specific aptamers to Vibrio parahaemolyticus using cell-SELEX powered by functionalized graphene oxide and rolling circle amplification. Anal. Chim. Acta 2019, 1052, 153–162. [Google Scholar] [CrossRef]
- Yan, W.; Gu, L.; Liu, S.; Ren, W.; Lyu, M.; Wang, S. Identification of a highly specific DNA aptamer for Vibrio vulnificus using systematic evolution of ligands by exponential enrichment coupled with asymmetric PCR. J. Fish. Dis. 2018, 41, 1821–1829. [Google Scholar] [CrossRef] [PubMed]
- Masoum, A.; Oloomi, M.; Yavari, A.; Bouzari, S. DNA aptamer idenfitication and characterization for E. coli O157 detetion using cell based SELEX method. Anal. Biochem. 2017, 536, 36–44. [Google Scholar] [CrossRef]
- Zou, Y.; Duan, N.; Wu, S.; Shen, M.; Wang, Z. Selection, Identification, and Binding Mechanism Studies of an ssDNA Aptamer Targeted to Different Stages of E. coli O157:H7. J. Agric. Food Chem. 2018, 66, 5677–5682. [Google Scholar] [CrossRef] [PubMed]
- Marton, S.; Cleto, F.; Krieger, M.A.; Cardoso, J. Isolation of an Aptamer that Binds Specifically to E. coli. PLoS ONE 2016, 11, 0153637. [Google Scholar] [CrossRef]
- Renders, M.; Miller, E.; Lam, C.H.; Perrin, D.M. Whole cell-SELEX of aptamers with a tyrosine-like side chain against live bacteria. Org. Biomol. Chem. 2017, 15, 1980–1989. [Google Scholar] [CrossRef]
- Hamula, C.L.A.; Peng, H.; Wang, Z.; Tyrrell, G.J.; Li, X.-F.; Le, X.C. An improved SELEX technique for selection of DNA aptamers binding to M-type 11 of Streptococcus pyogenes. Methods 2016, 51–57. [Google Scholar] [CrossRef]
- Alfavian, H.; Mousavi Gargari, S.L.; Rasoulinejad, S.; Medhat, A. Development of a DNA aptamer that binds specifically to group A Streptococcus serotype M3. Can. J. Microbiol. 2017, 63, 160–168. [Google Scholar] [CrossRef]
- Cui, W.; Liu, J.; Su, D.; Hu, D.; Hou, S.; Hu, T.; Yang, J.; Luo, Y.; Xi, Q.; Chu, B.; et al. Identification of ssDNA aptamers specific to clinical isolates of Streptococcus mutans strains with different cariogenicity. Acta. Biochim. Et. Biophys. Sin. 2016, 48, 563–572. [Google Scholar] [CrossRef]
- Graziani, A.C.; Stets, M.I.; Lopes, A.L.K.; Schluga, P.H.C.; Marton, S.; Ferreira, I.M.; De Andrade, A.S.R.; Krieger, M.A.; Cardoso, J. High efficiency binding aptamers for a wide range of bacterial sepsis agents. J. Microbiol. Biotechnol. 2017, 27, 838–843. [Google Scholar] [CrossRef]
- Sypabekova, M.; Bekmurzayeva, A.; Wang, R.; Li, Y.; Nogues, C.; Kanayeva, D. Selection, Characterization, and application of DNA aptamers for detection of Mycobacterium tuberculosis secreted protein MPT64. Tuberculosis 2017, 104, 70–78. [Google Scholar] [CrossRef]
- Mozioglu, E.; Gokmen, O.; Tamerler, C.; Kocagoz, Z.T.; Akgoz, M. Selection of Nucleic Acid Aptamers Specific for Mycobacterium tuberculosis. Appl. Biochem. Biotechnol. 2016, 178, 849–864. [Google Scholar] [CrossRef]
- Sun, X.; Pan, Q.; Yuan, C.; Wang, Q.; Tang, X.L.; Ding, K.; Zhou, X.; Zhang, X.L. A Single ssDNA Aptamer Binding to Mannose-Capped Lipoarabinomannan of Bacillus Calmette-Guérin Enhances Immunoprotective Effect against Tuberculosis. J. Am. Chem. Soc. 2016, 138, 11680–11689. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.-L.; Wu, S.-m.; Xie, Y.S.; Neng, G.Q.; Yuan, C.; Zhou, X.; Zhang, X.-L. Generation and application of ssDNA aptamers against glycolipid antigen ManLAM of mycobacterium tuberculosis for TB diagnosis. J. Infect. 2016, 72, 573–586. [Google Scholar] [CrossRef] [PubMed]
- Soundy, J.; Day, D. Selection of DNA aptamers specific for live Pseudomonas aeruginosa. PLoS ONE 2017, 12, e0185385. [Google Scholar] [CrossRef] [PubMed]
- Tsao, S.M.; Lai, J.C.; Horng, H.E.; Liu, T.C.; Hong, C.Y. Generation of aptamers from A primer-free randomized ssDNA library using magnetic-assisted rapid aptamer selection. Sci. Rep. 2017, 7, 45478. [Google Scholar] [CrossRef] [PubMed]
- Song, M.Y.; Nguyen, D.; Hong, S.W.; Kim, B.C. Broadly reactive aptamers targeting bacteria belonging to different genera using a sequential toggle cell-SELEX. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef]
