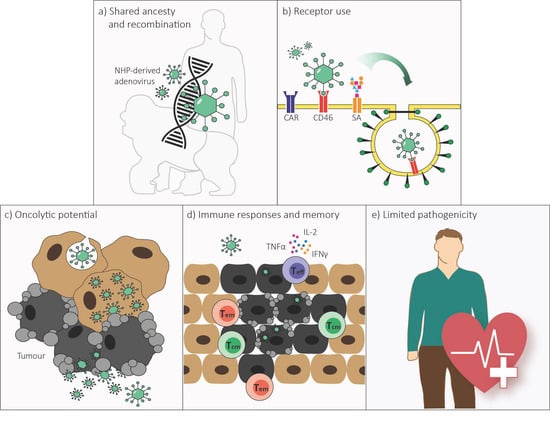
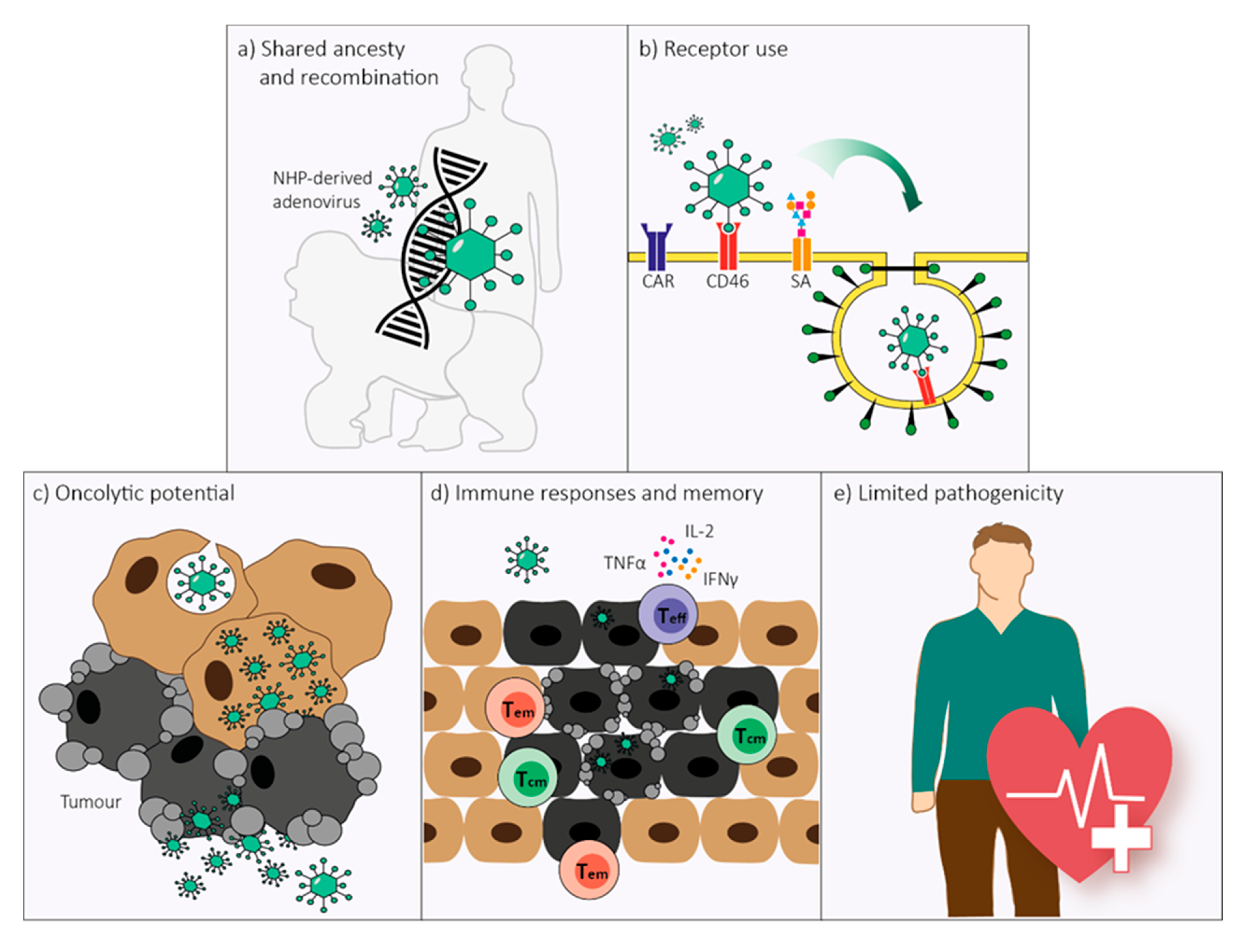
Non-Human Primate-Derived Adenoviruses for Future Use as Oncolytic Agents?
