Aggregation and Prion-Inducing Properties of the G-Protein Gamma Subunit Ste18 are Regulated by Membrane Association
Abstract
1. Introduction
2. Results
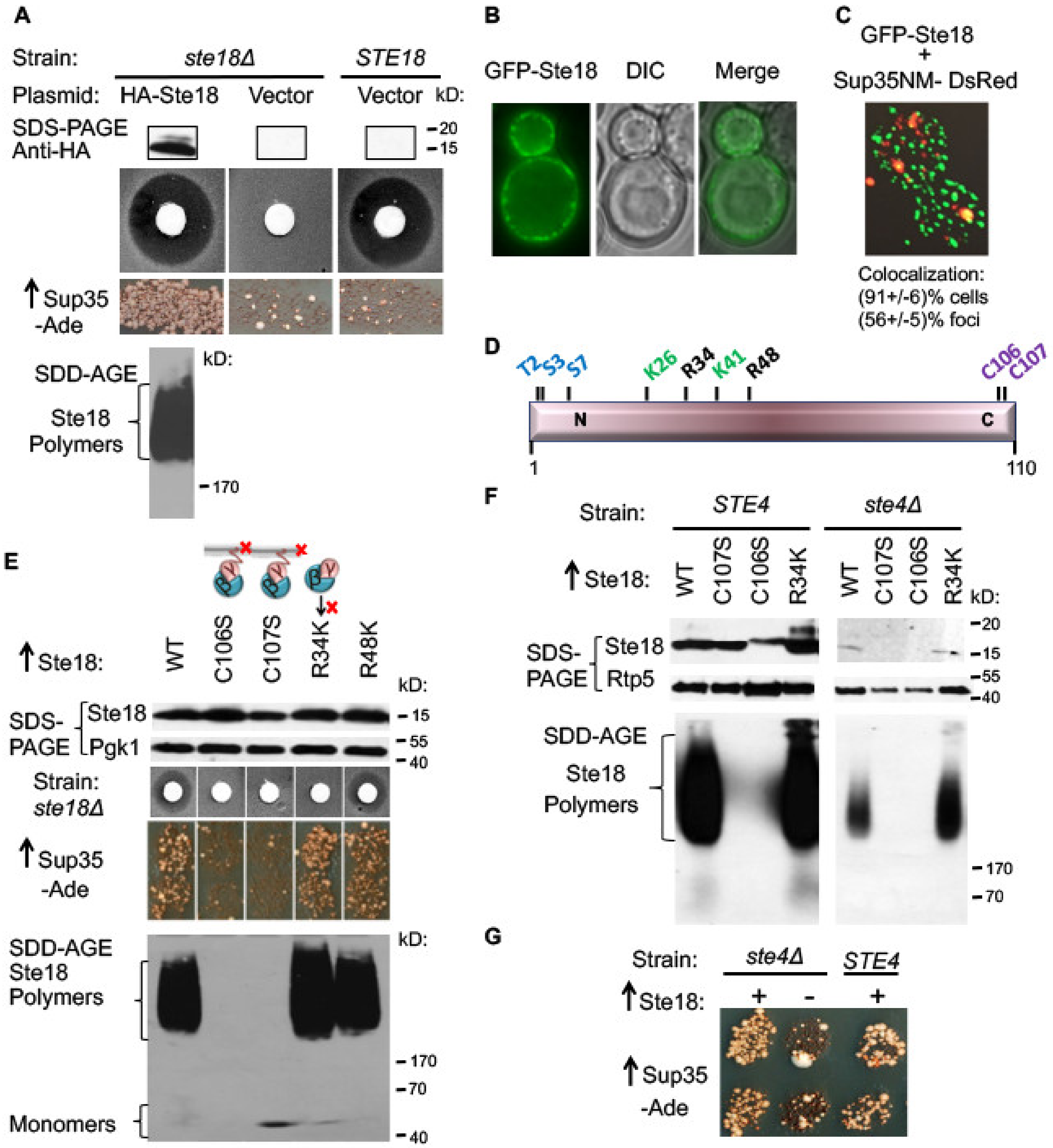
2.1. Overproduced Ste18 Promotes de novo [PSI+] Induction and Forms Aggregates that Are Transiently Associated with Aggregated Sup35
2.2. Ste18 Aggregation and Ste18-Mediated [PSI+] Induction Depend on the Membrane Association, but Do not Depend on Ste18 Function in the Signaling Pathway
2.3. Ste18 Ability to Induce Prions and Form Detergent-Resistant Aggregates Does not Depend on Association with Ste4
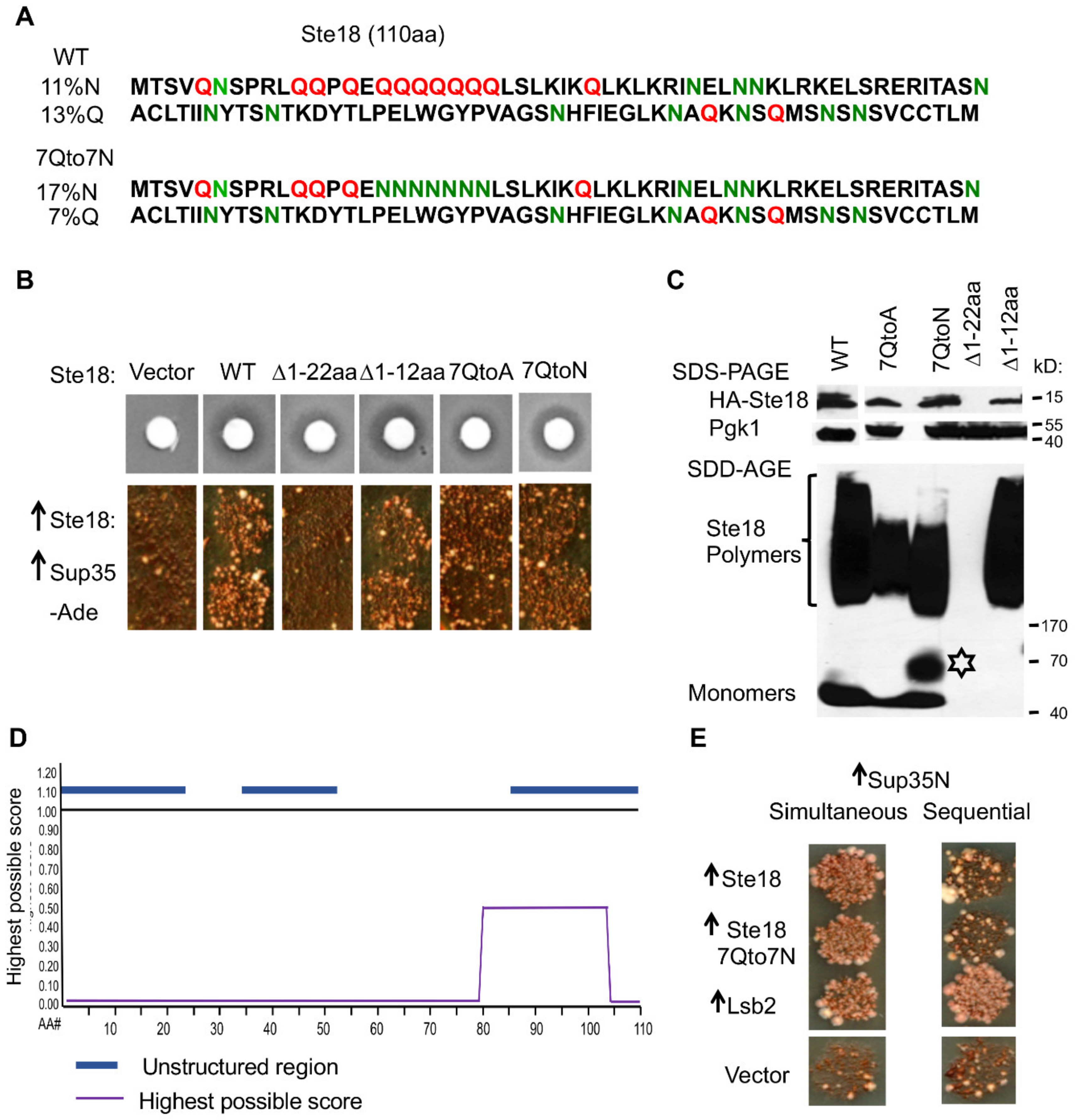
2.4. The Prion-Inducing Ability of Ste18 Does not Require the N-Terminal Q-Rich Stretch and Is not Affected by Glutamine to Asparagine Substitution
2.5. [PSI+] Induction is not Associated with the Formation of Detectable Heritable Prion State by Ste18
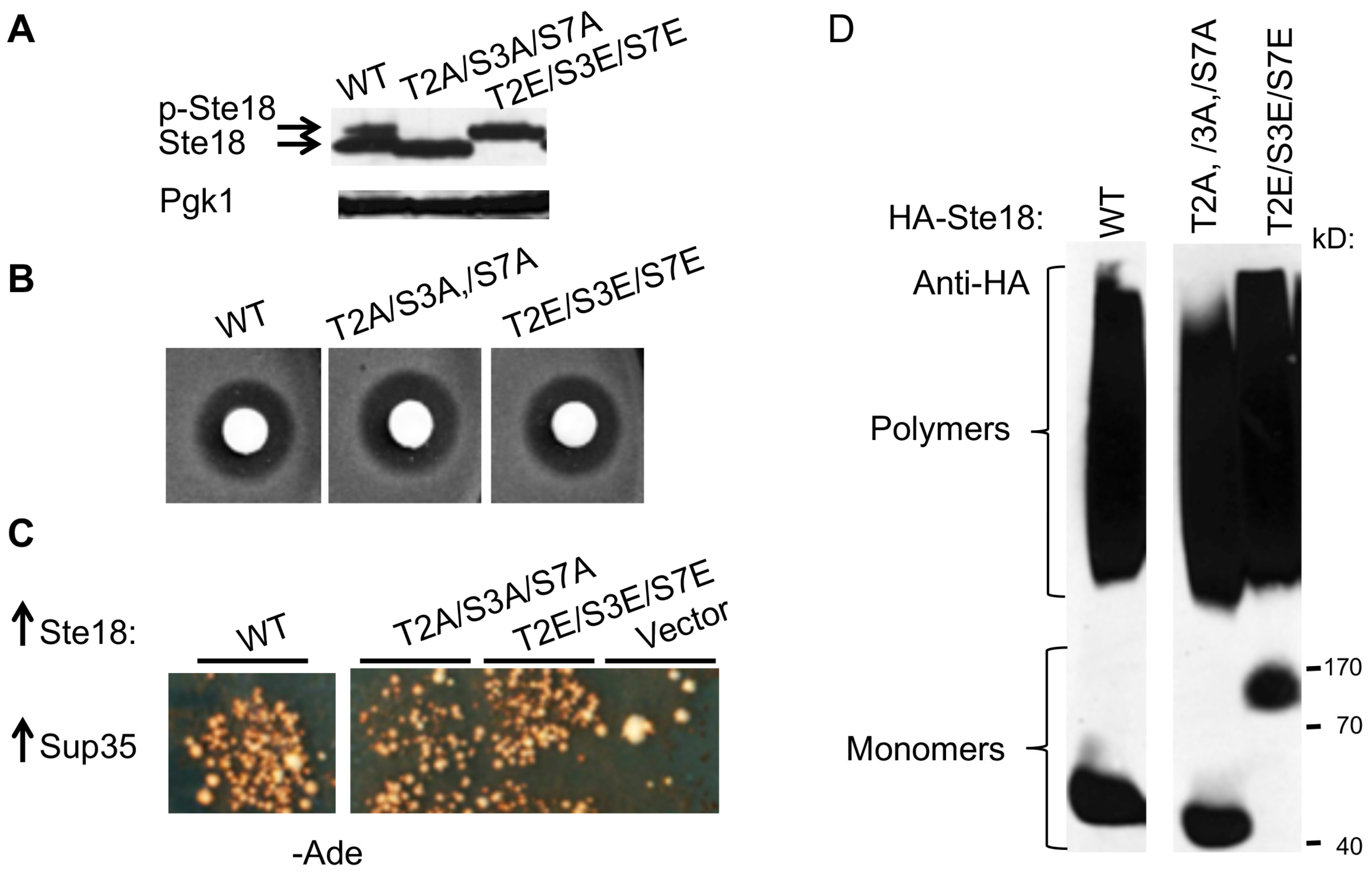
2.6. Posttranslational Modification of Ste18 by Phosphorylation Is not Essential for its Prion Inducing Ability
2.7. Ste18 is Ubiquitinated and Degraded in a Proteasome-Dependent Fashion
3. Discussion
3.1. Prion Inducing Properties of Ste18
3.2. Role of the Association of Ste18 with a Membrane in its Aggregation and Prion-Inducing Properties
3.3. Post-Translational Modifications Modulate Levels of Ste18 but not its Aggregation and Prion-Inducing Abilities
3.4. Protein Aggregation Based Memory of Environmental and Physiological Stimuli
4. Materials and Methods
4.1. Strains and Plasmids
4.2. Growth Conditions and Phenotype Detection
4.3. Protein Isolation and Analysis
4.4. Fluorescence Microscopy
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Prusiner, S.B. Prions. Proc. Natl. Acad. Sci. USA 1998, 95, 13363–13383. [Google Scholar] [CrossRef]
- Wickner, R.B. Yeast and Fungal Prions. Cold Spring Harb. Perspect. Biol. 2016, 8. [Google Scholar] [CrossRef]
- McGlinchey, R.P.; Kryndushkin, D.; Wickner, R.B. Suicidal [PSI+] is a lethal yeast prion. Proc. Natl. Acad. Sci. USA 2011, 108, 5337–5341. [Google Scholar] [CrossRef]
- Halfmann, R.; Jarosz, D.F.; Jones, S.K.; Chang, A.; Lancaster, A.K.; Lindquist, S. Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature 2012, 482, 363–368. [Google Scholar] [CrossRef]
- Jarosz, D.F.; Brown, J.C.S.; Walker, G.A.; Datta, M.S.; Ung, W.L.; Lancaster, A.K.; Rotem, A.; Chang, A.; Newby, G.A.; Weitz, D.A.; et al. Cross-kingdom chemical communication drives a heritable, mutually beneficial prion-based transformation of metabolism. Cell 2014, 158, 1083–1093. [Google Scholar] [CrossRef]
- Jarosz, D.F.; Lancaster, A.K.; Brown, J.C.; Lindquist, S. An evolutionarily conserved prion-like element converts wild fungi from metabolic specialists to generalists. Cell 2014, 158, 1072–1082. [Google Scholar] [CrossRef][Green Version]
- Newby, G.A.; Kayatekin, C. Microbial specialization by prions. Prion 2018, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Saupe, S.J. A short history of small s: A prion of the fungus Podospora anserina. Prion 2007, 1, 110–115. [Google Scholar] [CrossRef][Green Version]