- Lahousse, M.; Park, H.-C.; Lee, S.-C.; Ha, N.-R.; Jung, I.-P.; Schlesinger, S.R.; Shackelford, K.; Yoon, M.-Y.; Kim, S.-K. Inhibition of anthrax lethal factor by ssDNA aptamers. Arch. Biochem. Biophys. 2018, 646, 16–23. [Google Scholar] [CrossRef]
- Biondi, E.; Lane, J.D.; Das, D.; Dasgupta, S.; Piccirilli, J.A.; Hoshika, S.; Bradley, K.M.; Krantz, B.A.; Benner, S.A. Laboratory evolution of artificially expanded DNA gives redesignable aptamers that target the toxic form of anthrax protective antigen. Nucleic. Acids. Res. 2016, 44, 9565–9577. [Google Scholar] [CrossRef]
- Fischer, C.; Hünniger, T.; Jarck, J.H.; Frohnmeyer, E.; Kallinich, C.; Haase, I.; Hahn, U.; Fischer, M. Food Sensing: Aptamer-Based Trapping of Bacillus cereus Spores with Specific Detection via Real Time PCR in Milk. J. Agric. Food Chem. 2015, 63, 8050–8057. [Google Scholar] [CrossRef]
- Yan, W.; Gu, L.; Ren, W.; Ma, X.; Qin, M.; Lyu, M.; Wang, S. Recognition of Helicobacter pylori by protein-targeting aptamers. Helicobacter 2019, 24, 1–10. [Google Scholar] [CrossRef]
- Stoltenburg, R.; Schubert, T.; Strehlitz, B. In vitro selection and interaction studies of a DNA aptamer targeting Protein, A. PLoS ONE 2015, 10, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Gan, L.; Jiang, L.; Zhang, X.; Yang, X.; Chen, M.; Lan, X. Neutralization of staphylococcal enterotoxin B by an aptamer antagonist. Antimicrob. Agents. Chemother. 2015, 59, 2072–2077. [Google Scholar] [CrossRef] [PubMed]
- Ramlal, S.; Mondal, B.; Lavu, P.S.; Bhavanashri, N.; Kingston, J. Capture and detection of Staphylococcus aureus with dual labeled aptamers to cell surface components. Int. J. Food Microbiol. 2018, 265, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Stoltenburg, R.; Krafčiková, P.; Víglaský, V.; Strehlitz, B. G-quadruplex aptamer targeting Protein A and its capability to detect Staphylococcus aureus demonstrated by ELONA. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef]
- Stoltenburg, R.; Strehlitz, B. Refining the Results of a Classical SELEX Experiment by Expanding the Sequence Data Set of an Aptamer Pool Selected for Protein, A. Int. J. Mol. Sci. 2018, 19, 642. [Google Scholar] [CrossRef]
- Sedighian, H.; Halabian, R.; Amani, J.; Heiat, M.; Amin, M.; Fooladi, A.A.I. Staggered Target SELEX, a novel approach to isolate non-cross-reactive aptamer for detection of SEA by apta-qPCR. J. Biotechnol. 2018, 286, 45–55. [Google Scholar] [CrossRef]
- Sedighian, H.; Halabian, R.; Amani, J.; Heiat, M.; Taheri, R.A.; Imani Fooladi, A.A. Manufacturing of a novel double-function ssDNA aptamer for sensitive diagnosis and efficient neutralization of SEA. Anal. Biochem. 2018, 548, 69–77. [Google Scholar] [CrossRef]
- Huang, Y.; Chen, X.; Duan, N.; Wu, S.; Wang, Z.; Wei, X.; Wang, Y. Selection and characterization of DNA aptamers against Staphylococcus aureus enterotoxin C1. Food Chem. 2015, 166, 623–629. [Google Scholar] [CrossRef]
- Zhang, X.; Feng, Y.; Yao, Q.; He, F. Selection of a new Mycobacterium tuberculosis H37Rv aptamer and its application in the construction of a SWCNT/aptamer/Au-IDE MSPQC H37Rv sensor. Biosens. Bioelectron. 2017, 98, 261–266. [Google Scholar] [CrossRef]
- Aimaiti, R.; Qin, L.; Cao, T.; Yang, H.; Wang, J.; Lu, J.; Huang, X.; Hu, Z. Identification and application of ssDNA aptamers against H₃₇Rv in the detection of Mycobacterium tuberculosis. Appl. Microbiol. Biotechnol. 2015, 99, 9073–9083. [Google Scholar] [CrossRef]
- Ansari, N.; Ghazvini, K.; Ramezani, M.; Shahdordizadeh, M.; Yazdian-Robati, R.; Abnous, K.; Taghdisi, S.M. Selection of DNA aptamers against Mycobacterium tuberculosis Ag85A, and its application in a graphene oxide-based fluorometric assay. Microchim. Acta 2017, 185, 21. [Google Scholar] [CrossRef]
- Yu, X.; Chen, F.; Wang, R.; Li, Y. Whole-bacterium SELEX of DNA aptamers for rapid detection of E. coli O157:H7 using a QCM sensor. J. Biotechnol. 2018, 266, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Kaur, H.; Shorie, M.; Sharma, M.; Ganguli, A.K.; Sabherwal, P. Bridged Rebar Graphene functionalized aptasensor for pathogenic E. coli O78:K80:H11 detection. Biosens. Bioelectron. 2017, 98, 486–493. [Google Scholar] [CrossRef]
