Abstract
Adult neurogenesis is a multistage process by which neurons are generated and integrated into existing neuronal circuits. In the adult brain, neurogenesis is mainly localized in two specialized niches, the subgranular zone (SGZ) of the dentate gyrus and the subventricular zone (SVZ) adjacent to the lateral ventricles. Neurogenesis plays a fundamental role in postnatal brain, where it is required for neuronal plasticity. Moreover, perturbation of adult neurogenesis contributes to several human diseases, including cognitive impairment and neurodegenerative diseases. The interplay between extrinsic and intrinsic factors is fundamental in regulating neurogenesis. Over the past decades, several studies on intrinsic pathways, including transcription factors, have highlighted their fundamental role in regulating every stage of neurogenesis. However, it is likely that transcriptional regulation is part of a more sophisticated regulatory network, which includes epigenetic modifications, non-coding RNAs and metabolic pathways. Here, we review recent findings that advance our knowledge in epigenetic, transcriptional and metabolic regulation of adult neurogenesis in the SGZ of the hippocampus, with a special attention to the p53-family of transcription factors.
1. Introduction
The development of the mammalian central nervous system (CNS) is a spatial and temporal regulated process that evolves from a small number of cells that proliferate, acquire regional identities and give rise to different cell types [1]. These cells have been classified as neural stem cells (NSCs) and have the capability to produce identical NSCs progeny through symmetric cell division (self-renewal) and to differentiate into specialized brain cell types such as neurons, astrocytes and oligodendrocytes [2,3,4]. Under physiological conditions, adult neurogenesis is restricted in neurogenic niches that are localized in two different regions of the brain, the subventricular zone (SVZ) of the lateral ventricle where new neurons are generated and then migrate to the olfactory bulb (OB), and the subgranular zone (SGZ) of the dentate gyrus (DG) of the hippocampus [4,5,6].
Although the process of neurogenesis is complex and highly regulated, it can be divided into six different stages [7]. Stage 1 occurs 1–3 days after birth and is called the proliferation phase; during this, neuronal progenitor cells (NPCs) are capable of proliferation and multi-potential differentiation but are unable to self-renew. Stages 2–4 occur approximately 1 week after birth and are collectively called the differentiation phase; during this time, neuronal progenitors exit from the cell cycle and are committed to the neuronal lineage. After the commitment, immature neurons enter stage 5, known as the migration phase, to reach their final destination. This event occurs between 2 to 3 weeks after birth. Post-mitotic neurons start to extend their axonal projections and dendritic growth starts. Finally, stage 6 of adult neurogenesis occurs at approximately 4 weeks after birth and is the synaptic integration where newly generated neurons establish their synaptic contacts into the pre-existing circuits [8,9]. Overall, it takes about 2–4 months for indistinguishable adult-born neurons to fully integrate with surrounding cells and incorporate into the hippocampal circuits [10,11].
Multiple lines of evidence indicate that both extrinsic cues and intrinsic pathways are required for the regulation of this developmental process [8,12,13]. Among the intrinsic pathways, multilayered regulatory networks display connections between multiple complex transcription factors, epigenetic control, non-coding RNAs, signalling and metabolic pathways [14,15].
Understanding how neuronal differentiation is regulated in adulthood is crucial for understanding the pathogenesis of several disorders that affect the CNS including neurodegenerative diseases, psychiatric and neurodevelopmental disorders. The present review discusses and summarises the recent research on adult neurogenesis to improve our understanding of the transcriptional, epigenetic and metabolic regulation of neurogenesis in the human brain.
2. Epigenetic Regulation of Adult Neurogenesis in SGZ of the DG
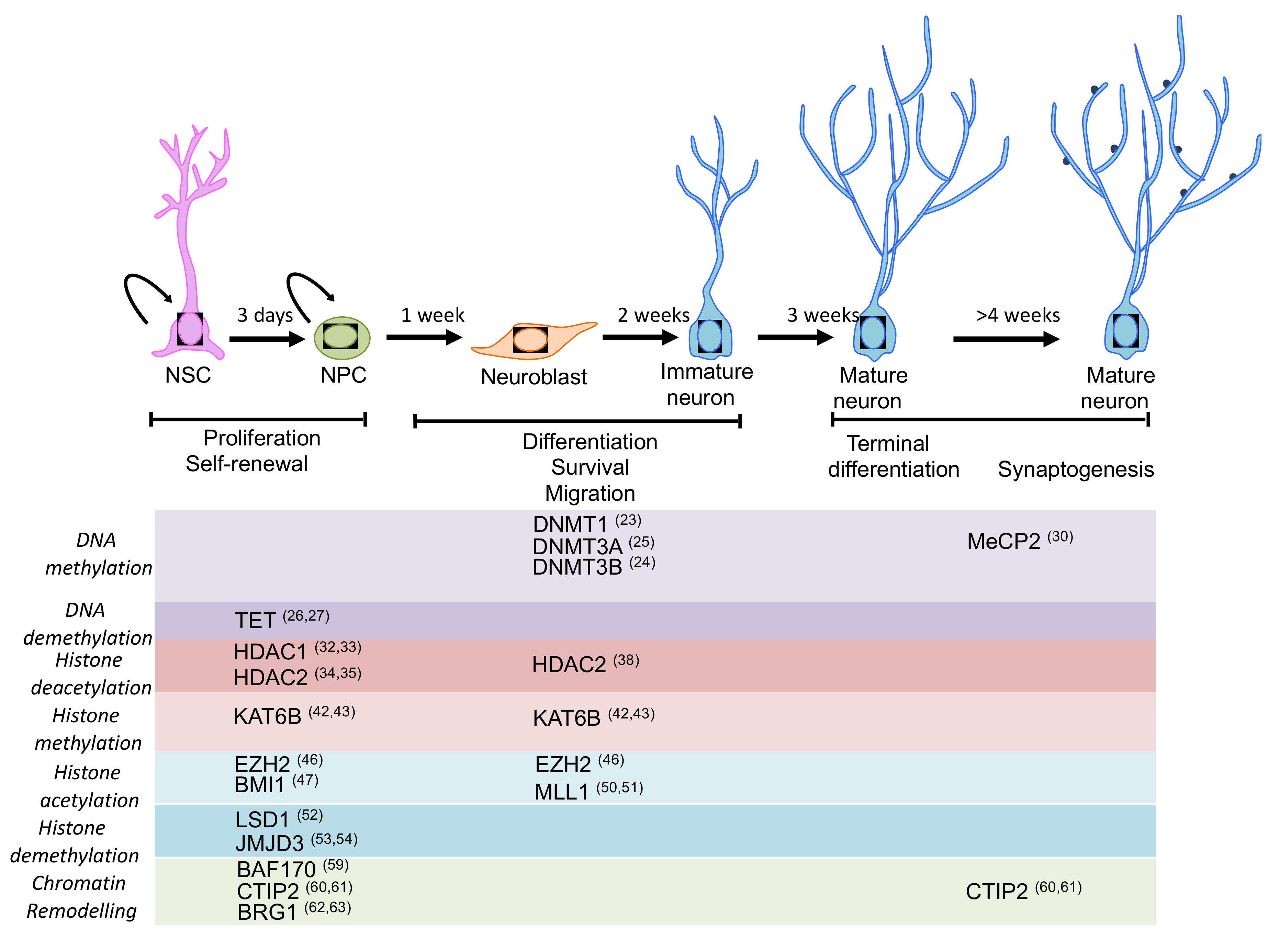
Epigenetic mechanisms such as DNA methylation, histone post-translational modifications and chromatin remodelling, are essential players in determining adult neuronal differentiation by regulation of gene expression [16]. Moreover, alterations (mutations of regulators and/or deregulations) of epigenetic mechanisms have critical implications in several human conditions, including neurological disorders, such as autism spectrum disorder, mental retardation and epilepsy. This highlights the important roles that epigenetic mechanisms exert for brain development and function [17,18]. See Figure 1 for a schematic view.
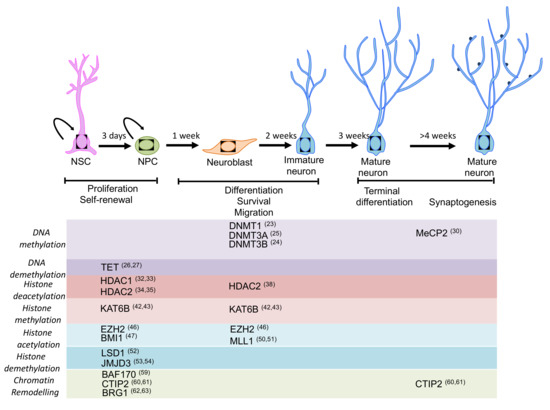
Figure 1.
Schematic representation of the main stages of neurogenesis and the epigenetic machinery involved in the regulation of neurogenesis. Expression pattern of the main epigenetic regulators for which a function in adult neurogenesis has been described or proposed. Most of the regulators play an important role in self-renewal, proliferation and fate specification during neurogenesis (please see main text for details).
2.1. DNA Methylation
Methylation of the cytosine (5mC) of the dinucleotide CpG islands within the promoters of genes has been the first described epigenetic modification [19,20]. CpG islands are short stretches of palindromic DNA with the sequence “CpG” that code for cytosine (C) and guanine (G) nucleotides with the “p” representing the linking phosphate. Typically, methylation of the CpG islands is associated with their transcription repression [21]. Moreover, 5mC is enzymatically mediated by DNA methyl transferases (DNMTs), while removal of the methyl group is triggered by the ten-eleven translocation (TET) family [21].
DNA methylation is a key process by which pluripotency genes are silenced during the neural induction of embryonic stem cells to NSCs, supporting the importance of DNA methylation during neurogenesis [22]. While DNMT1 controls the timing of astrogliogenesis through regulation of JAK-STAT signalling [23], DNMT3A and DNMT3B are crucial for neuron specification. DNMT3A is expressed in neurons and its expression increases postnatally. Ablation of DNMT3A in the brain (using Nestin-Cre system) results in an impaired postnatal neurogenesis, reduced number of neurons and mice death in early postnatal stages [24]. During neurogenesis, DNMT3B is expressed in NPCs and its deletion results in an accelerated neuronal maturation as shown by an upregulation of several neuronal genes including NeuroD1, Map2m and Ncam1 [25].
TET family enzymes are responsible for activating the erasure of the methyl group from 5mC. Deletion of Tet genes in mice using the Nestin-Cre ERT2 system leads to a reduction of the number of NPCs in the adult SGZ, and impairment in learning and memory [26]. At a molecular level, this phenotype is associated with the deregulation of several genes involved in NPCs proliferation such as, galanin (Gal), chondroitin sulphate proteoglycan 4 (Cspg4) and neuroglobin (Ngb). Moreover, neuronal activity in the adult mouse hippocampus can stimulate neurogenesis [27]. In this context, growth arrest and DNA-damage-inducible protein 45β (GADD45β) promote adult hippocampal neurogenesis by reducing the degree of DNA methylation of the promoter and therefore increasing the expression of key neuronal genes, including Bdnf and Fgf1. This results in promoting NPCs proliferation and the development of new neurons [28,29]. Moreover, deficiency of MeCp2 hinders maturation and impairs dendritic and spine morphogenesis of new neurons [30]. Hence, DNA methylation has a critical role in NPCs maintenance and fate specification in adult neurogenesis.
2.2. Histone Post-Translational Modifications
The chemical covalent modifications of the histone, including acetylation, methylation, ubiquitination, phosphorylation, ribosylation, and SUMOylation, play a key role in regulating the chromatin state [31]. In general, acetylation of specific amino acid residues and di- or tri-methylation of histone H3 lysine 4 (H3K4) are associated with active transcription. On contrary, di- or tri-methylation of H3K9 and H3K27 are stable repressive marks. Histone acetyltransferases (HATs) and histone deacetylases (HDACs) are the enzymes that regulate acetylation (transcriptional activation) or deacetylation (transcriptional silencing) of the histone, respectively.
HDACs are the best-studied histone modifiers in the context of neuronal differentiation [32]. A comprehensive gene expression analysis of the 11 isoforms of HDACs in rat brain show a ubiquitous expression of class I HDACs in the CNS, while class II HDACs show a tissue-specific pattern of expression, suggesting that HDAC isoforms expression may also be developmentally regulated [33]. Moreover, HDAC1 is mainly expressed in NSCs, while HDAC2 in mature neurons [34]. HDAC1 and HDAC2 have essential and redundant roles during brain development [35]. Indeed, deletion of HDAC1 or HDAC2 in neuronal precursors (using human glial fibrillary acidic protein (hGFAP)-Cre system) shows no obvious phenotypes, while combined deletion in developing neurons results in severe abnormalities in brain formation including, loss of hippocampal structure and disorganisation of cortical neurons. These abnormalities are mainly linked to a failure of neuronal precursors to differentiate into mature neurons and to excessive cell death [36,37]. HDAC2 has an important role in adult brain. Indeed, Nestin-Cre mediated deletion of HDAC2 shows that neurons derived from adult neurogenesis die at a specific maturation stage. However, brain development and adult stem cell fate are normal [38]. Nevertheless, embryonic neurogenesis is not affected by the loss of HDAC2 catalytic activity, possibly because HDAC1 can compensate for the lack of HDAC2 [35]. The class II HDAC3 and HDAC5 are highly expressed in neuronal stem/progenitor cells and have also been implicated in neuronal differentiation by regulating their proliferation and differentiation. Conditional deletion of HDAC3 in neurons (calcium/calmodulin-dependent protein kinase II (CaMKII)-Cre system) and glia (Nestin-Cre system) result in a disorganisation of the neocortex and cerebellum. These mice show also perinatal death [39]. Apart from deacetylation of histone proteins, HDAC family members (HDAC 1, 3, 6 and 9) by deacetylate components of the microtubule and actin cytoskeleton, regulate neurite formation, dendrite and axon growth [40,41].