Abstract
1. Introduction
2. Genome Organization and Recombination
3. Receptor Use
4. Replication and Cell Lysis
5. Immune Responses
6. Pathology in Humans
7. Future Perspectives
Supplementary Materials
Funding
Conflicts of Interest
Abbreviations
| NHP | Non-human primate |
| HAdV | Human adenovirus |
| SAdV | Simian adenovirus |
| APC | Antigen-presenting cell |
| DC | Dendritic cell |
| dLN | Draining Lymph Node |
| Teff | Effector T cell |
| Tem | Effector memory T cell |
| Tcm | Central memory T cell |
| IFN | Interferon |
| TNF | Tumor necrosis factor |
| IL | Interleukin |
| PBMC | Peripheral blood mononuclear cells |
| Trm | Tissue-resident memory T cell |
| Tfh | Follicular B cell helper T cell |
References
- Rowe, W.P.; Huebner, R.J.; Gilmore, L.K.; Parrott, R.H.; Ward, T.G. Isolation of a cytopathogenic agent from human adenoids undergoing spontaneous degeneration in tissue culture. Exp. Boil. Med. 1953, 84, 570–573. [Google Scholar] [CrossRef] [PubMed]
- Trentin, J.J.; Yabe, Y.; Taylor, G. The quest for human cancer viruses. Science 1962, 137, 835–841. [Google Scholar] [CrossRef] [PubMed]
- Huebner, R.J.; Rowe, W.P.; Lane, W.T. Oncogenic effects in hamsters of human adenovirus types 12 and 18. Proc. Natl. Acad. Sci. USA 1962, 48, 2051–2058. [Google Scholar] [CrossRef]
- Schenk-Braat, E.A.M.; Kaptein, L.C.M.; Hallemeesch, M.M.; Bangma, C.H.; Hoeben, R.C. Gene therapy in The Netherlands: Highlights from the Low Countries. J. Gene Med. 2007, 9, 895–903. [Google Scholar] [CrossRef] [PubMed]
- Hoeben, R.C.; Uil, T.G. Adenovirus DNA Replication. Cold Spring Harb. Perspect. Boil. 2013, 5, a013003. [Google Scholar] [CrossRef]
- King, A.M.Q.; Adams, M.J.; Carstens, E.B.; Lefkowitz, E.J. Virus Taxonomy: Classification and Nomenclature of Viruses; Ninth Report of the International Committee on Taxonomy of Viruses; Academic Press: London, UK; Waltham, MA, USA, 2012. [Google Scholar]
- Seto, D.; Chodosh, J.; Brister, J.R.; Jones, M.S. Members of the adenovirus research community using the whole-genome sequence to characterize and name human adenoviruses. J. Virol. 2011, 85, 5701–5702. [Google Scholar] [CrossRef]
- Aoki, K.; Benkö, M.; Davison, A.J.; Echavarria, M.; Erdman, D.D.; Harrach, B.; Kajon, A.E.; Schnurr, D.; Wadell, G. Members of the adenovirus research community toward an integrated human adenovirus designation system that utilizes molecular and serological data and serves both clinical and fundamental virology. J. Virol. 2011, 85, 5703–5704. [Google Scholar] [CrossRef]
- Wevers, D.; Metzger, S.; Babweteera, F.; Bieberbach, M.; Boesch, C.; Cameron, K.; Couacy-Hymann, E.; Cranfield, M.; Gray, M.; Harris, L.A.; et al. Novel adenoviruses in wild primates: A high level of genetic diversity and evidence of zoonotic transmissions. J. Virol. 2011, 85, 10774–10784. [Google Scholar] [CrossRef] [PubMed]
- Pantó, L.; Podgorski, I.I.; Jánoska, M.; Márkó, O.; Harrach, B. Taxonomy proposal for Old World monkey adenoviruses: Characterisation of several non-human, non-ape primate adenovirus lineages. Arch. Virol. 2015, 160, 3165–3177. [Google Scholar] [CrossRef] [PubMed]
- Lange, C.E.; Niama, F.R.; Cameron, K.; Olson, S.H.; Nina, R.A.; Ondzie, A.; Bounga, G.; Smith, B.R.; Pante, J.; Reed, P.; et al. First evidence of a new simian adenovirus clustering with Human mastadenovirus F viruses. Virol. J. 2019, 16, 147. [Google Scholar] [CrossRef]