- Bayraç, A.T.; Donmez, S.I. Selection of DNA aptamers to Streptococcus pneumonia and fabrication of graphene oxide based fluorescent assay. Anal. Biochem. 2018, 556, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Song, M.S.; Sekhon, S.S.; Shin, W.R.; Kim, H.C.; Min, J.; Ahn, J.Y.; Kim, Y.H. Detecting and discriminating shigella sonnei using an aptamer-based fluorescent biosensor platform. Molecules 2017, 22, 825. [Google Scholar] [CrossRef] [PubMed]
- Bayrac, C.; Eyidogan, F.; Oktem, H.A. DNA aptamer-based colorimetric detection platform for Salmonella Enteritidis. Biosens.Bioelectron. 2017, 98, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Mirzakhani, K.; Gargari, S.L.M.; Rasooli, I.; Rasoulinejad, S. Development of a DNA aptamer for screening Neisseria meningitidis serogroup b by cell SELEX. Iran. Biomed. J. 2018, 22, 193–201. [Google Scholar] [CrossRef]
- Chinnappan, R.; AlAmer, S.; Eissa, S.; Rahamn, A.A.; Abu Salah, K.M.; Zourob, M. Fluorometric graphene oxide-based detection of Salmonella enteritis using a truncated DNA aptamer. Microchim. Acta 2018, 185, 1–9. [Google Scholar] [CrossRef]
- Shin, W.R.; Sekhon, S.S.; Kim, S.G.; Rhee, S.J.; Yang, G.N.; Won, K.; Rhee, S.K.; Ryu, H.; Kim, K.; Min, J.; et al. Aptamer-based pathogen monitoring for salmonella enterica ser. Typhimurium. J. Biomed. Nanotechnol. 2018, 14, 1992–2002. [Google Scholar] [CrossRef]
- Rasoulinejad, S.; Gargari, S.L.M. Aptamer-nanobody based ELASA for specific detection of Acinetobacter baumannii isolates. J. Biotechnol. 2016, 231, 46–54. [Google Scholar] [CrossRef]
- Liu, M.; Yin, Q.; Brennan, J.D.; Li, Y. Selection and characterization of DNA aptamers for detection of glutamate dehydrogenase from Clostridium difficile. Biochimie 2018, 145, 151–157. [Google Scholar] [CrossRef]
- Frohnmeyer, E.; Frisch, F.; Falke, S.; Betzel, C.; Fischer, M. Highly affine and selective aptamers against cholera toxin as capture elements in magnetic bead-based sandwich ELAA. J. Biotechnol. 2018, 269, 35–42. [Google Scholar] [CrossRef]
- Lavu, P.S.; Mondal, B.; Ramlal, S.; Murali, H.S.; Batra, H.V. Selection and Characterization of Aptamers Using a Modified Whole Cell Bacterium SELEX for the Detection of Salmonella enterica Serovar Typhimurium. Acs. Comb. Sci. 2016, 18, 292–301. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Ahn, J.Y.; Lee, K.A.; Um, H.J.; Sekhon, S.S.; Sun Park, T.; Min, J.; Kim, Y.H. Analytical bioconjugates, aptamers, enable specific quantitative detection of Listeria monocytogenes. Biosens. Bioelectron. 2015, 68, 272–280. [Google Scholar] [CrossRef] [PubMed]
- Hong, K.L.; Yancey, K.; Battistella, L.; Williams, R.M.; Hickey, K.M.; Bostick, C.D.; Gannett, P.M.; Sooter, L.J. Selection of Single-Stranded DNA Molecular Recognition Elements against Exotoxin A Using a Novel Decoy-SELEX Method and Sensitive Detection of Exotoxin A in Human Serum. Biomed. Res. Int. 2015, 2015, 417641. [Google Scholar] [CrossRef] [PubMed]
- Shin, W.R.; Sekhon, S.S.; Rhee, S.K.; Ko, J.H.; Ahn, J.Y.; Min, J.; Kim, Y.H. Aptamer-Based Paper Strip Sensor for Detecting Vibrio fischeri. Acs. Comb. Sci. 2018, 20, 261–268. [Google Scholar] [CrossRef]
- Shin, H.S.; Gedi, V.; Kim, J.K.; Lee, D.k. Detection of Gram-negative bacterial outer membrane vesicles using DNA aptamers. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef]
- Hedayati Ch, M.; Amani, J.; Sedighian, H.; Amin, M.; Salimian, J.; Halabian, R.; Imani Fooladi, A.A. Isolation of a new ssDNA aptamer against staphylococcal enterotoxin B based on CNBr-activated sepharose-4B affinity chromatography. J. Mol. Recognit. 2016, 29, 436–445. [Google Scholar] [CrossRef]
- Fan, Y.; Cui, M.; Liu, Y.; Jin, M.; Zhao, H. Selection and characterization of DNA aptamers for constructing colorimetric biosensor for detection of PBP2a. Spectrochim. Acta A 2019. [Google Scholar] [CrossRef]
- Das, R.; Dhiman, A.; Mishra, S.K.; Haldar, S.; Sharma, N.; Bansal, A.; Ahmad, Y.; Kumar, A.; Tyagi, J.S.; Sharma, T.K. Structural switching electrochemical DNA aptasensor for the rapid diagnosis of tuberculous meningitis. Int. J. Nanomed. 2019, 14, 2103–2113. [Google Scholar] [CrossRef]
- Bai, L.; Chen, Y.; Bai, Y.; Chen, Y.; Zhou, J.; Huang, A. Fullerene-doped polyaniline as new redox nanoprobe and catalyst in electrochemical aptasensor for ultrasensitive detection of Mycobacterium tuberculosis MPT64 antigen in human serum. Biomaterials 2017, 133, 11–19. [Google Scholar] [CrossRef]