Opposite to the histone deacetylation, the reverse reaction (acetylation) that is catalysed by HAT is also involved in neurogenesis. KAT6B, which is highly expressed in the adult SVZ, plays an important role in adult neurogenesis [42]. KAT6B deficient mice (Querkopfgt/gt mutant mice) show reduction in the number of NSCs and in the migrating neuroblasts in the rostral migratory stream [43].
Histone methylation/demethylation is associated with either activation or repression of transcription, depending on the position of methylated residues and on the number of methyl groups added [44]. The reaction of histone methylation is catalysed by lysine methyltransferases (KMTs). The well-known PcG repressive complex (PRC) and Trithorax active complex (TRXG) are involved in the regulation of adult NPCs and differentiated neurons [45]. Specifically, the methyltransferase component of PcG Enhancer of zeste homologue 2 (Ezh2) is expressed both in the SVZ and SGZ of adult mice. Conditional deletion (using hGFAP-Cre system) of Ezh2 in cerebellar granule cell layer, hippocampal dentate gyrus and SVZ NSCs, results in a reduced neurogenesis in both the DG and olfactory bulb [46]. This, at least in part, is linked to reduced cell proliferation. Mechanistically, Ezh2 is critical for the repression of genes involved in self-renewal and neuronal differentiation such as, Ink4a/Arf. BMI1 is another component of the PRC1 complex that is required for the self-renewal of the NSC but not for their survival and differentiation [47]. At a molecular level, constitutive deletion of BMI1 in mice results in the deregulation of p21-Rb pathway in NSC [48,49].
Mixed-lineage leukaemia 1 (MLL1) is a histone methyltransferase, member of the TRXG complex that regulates neurogenesis in the mouse postnatal brain [50]. In particular, MLL1 depletion in NSCs, using hGFAP-Cre system results in a severe impairment of neuronal differentiation, which is due to the loss of distal-less homeobox 2 (Dlx2) expression. Interestingly, deletion of MLL1 is associated with an increase of H3K27me3 that impairs the proper activation of DLX2 gene. More recently, MLL1 mainly regulates neuronal genes, including brain-specific POU-box gene, Brn4 [51].
Lysine-specific demethylase 1 (LSD1) is expressed in NSCs and is an important regulator of proliferation. Indeed, pharmacological and genetic inhibition (siRNA-expressing lentivirus) of LSD1 leads to a dramatic reduction in NSC proliferation [52]. Mechanistically, the orphan nuclear receptor TLX, also known as NR2E1TLX, recruits both LSD1 and HDAC5 on its target genes and this complex mediates gene repression.
Jumonji domain-containing protein 3 (JMJD3) is an H3K27me3-specific demethylase that is required for neuronal commitment by directly regulating the expression of Pax6, Nestin and Sox1 [53]. Moreover, JMJD3 is expressed in the adult SVZ and its conditional deletion (using hGFAP-Cre system) leads to an impaired OB neurogenesis by controlling the expression of several neurogenic genes (Myt1, Slc32a1 and Gjb6). This is via interaction with the promoter regions as well as in conjunction with neurogenic enhancer elements (I12b), which in turn regulates Dlx2 [54].
Therefore, histone post-translational modifications play a critical role in neurodevelopment and embryonic and adult neurogenesis.
2.3. Chromatin Remodelling
Together with chemical modifications of histones and methylation of DNA, specific chromatin conformation is also required for the regulation of gene expression [55]. The presence of histones in the DNA poses a barrier to gene transcription, therefore the proper density and spacing of nucleosomes should be maintained. Specialized ATP-dependent chromatin-remodelling complexes including imitation switch (ISWI), chromodomain helicase DNA-binding (CHD), switch/sucrose non-fermentable (SWI/SNF also known as BRG1/BRM associated factor, BAF) and INO80 are responsible for nucleosome occupancy and composition. Chromatin remodelling complexes are formed by the assembly of different combinations of proteins and they have emerged as important regulators of neuronal development [56,57].
BAF is a multimeric complex comprised at least 10 to 15 subunits that bind to the promoter region of active genes involved in the maintenance of neuronal development by mainly influencing the expression of essential genes for neurogenesis, cell migration and functional integration of neurons [58]. In particular, components of BAF complex promote adult neurogenesis. Indeed, it has been shown that conditional deletion of the BAF170 subunit in adult brain by using hGFAP-Cre system results in the impairment of neuronal differentiation with premature generation of astrocytes [59].
Moreover, the Ctip2 subunit is mainly expressed in postmitotic granule neurons and is required for the proper development of the hippocampus. Conditional deletion (using Emx1-Cre system) of Ctip2 in adult hippocampus results in the reduction of the NPC pool and impairment of neuronal differentiation [60]. Moreover, Ctip2 subunit plays an important role in the regulation of neuronal survival and integration of new neurons into the pre-existing hippocampal circuits [61].
To further support the role of BAF complex during neurogenesis, the BRG1 subunit has been conditionally deleted (using Nestin-Cre system) in NSCs resulting in a premature neuronal differentiation [62]. Mechanistically, BRG1 interacts with Pax6, and the complex directly activates the neurogenic transcription factors such as Sox11, Nfib and Pou3f4 to Pax6 [63].
All together, these data indicate that epigenetic mechanisms are essential for maintaining neurogenesis throughout adult life.
3. Transcriptional Regulation of Adult Neurogenesis
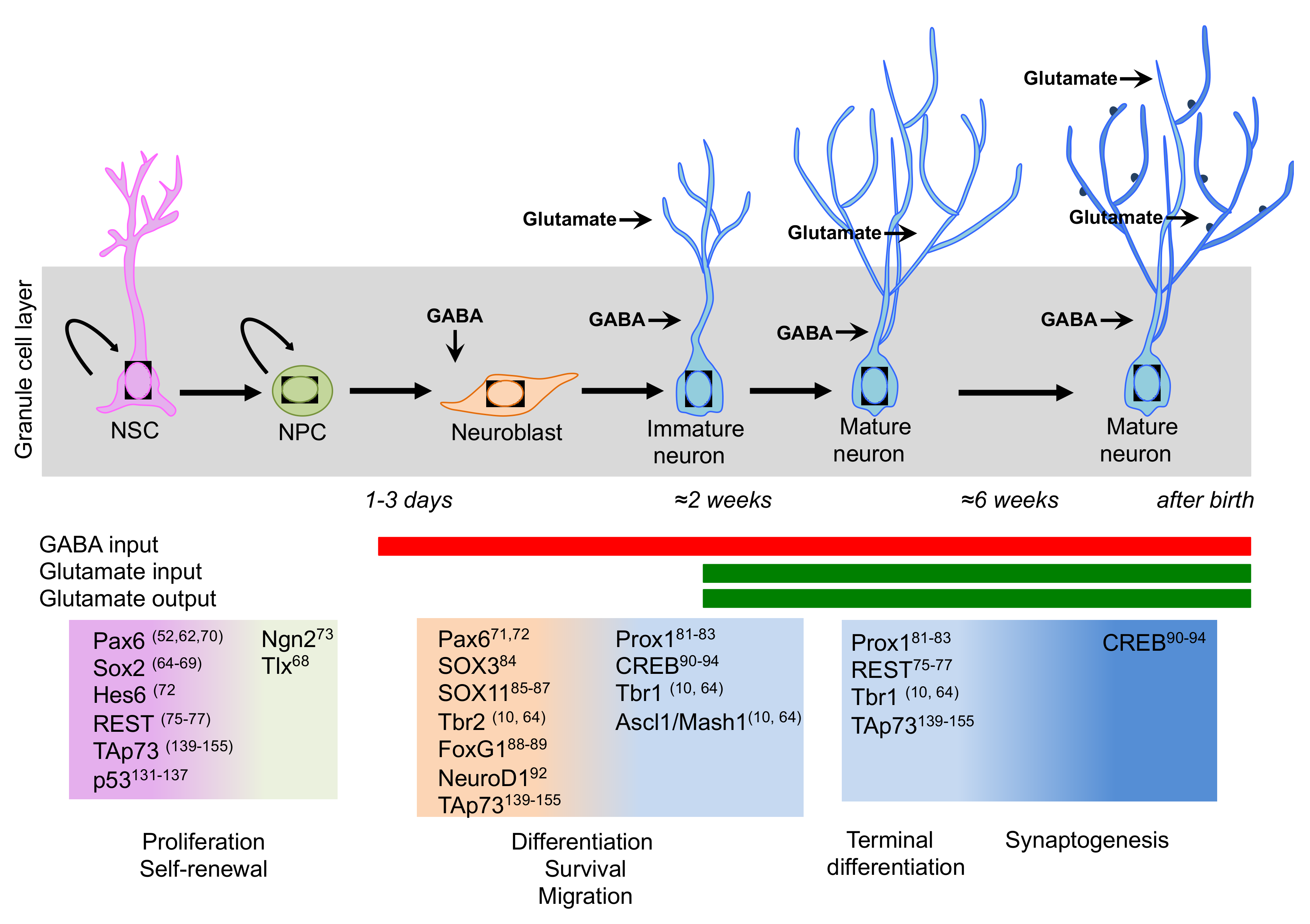
Among the intrinsic signals that regulate the sophisticated intracellular networks for NSC/NPC maintenance and neuronal differentiation, the transcription factors have been the most studied and investigated components. Progression through the different developmental stages during adult neurogenesis is accompanied by the up- and down-regulation of several transcription factors. Thus, transcription factors are the pivotal regulators of gene expression and potential mediators of extrinsic signals [64]. Interestingly, this network is almost conserved during neurogenesis in the developing and adult brain. Here, we will discuss some key neurogenic transcription factors, as visualized in Figure 2, and we will summarise recent work on the well-established role of a p53 family in regulating neurogenesis.
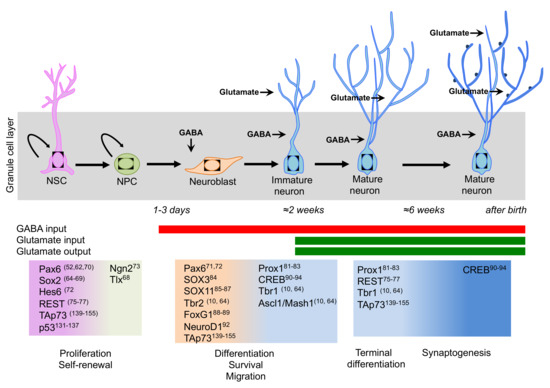
Figure 2.
Schematic representation of the transcription factor network involved in neurogenesis in the adult hippocampus. A sequential and coordinated expression pattern of the neurogenic transcription factors is fundamental for the proper progression from neural stem cells (NSCs) to mature neurons (Please see main text for details). During neurogenesis, a switch takes place from GABA excitatory to GABA inhibitory and glutamate excitatory inputs.
3.1. Transcriptional Control of Maintenance and Cell Fate Decision of NSCs in Adult Hippocampus
Among the transcription factors involved in the regulation of adult hippocampal neurogenesis, the Sox family is the most extensively investigated [65]. Sox2 has an active role in neural stem cells renewal. However, recent findings have shown that some neurons and glia cells keep high Sox2 expression, which is necessary for their function including cellular morphology and connectivity [66]. Conditional deletion (using Nestin-Cre system) of Sox2 during embryonic development results in a complete absence of the DG by post-natal day 7 [67]. To further support this, when Sox2 is deleted by retrovirus injection into an adult hippocampus, a decline of the positive type-1 cells population is observed suggesting that Sox2 is required for NPC maintenance in the dentate gyrus. Mechanistically, Sox2 directly regulates the expression of Sonic hedgehog (Shh) pathway. In parallel, Sox2 must repress the expression of NeuroD1 in order to maintain the self-renewal capacity of NSCs and therefore preventing the progression of neurogenesis [68]. Moreover, Sox2 controls the expression of the nuclear orphan receptor Tlx, which in turns supports proliferation and self-renewal of adult NSCs in the DG [69]. On the other hand, Sox2 expression is regulated by the Notch/RBPJk signalling, which is required for self-renewal and expansion of NSCs in the adult hippocampus [70].
The paired box protein, Pax6 is a transcription factor that was discovered to be essential for eye development; Pax6 is also crucial for the maintenance of NSCs pool [71]. It is mainly present in NSCs and its expression also persists to some extent in progenitor cells [72]. Pax6 directly regulates the expression of a cohort of genes important for self-renewal and neuronal differentiation including Hmga2, Cdk4, Gadd45g, Neurod1, Sstr2 and Hes6. Recently, the gene regulatory circuitry of Pax6 during neurogenesis has been mapped [73]. Among them, the pro-neural protein neurogenin 2 (Ngn2), regulated by Pax6, is expressed during DG formation and plays an essential role in neurogenesis [74]. Ngn2 constitutive knockout mice exhibit reduced cell proliferation and failure to form the infrapyramidal blade of the DG. Interestingly, many promoters bound by Pax6 are also occupied by Sox2, suggesting that they function together in the regulation of NSCs maintenance and proliferation.
In addition to Sox2, Tlx, Pax6 and Ngn2, an additional transcription factor that is fundamental for preserving the NSC pool is the restrictive element-1 silencing transcription factor (REST), also known as neuron-restrictive silencing factor (NRSF), which acts as a transcriptional repressor by recruiting other corepressor, such as mSin3A/B [75], N-CoR, CtBP [76] or CoREST, on the promoter of coding and non-coding target genes [77]. REST plays a fundamental role both at level of neurogenesis and in postmitotic neurons where it fine-tunes the expression of genes important for synaptic plasticity. During neurogenesis, REST is important for keeping the neuronal genes repressed and therefore preserves the undifferentiated state of the NSC pool [78]. Moreover, REST is also involved in shaping the synaptogenesis in adult neurons by regulating the expression of several synaptic proteins, including GluN2B and KCC2.