- Hoppe, E.; Pauly, M.; Gillespie, T.R.; Akoua-Koffi, C.; Hohmann, G.; Fruth, B.; Karhemere, S.; Madinda, N.F.; Mugisha, L.; Muyembe, J.-J.; et al. Multiple cross-species transmission events of human adenoviruses (HAdV) during hominine evolution. Mol. Boil. Evol. 2015, 32, 2072–2084. [Google Scholar] [CrossRef] [PubMed]
- Vogels, R.; Zuijdgeest, D.; Van Rijnsoever, R.; Hartkoorn, E.; Damen, I.; De Béthune, M.-P.; Kostense, S.; Penders, G.; Helmus, N.; Koudstaal, W.; et al. Replication-deficient human adenovirus type 35 vectors for gene transfer and vaccination: Efficient human cell infection and bypass of preexisting adenovirus immunity. J. Virol. 2003, 77, 8263–8271. [Google Scholar] [CrossRef] [PubMed]
- Nwanegbo, E.; Vardas, E.; Gao, W.; Whittle, H.; Sun, H.; Rowe, D.; Robbins, P.D.; Gambotto, A. Prevalence of neutralizing antibodies to adenoviral serotypes 5 and 35 in the adult populations of The Gambia, South Africa, and the United States. Clin. Diagn. Lab. Immunol. 2004, 11, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Xiang, Z.Q.; Li, Y.; Kurupati, R.K.; Jia, B.; Bian, A.; Zhou, D.M.; Hutnick, N.; Yuan, S.; Gray, C.; et al. Adenovirus-based vaccines: Comparison of vectors from three species of adenoviridae. J. Virol. 2010, 84, 10522–10532. [Google Scholar] [CrossRef]
- Roy, S.; Medina-Jaszek, A.; Wilson, M.J.; Sandhu, A.; Calcedo, R.P.; Lin, J.; Wilson, J.M. Creation of a panel of vectors based on ape adenovirus isolates. J. Gene Med. 2010, 13, 17–25. [Google Scholar] [CrossRef]
- Quinn, K.M.; Da Costa, A.; Yamamoto, A.; Berry, D.; Lindsay, R.W.B.; Darrah, P.A.; Wang, L.; Cheng, C.; Kong, W.-P.; Gall, J.G.D.; et al. Comparative analysis of the magnitude, quality, phenotype, and protective capacity of simian immunodeficiency virus gag-specific CD8+ T cells following human-, simian-, and chimpanzee-derived recombinant adenoviral vector immunization. J. Immunol. 2013, 190, 2720–2735. [Google Scholar] [CrossRef]
- Capasso, C.; Hirvinen, M.; Garofalo, M.; Romaniuk, D.; Kuryk, L.; Sarvela, T.; Vitale, A.; Antopolsky, M.; Magarkar, A.; Viitala, T.; et al. Oncolytic adenoviruses coated with MHC-I tumor epitopes increase the antitumor immunity and efficacy against melanoma. OncoImmunology 2015, 5, e1105429. [Google Scholar] [CrossRef]
- Kaján, G.L.; Lipiec, A.; Bartha, D.; Allard, A.; Arnberg, N. A multigene typing system for human adenoviruses reveals a new genotype in a collection of Swedish clinical isolates. PLoS ONE 2018, 13, e0209038. [Google Scholar] [CrossRef]
- Dehghan, S.; Seto, J.; Liu, E.B.; Ismail, A.M.; Madupu, R.; Heim, A.; Jones, M.S.; Dyer, D.W.; Chodosh, J.; Seto, D. A zoonotic adenoviral human pathogen emerged through genomic recombination among human and nonhuman simian hosts. J. Virol. 2019, 93, e00564-19. [Google Scholar] [CrossRef]
- Wevers, D.; Leendertz, F.H.; Scuda, N.; Boesch, C.; Robbins, M.M.; Head, J.; Ludwig, C.; Kühn, J.; Ehlers, B. A novel adenovirus of Western lowland gorillas (Gorilla gorilla gorilla). Virol. J. 2010, 7, 303. [Google Scholar] [CrossRef]