- Sypabekova, M.; Jolly, P.; Estrela, P.; Kanayeva, D. Electrochemical aptasensor using optimized surface chemistry for the detection of Mycobacterium tuberculosis secreted protein MPT64 in human serum. Biosens. Bioelectron. 2019, 123, 141–151. [Google Scholar] [CrossRef]
- Lavania, S.; Das, R.; Dhiman, A.; Myneedu, V.P.; Verma, A.; Singh, N.; Sharma, T.K.; Tyagi, J.S. Aptamer-Based TB Antigen Tests for the Rapid Diagnosis of Pulmonary Tuberculosis: Potential Utility in Screening for Tuberculosis. Acs. Infect. Dis. 2018, 4, 1718–1726. [Google Scholar] [CrossRef]
- Zhang, X.; Feng, Y.; Duan, S.; Su, L.; Zhang, J.; He, F. Mycobacterium tuberculosis strain H37Rv Electrochemical Sensor Mediated by Aptamer and AuNPs-DNA. Acs. Sens. 2019, 4, 849–855. [Google Scholar] [CrossRef]
- Borsa, B.A.; Tuna, B.G.; Hernandez, F.J.; Hernandez, L.I.; Bayramoglu, G.; Arica, M.Y.; Ozalp, V.C. Staphylococcus aureus detection in blood samples by silica nanoparticle-oligonucleotides conjugates. Biosens. Bioelectron. 2016, 86, 27–32. [Google Scholar] [CrossRef]
- Cheng, D.; Yu, M.; Fu, F.; Han, W.; Li, G.; Xie, J.; Song, Y.; Swihart, M.T.; Song, E. Dual Recognition Strategy for Specific and Sensitive Detection of Bacteria Using Aptamer-Coated Magnetic Beads and Antibiotic-Capped Gold Nanoclusters. Anal. Chem. 2016, 88, 820–825. [Google Scholar] [CrossRef]
- Gao, W.; Li, B.; Yao, R.; Li, Z.; Wang, X.; Dong, X.; Qu, H.; Li, Q.; Li, N.; Chi, H.; et al. Intuitive Label-Free SERS Detection of Bacteria Using Aptamer-Based in Situ Silver Nanoparticles Synthesis. Anal. Chem. 2017, 89, 9836–9842. [Google Scholar] [CrossRef]
- Pang, Y.; Wan, N.; Shi, L.; Wang, C.; Sun, Z.; Xiao, R.; Wang, S. Dual-recognition surface-enhanced Raman scattering(SERS)biosensor for pathogenic bacteria detection by using vancomycin-SERS tags and aptamer-Fe3O4@Au. Anal. Chim. Acta 2019, 1077, 288–296. [Google Scholar] [CrossRef]
- Lian, Y.; He, F.; Wang, H.; Tong, F. A new aptamer/graphene interdigitated gold electrode piezoelectric sensor for rapid and specific detection of Staphylococcus aureus. Biosens. Bioelectron. 2015, 65, 334–339. [Google Scholar] [CrossRef]
- Sundararaj, N.; Kalagatur, N.K.; Mudili, V.; Krishna, K.; Antonysamy, M. Isolation and identification of enterotoxigenic Staphylococcus aureus isolates from Indian food samples: Evaluation of in-house developed aptamer linked sandwich ELISA (ALISA) method. J. Food Sci. Technol. 2019, 56, 1016–1026. [Google Scholar] [CrossRef]
- Ranjbar, S.; Shahrokhian, S. Designa nd fabrication of an electrochemical aptasensor using Au nanoparticles/carbon nanoparticles/cellulose nanofibers nanocomposite for rapid and sensitive detection of Staphyloccus aureus. Bioelectrochemistry 2018, 123, 70–76. [Google Scholar] [CrossRef]
- Wang, S.; Deng, W.; Yang, L.; Tan, Y.; Xie, Q.; Yao, S. Copper-Based Metal-Organic Framework Nanoparticles with Peroxidase-Like Activity for Sensitive Colorimetric Detection of Staphylococcus aureus. ACS Appl. Mater. Interfaces 2017, 9, 24440–24445. [Google Scholar] [CrossRef]
- Li, J.; Jiang, H.; Rao, X.; Liu, Z.; Zhu, H.; Xu, Y. Point-of-Care Testing of Pathogenic Bacteria at the Single-Colony Level via Gas Pressure Readout Using Aptamer-Coated Magnetic CuFe 2 O 4 and Vancomycin-Capped Platinum Nanoparticles. Anal. Chem. 2019, 91, 1494–1500. [Google Scholar] [CrossRef]
- Yu, T.; Xu, H.; Zhao, Y.; Han, Y.; Zhang, Y.; Zhang, J.; Xu, C.; Wang, W.; Guo, Q.; Ge, J. Aptamer based high throughput colorimetric biosensor for detection of staphylococcus aureus. Sci. Rep. 2020, 10, 1–6. [Google Scholar] [CrossRef]
- Xu, J.; Guo, J.; Maina, S.W.; Yang, Y.; Hu, Y.; Li, X.; Qiu, J.; Xin, Z. An aptasensor for staphylococcus aureus based on nicking enzyme amplification reaction and rolling circle amplification. Anal. Biochem. 2018, 549, 136–142. [Google Scholar] [CrossRef]
- Wang, J.; Wu, X.; Wang, C.; Shao, N.; Dong, P.; Xiao, R.; Wang, S. Magnetically Assisted Surface-Enhanced Raman Spectroscopy for the Detection of Staphylococcus aureus Based on Aptamer Recognition. ACS Appl. Mater. Interfaces 2015, 7, 20919–20929. [Google Scholar] [CrossRef]