- Caudron, F.; Barral, Y. A super-assembly of Whi3 encodes memory of deceptive encounters by single cells during yeast courtship. Cell 2013, 155, 1244–1257. [Google Scholar] [CrossRef] [PubMed]
- Liebman, S.W.; Chernoff, Y.O. Prions in yeast. Genetics 2012, 191, 1041–1072. [Google Scholar] [CrossRef]
- Tuite, M.F.; Serio, T.R. The prion hypothesis: From biological anomaly to basic regulatory mechanism. Nat. Rev. Mol. Cell Biol. 2010, 11, 823–833. [Google Scholar] [CrossRef] [PubMed]
- Killian, A.N.; Miller, S.C.; Hines, J.K. Impact of Amyloid Polymorphism on Prion-Chaperone Interactions in Yeast. Viruses 2019, 11. [Google Scholar] [CrossRef] [PubMed]
- Chernova, T.A.; Wilkinson, K.D.; Chernoff, Y.O. Prions, Chaperones, and Proteostasis in Yeast. Cold Spring Harb. Perspect. Biol. 2017, 9. [Google Scholar] [CrossRef]
- Chernova, T.A.; Wilkinson, K.D.; Chernoff, Y.O. Physiological and environmental control of yeast prions. FEMS Microbiol. Rev. 2014, 38, 326–344. [Google Scholar] [CrossRef] [PubMed]
- Chernova, T.A.; Chernoff, Y.O.; Wilkinson, K.D. Yeast Models for Amyloids and Prions: Environmental. Modul. Drug Discov. Mol. 2019, 24. [Google Scholar] [CrossRef]
- Tuite, M.F. The natural history of yeast prions. Adv. Appl. Microbiol. 2013, 84, 85–137. [Google Scholar] [CrossRef] [PubMed]
- Derkatch, I.L.; Bradley, M.E.; Hong, J.Y.; Liebman, S.W. Prions affect the appearance of other prions: The story of [PIN(+)]. Cell 2001, 106, 171–182. [Google Scholar] [CrossRef]
- Osherovich, L.Z.; Weissman, J.S. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI(+)] prion. Cell 2001, 106, 183–194. [Google Scholar] [CrossRef]
- Derkatch, I.L.; Uptain, S.M.; Outeiro, T.F.; Krishnan, R.; Lindquist, S.L.; Liebman, S.W. Effects of Q/N-rich, polyQ, and non-polyQ amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro. Proc. Natl. Acad. Sci. USA 2004, 101, 12934–12939. [Google Scholar] [CrossRef]
- Yang, Z.; Hong, J.Y.; Derkatch, I.L.; Liebman, S.W. Heterologous gln/asn-rich proteins impede the propagation of yeast prions by altering chaperone availability. PLoS Genet. 2013, 9, e1003236. [Google Scholar] [CrossRef]
- Li, X.; Rayman, J.B.; Kandel, E.R.; Derkatch, I.L. Functional role of Tia1/Pub1 and Sup35 prion domains: Directing protein synthesis machinery to the tubulin cytoskeleton. Mol. Cell 2014, 55, 305–318. [Google Scholar] [CrossRef] [PubMed]
- Gokhale, K.C.; Newnam, G.P.; Sherman, M.Y.; Chernoff, Y.O. Modulation of prion-dependent polyglutamine aggregation and toxicity by chaperone proteins in the yeast model. J. Biol. Chem. 2005, 280, 22809–22818. [Google Scholar] [CrossRef]
- Gong, H.; Romanova, N.V.; Allen, K.D.; Chandramowlishwaran, P.; Gokhale, K.; Newnam, G.P.; Mieczkowski, P.; Sherman, M.Y.; Chernoff, Y.O. Polyglutamine toxicity is controlled by prion composition and gene dosage in yeast. PLoS Genet. 2012, 8, e1002634. [Google Scholar] [CrossRef]
- Meriin, A.B.; Zhang, X.; He, X.; Newnam, G.P.; Chernoff, Y.O.; Sherman, M.Y. Huntington toxicity in yeast model depends on polyglutamine aggregation mediated by a prion-like protein Rnq1. J. Cell Biol. 2002, 157, 997–1004. [Google Scholar] [CrossRef]