Overall, the NSCs pool in the adult hippocampus is maintained in part by the integration of extrinsic signals including Notch and the canonical Wnt pathway, which both directly control the expression of Sox2. Sox2 on the other hand, exerts dual gene regulatory impacts, specifically by activating the expression of Tlx and Shh pathway and repressing the expression of the prodifferentiation genes, such as NeuroD1.
3.2. Transcriptional Control of Differentiation, Survival and Integration of Dentate Granule Neurons
As intermediate progenitors exit from the cell cycle, they commit to neuronal lineage and then differentiate into glutamatergic granule neurons; there is a switch in the transcriptional program to control the later stages of adult neurogenesis. One of the key transcription factors that regulates this transition is NeuroD1. NeuroD1 is expressed at high levels in several areas of the adult brain including hippocampus, OB and cerebellum. NeuroD1 expression is both under the control of canonical Wnt signalling and GABA neurotransmitter and its expression promotes neuronal differentiation of NSCs in vivo [79]. Generation of the first NeuroD1 constitutive knockout mouse shows a disorganization of the DG indicating the essential role of NeuroD1 in regulating hippocampal development [80]. This first evidence was also confirmed by the conditional deletion NeuroD1 in the adult DG [81]. In these mice, newborn granule neurons are markedly reduced due to failing in survive and integrate into the pre-existing network.
In immature granule neurons, NeuroD1 is co-expressed with the prospero related homeobox gene Prox1 [82] and its overexpression promotes neuronal differentiation of NSCs. Moreover, conditional ablation of Prox1 results in the alteration of DG structure. In particular, the infrapyramidal blade fails to form and the size of the suprapyramidal blade is reduced [83]. Although most of the transcription factors are transiently expressed during neurogenesis, Prox1 expression seems to persist also in mature granule neurons. Indeed, conditional deletion of Prox1 in newly generated mature neurons affects Calbindin expression [84]. Overall, NeuroD1 and Prox1 appear to be a key player in regulating the differentiation of dentate granule neurons.
Sox family may also participate in the regulation of the transition from NPCs to immature neurons. Sox3 is closely related to Sox1 and Sox2 gene and is expressed in the early CNS. Consistent with this pattern of expression, constitutive deletion of Sox3 in mice results in congenital abnormalities in the hippocampus and corpus callosum [85]. Sox11 is mainly expressed in neurogenic niches of the adult mammalian brain and its ectopic expression promotes neuronal differentiation of NPCs [86]. In addition, in vivo conditional ablation of Sox11, by stereotactic injection of the Moloney Murine Leukemia Virus (M-MLV) CAG–GFP–IRES–Cre into the dentate gyrus of adult Sox4/Sox11dcKO mice inhibits neurogenesis without affecting proliferation and survival of NPCs. Mechanistically, Sox11 directly regulates the expression of the neuronal lineage-specific gene DCX [87]. In addition, Sox11 also modulates dendrites development in adult DG neurons [88].
Moreover, FoxG1 is a Winged-Helix transcriptional repressor mainly expressed in telencephalon and in the adult DG, and plays an important role in postnatal hippocampus neurogenesis. In fact, FoxG1 haploinsufficiency results in the reduction of hippocampal volume and granule cell number with memory deficits [89]. Moreover, conditional deletion of Foxg1 leads to developmental defects in the adult DG, characterized by the loss of the SGZ [90]. These abnormalities are caused by a defect in self-renewal, differentiation and migration of newly generated neurons.
The last stage of hippocampal neurogenesis, maturation ad integration of granule neurons is mainly under the control of the transcription factor cAMP response-element binding protein (CREB1). CREB1 is a member of the bZIP superfamily of transcription factors that plays an important role in modulating neuronal survival, maturation and plasticity in the adult DG [91]. In particular, GABA-mediated excitation increases CREB signalling and results in the enhancement of dendrite length and an increase in dendritic branching [92]. Conversely, when CREB function is lost by in vivo retrovirus-mediated genetic manipulation into adult brain, maturation and complexity of dendritic tree is impaired and the expression of the neurogenic transcription factor NeuroD1 is decreased [93]. In addition, it has been shown that CREB regulates the survival of newborn neurons. However, it should be noted that CREB mutant mice have increased levels of hippocampal neurogenesis [94,95].
3.3. The Well-Established Role of p53-Family During Hippocampal Neurogenesis
The family of transcriptional factors p53, p63 and p73 [96,97,98,99,100,101,102] are well known and characterised for their role in the control of the cell cycle arrest and apoptosis [103,104,105,106,107,108,109,110,111]. Together with the frequent p53 mutations in cancer [112,113,114,115,116,117,118,119,120,121,122], this signalling leads to associate their function to oncosuppression [123,124,125,126]. There is, however, strong in vivo and in vitro evidence that the family is also implicated in the regulation of the CNS functions [127,128,129,130,131]. Mechanistically, the p53-family proteins control NSCs survival, self-renewal and terminal differentiation via a complex transcriptional regulatory network by binding to specific DNA sequences to regulate the expression of coding- and non-coding genes.
p53 mRNA expression is mainly confined to an in area of the developing brain that does not undergo apoptosis, suggesting that p53 participates in neuronal differentiation [132,133,134]. The role of p53 in neuronal differentiation is further supported by in vivo studies. p53-null mice, in particular female, display exencephaly due to an overgrowth of neural tissue, which in turn, causes failure of the neural tube to close during brain development [135]. p53 controls proliferation and differentiation of NPCs both in vitro and in vivo mainly by regulating the expression of several cell-cycle regulators without affecting the expression of the canonical neuronal markers [136,137]. In post-mitotic neurons beyond the regulation of neuronal apoptosis, post-translational modifications of p53 (acetylation) might promote neuronal maturation and axonal regeneration [138]. p53 has also been associated to phosphorylation of tau protein in the pathogenesis of the Alzheimer’s disease [139].
Among the members of the p53 family, p73 is a key player in the regulation of CNS development and function by modulating NSC self-renewal and differentiation as well as promoting terminal neuronal differentiation [140,141,142]. The Trp73 gene, codifying the p73 proteins, has a complex structure, including two alternative promoters that regulate expression of the N-terminal full-term isoform, TAp73, and the N-terminal truncated ΔNp73 isoform. The crucial role of p73 in the CNS has been highlighted by the phenotype of the several knockout mice for p73: global p73KO mice and the isoform-specific TAp73 or ΔNp73 models [101,143] [144,145,146,147]. Indeed, deletion of both isoforms results in hippocampal dysgenesis that is characterized by the partial or total loss of the lower blade of the DG and by an impaired organization of CA1 and CA3 regions [140]. Moreover, p73 is essential for maintaining the neurogenic pool in SVZ and SGZ by promoting self-renewal and proliferation and inhibiting premature senescence of NSCs and/or NPCs [148,149,150]. Mechanistically, TAp73 either directly or indirectly regulates the expression of genes involved in metabolism [151,152,153] and NSCs maintenance including, Sox2, Sox3, TRIM32 and Notch signalling pathway. Similarly, TAp73 regulates NSCs maintenance in the olfactory bulb by transcriptionally regulating the expression of Hey-2 [154]. TAp73 is also implicated in the regulation of post-mitotic neurons, by modulating expression of the neurotrophin receptor p75 (p75NTR), which is implicated in axonal growth and dendritic arborisation [155,156]. Moreover, TAp73 controls neuronal terminal differentiation by regulating the expression of synaptic proteins such as synaptotagmin-1 and syntaxin-1A via miR-34a [157,158,159].
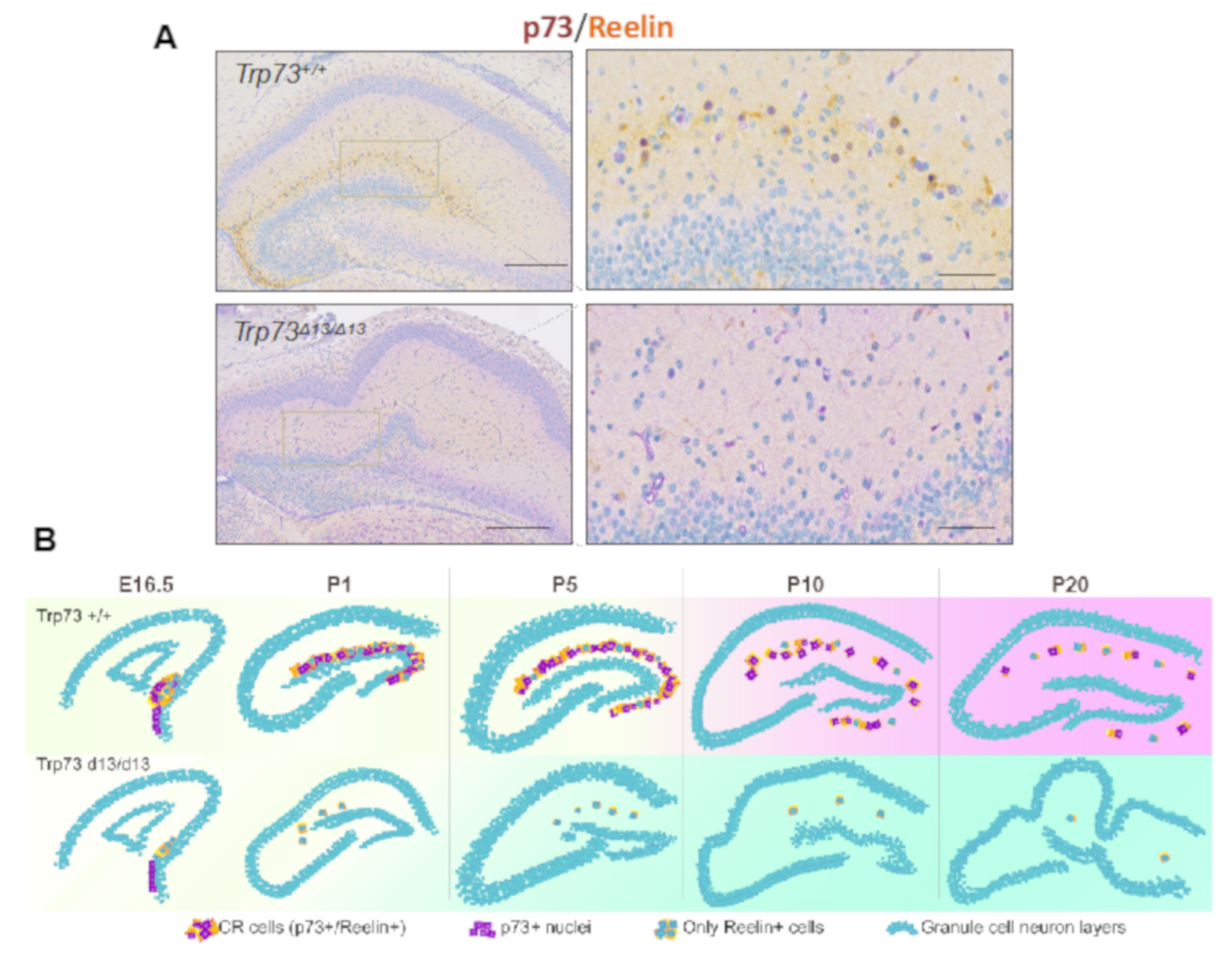
The recently described Trp73d13/d13 mice, lacking exon 13 in the p73 gene, have elucidated p73 C-terminus contribution to brain development. p73 C-terminus can undergo a complex alternative splicing, which can give rise up to 7 different isoforms (α, β, γ, σ, ε, ζ, η) [160,161]. Deletion of exon 13 produces a switch of the longest and most expressed isoform α into the isoform β, which in contrast to α does not contain the sterile alpha-motif (SAM) domain. Replacement of α with β substantially affects brain development, producing hippocampal dysgenesis, which largely recapitulates the phenotype of the global p73KO and the selective TAp73 KO. In particular, Trp73d13/d13 mouse developing brain displays a progressive depauperation of Cajal–Retzius (CR) cells [162] (Figure 3). Thus, the hippocampal dysgenesis appears to be a consequence of deprivation of the CR cells, which are physiologically deputed to direct brain architecture during embryonic development.
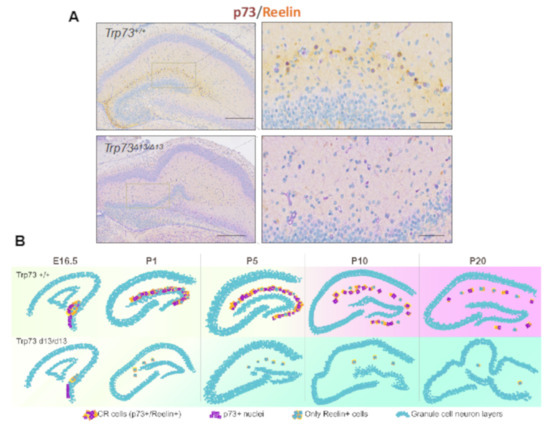
Figure 3.
Trp73D13/D13 mice display hippocampal dysgenesis and CR cells depletion. (A) Immunohistochemistry of postnatal day 5 (P5) mouse hippocampus displays disrupted morphology and reduced presence of Reelin+ CR cells in Trp73D13/D13 genotype. Scale bars indicate 500 µm and 50 m.(B) Representative summary of morphological developmental progression in the hippocampus of Trp73+/+ and Trp73D13/D13 from embryonic 16.5 (E16.5) to postnatal day 20 (P20) stage. Adapted with modifications from Amelio et al. [162].