- Cai, R.; Yin, F.; Chen, H.; Tian, Y.; Zhou, N. A fluorescent aptasensor for Staphylococcus aureus based on strand displacement amplification and self-assembled DNA hexagonal structure. Microchim. Acta 2020, 187, 604–620. [Google Scholar] [CrossRef]
- Qiao, J.; Meng, X.; Sun, Y.; Li, Q.; Zhao, R.; Zhang, Y.; Wang, J.; Yi, Z. Aptamer-based fluorometric assay for direct identification of methicillin-resistant Staphylococcus aureus from clinical samples. J. Microbiol. Methods 2018, 153, 92–98. [Google Scholar] [CrossRef]
- Oscoy, I.; Yusuflbeyoglu, S.; Yilmaz, V.; McLamore, E.S.; Ildiz, N.; Ulgen, A. DNA aptamer functionalized gold nanostructores for molecular recognition and photothermal inactivation of methicillin-Resistant Satphylococcus aureus. Colloids Surf. B 2017, 159, 16–22. [Google Scholar]
- Xu, Y.; Wang, H.; Luan, C.; Liu, Y.; Chen, B.; Zhao, Y. Aptamer-based hydrogel barcodes for the capture and detection of multiple types of pathogenic bacteria. Biosens. Bioelectron. 2018, 100, 404–410. [Google Scholar] [CrossRef]
- Jin, B.; Wang, S.; Lin, M.; Jin, Y.; Zhang, S.; Cui, X.; Gong, Y.; Li, A.; Xu, F.; Lu, T.J. Upconversion nanoparticles based FRET aptasensor for rapid and ultrasensitive bacteria detection. Biosens. Bioelectron. 2017, 90, 525–533. [Google Scholar] [CrossRef]
- Jo, N.; Kim, H.; Lee, S.-M.; Oh, J.; Park, I.H.; Lim, K.J.; Shin, J.-s.; Yoo, K.H. Aptamer-functionalized capacitance sensors for real-time monitoring of bacterial growna dn antibiotics susceptibility. Biosensors 2018, 102, 164–170. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, X.; Yang, Q.; Jiang, X.; Li, Y.; Zhao, J.; Qu, K. Conductometric sensor for viable Escherichia coli and Staphylococcus aureus based on magnetic analyte separation via aptamer. Microchim. Acta 2020, 187, 43. [Google Scholar] [CrossRef]
- Wu, W.; Zhao, S.; Mao, Y.; Fang, Z.; Lu, X.; Zeng, L. A sensitive lateral flow biosensor for Escherichia coli O157: H7 detection based on aptamer mediated strand displacement amplification. Anal. Chim. Acta 2015, 861, 62–68. [Google Scholar] [CrossRef]
- Khang, J.; Kim, D.; Chung, K.W.; Lee, J.H. Chemiluminescent aptasensor capable of rapidly quantifying Escherichia coli O157:H7. Talanta 2016, 147, 177–183. [Google Scholar] [CrossRef]
- Shahrokhian, S.; Ranjbar, S. Aptamer immobilization on amino-functionalized metal-organic frameworks: An ultrasensitive platform for the electrochemical diagnostic of Escherichia coli O157:H7. Analyst 2018, 143, 3191–3201. [Google Scholar] [CrossRef]
- Bu, S.; Wang, K.; Li, Z.; Wang, C.; Hao, Z.; Liu, W.; Wan, J. An electrochemical biosensor based on methylene blue-loaded nanocomposites as signal-amplifying tags to detect pathogenic bacteria. Analyst 2020. [Google Scholar] [CrossRef]
- Dong, X.; Shi, Z.; Xu, C.; Yang, C.; Chen, F.; Lei, M. CdS quantum dots/Au nanoparticles/ZnO nanowire array for self-powered photoelectrochemical detection of Escherichia coli O157:H7. Biosens. Bioelectron. 2020, 149, 111843. [Google Scholar] [CrossRef]
- Ling, M.; Peng, Z.; Cheng, L.; Deng, L. Rapid fluorescent detection of enterotoxigenic Escherichia coli (ETEC) K88 based on graphene oxide-dependent nanoquencher and Klenow fragment-triggered target cyclic amplification. Appl. Spectrosc. 2015, 69, 1175–1181. [Google Scholar] [CrossRef]
- Wang, Z.; Lu, Q.; Xu, T.; Wang, F.; Huang, F.; Peng, Y.; Deng, L. G-quadruplex-based assay combined with aptamer and gold nanoparticles for Escherichia coli K88 determination. Microchim. Acta 2020, 187, 604–620. [Google Scholar] [CrossRef]
- Hao, N.; Zhang, X.; Zhou, Z.; Hua, R.; Zhang, Y.; Liu, Q.; Qian, J.; Li, H.; Wang, K. AgBr nanoparticles/3D nitrogen-doped graphene hydrogel for fabricating all-solid-state luminol-electrochemiluminescence Escherichia coli aptasensors. Biosens. Bioelectron. 2017, 97, 377–383. [Google Scholar] [CrossRef]
- Hua, R.; Hao, N.; Lu, J.; Qian, J.; Liu, Q.; Li, H.; Wang, K. A sensitive Potentiometric resolved ratiometric Photoelectrochemical aptasensor for Escherichia coli detection fabricated with non-metallic nanomaterials. Biosens. Bioelectron. 2018, 106, 57–63. [Google Scholar] [CrossRef]