- Jucker, M.; Walker, L.C. Pathogenic protein seeding in Alzheimer disease and other neurodegenerative disorders. Ann. Neurol. 2011, 70, 532–540. [Google Scholar] [CrossRef] [PubMed]
- Walker, L.C.; LeVine, H., 3rd. Corruption and spread of pathogenic proteins in neurodegenerative diseases. J. Biol. Chem. 2012, 287, 33109–33115. [Google Scholar] [CrossRef]
- Liu, B.; Larsson, L.; Caballero, A.; Hao, X.; Oling, D.; Grantham, J.; Nystrom, T. The polarisome is required for segregation and retrograde transport of protein aggregates. Cell 2010, 140, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Toret, C.P.; Drubin, D.G. The budding yeast endocytic pathway. J. Cell Sci. 2006, 119, 4585–4587. [Google Scholar] [CrossRef] [PubMed]
- Ganusova, E.E.; Ozolins, L.N.; Bhagat, S.; Newnam, G.P.; Wegrzyn, R.D.; Sherman, M.Y.; Chernoff, Y.O. Modulation of prion formation, aggregation, and toxicity by the actin cytoskeleton in yeast. Mol. Cell. Biol. 2006, 26, 617–629. [Google Scholar] [CrossRef]
- Bailleul, P.A.; Newnam, G.P.; Steenbergen, J.N.; Chernoff, Y.O. Genetic study of interactions between the cytoskeletal assembly protein sla1 and prion-forming domain of the release factor Sup35 (eRF3) in Saccharomyces cerevisiae. Genetics 1999, 153, 81–94. [Google Scholar] [PubMed]
- Chernova, T.A.; Romanyuk, A.V.; Karpova, T.S.; Shanks, J.R.; Ali, M.; Moffatt, N.; Howie, R.L.; O’Dell, A.; McNally, J.G.; Liebman, S.W.; et al. Prion induction by the short-lived, stress-induced protein Lsb2 is regulated by ubiquitination and association with the actin cytoskeleton. Mol. Cell 2011, 43, 242–252. [Google Scholar] [CrossRef] [PubMed]
- Dowell, S.J.; Brown, A.J. Yeast assays for G protein-coupled receptors. Methods Mol. Biol. 2009, 552, 213–229. [Google Scholar] [CrossRef] [PubMed]
- Cattaneo, F.; Guerra, G.; Parisi, M.; De Marinis, M.; Tafuri, D.; Cinelli, M.; Ammendola, R. Cell-Surface Receptors Transactivation Mediated by G Protein-Coupled Receptors. Int. J. Mol. Sci. 2014, 15, 19700–19728. [Google Scholar] [CrossRef] [PubMed]
- Dohlman, H.G.; Thorner, J.W. Regulation of G protein-initiated signal transduction in yeast: Paradigms and principles. Annu. Rev. Biochem. 2001, 70, 703–754. [Google Scholar] [CrossRef] [PubMed]
- Alvaro, C.G.; O’Donnell, A.F.; Prosser, D.C.; Augustine, A.A.; Goldman, A.; Brodsky, J.L.; Cyert, M.S.; Wendland, B.; Thorner, J. Specific alpha-arrestins negatively regulate Saccharomyces cerevisiae pheromone response by down-modulating the G-protein-coupled receptor Ste2. Mol. Cell. Biol. 2014, 34, 2660–2681. [Google Scholar] [CrossRef] [PubMed]
- Arkowitz, R.A. Chemical gradients and chemotropism in yeast. Cold Spring Harb. Perspect. Biol. 2009, 1, a001958. [Google Scholar] [CrossRef]
- Dewhurst, H.M.; Choudhury, S.; Torres, M.P. Structural Analysis of PTM Hotspots (SAPH-ire)--A Quantitative Informatics Method Enabling the Discovery of Novel Regulatory Elements in Protein Families. Mol. Cell Proteomics 2015, 14, 2285–2297. [Google Scholar] [CrossRef]