4. Metabolic Regulation of Adult Neurogenesis
The mammalian neurons strictly depend on glucose as the main source of energy and energy metabolism is tightly regulated during neuronal differentiation [163,164,165,166] and degeneration [167]. Neurons rely on oxidative phosphorylation (OXPHOS) to meet energy demands, e OXPHOS, neurons metabolise one glucose molecule to obtain 30–32 ATP molecules of energy. Therefore, mitochondria play a key role during neurodevelopment and adult neurogenesis for cytoskeletal remodelling, outgrowth of axons, dendrites and synaptic activity [168,169]. Peroxisome proliferator-activated receptor gamma coactivator 1 (PGC-1α) is a master regulator of mitochondrial biogenesis by activating the expression of nuclear respiratory factor 1 (NRF1), which controls the expression of mitochondrial proteins including cytochrome-c (cyt-c), and mitochondrial transcription factor A (TFAM). PGC-1α regulates the formation and maintenance of the synapsis in developing and adult hippocampal neurons [170].
Nuclear-encoded mitochondrial proteins are regulated by several factors such as oncogenes and tumour suppressor proteins [171], including p73, oestrogen-related receptor (ERRα), nuclear respiratory factors NRF1 and NRF2 and the yin yang 1 transcription factor (YY1). Indeed, TAp73 directly regulates the expression of the cytochrome C oxidase subunit 4 (COX4I1), a mitochondrial protein from the complex IV, which is essential for energy supply in neurons [172,173]. NRF2 deletion affects the proliferation capacity of NSCs from SGZ and impairs neuronal differentiation [174] and the psychiatric susceptibility gene Cacna1c also promotes mitochondrial resilience to oxidative stress in neurons [175].
Post-translation modification also regulates mitochondria function during neuronal differentiation. Cytoplasmic polyadenylation element-binding protein 1 (CPEB1) controls translation of NDUFV2 and complex I activity. Indeed, CPEB1 deficient neurons show reduced ATP production and impaired dendrite branching. This phenotype can be rescued by overexpressing NADH:Ubiquinone Oxidoreductase Core Subunit V2 (NDUFV2) [176].
Neuronal differentiation implies a substantial metabolic reprograming [177,178]: the switch from aerobic glycolysis to OXPHOS characterizes neuronal differentiation of NPCs [179]. In particular, the transition from NPCs to neurons is characterized by a reduced expression of hexokinase (HK2) and lactate dehydrogenase (LDHA), together with a switch in pyruvate kinase splicing from PKM2 to PKM1. In addition, neuronal differentiation is associated with the upregulation of the master regulators of mitochondrial biogenesis, PGC-1α, ERRγ and TFAM. Since mitochondrial mass increases proportionally with neuronal mass growth, the upregulation of mitochondrial biogenesis could in part support cell growth, i.e., axonal growth and dendritic development [180]. During terminal differentiation of cortical neurons, an increase in glucose metabolism is associated with a rise in glucose uptake and enhanced GLUT3 expression. Aerobic glycolysis in neurons sustains axonal elongation and synaptogenesis [181], regardless of the efficiency of the related energy production. This metabolic reprogramming is secondary to activation of the PI3K/mTOR axis [180]. Indeed, pharmacological inhibition by Rapamycin of the PI3K/mTOR axis results in a reduction of mitochondrial biogenesis and glucose metabolism in neurons that are associated with a reduction in terminal neuronal differentiation. Therefore, mTOR signalling plays a role in the regulation of mitochondrial mass and functions together with the regulation of glycolysis, which in turn is required for dendrite and synapse formation.
Glutamine is also an amino acid linked to cellular energy homeostasis. Glutamine can be converted to glutamate and then into α-ketoglutarate and thus be oxidized in the tricarboxylic acid (TCA) cycle to synthetize ATP. Terminal neuronal differentiation is also associated with increased glutamine metabolism, resulting in increased expression of neurotransmitters such as glutamate and GABA, and increased TCA cycle activity [180].
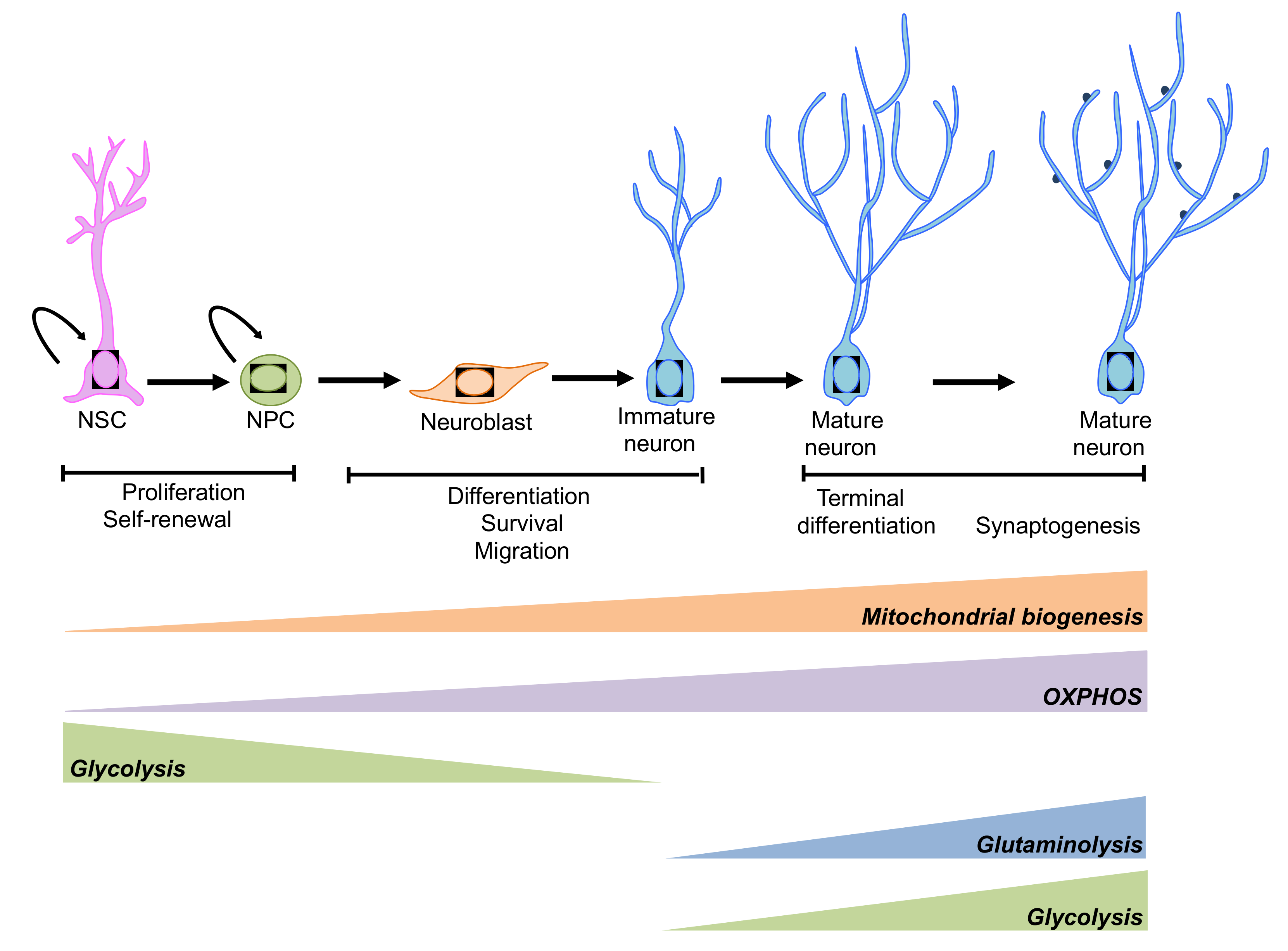
Overall, Figure 4, aerobic glycolysis and OXPHOS are the two main pathways providing metabolic precursors for biosynthesis and energy production. The activities of these pathways are tightly regulated to guarantee optimal resource supply, conforming to cellular function. The balance between aerobic glycolysis and OXPHOS is vital for neuronal development [182].
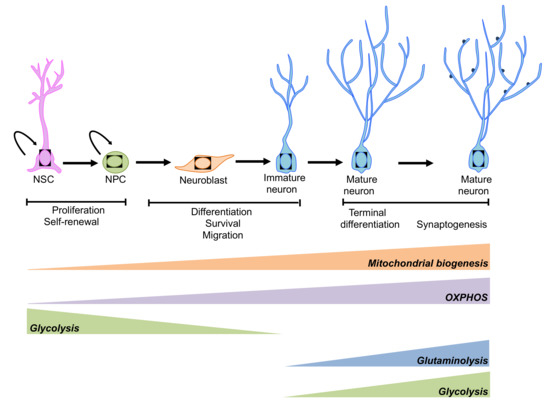
Figure 4.
Schematic representation of the metabolic pathways involved in the regulation of neurogenesis. An extensive reprogramming of cell metabolism is associated with neurogenesis. NSCs and neuronal progenitor cells (NPCs) rely mainly on glycolysis, while mature neurons preferentially use OXPHOS. However, under some circumstances aerobic glycolysis and glutaminolysis has been observed during neuronal terminal differentiation (please see main text for details).
All together, these observations support a crosstalk between signalling and metabolism.
In the past 20 years, many significant questions have been addressed and some basic principles have emerged in the field of adult neurogenesis. However, more recently, the persistency of adult neurogenesis in adult human brain has been questioned [183]. The main challenge in studying human adult neurogenesis is the access to samples. Since the preservation of human brain will affect the labelling of new adult-born neurons, the creation of brain banks with standardised tissues collection and preservation is a main priority for studying adult neurogenesis. In addition, we need to have unanimous criteria for how to identify NSCs from progenitor cells. Moreover, single-cell RNA sequencing will also give fundamental information for understanding the cellular components in neurogenic niches.
5. Conclusions
A large body of evidence indicates that the alteration of neurogenesis is associated with several human pathologies and with age-associated decline in cognitive function [184]. Therefore, understanding the epigenetic, transcriptional and metabolic mechanisms underlying neuronal differentiation, and how these processes are deregulated, could open novel therapeutic strategies for treating brain disorders. Several HDAC inhibitors are neuroprotective in many mouse models of neurodegeneration [185] and several clinical trials are either ongoing or completed (NCT03056495 and NCT02124083). Moreover, modulation of metabolic pathways with small molecules might be an alternative possible novel therapeutic strategy in the near future. Indeed, mTOR signalling by regulating cell growth and metabolism is emerging as a crucial player in proliferation, differentiation, and neurite outgrowth and synaptic formation and its activity can be modulated by rapamycin/rapalogs [186,187]. Failures in mitochondria OXPHOS lead to several neurological disorders including Leigh syndrome, a severe childhood neurological disorder [188]. Rapamycin treatment of a mouse model of the syndrome delays onset of neurological symptoms reduces neuroinflammation and prevents brain lesions [189].
Author Contributions
Conceptualization, G.M. Writing—original draft preparation, M.V.N.-C., M.A., I.A.; Writing—review and editing, M.V.N.-C., M.A., I.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Medical Research Council (to GM), Associazione Italiana per la Ricerca contro il Cancro (AIRC) to GM (IG#20473; 2018–2022), to IA (AIRC Start-Up ID 23219; 2020–2024), Ministry of Health and MAECI Italy-China Science and Technology Cooperation (#PGR00961) to G.M. Work has been also supported by Regione Lazio through LazioInnova Progetto Gruppo di Ricerca n 85-2017-14986, Children with Cancer UK fellowship (2014-178), Life Sciences Seed Fund-QMUL and Bart Charity Grant (MGU0473) to MVNC.