- Guo, Y.; Wang, Y.; Liu, S.; Yu, J.; Wang, H.; Wang, Y.; Huang, J. Label-free and highly sensitive electrochemical detection of E. coli based on rolling circle amplifications coupled peroxidase-mimicking DNAzyme amplification. Biosens. Bioelectron. 2016, 75, 315–319. [Google Scholar] [CrossRef]
- Wu, G.; Dai, Z.; Tang, X.; Lin, Z.; Lo, P.K.; Meyyappan, M.; Lai, K.W.C. Graphene Field-Effect Transistors for the Sensitive and Selective Detection of Escherichia coli Using Pyrene-Tagged DNA Aptamer. Adv. Healthc. Mater. 2017, 6. [Google Scholar] [CrossRef]
- Abdelrasoul, G.N.; Anwar, A.; MacKay, S.; Tamura, M.; Shah, M.A.; Khasa, D.P.; Montgomery, R.R.; Ko, A.I.; Chen, J. DNA aptamer-based non-faradaic impedance biosensor for detecting E. coli. Anal. Chim. Acta 2020, 1107, 135–144. [Google Scholar] [CrossRef]
- Zhao, L.; Liu, Y.; Zhang, Z.; Wei, J.; Xie, S.; Li, X. Fibrous testing papers for fluorescence trace sensing and photodynamic destruction of antibiotic-resistant bacteria. J. Mater. Chem. B 2020, 8, 2709–2718. [Google Scholar] [CrossRef]
- Bruno, J.G.; Sivils, J.C.; Phillips, T. Aptamer-magnetic bead quantum dot sandwich assays for foodborne pathogen detection: Pros, cons, and lessons learned. J. Aoac. Int. 2017, 100, 895–899. [Google Scholar] [CrossRef]
- Yoo, S.M.; Kim, D.K.; Lee, S.Y. Aptamer-functionalized localized surface plasmon resonance sensor for the multiplexed detection of different bacterial species. Talanta 2015, 132, 112–117. [Google Scholar] [CrossRef]
- Das, R.; Dhiman, A.; Kapil, A.; Bansal, V.; Sharma, T.K. Aptamer-mediated colorimetric and electrochemical detection of Pseudomonas aeruginosa utilizing peroxidase-mimic activity of gold NanoZyme. Anal. Bioanal. Chem. 2019, 411, 1229–1238. [Google Scholar] [CrossRef]
- Zhong, Z.; Gao, X.; Gao, R.; Jia, L. Selective capture and sensitive fluorometric determination of Pseudomonas aeruginosa by using aptamer modified magnetic nanoparticles. Mikrochim. Acta 2018, 185, 377. [Google Scholar] [CrossRef]
- Wu, Z.; He, D.; Cui, B.; Jin, Z. A bimodal (SERS and colorimetric) aptasensor for the detection of Pseudomonas aeruginosa. Microchim. Acta 2018, 185, 1–7. [Google Scholar] [CrossRef]
- Hu, J.; Fu, K.; Bohn, P.W. Whole-Cell Pseudomonas aeruginosa Localized Surface Plasmon Resonance Aptasensor. Anal. Chem. 2018, 90, 2326–2332. [Google Scholar] [CrossRef]
- Zhang, X.; Xie, G.; Gou, D.; Luo, P.; Yao, Y.; Chen, H. A novel enzyme-free electrochemical biosensor for rapid detection of Pseudomonas aeruginosa based on high catalytic Cu-ZrMOF and conductive Super, P. Biosens. Bioelectron. 2019, 142. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, J.; Estévez, M.C.; Fernández-Gavela, A.; González-López, J.J.; González-Guerrero, A.B.; Lechuga, L.M. Label-free detection of nosocomial bacteria using a nanophotonic interferometric biosensor. Analyst 2020, 145, 497–506. [Google Scholar] [CrossRef]
- Wu, J.H.; Wang, C.H.; Ma, Y.D.; Lee, G.B. A nitrocellulose membrane-based integrated microfluidic system for bacterial detection utilizing magnetic-composite membrane microdevices and bacteria-specific aptamers. Lab. Chip. 2018, 18, 1633–1640. [Google Scholar] [CrossRef] [PubMed]
- Su, C.H.; Tsai, M.H.; Lin, C.Y.; Ma, Y.D.; Wang, C.H.; Chung, Y.D.; Lee, G.B. Dual aptamer assay for detection of Acinetobacter baumannii on an electromagnetically-driven microfluidic platform. Biosens. Bioelectron. 2020, 159, 112148. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Guo, Y.; Fan, J.; Yang, Y.; Zuo, C.; Bai, S.; Sheng, S.; Li, J.; Xie, G. A fluorometric assay for rapid enrichment and determination of bacteria by using zirconium-metal organic frameworks as both capture surface and signal amplification tag. Microchim. Acta 2020, 187, 604–620. [Google Scholar] [CrossRef] [PubMed]
- Hills, K.D.; Oliveira, D.A.; Cavallaro, N.D.; Gomes, C.L.; McLamore, E.S. Actuation of chitosan-aptamer nanobrush borders for pathogen sensing. Analyst 2018, 143, 1650–1661. [Google Scholar] [CrossRef]
- Pei, Q.; Song, X.; Liu, S.; Wang, J.; Leng, X.; Cui, X.; Yu, J.; Wang, Y.; Huang, J. A facile signal-on electrochemical DNA sensing platform for ultrasensitive detection of pathogenic bacteria based on Exo III-assisted autonomous multiple-cycle amplification. Analyst 2019, 144, 3023–3029. [Google Scholar] [CrossRef]