- Choudhury, S.; Baradaran-Mashinchi, P.; Torres, M.P. Negative Feedback Phosphorylation of Ggamma Subunit Ste18 and the Ste5 Scaffold Synergistically Regulates MAPK Activation in Yeast. Cell Rep. 2018, 23, 1504–1515. [Google Scholar] [CrossRef]
- Hirschman, J.E.; Jenness, D.D. Dual lipid modification of the yeast ggamma subunit Ste18p determines membrane localization of Gbetagamma. Mol. Cell. Biol. 1999, 19, 7705–7711. [Google Scholar] [CrossRef][Green Version]
- Manahan, C.L.; Patnana, M.; Blumer, K.J.; Linder, M.E. Dual lipid modification motifs in G(alpha) and G(gamma) subunits are required for full activity of the pheromone response pathway in Saccharomyces cerevisiae. Mol. Biol. Cell 2000, 11, 957–968. [Google Scholar] [CrossRef]
- Swaney, D.L.; Beltrao, P.; Starita, L.; Guo, A.; Rush, J.; Fields, S.; Krogan, N.J.; Villen, J. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat. Methods 2013, 10, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Chernoff, Y.O.; Uptain, S.M.; Lindquist, S.L. Analysis of prion factors in yeast. Methods Enzymol. 2002, 351, 499–538. [Google Scholar] [PubMed]
- Bagriantsev, S.N.; Kushnirov, V.V.; Liebman, S.W. Analysis of amyloid aggregates using agarose gel electrophoresis. Methods Enzymol. 2006, 412, 33–48. [Google Scholar] [CrossRef] [PubMed]
- Grishin, A.V.; Weiner, J.L.; Blumer, K.J. Biochemical and genetic analysis of dominant-negative mutations affecting a yeast G-protein gamma subunit. Mol. Cell. Biol. 1994, 14, 4571–4578. [Google Scholar] [CrossRef][Green Version]
- Hirschman, J.E.; De Zutter, G.S.; Simonds, W.F.; Jenness, D.D. The G beta gamma complex of the yeast pheromone response pathway. Subcellular fractionation and protein-protein interactions. J. Biol. Chem. 1997, 272, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.B.; Znassi, N.; Chateau, M.T.; Kajava, A.V. A structure-based approach to predict predisposition to amyloidosis. Alzheimers Dement. 2015, 11, 681–690. [Google Scholar] [CrossRef]
- Halfmann, R.; Alberti, S.; Krishnan, R.; Lyle, N.; O’Donnell, C.W.; King, O.D.; Berger, B.; Pappu, R.V.; Lindquist, S. Opposing effects of glutamine and asparagine govern prion formation by intrinsically disordered proteins. Mol. Cell 2011, 43, 72–84. [Google Scholar] [CrossRef] [PubMed]
- Chernova, T.A.; Kiktev, D.A.; Romanyuk, A.V.; Shanks, J.R.; Laur, O.; Ali, M.; Ghosh, A.; Kim, D.; Yang, Z.; Mang, M.; et al. Yeast Short-Lived Actin-Associated Protein Forms a Metastable Prion in Response to Thermal Stress. Cell Rep. 2017, 18, 751–761. [Google Scholar] [CrossRef]
- Toombs, J.A.; McCarty, B.R.; Ross, E.D. Compositional determinants of prion formation in yeast. Mol. Cell. Biol. 2010, 30, 319–332. [Google Scholar] [CrossRef]
- Derkatch, I.L.; Bradley, M.E.; Zhou, P.; Chernoff, Y.O.; Liebman, S.W. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics 1997, 147, 507–719. [Google Scholar]
- Allen, K.D.; Chernova, T.A.; Tennant, E.P.; Wilkinson, K.D.; Chernoff, Y.O. Effects of ubiquitin system alterations on the formation and loss of a yeast prion. J. Biol. Chem. 2007, 282, 3004–3013. [Google Scholar] [CrossRef]