- Podgorski, I.I.; Pantó, L.; Papp, T.; Harrach, B.; Benkő, M. Genome analysis of four Old World monkey adenoviruses supports the proposed species classification of primate adenoviruses and reveals signs of possible homologous recombination. J. Gen. Virol. 2016, 97, 1604–1614. [Google Scholar] [CrossRef] [PubMed]
- Han, J.-W.; La, T.-M.; Kim, J.-H.; Choi, I.-S.; Song, C.-S.; Park, S.-Y.; Lee, J.-B.; Lee, S.-W. The possible origin of human adenovirus type 3: Evidence of natural genetic recombination between human and simian adenovirus. Infect. Genet. Evol. 2018, 65, 380–384. [Google Scholar] [CrossRef] [PubMed]
- Hoppe, E.; Pauly, M.; Robbins, M.; Gray, M.; Kujirakwinja, D.; Nishuli, R.; Mungu-Akonkwa, D.-D.B.; Leendertz, F.H.; Ehlers, B. Phylogenomic evidence for recombination of adenoviruses in wild gorillas. J. Gen. Virol. 2015, 96, 3090–3098. [Google Scholar] [CrossRef] [PubMed]
- Dhingra, A.; Hage, E.; Ganzenmueller, T.; Böttcher, S.; Hofmann, J.; Hamprecht, K.; Obermeier, P.; Rath, B.; Hausmann, F.; Dobner, T.; et al. Molecular evolution of human adenovirus (HAdV) species C. Sci. Rep. 2019, 9, 1039. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Martin, N.; Ramani, S.R.; Hackney, J.A.; Tom, I.; Wranik, B.J.; Chan, M.; Wu, J.; Paluch, M.T.; Takeda, K.; Hass, P.E.; et al. The extracellular interactome of the human adenovirus family reveals diverse strategies for immunomodulation. Nat. Commun. 2016, 7, 11473. [Google Scholar] [CrossRef]
- Calcedo, R.; Vandenberghe, L.H.; Roy, S.; Somanathan, S.; Wang, L.; Wilson, J.M. Host immune responses to chronic adenovirus infections in human and nonhuman primates. J. Virol. 2008, 83, 2623–2631. [Google Scholar] [CrossRef]
- Suzuki, K.; Alemany, R.; Yamamoto, M.; Curiel, D.T. The presence of the adenovirus E3 region improves the oncolytic potency of conditionally replicative adenoviruses. Clin. Cancer Res. 2002, 8, 3348–3359. [Google Scholar]
- Smith, T.; Waterman, M. Identification of common molecular subsequences. J. Mol. Boil. 1981, 147, 195–197. [Google Scholar] [CrossRef]
- Katoh, K. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Lole, K.S.; Bollinger, R.C.; Paranjape, R.S.; Gadkari, D.; Kulkarni, S.S.; Novak, N.G.; Ingersoll, R.; Sheppard, H.W.; Ray, S.C. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 1999, 73, 152–160. [Google Scholar] [CrossRef]
- Arnberg, N. Adenovirus receptors: Implications for tropism, treatment and targeting. Rev. Med. Virol. 2009, 19, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, Z.-Y.; Liu, Y.; Persson, J.; Beyer, I.; Möller, T.; Koyuncu, D.; Drescher, M.R.; Strauss, R.; Zhang, X.-B.; et al. Desmoglein 2 is a receptor for adenovirus serotypes 3, 7, 11 and 14. Nat. Med. 2010, 17, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.-Y.; Wang, X.; Han, Y.; Li, G.; Wang, H.-J.; Wang, S.-B.; Chen, X.-Y.; Liu, F.-L.; He, X.-L.; Tong, X.-M.; et al. Loss of coxsackie and adenovirus receptor expression in human colorectal cancer: A potential impact on the efficacy of adenovirus-mediated gene therapy in Chinese Han population. Mol. Med. Rep. 2016, 14, 2541–2547. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yang, X.; Li, S.; Wang, H.; Chen, W.; Mou, X.; Wang, S. Expression of coxsackie and adenovirus receptor is correlated with inferior prognosis in liver cancer patients. Oncol. Lett. 2018, 17, 2485–2490. [Google Scholar] [CrossRef] [PubMed]