- Park, K.S.; Charles, R.C.; Ryan, E.T.; Weissleder, R.; Lee, H. Fluorescence polarization-based nucleic acid testing for rapid and cost-effective diagnosis of infectious disease. Chemistry 2015, 21. [Google Scholar] [CrossRef]
- Wei, X.; Zhou, W.; Sanjay, S.T.; Zhang, J.; Jin, Q.; Xu, F.; Dominguez, D.C.; Li, X. Multiplexed Instrument-Free Bar-Chart SpinChip Integrated with Nanoparticle-Mediated Magnetic Aptasensors for Visual Quantitative Detection of Multiple Pathogens. Anal. Chem. 2018, 90, 9888–9896. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ma, X.; Liu, Y.; Duan, N.; Wu, S.; Wang, Z.; Xu, B. Gold nanoparticles enhanced SERS aptasensor for the simultaneous detection of Salmonella typhimurium and Staphylococcus aureus. Biosens. Bioelectron. 2015, 74, 872–877. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Zhou, X.; Shan, Y.; Yue, H.; Huang, R.; Hu, J.; Xing, D. Sensitive detection of a bacterial pathogen using allosteric probe-initiated catalysis and CRISPR-Cas13a amplification reaction. Nat. Commun. 2020, 11, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Bruno, J.G.; Phillips, T.; Montez, T.; Garcia, A.; Sivils, J.C.; Mayo, M.W.; Greis, A. Development of a fluorescent enzyme-linked DNA aptamer-magnetic bead sandwich assay and portable fluorometer for sensitive and rapid listeria Detection. J. Fluoresc. 2015, 25, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Wen, L.x.; Lv, J.j.; Chen, L.; Li, S.b.; Mou, X.j.; Xu, Y. A fluorescent probe composed of quantum dot labeled aptamer and graphene oxide for the determination of the lipopolysaccharide endotoxin. Microchim. Acta 2019, 186, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Oueslati, R.; Cheng, C.; Zhao, L.; Chen, J.; Almeida, R.; Wu, J. Rapid, highly sensitive detection of Gram-negative bacteria with lipopolysaccharide absed disposable aptasensor. Biosens. Bioelectron. 2018, 112, 48–53. [Google Scholar] [CrossRef]
- Niu, J.; Hu, X.; Ouyang, W.; Chen, Y.; Liu, S.; Han, J.; Liu, L. Femtomolar Detection of Lipopolysaccharide in Injectables and Serum Samples Using Aptamer-Coupled Reduced Graphene Oxide in a Continuous Injection-Electrostacking Biochip. Anal. Chem. 2019, 91, 2360–2367. [Google Scholar] [CrossRef]
- Zhu, L.; Li, S.; Shao, X.; Feng, Y.; Xie, P.; Luo, Y.; Huang, K.; Xu, W. Colorimetric detection and typing of E. coli lipopolysaccharides based on a dual aptamer-functionalized gold nanoparticle probe. Microchim. Acta 2019, 186, 6–11. [Google Scholar] [CrossRef]
- Posha, B.; Sandhyarani, N. Highly sensitive endotoxin detection using a gold nanoparticle loaded layered molybdenum disulfide-polyacrylic acid nanocomposite. Analyst 2020, 145, 3939–3947. [Google Scholar] [CrossRef]
- Pourmadadi, M.; Shayeh, J.S.; Omidi, M.; Yazdian, F.; Alebouyeh, M. A glassy carbon electrode modified with reduced graphene oxide and gold nanoparticles for electrochemical aptasensing of lipopolysaccharides from Escherichia coli bacteria. Microchim. Acta 2019, 186, 787. [Google Scholar] [CrossRef]
- Xie, P.; Zhu, L.; Shao, X.; Huang, K.; Tian, J.; Xu, W. Highly sensitive detection of lipopolysaccharides using an aptasensor based on hybridization chain reaction. Sci. Rep. 2016, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Duan, N.; Gu, H.; Wang, H.; Wang, Z. Fluorometric determination of lipopolysaccharides via changes of the graphene oxide-enhanced fluorescence polarization caused by truncated aptamers. Microchim. Acta 2019, 186, 3–10. [Google Scholar] [CrossRef]
- Mazzaracchio, V.; Neagu, D.; Porchetta, A.; Marcoccio, E.; Pomponi, A.; Faggioni, G.; D’Amore, N.; Notargiacomo, A.; Pea, M.L.i.; Moscone, D.; et al. A label-free impedimetric aptasensor for the detection of Bacillus anthracis spore simulant. Biosens. Bioelectron. 2019, 126, 640–646. [Google Scholar] [CrossRef]