- Chernova, T.A.; Allen, K.D.; Wesoloski, L.M.; Shanks, J.R.; Chernoff, Y.O.; Wilkinson, K.D. Pleiotropic effects of Ubp6 loss on drug sensitivities and yeast prion are due to depletion of the free ubiquitin pool. J. Biol. Chem. 2003, 278, 52102–52115. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Hochstrasser, M. Autocatalytic subunit processing couples active site formation in the 20S proteasome to completion of assembly. Cell 1996, 86, 961–972. [Google Scholar] [CrossRef]
- Buchanan, B.W.; Lloyd, M.E.; Engle, S.M.; Rubenstein, E.M. Cycloheximide Chase Analysis of Protein Degradation in Saccharomyces cerevisiae. J. Vis. Exp. 2016. [Google Scholar] [CrossRef]
- Alberti, S.; Halfmann, R.; King, O.; Kapila, A.; Lindquist, S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 2009, 137, 146–158. [Google Scholar] [CrossRef] [PubMed]
- Bondarev, S.A.; Bondareva, O.V.; Zhouravleva, G.A.; Kajava, A.V. BetaSerpentine: A bioinformatics tool for reconstruction of amyloid structures. Bioinformatics 2018, 34, 599–608. [Google Scholar] [CrossRef]
- Roche, D.B.; Villain, E.; Kajava, A.V. Usage of a dataset of NMR resolved protein structures to test aggregation versus solubility prediction algorithms. Sci. A Publ. Protein Soc. 2017, 26, 1864–1869. [Google Scholar] [CrossRef]
- Suzuki, G.; Tanaka, M. Expanding the yeast prion world: Active prion conversion of non-glutamine/asparagine-rich Mod5 for cell survival. Prion 2013, 7, 109–113. [Google Scholar] [CrossRef]
- Maddelein, M.L.; Dos Reis, S.; Duvezin-Caubet, S.; Coulary-Salin, B.; Saupe, S.J. Amyloid aggregates of the HET-s prion protein are infectious. Proc. Natl. Acad. Sci. USA 2002, 99, 7402–7407. [Google Scholar] [CrossRef]
- Sondheimer, N.; Lindquist, S. Rnq1: An epigenetic modifier of protein function in yeast. Mol. Cell 2000, 5, 163–172. [Google Scholar] [CrossRef]
- Suzuki, G.; Shimazu, N.; Tanaka, M. A yeast prion, Mod5, promotes acquired drug resistance and cell survival under environmental stress. Science 2012, 336, 355–359. [Google Scholar] [CrossRef] [PubMed]
- Schlissel, G.; Krzyzanowski, M.K.; Caudron, F.; Barral, Y.; Rine, J. Aggregation of the Whi3 protein, not loss of heterochromatin, causes sterility in old yeast cells. Science 2017, 355, 1184–1187. [Google Scholar] [CrossRef]
- Sarnataro, D. Attempt to Untangle the Prion-Like Misfolding Mechanism for Neurodegenerative Diseases. Int. J. Mol. Sci. 2018, 19. [Google Scholar] [CrossRef] [PubMed]
- Pepe, A.; Avolio, R.; Matassa, D.S.; Esposito, F.; Nitsch, L.; Zurzolo, C.; Paladino, S.; Sarnataro, D. Regulation of sub-compartmental targeting and folding properties of the Prion-like protein Shadoo. Sci. Rep. 2017, 7, 3731. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Derkatch, I.L.; Liebman, S.W. The relationship between visible intracellular aggregates that appear after overexpression of Sup35 and the yeast prion-like elements [PSI (+)] and [PIN (+)]. Mol. Microbiol. 2001, 39, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Mathur, V.; Taneja, V.; Sun, Y.; Liebman, S.W. Analyzing the birth and propagation of two distinct prions, [PSI+] and [Het-s] (y), in yeast. Mol. Biol. Cell 2010, 21, 1449–1461. [Google Scholar] [CrossRef][Green Version]