Acknowledgments
We would like to thank Tina Nath Varma and E. Panatta for helpful comments and suggestions.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| CaMKII | Calcium/Calmodulin-Dependent Protein Kinase II |
| CHD | Chromodomain Helicase DNA-Binding |
| CNS | Central Nervous System |
| COX4I1 | Cytochrome C Oxidase Subunit 4 |
| CPEB1 | Cytoplasmic Polyadenylation Element-Binding Protein 1 |
| Cspg4 | Chondroitin Sulphate Proteoglycan 4 |
| cyt-c | Cytochrome-C |
| DG | Dentate Gyrus |
| Dlx2 | Distal-Less Homeobox 2 |
| DNMTs | Dna Methyl Transferases |
| ERRα | Oestrogen-Related Receptor Alpha |
| Ezh2 | Zeste Homologue 2 |
| GADD45β | DNA-Damage-Inducible Protein 45β |
| Gal | Galanin |
| HATs | Histone Acetyltransferases |
| HDACs | Histone Deacetylases |
| hGFAP | Human Glial Fibrillary Acidic Protein |
| HK2 | hexokinase |
| JMJD3 | Jumonji Domain-Containing Protein 3 |
| KMTs | Lysine Methyltransferases |
| LDHA | lactate dehydrogenase |
| LSD1 | Lysine-Specific Demethylase 1 |
| MLL1 | Mixed-Lineage Leukaemia 1 |
| M-MLV | Moloney Murine Leukemia Virus |
| Ngb | Neuroglobin |
| NPCs | Neuronal Progenitor Cells |
| NRSF | Neuron-Restrictive Silencing Factor |
| NSCs | Neural Stem Cells |
| OB | Olfactory Bulb |
| OXPHOS | oxidative phosphorylation |
| PRC | Pcg Repressive Complex |
| REST | Restrictive Element-1 Silencing Transcription Factor |
| SAM | sterile alpha-motif |
| SGZ | Subgranular Zone |
| Shh | Sonic hedgehog |
| SVZ | Subventricular Zone |
| TET | Ten-Eleven Translocation |
| TRXG | Trithorax Active Complex |
| YY1 | Yin Yang 1 Transcription Factor |
References
- Ming, G.L.; Song, H. Adult neurogenesis in the mammalian brain: Significant answers and significant questions. Neuron 2011, 70, 687–702. [Google Scholar] [CrossRef] [PubMed]
- Yadirgi, G.; Marino, S. Adult neural stem cells and their role in brain pathology. J. Pathol. 2009, 217, 242–253. [Google Scholar] [CrossRef] [PubMed]
- Kole, A.J.; Annis, R.P.; Deshmukh, M. Mature neurons: Equipped for survival. Cell Death Dis. 2013, 4, e689. [Google Scholar] [CrossRef] [PubMed]
- Ming, G.L.; Song, H. Adult neurogenesis in the mammalian central nervous system. Annu. Rev. Neurosci. 2005, 28, 223–250. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Deng, W.; Gage, F.H. Mechanisms and functional implications of adult neurogenesis. Cell 2008, 132, 645–660. [Google Scholar] [CrossRef]
- Feliciano, D.M.; Bordey, A.; Bonfanti, L. Noncanonical Sites of Adult Neurogenesis in the Mammalian Brain. Cold Spring Harb. Perspect. Biol. 2015, 7, a018846. [Google Scholar] [CrossRef]
- Kempermann, G.; Jessberger, S.; Steiner, B.; Kronenberg, G. Milestones of neuronal development in the adult hippocampus. Trends Neurosci. 2004, 27, 447–452. [Google Scholar] [CrossRef]
- De la Torre-Ubieta, L.; Bonni, A. Transcriptional regulation of neuronal polarity and morphogenesis in the mammalian brain. Neuron 2011, 72, 22–40. [Google Scholar] [CrossRef]
- Kristiansen, M.; Ham, J. Programmed cell death during neuronal development: The sympathetic neuron model. Cell Death Differ. 2014, 21, 1025–1035. [Google Scholar] [CrossRef]
- Goncalves, J.T.; Schafer, S.T.; Gage, F.H. Adult Neurogenesis in the Hippocampus: From Stem Cells to Behavior. Cell 2016, 167, 897–914. [Google Scholar] [CrossRef]
- Gage, F.H. Adult neurogenesis in mammals. Science 2019, 364, 827–828. [Google Scholar] [CrossRef] [PubMed]
- Avila, A.; Vidal, P.M.; Tielens, S.; Morelli, G.; Laguesse, S.; Harvey, R.J.; Rigo, J.M.; Nguyen, L. Glycine receptors control the generation of projection neurons in the developing cerebral cortex. Cell Death Differ. 2014, 21, 1696–1708. [Google Scholar] [CrossRef] [PubMed]
- Secondo, A.; Esposito, A.; Petrozziello, T.; Boscia, F.; Molinaro, P.; Teadeschi, V.; Pannaccione, A.; Ciccone, R.; Guida, N.; Di Renzo, G.; et al. Na(+)/Ca(2+) exchanger 1 on nuclear envelope controls PTEN/Akt pathway via nucleoplasmic Ca(2+) regulation during neuronal differentiation. Cell Death Discov. 2018, 4, 12. [Google Scholar] [CrossRef] [PubMed]
- Stappert, L.; Klaus, F.; Brustle, O. MicroRNAs Engage in Complex Circuits Regulating Adult Neurogenesis. Front. Neurosci. 2018, 12, 707. [Google Scholar] [CrossRef]
- Mira, H.; Lie, D.C. Regulation of Adult Neurogenesis 2.0—Beyond Signaling Pathways and Transcriptional Regulators. Brain Plast 2017, 3, 1–3. [Google Scholar] [CrossRef]
- Yao, B.; Christian, K.M.; He, C.; Jin, P.; Ming, G.L.; Song, H. Epigenetic mechanisms in neurogenesis. Nat. Rev. Neurosci. 2016, 17, 537–549. [Google Scholar] [CrossRef]
- Gabriele, M.; Lopez Tobon, A.; D’Agostino, G.; Testa, G. The chromatin basis of neurodevelopmental disorders: Rethinking dysfunction along the molecular and temporal axes. Prog. Neuropsychopharmacol. Biol. Psychiatry 2018, 84, 306–327. [Google Scholar] [CrossRef]
- Qureshi, I.A.; Mehler, M.F. Epigenetic mechanisms underlying human epileptic disorders and the process of epileptogenesis. Neurobiol. Dis. 2010, 39, 53–60. [Google Scholar] [CrossRef]
- Li, E.; Beard, C.; Jaenisch, R. Role for DNA methylation in genomic imprinting. Nature 1993, 366, 362–365. [Google Scholar] [CrossRef]
- Peters, J. The role of genomic imprinting in biology and disease: An expanding view. Nat. Rev. Genet. 2014, 15, 517–530. [Google Scholar] [CrossRef]
- Schubeler, D. Function and information content of DNA methylation. Nature 2015, 517, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Mohn, F.; Weber, M.; Rebhan, M.; Roloff, T.C.; Richter, J.; Stadler, M.B.; Bibel, M.; Schübeler, D. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol. Cell 2008, 30, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Fan, G.; Martinowich, K.; Chin, M.H.; He, F.; Fouse, S.D.; Hutnick, L.; Hattori, D.; Ge, W.; Shen, Y.; Wu, H.; et al. DNA methylation controls the timing of astrogliogenesis through regulation of JAK-STAT signaling. Development 2005, 132, 3345–3356. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, S.; Meletis, K.; Fu, D.; Jhaveri, S.; Jaenisch, R. Ablation of de novo DNA methyltransferase Dnmt3a in the nervous system leads to neuromuscular defects and shortened lifespan. Dev. Dyn. 2007, 236, 1663–1676. [Google Scholar] [CrossRef] [PubMed]
- Martins-Taylor, K.; Schroeder, D.I.; LaSalle, J.M.; Lalande, M.; Xu, R.H. Role of DNMT3B in the regulation of early neural and neural crest specifiers. Epigenetics 2012, 7, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.R.; Cui, Q.Y.; Murai, K.; Lim, Y.C.; Smith, Z.D.; Jin, S.; Ye, P.; Rosa, L.; Lee, Y.K.; Wu, H.P.; et al. Tet1 regulates adult hippocampal neurogenesis and cognition. Cell Stem Cell 2013, 13, 237–245. [Google Scholar] [CrossRef]
- Kempermann, G. Activity Dependency and Aging in the Regulation of Adult Neurogenesis. Cold Spring Harb. Perspect. Biol. 2015, 7, a018929. [Google Scholar] [CrossRef]
- Ma, D.K.; Jang, M.H.; Guo, J.U.; Kitabatake, Y.; Chang, M.L.; Pow-Anpongkul, N.; Flavell, R.A.; Lu, B.; Ming, G.L.; Song, H. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science 2009, 323, 1074–1077. [Google Scholar] [CrossRef]
- Barreto, G.; Schafer, A.; Marhold, J.; Stach, D.; Swaminathan, S.K.; Handa, V.; Döderlien, G.; Maltry, N.; Wu, W.; Lyko, F. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature 2007, 445, 671–675. [Google Scholar] [CrossRef]
- Smrt, R.D.; Eaves-Egenes, J.; Barkho, B.Z.; Santistevan, N.J.; Zhao, C.; Aimone, J.B.; Gage, F.H.; Zhao, X. Mecp2 deficiency leads to delayed maturation and altered gene expression in hippocampal neurons. Neurobiol. Dis. 2007, 27, 77–89. [Google Scholar] [CrossRef]
- Bernstein, B.E.; Meissner, A.; Lander, E.S. The mammalian epigenome. Cell 2007, 128, 669–681. [Google Scholar] [CrossRef] [PubMed]
- Kazantsev, A.G.; Thompson, L.M. Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat. Rev. Drug Discov. 2008, 7, 854–868. [Google Scholar] [CrossRef]
- Haberland, M.; Montgomery, R.L.; Olson, E.N. The many roles of histone deacetylases in development and physiology: Implications for disease and therapy. Nat. Rev. Genet. 2009, 10, 32–42. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, J.L.; Roskams, A.J. Histone deacetylases 1 and 2 are expressed at distinct stages of neuro-glial development. Dev. Dyn. 2008, 237, 2256–2267. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, R.L.; Hsieh, J.; Barbosa, A.C.; Richardson, J.A.; Olson, E.N. Histone deacetylases 1 and 2 control the progression of neural precursors to neurons during brain development. Proc. Natl. Acad. Sci USA 2009, 106, 7876–7881. [Google Scholar] [CrossRef]
- Galluzzi, L.; Vitale, I.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; Amelio, I.; Andrews, D.W. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018, 25, 486–541. [Google Scholar] [CrossRef]
- Melino, G.; Knight, R.A.; Nicotera, P. How many ways to die? How many different models of cell death? Cell Death Differ. 2005, 12 (Suppl. 2), 1457–1462. [Google Scholar] [CrossRef][Green Version]
- Hagelkruys, A.; Lagger, S.; Krahmer, J.; Leopoldi, A.; Artaker, M.; Pusch, O.; Zezula, J.; Weissmann, S.; Xie, Y.; Schöfer, C. A single allele of Hdac2 but not Hdac1 is sufficient for normal mouse brain development in the absence of its paralog. Development 2014, 141, 604–616. [Google Scholar] [CrossRef]
- Norwood, J.; Franklin, J.M.; Sharma, D.; D’Mello, S.R. Histone deacetylase 3 is necessary for proper brain development. J. Biol. Chem. 2014, 289, 34569–34582. [Google Scholar] [CrossRef]
- Ageta-Ishihara, N.; Miyata, T.; Ohshima, C.; Watanabe, M.; Sato, Y.; Hamamura, Y.; Higashiyama, T.; Mazitschek, R.; Bito, H.; Kinoshita, M. Septins promote dendrite and axon development by negatively regulating microtubule stability via HDAC6-mediated deacetylation. Nat. Commun. 2013, 4, 2532. [Google Scholar] [CrossRef]
- Zhang, S.; Fujita, Y.; Matsuzaki, R.; Yamashita, T. Class I histone deacetylase (HDAC) inhibitor CI-994 promotes functional recovery following spinal cord injury. Cell Death Dis. 2018, 9, 460. [Google Scholar] [CrossRef] [PubMed]
- Sheikh, B.N.; Dixon, M.P.; Thomas, T.; Voss, A.K. Querkopf is a key marker of self-renewal and multipotency of adult neural stem cells. J. Cell Sci. 2012, 125, 295–309. [Google Scholar] [CrossRef] [PubMed]
- Merson, T.D.; Dixon, M.P.; Collin, C.; Rietze, R.L.; Bartlett, P.F.; Thomas, T.; Voss, A.K. The transcriptional coactivator Querkopf controls adult neurogenesis. J. Neurosci. 2006, 26, 11359–11370. [Google Scholar] [CrossRef] [PubMed]
- Hyun, K.; Jeon, J.; Park, K.; Kim, J. Writing, erasing and reading histone lysine methylations. Exp. Mol. Med. 2017, 49, e324. [Google Scholar] [CrossRef]
- Corley, M.; Kroll, K.L. The roles and regulation of Polycomb complexes in neural development. Cell Tissue Res. 2015, 359, 65–85. [Google Scholar] [CrossRef]
- Hwang, W.W.; Salinas, R.D.; Siu, J.J.; Kelley, K.W.; Delgado, R.N.; Paredes, M.F.; Alvarez-Buylla, A.; Oldham, M.C.; Lim, D.A. Distinct and separable roles for EZH2 in neurogenic astroglia. eLife 2014, 3, e02439. [Google Scholar] [CrossRef]
- Molofsky, A.V.; Pardal, R.; Iwashita, T.; Park, I.K.; Clarke, M.F.; Morrison, S.J. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature 2003, 425, 962–967. [Google Scholar] [CrossRef]