- Wu, Z.; He, D.; Cui, B. A fluorometric assay for staphylococcal enterotoxin B by making use of platinum coated gold nanorods and of upconversion nanoparticles. Microchim. Acta 2018, 185, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Mousavi Nodoushan, S.; Nasirizadeh, N.; Amani, J.; Halabian, R.; Imani Fooladi, A.A. An electrochemical aptasensor for staphylococcal enterotoxin B detection based on reduced graphene oxide and gold nano-urchins. Biosens. Bioelectron. 2019, 127, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Mondal, B.; Ramlal, S.; Lavu, P.S.; Bhavanashri, N.; Kingston, J. Highly sensitive colorimetric biosensor for staphylococcal enterotoxin B by a label-free aptamer and gold nanoparticles. Front. Microbiol. 2018, 9, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zhang, H.; Chen, X.; Wang, X.; Duan, N.; Wu, S.; Xu, B.; Wang, Z. A multicolor time-resolved fluorescence aptasensor for the simultaneous detection of multiplex Staphylococcus aureus enterotoxins in the milk. Biosens. Bioelectron. 2015, 74, 170–176. [Google Scholar] [CrossRef]
- Zhang, L.; Qi, Z.; Zou, Y.; Zhang, J.; Xia, W.; Zhang, R.; He, Z.; Cai, X.; Lin, Y.; Duan, S.-Z.; et al. Engineering DNA − Nanozyme Interfaces for Rapid Detection of Dental Bacteria. ACS Appl. Mater. Interfaces 2019, 11, 30640–30647. [Google Scholar] [CrossRef]
- Zhao, G.; Ding, J.; Yu, H.; Yin, T.; Qin, W. Potentiometric aptasensing of Vibrio alginolyticus based on DNA nanostructure-modified magnetic beads. Sensors 2016, 16, 2052. [Google Scholar] [CrossRef]
- Sun, Y.; Duan, N.; Ma, P.; Liang, Y.; Zhu, X.; Wang, Z. Colorimetric Aptasensor Based on Truncated Aptamer and Trivalent DNAzyme for Vibrio parahemolyticus Determination. J. Agric. Food Chem. 2019, 67, 2313–2320. [Google Scholar] [CrossRef]
- Oroval, M.; Coronado-Puchau, M.; Langer, J.; Sanz-Ortiz, M.N.; Ribes, Á.; Aznar, E.; Coll, C.; Marcos, M.D.; Sancenón, F.; Liz-Marzán, L.M.; et al. Surface Enhanced Raman Scattering and Gated Materials for Sensing Applications: The Ultrasensitive Detection of Mycoplasma and Cocaine. Chem. A. Eur. J. 2016, 22, 13488–13495. [Google Scholar] [CrossRef] [PubMed]
- Reich, P.; Stoltenburg, R.; Strehlitz, B.; Frense, D.; Beckmann, D. Development of An Impedimetric Aptasensor for the Detection of Staphylococcus aureus. Int. J. Mol. Sci. 2017, 18, 2484. [Google Scholar] [CrossRef] [PubMed]
- Urmann, K.; Reich, P.; Walter, J.G.; Beckmann, D.; Segal, E.; Scheper, T. Rapid and label-free detection of protein a by aptamer-tethered porous silicon nanostructures. J. Biotechnol. 2017, 257, 171–177. [Google Scholar] [CrossRef]
- Oliveira, D.; Borges, A.; Simões, M. Staphylococcus aureus Toxins and Their Molecular Activity in Infectious Diseases. Toxins 2018, 10, 252. [Google Scholar] [CrossRef]
- Lee, A.S.; de Lencastre, H.; Garau, J.; Kluytmans, J.; Malhotra-Kumar, S.; Peschel, A.; Harbarth, S. Methicillin-resistant Staphylococcus aureus. Nat. Rev. Dis. Primers 2018, 4, 18033. [Google Scholar] [CrossRef]
- Ehrt, S.; Schnappinger, D.; Rhee, K.Y. Metabolic principles of persistence and pathogenicity in Mycobacterium tuberculosis. Nat. Rev. Microbiol. 2018, 16, 496–507. [Google Scholar] [CrossRef]
- Hartzell, J.D.; Kim, A.S.; Kortepeter, M.G.; Moran, K.A. Acinetobacter pneumonia: A review. Med. Gen. Med. 2007, 9, 4. [Google Scholar]
- Weiser, J.N.; Ferreira, D.M.; Paton, J.C. Streptococcus pneumoniae: Transmission, colonization and invasion. Nat. Rev. Microbiol. 2018, 16, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Moradali, M.F.; Ghods, S.; Rehm, B.H.A. Pseudomonas aeruginosa Lifestyle: A Paradigm for Adaptation, Survival, and Persistence. Front. Cell Infect. Microbiol. 2017, 7, 39. [Google Scholar] [CrossRef]
- Ehling-Schulz, M.; Lereclus, D.; Koehler, T.M. The Bacillus cereus Group: Bacillus Species with Pathogenic Potential. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef]
- Gao, R.; Li, D.; Lin, Y.; Lin, J.; Xia, X.; Wang, H.; Bi, L.; Zhu, J.; Hassan, B.; Wang, S.; et al. Structural and Functional Characterization of the FadR Regulatory Protein from Vibrio alginolyticus. Front. Cell Infect. Microbiol. 2017, 7, 513. [Google Scholar] [CrossRef] [PubMed]
- Wan, Q.; Liu, X.; Zeng, Z.; Chen, Z.; Liu, Y.; Zu, Y. Aptamer Cocktail to Detect Multiple Species of Mycoplasma in Cell Culture. Int. J. Mol. Sci. 2020, 21, 3784. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).