- Finegold, A.A.; Schafer, W.R.; Rine, J.; Whiteway, M.; Tamanoi, F. Common modifications of trimeric G proteins and ras protein: Involvement of polyisoprenylation. Science 1990, 249, 165–169. [Google Scholar] [CrossRef]
- Ardito, F.; Giuliani, M.; Perrone, D.; Troiano, G.; Lo Muzio, L. The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy (Review). Int. J. Mol. Med. 2017, 40, 271–280. [Google Scholar] [CrossRef]
- Liu, Z.; Li, T.; Li, P.; Wei, N.; Zhao, Z.; Liang, H.; Ji, X.; Chen, W.; Xue, M.; Wei, J. The Ambiguous Relationship of Oxidative Stress, Tau Hyperphosphorylation, and Autophagy Dysfunction in Alzheimer’s Disease. Oxid. Med. Cell. Longev. 2015, 2015, 352723. [Google Scholar] [CrossRef]
- Marcelli, S.; Corbo, M.; Iannuzzi, F.; Negri, L.; Blandini, F.; Nistico, R.; Feligioni, M. The Involvement of Post-Translational Modifications in Alzheimer’s Disease. Curr. Alzheimer Res. 2018, 15, 313–335. [Google Scholar] [CrossRef]
- Li, H.Y.; Yeh, P.A.; Chiu, H.C.; Tang, C.Y.; Tu, B.P. Hyperphosphorylation as a defense mechanism to reduce TDP-43 aggregation. PLoS ONE 2011, 6, e23075. [Google Scholar] [CrossRef] [PubMed]
- Nonaka, T.; Suzuki, G.; Tanaka, Y.; Kametani, F.; Hirai, S.; Okado, H.; Miyashita, T.; Saitoe, M.; Akiyama, H.; Masai, H.; et al. Phosphorylation of TAR DNA-binding Protein of 43 kDa (TDP-43) by Truncated Casein Kinase 1delta Triggers Mislocalization and Accumulation of TDP-43. J. Biol. Chem. 2016, 291, 5473–5483. [Google Scholar] [CrossRef] [PubMed]
- Rhoads, S.N.; Monahan, Z.T.; Yee, D.S.; Shewmaker, F.P. The Role of Post-Translational Modifications on Prion-Like Aggregation and Liquid-Phase Separation of FUS. Int. J. Mol. Sci. 2018, 19. [Google Scholar] [CrossRef] [PubMed]
- Gnad, F.; de Godoy, L.M.; Cox, J.; Neuhauser, N.; Ren, S.; Olsen, J.V.; Mann, M. High-accuracy identification and bioinformatic analysis of in vivo protein phosphorylation sites in yeast. Proteomics 2009, 9, 4642–4652. [Google Scholar] [CrossRef]
- Rangarajan, N.; Gordy, C.L.; Askew, L.; Bevill, S.M.; Elston, T.C.; Errede, B.; Hurst, J.H.; Kelley, J.B.; Sheetz, J.B.; Suzuki, S.K.; et al. Systematic analysis of F-box proteins reveals a new branch of the yeast mating pathway. J. Biol. Chem. 2019, 294, 14717–14731. [Google Scholar] [CrossRef] [PubMed]
- Caudron, F.; Barral, Y. Mnemons: Encoding memory by protein super-assembly. Microb. Cell 2014, 1, 100–102. [Google Scholar] [CrossRef] [PubMed]
- Majumdar, A.; Cesario, W.C.; White-Grindley, E.; Jiang, H.; Ren, F.; Khan, M.R.; Li, L.; Choi, E.M.; Kannan, K.; Guo, F.; et al. Critical role of amyloid-like oligomers of Drosophila Orb2 in the persistence of memory. Cell 2012, 148, 515–529. [Google Scholar] [CrossRef]
- Fioriti, L.; Myers, C.; Huang, Y.Y.; Li, X.; Stephan, J.S.; Trifilieff, P.; Colnaghi, L.; Kosmidis, S.; Drisaldi, B.; Pavlopoulos, E.; et al. The Persistence of Hippocampal-Based Memory Requires Protein Synthesis Mediated by the Prion-like Protein CPEB3. Neuron 2015, 86, 1433–1448. [Google Scholar] [CrossRef]