- Fasano, C.A.; Dimos, J.T.; Ivanova, N.B.; Lowry, N.; Lemischka, I.R.; Temple, S. shRNA knockdown of Bmi-1 reveals a critical role for p21-Rb pathway in NSC self-renewal during development. Cell Stem Cell 2007, 1, 87–99. [Google Scholar] [CrossRef]
- Li, Y.Q.; Wong, C.S. Effects of p21 on adult hippocampal neuronal development after irradiation. Cell Death Discov. 2018, 4, 15. [Google Scholar] [CrossRef]
- Lim, D.A.; Huang, Y.C.; Swigut, T.; Mirick, A.L.; Garcia-Verdugo, J.M.; Wysocka, J.; Ernest, P.; Alvarez-Buylla, A. Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature 2009, 458, 529–533. [Google Scholar] [CrossRef]
- Potts, M.B.; Siu, J.J.; Price, J.D.; Salinas, R.D.; Cho, M.J.; Ramos, A.D.; Hahn, J.; Margeta, M.; Oldman, M.C.; Lim, D.A. Analysis of Mll1 deficiency identifies neurogenic transcriptional modules and Brn4 as a factor for direct astrocyte-to-neuron reprogramming. Neurosurgery 2014, 75, 472–482; discussion 482. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Alzayady, K.; Stewart, R.; Ye, P.; Yang, S.; Li, W.; Shi, Y. Histone demethylase LSD1 regulates neural stem cell proliferation. Mol. Cell. Biol. 2010, 30, 1997–2005. [Google Scholar] [CrossRef]
- Burgold, T.; Spreafico, F.; De Santa, F.; Totaro, M.G.; Prosperini, E.; Natoli, G.; Testa, G. The histone H3 lysine 27-specific demethylase Jmjd3 is required for neural commitment. PLoS ONE 2008, 3, e3034. [Google Scholar] [CrossRef] [PubMed]
- Park, D.H.; Hong, S.J.; Salinas, R.D.; Liu, S.J.; Sun, S.W.; Sgualdino, J.; Testa, G.; Matzuk, M.M.; Iwamori, N.; Lim, D.A. Activation of neuronal gene expression by the JMJD3 demethylase is required for postnatal and adult brain neurogenesis. Cell Rep. 2014, 8, 1290–1299. [Google Scholar] [CrossRef] [PubMed]
- Clapier, C.R.; Iwasa, J.; Cairns, B.R.; Peterson, C.L. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 2017, 18, 407–422. [Google Scholar] [CrossRef] [PubMed]
- Sokpor, G.; Castro-Hernandez, R.; Rosenbusch, J.; Staiger, J.F.; Tuoc, T. ATP-Dependent Chromatin Remodeling During Cortical Neurogenesis. Front. Neurosci. 2018, 12, 226. [Google Scholar] [CrossRef]
- Narayanan, R.; Tuoc, T.C. Roles of chromatin remodeling BAF complex in neural differentiation and reprogramming. Cell Tissue Res. 2014, 356, 575–584. [Google Scholar] [CrossRef]
- Sokpor, G.; Xie, Y.; Rosenbusch, J.; Tuoc, T. Chromatin Remodeling BAF (SWI/SNF) Complexes in Neural Development and Disorders. Front. Mol. Neurosci. 2017, 10, 243. [Google Scholar] [CrossRef]
- Tuoc, T.; Dere, E.; Radyushkin, K.; Pham, L.; Nguyen, H.; Tonchev, A.B.; Sun, G.; Ronnenberg, A.; Shi, Y.; Satiger, J.F.; et al. Ablation of BAF170 in Developing and Postnatal Dentate Gyrus Affects Neural Stem Cell Proliferation, Differentiation, and Learning. Mol. Neurobiol. 2017, 54, 4618–4635. [Google Scholar] [CrossRef]
- Simon, R.; Brylka, H.; Schwegler, H.; Venkataramanappa, S.; Andratschke, J.; Wiegreffe, C.; Liu, P.; Fuchs, E.; Jenkins, N.A.; Copeland, N.G.; et al. A dual function of Bcl11b/Ctip2 in hippocampal neurogenesis. EMBO J. 2012, 31, 2922–2936. [Google Scholar] [CrossRef]
- Simon, R.; Baumann, L.; Fischer, J.; Seigfried, F.A.; De Bruyckere, E.; Liu, P.; Jenkins, N.A.; Schwegler, H.; Britisch, S. Structure-function integrity of the adult hippocampus depends on the transcription factor Bcl11b/Ctip2. Genes Brain Behav. 2016, 15, 405–419. [Google Scholar] [CrossRef]
- Matsumoto, S.; Banine, F.; Struve, J.; Xing, R.; Adams, C.; Liu, Y.; Metzger, D.; Chambon, P.; Rao, M.S.; Sherman, L.S.; et al. Brg1 is required for murine neural stem cell maintenance and gliogenesis. Dev. Biol. 2006, 289, 372–383. [Google Scholar] [CrossRef] [PubMed]
- Ninkovic, J.; Steiner-Mezzadri, A.; Jawerka, M.; Akinci, U.; Masserdotti, G.; Petricca, S.; Fisher, J.; von Holst, A.; Beckers, J.; Lie, C.D.; et al. The BAF complex interacts with Pax6 in adult neural progenitors to establish a neurogenic cross-regulatory transcriptional network. Cell Stem Cell 2013, 13, 403–418. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, J. Orchestrating transcriptional control of adult neurogenesis. Genes Dev. 2012, 26, 1010–1021. [Google Scholar] [CrossRef] [PubMed]
- Wegner, M. SOX after SOX: SOXession regulates neurogenesis. Genes Dev. 2011, 25, 2423–2428. [Google Scholar] [CrossRef]
- Mercurio, S.; Serra, L.; Nicolis, S.K. More than just Stem Cells: Functional Roles of the Transcription Factor Sox2 in Differentiated Glia and Neurons. Int. J. Mol. Sci. 2019, 20, 4540. [Google Scholar] [CrossRef]
- Favaro, R.; Valotta, M.; Ferri, A.L.; Latorre, E.; Mariani, J.; Giachino, C.; Lanchini, C.; Tosetti, V.; Ottolenghi, S.; Taylor, V.; et al. Hippocampal development and neural stem cell maintenance require Sox2-dependent regulation of Shh. Nat. Neurosci. 2009, 12, 1248–1256. [Google Scholar] [CrossRef]
- Kuwabara, T.; Hsieh, J.; Muotri, A.; Yeo, G.; Warashina, M.; Lie, D.C.; Moore, L.; Nakashima, K.; Asashima, M.; Gage, F.H.; et al. Wnt-mediated activation of NeuroD1 and retro-elements during adult neurogenesis. Nat. Neurosci. 2009, 12, 1097–1105. [Google Scholar] [CrossRef]
- Shimozaki, K.; Zhang, C.L.; Suh, H.; Denli, A.M.; Evans, R.M.; Gage, F.H. SRY-box-containing gene 2 regulation of nuclear receptor tailless (Tlx) transcription in adult neural stem cells. J. Biol. Chem. 2012, 287, 5969–5978. [Google Scholar] [CrossRef]
- Ehm, O.; Goritz, C.; Covic, M.; Schaffner, I.; Schwarz, T.J.; Karaca, E.; Kempkes, B.; Kremmer, E.; Pfrieger, F.W.; Espinosa, L.; et al. RBPJkappa-dependent signaling is essential for long-term maintenance of neural stem cells in the adult hippocampus. J. Neurosci. 2010, 30, 13794–13807. [Google Scholar] [CrossRef]
- Sansom, S.N.; Griffiths, D.S.; Faedo, A.; Kleinjan, D.J.; Ruan, Y.; Smith, J.; van Heyningen, V.; Rubenstein, J.L.; Livesey, F.J. The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self-renewal and neurogenesis. PLoS Genet. 2009, 5, e1000511. [Google Scholar] [CrossRef] [PubMed]
- Nacher, J.; Varea, E.; Blasco-Ibanez, J.M.; Castillo-Gomez, E.; Crespo, C.; Martinez-Guijarro, F.J.; McEwen, B.S. Expression of the transcription factor Pax 6 in the adult rat dentate gyrus. J. Neurosci. Res. 2005, 81, 753–761. [Google Scholar] [CrossRef] [PubMed]
- Thakurela, S.; Tiwari, N.; Schick, S.; Garding, A.; Ivanek, R.; Berninger, B.; Tiwari, V.K. Mapping gene regulatory circuitry of Pax6 during neurogenesis. Cell Discov. 2016, 2, 15045. [Google Scholar] [CrossRef]
- Galichet, C.; Guillemot, F.; Parras, C.M. Neurogenin 2 has an essential role in development of the dentate gyrus. Development 2008, 135, 2031–2041. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Naruse, Y.; Aoki, T.; Kojima, T.; Mori, N. Neural restrictive silencer factor recruits mSin3 and histone deacetylase complex to repress neuron-specific target genes. Proc. Natl. Acad. Sci. USA 1999, 96, 13691–13696. [Google Scholar] [CrossRef] [PubMed]
- Garriga-Canut, M.; Schoenike, B.; Qazi, R.; Bergendahl, K.; Daley, T.J.; Pfender, R.M.; Morrison, J.F.; Ockuly, J.; Stafstrom, C.; Sutula, T.; et al. 2-Deoxy-D-glucose reduces epilepsy progression by NRSF-CtBP-dependent metabolic regulation of chromatin structure. Nat. Neurosci. 2006, 9, 1382–1387. [Google Scholar] [CrossRef] [PubMed]
- Ballas, N.; Mandel, G. The many faces of REST oversee epigenetic programming of neuronal genes. Curr. Opin. Neurobiol. 2005, 15, 500–506. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Ure, K.; Ding, P.; Nashaat, M.; Yuan, L.; Ma, J.; Hammer, R.E.; Hsieh, J. The master negative regulator REST/NRSF controls adult neurogenesis by restraining the neurogenic program in quiescent stem cells. J. Neurosci. 2011, 31, 9772–9786. [Google Scholar] [CrossRef] [PubMed]
- Tozuka, Y.; Fukuda, S.; Namba, T.; Seki, T.; Hisatsune, T. GABAergic excitation promotes neuronal differentiation in adult hippocampal progenitor cells. Neuron 2005, 47, 803–815. [Google Scholar] [CrossRef]
- Miyata, T.; Maeda, T.; Lee, J.E. NeuroD is required for differentiation of the granule cells in the cerebellum and hippocampus. Genes Dev. 1999, 13, 1647–1652. [Google Scholar] [CrossRef]
- Gao, Z.; Ure, K.; Ables, J.L.; Lagace, D.C.; Nave, K.A.; Goebbels, S.; Eisch, A.J.; Hsieh, J. Neurod1 is essential for the survival and maturation of adult-born neurons. Nat. Neurosci. 2009, 12, 1090–1092. [Google Scholar] [CrossRef]
- Jessberger, S.; Toni, N.; Clemenson, G.D., Jr.; Ray, J.; Gage, F.H. Directed differentiation of hippocampal stem/progenitor cells in the adult brain. Nat. Neurosci. 2008, 11, 888–893. [Google Scholar] [CrossRef]
- Lavado, A.; Lagutin, O.V.; Chow, L.M.; Baker, S.J.; Oliver, G. Prox1 is required for granule cell maturation and intermediate progenitor maintenance during brain neurogenesis. PLoS Biol. 2010, 8, e1000460. [Google Scholar] [CrossRef] [PubMed]
- Iwano, T.; Masuda, A.; Kiyonari, H.; Enomoto, H.; Matsuzaki, F. Prox1 postmitotically defines dentate gyrus cells by specifying granule cell identity over CA3 pyramidal cell fate in the hippocampus. Development 2012, 139, 3051–3062. [Google Scholar] [CrossRef] [PubMed]
- Rizzoti, K.; Brunelli, S.; Carmignac, D.; Thomas, P.Q.; Robinson, I.C.; Lovell-Badge, R. SOX3 is required during the formation of the hypothalamo-pituitary axis. Nat. Genet. 2004, 36, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Haslinger, A.; Schwarz, T.J.; Covic, M.; Lie, D.C. Expression of Sox11 in adult neurogenic niches suggests a stage-specific role in adult neurogenesis. Eur. J. Neurosci. 2009, 29, 2103–2114. [Google Scholar] [CrossRef]
- Mu, L.; Berti, L.; Masserdotti, G.; Covic, M.; Michaelidis, T.M.; Doberauer, K.; Merz, K.; Rehfeld, F.; Haslinger, A.; Wegner, M.; et al. SoxC transcription factors are required for neuronal differentiation in adult hippocampal neurogenesis. J. Neurosci. 2012, 32, 3067–3080. [Google Scholar] [CrossRef] [PubMed]
- Balta, E.A.; Schaffner, I.; Wittmann, M.T.; Sock, E.; von Zweydorf, F.; von Wittgenstein, J.; Steib, K.; Heim, B.; Kremmer, E.; Häberle, B.; et al. Phosphorylation of the neurogenic transcription factor SOX11 on serine 133 modulates neuronal morphogenesis. Sci. Rep. 2018, 8, 16196. [Google Scholar] [CrossRef]
- Shen, L.; Nam, H.S.; Song, P.; Moore, H.; Anderson, S.A. FoxG1 haploinsufficiency results in impaired neurogenesis in the postnatal hippocampus and contextual memory deficits. Hippocampus 2006, 16, 875–890. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Gong, Y.; Yang, Y.; Shen, W.; Wang, K.; Liu, J.; Xu, B.; Zhao, J.; Zhao, C. Foxg1 has an essential role in postnatal development of the dentate gyrus. J. Neurosci. 2012, 32, 2931–2949. [Google Scholar] [CrossRef] [PubMed]
- Lonze, B.E.; Ginty, D.D. Function and regulation of CREB family transcription factors in the nervous system. Neuron 2002, 35, 605–623. [Google Scholar] [CrossRef]
- Giachino, C.; De Marchis, S.; Giampietro, C.; Parlato, R.; Perroteau, I.; Schutz, G.; Fasolo, A.; Peretto, P. cAMP response element-binding protein regulates differentiation and survival of newborn neurons in the olfactory bulb. J. Neurosci. 2005, 25, 10105–10118. [Google Scholar] [CrossRef] [PubMed]
- Jagasia, R.; Steib, K.; Englberger, E.; Herold, S.; Faus-Kessler, T.; Saxe, M.; Gage, F.H.; Song, H.; Lie, D.C. GABA-cAMP response element-binding protein signaling regulates maturation and survival of newly generated neurons in the adult hippocampus. J. Neurosci. 2009, 29, 7966–7977. [Google Scholar] [CrossRef] [PubMed]