- Gustin, M.C.; Albertyn, J.; Alexander, M.; Davenport, K. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1998, 62, 1264–1300. [Google Scholar] [CrossRef]
- Mendenhall, M.D.; Hodge, A.E. Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1998, 62, 1191–1243. [Google Scholar] [CrossRef]
- Merlini, L.; Dudin, O.; Martin, S.G. Mate and fuse: How yeast cells do it. Open Biol. 2013, 3, 130008. [Google Scholar] [CrossRef] [PubMed]
- Butty, A.C.; Pryciak, P.M.; Huang, L.S.; Herskowitz, I.; Peter, M. The role of Far1p in linking the heterotrimeric G protein to polarity establishment proteins during yeast mating. Science 1998, 282, 1511–1516. [Google Scholar] [CrossRef] [PubMed]
- Nern, A.; Arkowitz, R.A. A Cdc24p-Far1p-Gbetagamma protein complex required for yeast orientation during mating. J. Cell Biol. 1999, 144, 1187–1202. [Google Scholar] [CrossRef] [PubMed]
- Nern, A.; Arkowitz, R.A. G proteins mediate changes in cell shape by stabilizing the axis of polarity. Mol. Cell 2000, 5, 853–864. [Google Scholar] [CrossRef]
- Wang, X.; Tian, W.; Banh, B.T.; Statler, B.M.; Liang, J.; Stone, D.E. Mating yeast cells use an intrinsic polarity site to assemble a pheromone-gradient tracking machine. J. Cell Biol. 2019, 218, 3730–3752. [Google Scholar] [CrossRef] [PubMed]
- Madania, A.; Dumoulin, P.; Grava, S.; Kitamoto, H.; Scharer-Brodbeck, C.; Soulard, A.; Moreau, V.; Winsor, B. The Saccharomyces cerevisiae homologue of human Wiskott-Aldrich syndrome protein Las17p interacts with the Arp2/3 complex. Mol. Biol. Cell 1999, 10, 3521–3538. [Google Scholar] [CrossRef] [PubMed]
- Chernova, T.A.; Chernoff, Y.O.; Wilkinson, K.D. Prion-based memory of heat stress in yeast. Prion 2017, 11, 151–161. [Google Scholar] [CrossRef]
- Nagiec, M.J.; Dohlman, H.G. Checkpoints in a yeast differentiation pathway coordinate signaling during hyperosmotic stress. PLoS Genet. 2012, 8, e1002437. [Google Scholar] [CrossRef]
- Dupre, D.J.; Robitaille, M.; Rebois, R.V.; Hebert, T.E. The role of Gbetagamma subunits in the organization, assembly, and function of GPCR signaling complexes. Annu. Rev. Pharmacol. Toxicol. 2009, 49, 31–56. [Google Scholar] [CrossRef]
- Campanale, J.P.; Sun, T.Y.; Montell, D.J. Development and dynamics of cell polarity at a glance. J. Cell Sci. 2017, 130, 1201–1207. [Google Scholar] [CrossRef]
- Chen, P.; Johnson, P.; Sommer, T.; Jentsch, S.; Hochstrasser, M. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MAT alpha 2 repressor. Cell 1993, 74, 357–369. [Google Scholar] [CrossRef]
- Longtine, M.S.; McKenzie, A., 3rd; Demarini, D.J.; Shah, N.G.; Wach, A.; Brachat, A.; Philippsen, P.; Pringle, J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 1998, 14, 953–961. [Google Scholar] [CrossRef]
- Sikorski, R.S.; Hieter, P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 1989, 122, 19–27. [Google Scholar]
- Kiktev, D.A.; Patterson, J.C.; Muller, S.; Bariar, B.; Pan, T.; Chernoff, Y.O. Regulation of chaperone effects on a yeast prion by cochaperone Sgt2. Mol. Cell. Biol. 2012, 32, 4960–4970. [Google Scholar] [CrossRef] [PubMed]
- Sherman, F. Getting started with yeast. Methods Enzymol. 2002, 350, 3–41. [Google Scholar] [PubMed]
- Dohlman, H.G.; Song, J.; Ma, D.; Courchesne, W.E.; Thorner, J. Sst2, a negative regulator of pheromone signaling in the yeast Saccharomyces cerevisiae: Expression, localization, and genetic interaction and physical association with Gpa1 (the G-protein alpha subunit). Mol. Cell. Biol. 1996, 16, 5194–5209. [Google Scholar] [CrossRef] [PubMed]
- Allen, K.D.; Wegrzyn, R.D.; Chernova, T.A.; Muller, S.; Newnam, G.P.; Winslett, P.A.; Wittich, K.B.; Wilkinson, K.D.; Chernoff, Y.O. Hsp70 chaperones as modulators of prion life cycle: Novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+]. Genetics 2005, 169, 1227–1242. [Google Scholar] [CrossRef] [PubMed]
- Kryndushkin, D.S.; Alexandrov, I.M.; Ter-Avanesyan, M.D.; Kushnirov, V.V. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J. Biol. Chem. 2003, 278, 49636–49643. [Google Scholar] [CrossRef]
- Drozdova, P.B.; Barbitoff, Y.A.; Belousov, M.V.; Skitchenko, R.K.; Rogoza, T.M.; Leclercq, J.Y.; Kajava, A.V.; Matveenko, A.G.; Zhouravleva, G.A.; Bondarev, S.A. Estimation of amyloid aggregate sizes with semi-denaturing detergent agarose gel electrophoresis and its limitations. Prion 2020, 14, 118–128. [Google Scholar] [CrossRef]

| Strain | Genotype | Source |
|---|---|---|
| GT409 (WTY222) | MATa ade1–14 (UGA) his3-∆200 leu2–3,112 lys2–801 trp1 ura3–52 [psi− pin−] | [51] |
| WTY775 | ste18Δ::kanMX6 disruption in GT409 | This study |
| WTY770 | ste4Δ::kanMX6 disruption in GT409 | This study |
| MHY501 | MATα his3-Δ200 leu2-3,112 lys2-801 trp1 ura3-52 | [91] |
| MHY3646 | doa3-1 derivative of MHY501 | [91] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chernova, T.A.; Yang, Z.; Karpova, T.S.; Shanks, J.R.; Shcherbik, N.; Wilkinson, K.D.; Chernoff, Y.O. Aggregation and Prion-Inducing Properties of the G-Protein Gamma Subunit Ste18 are Regulated by Membrane Association. Int. J. Mol. Sci. 2020, 21, 5038. https://doi.org/10.3390/ijms21145038
Chernova TA, Yang Z, Karpova TS, Shanks JR, Shcherbik N, Wilkinson KD, Chernoff YO. Aggregation and Prion-Inducing Properties of the G-Protein Gamma Subunit Ste18 are Regulated by Membrane Association. International Journal of Molecular Sciences. 2020; 21(14):5038. https://doi.org/10.3390/ijms21145038
Chicago/Turabian StyleChernova, Tatiana A., Zhen Yang, Tatiana S. Karpova, John R. Shanks, Natalia Shcherbik, Keith D. Wilkinson, and Yury O. Chernoff. 2020. "Aggregation and Prion-Inducing Properties of the G-Protein Gamma Subunit Ste18 are Regulated by Membrane Association" International Journal of Molecular Sciences 21, no. 14: 5038. https://doi.org/10.3390/ijms21145038
APA StyleChernova, T. A., Yang, Z., Karpova, T. S., Shanks, J. R., Shcherbik, N., Wilkinson, K. D., & Chernoff, Y. O. (2020). Aggregation and Prion-Inducing Properties of the G-Protein Gamma Subunit Ste18 are Regulated by Membrane Association. International Journal of Molecular Sciences, 21(14), 5038. https://doi.org/10.3390/ijms21145038