- Gur, T.L.; Conti, A.C.; Holden, J.; Bechtholt, A.J.; Hill, T.E.; Lucki, I.; Malberg, J.E.; Blendly, J.A. cAMP response element-binding protein deficiency allows for increased neurogenesis and a rapid onset of antidepressant response. J. Neurosci. 2007, 27, 7860–7868. [Google Scholar] [CrossRef]
- Chong, C.M.; Ke, M.; Tan, Y.; Huang, Z.; Zhang, K.; Ai, N.; Ge, W.; Qin, D.; Lu, J.H.; Su, H. Presenilin 1 deficiency suppresses autophagy in human neural stem cells through reducing gamma-secretase-independent ERK/CREB signaling. Cell Death Dis. 2018, 9, 879. [Google Scholar] [CrossRef]
- Sullivan, K.D.; Galbraith, M.D.; Andrysik, Z.; Espinosa, J.M. Mechanisms of transcriptional regulation by p53. Cell Death Differ. 2018, 25, 133–143. [Google Scholar] [CrossRef]
- Basu, S.; Murphy, M.E. p53 family members regulate cancer stem cells. Cell Cycle 2016, 15, 1403–1404. [Google Scholar] [CrossRef][Green Version]
- Levrero, M.; De Laurenzi, V.; Costanzo, A.; Gong, J.; Melino, G.; Wang, J.Y. Structure, function and regulation of p63 and p73. Cell Death Differ. 1999, 6, 1146–1153. [Google Scholar] [CrossRef]
- De Laurenzi, V.; Melino, G. Evolution of functions within the p53/p63/p73 family. Ann. N. Y. Acad. Sci. 2000, 926, 90–100. [Google Scholar] [CrossRef]
- Candi, E.; Agostini, M.; Melino, G.; Bernassola, F. How the TP53 family proteins TP63 and TP73 contribute to tumorigenesis: Regulators and effectors. Hum. Mutat. 2014, 35, 702–714. [Google Scholar] [CrossRef]
- Tomasini, R.; Mak, T.W.; Melino, G. The impact of p53 and p73 on aneuploidy and cancer. Trends Cell Biol. 2008, 18, 244–252. [Google Scholar] [CrossRef]
- Tomasini, R.; Tsuchihara, K.; Tsuda, C.; Lau, S.K.; Wilhelm, M.; Rufini, A.; Tsao, M.S.; Iovanna, J.L.; Jurisicova, A.; Melino, G.; et al. TAp73 regulates the spindle assembly checkpoint by modulating BubR1 activity. Proc. Natl. Acad. Sci. USA 2009, 106, 797–802. [Google Scholar] [CrossRef]
- Engeland, K. Cell cycle arrest through indirect transcriptional repression by p53: I have a DREAM. Cell Death Differ. 2018, 25, 114–132. [Google Scholar] [CrossRef]
- Gomes, S.; Leao, M.; Raimundo, L.; Ramos, H.; Soares, J.; Saraiva, L. p53 family interactions and yeast: Together in anticancer therapy. Drug Discov. Today 2016, 21, 616–624. [Google Scholar] [CrossRef]
- Wu, D.; Prives, C. Relevance of the p53-MDM2 axis to aging. Cell Death Differ. 2018, 25, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Billant, O.; Leon, A.; Le Guellec, S.; Friocourt, G.; Blondel, M.; Voisset, C. The dominant-negative interplay between p53, p63 and p73: A family affair. Oncotarget 2016, 7, 69549–69564. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Garcia-Barros, M.; Wen, S.; Li, F.; Lin, C.L.; Hannun, Y.A.; Obeid, L.M.; Mao, C. Tumor suppressor p53 links ceramide metabolism to DNA damage response through alkaline ceramidase 2. Cell Death Differ. 2018, 25, 841–856. [Google Scholar] [CrossRef] [PubMed]
- Ferraiuolo, M.; Di Agostino, S.; Blandino, G.; Strano, S. Oncogenic Intra-p53 Family Member Interactions in Human Cancers. Front. Oncol. 2016, 6, 77. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.M.; Jung, C.H.; Kim, J.; Hwang, S.G.; Park, J.K.; Um, H.D. The p53/p21 Complex Regulates Cancer Cell Invasion and Apoptosis by Targeting Bcl-2 Family Proteins. Cancer Res. 2017, 77, 3092–3100. [Google Scholar] [CrossRef]
- Min, S.; Kim, K.; Kim, S.G.; Cho, H.; Lee, Y. Chromatin-remodeling factor, RSF1, controls p53-mediated transcription in apoptosis upon DNA strand breaks. Cell Death Dis. 2018, 9, 1079. [Google Scholar] [CrossRef]
- Contadini, C.; Monteonofrio, L.; Virdia, I.; Prodosmo, A.; Valente, D.; Chessa, L.; Musio, A.; Fava, L.L.; Rinaldo, C.; Di Rocco, G.; et al. p53 mitotic centrosome localization preserves centrosome integrity and works as sensor for the mitotic surveillance pathway. Cell Death Dis. 2019, 10, 850. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, F.; Collavin, L.; Del Sal, G. Mutant p53 as a guardian of the cancer cell. Cell Death Differ. 2019, 26, 199–212. [Google Scholar] [CrossRef] [PubMed]
- Parrales, A.; Thoenen, E.; Iwakuma, T. The interplay between mutant p53 and the mevalonate pathway. Cell Death Differ. 2018, 25, 460–470. [Google Scholar] [CrossRef]
- Post, S.M.; Kornblau, S.M.; Quintas-Cardama, A. p53 pathway dysfunction in AML: Beyond TP53 mutations. Oncotarget 2017, 8, 108288–108289. [Google Scholar] [CrossRef] [PubMed]
- Sorrentino, G.; Mantovani, F.; Del Sal, G. The stiff RhoAd from mevalonate to mutant p53. Cell Death Differ. 2018, 25, 645–647. [Google Scholar] [CrossRef]
- Goel, S.; Hall, J.; Pradhan, K.; Hirsch, C.; Przychodzen, B.; Shastri, A.; Mantzaris, I.; Jankiram, M.; Battini, R.; Kornblum, N.; et al. High prevalence and allele burden-independent prognostic importance of p53 mutations in an inner-city MDS/AML cohort. Leukemia 2016, 30, 1793–1795. [Google Scholar] [CrossRef]
- Kim, M.P.; Lozano, G. Mutant p53 partners in crime. Cell Death Differ. 2018, 25, 161–168. [Google Scholar] [CrossRef]
- Bailey, J.M.; Hendley, A.M.; Lafaro, K.J.; Pruski, M.A.; Jones, N.C.; Alsina, J.; Younes, M.; Maitra, A.; McAlister, F.; Iacobuzio-Donahue, C.A.; et al. p53 mutations cooperate with oncogenic Kras to promote adenocarcinoma from pancreatic ductal cells. Oncogene 2016, 35, 4282–4288. [Google Scholar] [CrossRef]
- Pitolli, C.; Wang, Y.; Mancini, M.; Shi, Y.; Melino, G.; Amelio, I. Do Mutations Turn p53 into an Oncogene? Int. J. Mol. Sci. 2019, 20, 6241. [Google Scholar] [CrossRef]
- Amelio, I.; Melino, G. The p53 family and the hypoxia-inducible factors (HIFs): Determinants of cancer progression. Trends Biochem. Sci. 2015, 40, 425–434. [Google Scholar] [CrossRef]
- Amelio, I.; Mancini, M.; Petrova, V.; Cairns, R.A.; Vikhreva, P.; Nicolai, S.; Marini, A.; Antonov, A.A.; Le Quesne, J.; Baena Acevedo, J.D.; et al. p53 mutants cooperate with HIF-1 in transcriptional regulation of extracellular matrix components to promote tumor progression. Proc. Natl. Acad. Sci. USA 2018, 115, E10869–E10878. [Google Scholar] [CrossRef] [PubMed]
- Amelio, I. How mutant p53 empowers Foxh1 fostering leukaemogenesis? Cell Death Discov. 2019, 5, 108. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, A.M.; Attardi, L.D. Deconstructing networks of p53-mediated tumor suppression in vivo. Cell Death Differ. 2018, 25, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Manzella, L.; Stella, S.; Pennisi, M.S.; Tirro, E.; Massimino, M.; Romano, C.; Puma, A.; Tavarelli, M.; Vigneri, P. New Insights in Thyroid Cancer and p53 Family Proteins. Int. J. Mol. Sci. 2017, 18, 1325. [Google Scholar] [CrossRef]
- Amelio, I.; Inoue, S.; Markert, E.K.; Levine, A.J.; Knight, R.A.; Mak, T.W.; Melino, G. TAp73 opposes tumor angiogenesis by promoting hypoxia-inducible factor 1alpha degradation. Proc. Natl. Acad. Sci. USA 2015, 112, 226–231. [Google Scholar] [CrossRef]
- Pitolli, C.; Wang, Y.; Candi, E.; Shi, Y.; Melino, G.; Amelio, I. p53-Mediated Tumor Suppression: DNA-Damage Response and Alternative Mechanisms. Cancers (Basel) 2019, 11, 1983. [Google Scholar] [CrossRef]
- Agostini, M.; Melino, G.; Bernassola, F. The p53 Family in Brain Disease. Antioxid. Redox Signal 2018, 29, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, W.B.; Kaplan, D.R.; Miller, F.D. The p53 family in nervous system development and disease. J. Neurochem. 2006, 97, 1571–1584. [Google Scholar] [CrossRef] [PubMed]
- Nemajerova, A.; Amelio, I.; Gebel, J.; Dotsch, V.; Melino, G.; Moll, U.M. Non-oncogenic roles of TAp73: From multiciliogenesis to metabolism. Cell Death Differ. 2018, 25, 144–153. [Google Scholar] [CrossRef]
- Wang, Q.; Zou, Y.; Nowotschin, S.; Kim, S.Y.; Li, Q.V.; Soh, C.L.; Su, C.; Zhang, C.; Shu, W.; Xi, Q.; et al. The p53 Family Coordinates Wnt and Nodal Inputs in Mesendodermal Differentiation of Embryonic Stem Cells. Cell Stem Cell 2017, 20, 70–86. [Google Scholar] [CrossRef]
- Billon, N.; Terrinoni, A.; Jolicoeur, C.; McCarthy, A.; Richardson, W.D.; Melino, G.; Raff, M. Roles for p53 and p73 during oligodendrocyte development. Development 2004, 131, 1211–1220. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rogel, A.; Popliker, M.; Webb, C.G.; Oren, M. p53 cellular tumor antigen: Analysis of mRNA levels in normal adult tissues, embryos, and tumors. Mol. Cell. Biol. 1985, 5, 2851–2855. [Google Scholar] [CrossRef]
- Komarova, E.A.; Chernov, M.V.; Franks, R.; Wang, K.; Armin, G.; Zelnick, C.R.; Chin, D.M.; Bacus, C.S.; Stark, G.R.; Gudkov, A.V.; et al. Transgenic mice with p53-responsive lacZ: p53 activity varies dramatically during normal development and determines radiation and drug sensitivity in vivo. EMBO J. 1997, 16, 1391–1400. [Google Scholar] [CrossRef]
- Van Lookeren Campagne, M.; Gill, R. Tumor-suppressor p53 is expressed in proliferating and newly formed neurons of the embryonic and postnatal rat brain: Comparison with expression of the cell cycle regulators p21Waf1/Cip1, p27Kip1, p57Kip2, p16Ink4a, cyclin G1, and the proto-oncogene Bax. J. Comp. Neurol. 1998, 397, 181–198. [Google Scholar] [CrossRef]
- Sah, V.P.; Attardi, L.D.; Mulligan, G.J.; Williams, B.O.; Bronson, R.T.; Jacks, T. A subset of p53-deficient embryos exhibit exencephaly. Nat. Genet. 1995, 10, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Armesilla-Diaz, A.; Bragado, P.; Del Valle, I.; Cuevas, E.; Lazaro, I.; Martin, C.; Cigudosa, J.C.; Silva, A. p53 regulates the self-renewal and differentiation of neural precursors. Neuroscience 2009, 158, 1378–1389. [Google Scholar] [CrossRef] [PubMed]
- Meletis, K.; Wirta, V.; Hede, S.M.; Nister, M.; Lundeberg, J.; Frisen, J. p53 suppresses the self-renewal of adult neural stem cells. Development 2006, 133, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Tedeschi, A.; Di Giovanni, S. The non-apoptotic role of p53 in neuronal biology: Enlightening the dark side of the moon. EMBO Rep. 2009, 10, 576–583. [Google Scholar] [CrossRef]
- Hooper, C.; Meimaridou, E.; Tavassoli, M.; Melino, G.; Lovestone, S.; Killick, R. p53 is upregulated in Alzheimer’s disease and induces tau phosphorylation in HEK293a cells. Neurosci. Lett. 2007, 418, 34–37. [Google Scholar] [CrossRef]
- Yang, A.; Walker, N.; Bronson, R.; Kaghad, M.; Oosterwegel, M.; Bonnin, J.; Vagner, C.; Bonnet, H.; Dikkes, P.; Sharpe, A.; et al. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature 2000, 404, 99–103. [Google Scholar] [CrossRef]
- Pozniak, C.D.; Barnabe-Heider, F.; Rymar, V.V.; Lee, A.F.; Sadikot, A.F.; Miller, F.D. p73 is required for survival and maintenance of CNS neurons. J. Neurosci. 2002, 22, 9800–9809. [Google Scholar] [CrossRef]
- Fuertes-Alvarez, S.; Maeso-Alonso, L.; Villoch-Fernandez, J.; Wildung, M.; Martin-Lopez, M.; Marshall, C.; Villena-Cortes, A.J.; Diez-Prieto, I.; Pietenpol, J.A.; Tissir, F.; et al. p73 regulates ependymal planar cell polarity by modulating actin and microtubule cytoskeleton. Cell Death Dis. 2018, 9, 1183. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, M.T.; Rufini, A.; Wetzel, M.K.; Tsuchihara, K.; Inoue, S.; Tomasini, R.; Itie-Youten, A.; Wakeham, A.; Arsenian-Henriksson, M.; Melino, G.; et al. Isoform-specific p73 knockout mice reveal a novel role for delta Np73 in the DNA damage response pathway. Genes Dev. 2010, 24, 549–560. [Google Scholar] [CrossRef] [PubMed]
- Niklison-Chirou, M.V.; Killick, R.; Knight, R.A.; Nicotera, P.; Melino, G.; Agostini, M. How Does p73 Cause Neuronal Defects? Mol. Neurobiol. 2016, 53, 4509–4520. [Google Scholar] [CrossRef] [PubMed]
- Killick, R.; Niklison-Chirou, M.; Tomasini, R.; Bano, D.; Rufini, A.; Grespi, F.; Gallagher, E.; Nicotera, P.; Mak, T.W.; Melino, G.; et al. p73: A multifunctional protein in neurobiology. Mol. Neurobiol. 2011, 43, 139–146. [Google Scholar] [CrossRef]
- Nemajerova, A.; Moll, U.M. Tissue-specific roles of p73 in development and homeostasis. J. Cell Sci. 2019, 132, jcs233338. [Google Scholar] [CrossRef] [PubMed]
- Sayan, B.S.; Yang, A.L.; Conforti, F.; Tucci, P.; Piro, M.C.; Browne, G.J.; Agostini, M.; Bernardini, S.; Knight, R.A.; Mak, T.W.; et al. Differential control of TAp73 and DeltaNp73 protein stability by the ring finger ubiquitin ligase PIR2. Proc. Natl. Acad. Sci. USA 2010, 107, 12877–12882. [Google Scholar] [CrossRef]
- Agostini, M.; Tucci, P.; Chen, H.; Knight, R.A.; Bano, D.; Nicotera, P.; McKeon, F.; Melino, G. p73 regulates maintenance of neural stem cell. Biochem. Biophys. Res. Commun. 2010, 403, 13–17. [Google Scholar] [CrossRef]
- Talos, F.; Abraham, A.; Vaseva, A.V.; Holembowski, L.; Tsirka, S.E.; Scheel, A.; Bode, D.; Dobbelstien, M.; Bruck, W.; Moll, U.M. p73 is an essential regulator of neural stem cell maintenance in embryonal and adult CNS neurogenesis. Cell Death Differ. 2010, 17, 1816–1829. [Google Scholar] [CrossRef]
- Gonzalez-Cano, L.; Herreros-Villanueva, M.; Fernandez-Alonso, R.; Ayuso-Sacido, A.; Meyer, G.; Garcia-Verdugo, J.M.; Silva, A.; Marques, M.M.; Marin, M.C. p73 deficiency results in impaired self renewal and premature neuronal differentiation of mouse neural progenitors independently of p53. Cell Death Dis. 2010, 1, e109. [Google Scholar] [CrossRef]
- Agostini, M.; Niklison-Chirou, M.V.; Annicchiarico-Petruzzelli, M.M.; Grelli, S.; Di Daniele, N.; Pestlikis, I.; Knight, R.A.; Melino, G.; Rufini, A. p73 Regulates Primary Cortical Neuron Metabolism: A Global Metabolic Profile. Mol. Neurobiol. 2018, 55, 3237–3250. [Google Scholar] [CrossRef]
- Amelio, I.; Cutruzzola, F.; Antonov, A.; Agostini, M.; Melino, G. Serine and glycine metabolism in cancer. Trends Biochem. Sci. 2014, 39, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Amelio, I.; Antonov, A.A.; Catani, M.V.; Massoud, R.; Bernassola, F.; Knight, R.A. TAp73 promotes anabolism. Oncotarget 2014, 5, 12820–12934. [Google Scholar] [CrossRef]
- Fujitani, M.; Cancino, G.I.; Dugani, C.B.; Weaver, I.C.; Gauthier-Fisher, A.; Paquin, A.; Mak, T.W.; Wojtowicz, M.J.; Miller, F.D.; Kaplan, D.R. TAp73 acts via the bHLH Hey2 to promote long-term maintenance of neural precursors. Curr. Biol. 2010, 20, 2058–2065. [Google Scholar] [CrossRef] [PubMed]
- Dechant, G.; Barde, Y.A. The neurotrophin receptor p75(NTR): Novel functions and implications for diseases of the nervous system. Nat. Neurosci. 2002, 5, 1131–1136. [Google Scholar] [CrossRef] [PubMed]
- Niklison-Chirou, M.V.; Steinert, J.R.; Agostini, M.; Knight, R.A.; Dinsdale, D.; Cattaneo, A.; Mak, T.W.; Melino, G. TAp73 knockout mice show morphological and functional nervous system defects associated with loss of p75 neurotrophin receptor. Proc. Natl. Acad. Sci. USA 2013, 110, 18952–18957. [Google Scholar] [CrossRef]
- Agostini, M.; Tucci, P.; Killick, R.; Candi, E.; Sayan, B.S.; Rivetti di Val Cervo, P.; Nicotera, P.; McKeon, F.; Knight, R.A.; Mak, T.W.; et al. Neuronal differentiation by TAp73 is mediated by microRNA-34a regulation of synaptic protein targets. Proc. Natl. Acad. Sci. USA 2011, 108, 21093–21098. [Google Scholar] [CrossRef]
- Agostini, M.; Tucci, P.; Steinert, J.R.; Shalom-Feuerstein, R.; Rouleau, M.; Aberdam, D.; Forsythe, I.D.; Young, K.W.; Ventura, A.; Concepcion, C.P.; et al. microRNA-34a regulates neurite outgrowth, spinal morphology, and function. Proc. Natl. Acad. Sci. USA 2011, 108, 21099–21104. [Google Scholar] [CrossRef]
- Chen, H.; Shalom-Feuerstein, R.; Riley, J.; Zhang, S.D.; Tucci, P.; Agostini, M.; Aberdam, D.; Knight, R.A.; Genchi, G.; Nicotera, P.; et al. miR-7 and miR-214 are specifically expressed during neuroblastoma differentiation, cortical development and embryonic stem cells differentiation, and control neurite outgrowth in vitro. Biochem. Biophys. Res. Commun. 2010, 394, 921–927. [Google Scholar] [CrossRef]
- Vikhreva, P.; Melino, G.; Amelio, I. p73 Alternative Splicing: Exploring a Biological Role for the C-Terminal Isoforms. J. Mol. Biol. 2018, 430, 1829–1838. [Google Scholar] [CrossRef]
- Grespi, F.; Amelio, I.; Tucci, P.; Annicchiarico-Petruzzelli, M.; Melino, G. Tissue-specific expression of p73 C-terminal isoforms in mice. Cell Cycle 2012, 11, 4474–4483. [Google Scholar] [CrossRef] [PubMed]
- Amelio, I.; Panatta, E.; Niklison-Chirou, M.V.; Steinert, J.R.; Agostini, M.; Morone, N.; Knight, R.A.; Melino, G. The C terminus of p73 is essential for hippocampal development. Proc. Natl. Acad. Sci. USA 2020, 117, 15694–15701. [Google Scholar] [CrossRef] [PubMed]
- Maffezzini, C.; Calvo-Garrido, J.; Wredenberg, A.; Freyer, C. Metabolic regulation of neurodifferentiation in the adult brain. Cell. Mol. Life Sci. 2020, 77, 2483–2496. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.T.; Mahmood, S.; Patel, M.S. Intermediary metabolism and energetics during murine early embryogenesis. J. Biol. Chem. 2003, 278, 31457–31460. [Google Scholar] [CrossRef]
- Shyh-Chang, N.; Ng, H.H. The metabolic programming of stem cells. Genes Dev. 2017, 31, 336–346. [Google Scholar] [CrossRef] [PubMed]
- Uittenbogaard, M.; Brantner, C.A.; Chiaramello, A. Epigenetic modifiers promote mitochondrial biogenesis and oxidative metabolism leading to enhanced differentiation of neuroprogenitor cells. Cell Death Dis. 2018, 9, 360. [Google Scholar] [CrossRef]
- Bourgognon, J.M.; Spiers, J.G.; Scheiblich, H.; Antonov, A.; Bradley, S.J.; Tobin, A.B.; Steiner, J.R. Alterations in neuronal metabolism contribute to the pathogenesis of prion disease. Cell Death Differ. 2018, 25, 1408–1425. [Google Scholar] [CrossRef]
- Raefsky, S.M.; Mattson, M.P. Adaptive responses of neuronal mitochondria to bioenergetic challenges: Roles in neuroplasticity and disease resistance. Free Radic. Biol. Med. 2017, 102, 203–216. [Google Scholar] [CrossRef]
- Garrido-Maraver, J.; Celardo, I.; Costa, A.C.; Lehmann, S.; Loh, S.H.Y.; Martins, L.M. Enhancing folic acid metabolism suppresses defects associated with loss of Drosophila mitofusin. Cell Death Dis. 2019, 10, 288. [Google Scholar] [CrossRef]
- Cheng, A.; Wan, R.; Yang, J.L.; Kamimura, N.; Son, T.G.; Ouyang, X.; Luo, Y.; Okun, E.; Mattson, M.P. Involvement of PGC-1alpha in the formation and maintenance of neuronal dendritic spines. Nat. Commun. 2012, 3, 1250. [Google Scholar] [CrossRef]
- Scarpulla, R.C. Nucleus-encoded regulators of mitochondrial function: Integration of respiratory chain expression, nutrient sensing and metabolic stress. Biochim. Biophys. Acta 2012, 1819, 1088–1097. [Google Scholar] [CrossRef] [PubMed]
- Rufini, A.; Niklison-Chirou, M.V.; Inoue, S.; Tomasini, R.; Harris, I.S.; Marino, A.; Federici, M.; Dinsdale, D.; Knight, R.A.; Melino, G.; et al. TAp73 depletion accelerates aging through metabolic dysregulation. Genes Dev. 2012, 26, 2009–2014. [Google Scholar] [CrossRef]
- Wong-Riley, M.T. Cytochrome oxidase: An endogenous metabolic marker for neuronal activity. Trends Neurosci. 1989, 12, 94–101. [Google Scholar] [CrossRef]
- Robledinos-Anton, N.; Rojo, A.I.; Ferreiro, E.; Núñez, Á.; Krause, K.-H.; Jaquet, V.; Cuardo, A. Transcription factor NRF2 controls the fate of neural stem cells in the subgranular zone of the hippocampus. Redox Biol. 2017, 13, 393–401. [Google Scholar] [CrossRef]
- Michels, S.; Ganjam, G.K.; Martins, H.; Schratt, G.M.; Wöhr, M.; Schwarting, R.K.W.; Culmsee, C. Downregulation of the psychiatric susceptibility gene Cacna1c promotes mitochondrial resilience to oxidative stress in neuronal cells. Cell Death Discov. 2018, 4, 54. [Google Scholar] [CrossRef] [PubMed]
- Oruganty-Das, A.; Ng, T.; Udagawa, T.; Goh, E.L.; Richter, J.D. Translational control of mitochondrial energy production mediates neuron morphogenesis. Cell Metab. 2012, 16, 789–800. [Google Scholar] [CrossRef] [PubMed]
- Wanet, A.; Arnould, T.; Najimi, M.; Renard, P. Connecting Mitochondria, Metabolism, and Stem Cell Fate. Stem Cells Dev. 2015, 24, 1957–1971. [Google Scholar] [CrossRef]
- Khacho, M.; Slack, R.S. Mitochondrial activity in the regulation of stem cell self-renewal and differentiation. Curr. Opin. Cell Biol. 2017, 49, 1–8. [Google Scholar] [CrossRef]
- Zheng, X.; Boyer, L.; Jin, M.; Mertens, J.; Kim, Y.; Ma, L.; Hamm, M.; Gage, F.H.; Hunter, T. Metabolic reprogramming during neuronal differentiation from aerobic glycolysis to neuronal oxidative phosphorylation. eLife 2016, 5, e13374. [Google Scholar] [CrossRef]
- Agostini, M.; Romeo, F.; Inoue, S.; Niklison-Chirou, M.V.; Elia, A.J.; Dinsdale, D.; Morone, N.; Knight, R.A.; Mak, T.W.; Melino, G. Metabolic reprogramming during neuronal differentiation. Cell Death Differ. 2016, 23, 1502–1514. [Google Scholar] [CrossRef]
- Bauernfeind, A.; Barks, S.K.; Duka, T.; Grossman, L.I.; Hof, P.R.; Sherwood, C.C. Aerobic glycolysis in the primate brain: Reconsidering the implications for growth and maintenance. Brain Struct. Funct. 2014, 219, 1149–1167. [Google Scholar] [CrossRef] [PubMed]
- Teslaa, T.; Teitell, M.A. Pluripotent stem cell energy metabolism: An update. EMBO J. 2015, 34, 138–153. [Google Scholar] [CrossRef]
- Orrells, S.F.; Paredes, M.F.; Cebrian-Silla, A.; Sandoval, K.; Qi, D.; Kelley, K.W.; James, D.; Mayer, S.; Chang, J.; Auguste, K.I.; et al. Human hippocampal neurogenesis drops sharply in children to undetectable levels in adults. Nature 2018, 555, 377–381. [Google Scholar] [CrossRef] [PubMed]
- Apple, D.M.; Solano-Fonseca, R.; Kokovay, E. Neurogenesis in the aging brain. Biochem. Pharmacol. 2017, 141, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Ziemka-Nalecz, M.; Jaworska, J.; Sypecka, J.; Zalewska, T. Histone Deacetylase Inhibitors: A Therapeutic Key in Neurological Disorders? J. Neuropathol. Exp. Neurol. 2018, 77, 855–870. [Google Scholar] [CrossRef]
- Tee, A.R.; Sampson, J.R.; Pal, D.K.; Bateman, J.M. The role of mTOR signalling in neurogenesis, insights from tuberous sclerosis complex. Semin. Cell Dev. Biol. 2016, 52, 12–20. [Google Scholar]
- Bove, J.; Martinez-Vicente, M.; Vila, M. Fighting neurodegeneration with rapamycin: Mechanistic insights. Nat. Rev. Neurosci. 2011, 12, 437–452. [Google Scholar] [CrossRef] [PubMed]
- Finsterer, J. Leigh and Leigh-like syndrome in children and adults. Pediatr. Neurol. 2008, 39, 223–235. [Google Scholar] [CrossRef]
- Johnson, S.C.; Yanos, M.E.; Kayser, E.-B.; Quintana, A.; Sangesland, M.; Castanza, A.; Uhde, L.; Hui, J.; Wall, V.Z.; Gagnidze, A.; et al. mTOR inhibition alleviates mitochondrial disease in a mouse model of Leigh syndrome. Science 2013, 342, 1524–1528. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).