- Cohen, C.J.; Xiang, Z.Q.; Gao, G.; Ertl, H.C.J.; Wilson, J.M.; Bergelson, J.M. Chimpanzee adenovirus CV-68 adapted as a gene delivery vector interacts with the coxsackievirus and adenovirus receptor. J. Gen. Virol. 2002, 83, 151–155. [Google Scholar] [CrossRef]
- Tatsis, N.; Blejer, A.; Lásaro, M.O.; Hensley, S.E.; Cun, A.; Tesema, L.; Li, Y.; Gao, G.; Xiang, Z.Q.; Zhou, D.; et al. A CD46-binding chimpanzee adenovirus vector as a vaccine carrier. Mol. Ther. 2007, 15, 608–617. [Google Scholar] [CrossRef]
- Abbink, P.; Kirilova, M.; Boyd, M.; Mercado, N.; Li, Z.; Nityanandam, R.; Nanayakkara, O.; Peterson, R.; LaRocca, R.A.; Aïd, M.; et al. Rapid cloning of novel rhesus adenoviral vaccine vectors. J. Virol. 2018, 92, e01924-17. [Google Scholar] [CrossRef]
- Baker, A.T.; Mundy, R.; Davies, J.; Rizkallah, P.J.; Parker, A.L. Human adenovirus type 26 uses sialic acid–bearing glycans as a primary cell entry receptor. Sci. Adv. 2019, 5, eaax3567. [Google Scholar] [CrossRef]
- Lenman, A.; Liaci, A.M.; Liu, Y.; Årdahl, C.; Rajan, A.; Nilsson, E.; Bradford, W.; Kaeshammer, L.; Jones, M.S.; Frängsmyr, L.; et al. Human adenovirus 52 uses sialic acid-containing glycoproteins and the coxsackie and adenovirus receptor for binding to target cells. PLoS Pathog. 2015, 11, e1004657. [Google Scholar] [CrossRef]
- Dicks, M.D.J.; Spencer, A.; Coughlan, L.; Bauza, K.; Gilbert, S.C.; Hill, A.V.S.; Cottingham, M. Differential immunogenicity between HAdV-5 and chimpanzee adenovirus vector ChAdOx1 is independent of fiber and penton RGD loop sequences in mice. Sci. Rep. 2015, 5, 16756. [Google Scholar] [CrossRef]
- Stasiak, A.C.; Stehle, T. Human adenovirus binding to host cell receptors: A structural view. Med. Microbiol. Immunol. 2019, 209, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Yamano, T.; Kubo, S.; Fukumoto, M.; Yano, A.; Mawatari-Furukawa, Y.; Okamura, H.; Tomita, N. Whole cell vaccination using immunogenic cell death by an oncolytic adenovirus is effective against a colorectal cancer model. Mol. Oncolytics 2016, 3, 16031. [Google Scholar] [CrossRef]
- Wu, C.; Öberg, D.; Rashid, A.; Gupta, R.; Mignardi, M.; Johansson, S.; Akusjärvi, G.; Svensson, C. A mouse mammary epithelial cell line permissive for highly efficient human adenovirus growth. Virology 2013, 435, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.; Jacobus, E.J.; Taverner, W.; Fisher, K.; Hemmi, S.; West, K.; Slater, L.; Lilley, F.; Brown, A.C.; Champion, B.; et al. Expression of human CD46 and trans-complementation by murine adenovirus 1 fails to allow productive infection by a group B oncolytic adenovirus in murine cancer cells. J. Immunother. Cancer 2018, 6, 55. [Google Scholar] [CrossRef]
- Bots, S.T.F.; Kemp, V.; Lamfers, M.L.M.; Van der Pluijm, G.; Hoeben, R.C. New non-human primate adenoviruses for use as oncolytic agents. In Proceedings of the International Oncolytic Virus Conference, Rochester, MN, USA, 9–12 October 2019; p. 31. [Google Scholar]
- Cheng, T.; Song, Y.; Zhang, Y.; Zhang, C.; Yin, J.; Chi, Y.; Zhou, D. A novel oncolytic adenovirus based on simian adenovirus serotype 24. Oncotarget 2017, 8, 26871–26885. [Google Scholar] [CrossRef]
- Dyer, A.; Di, Y.; Calderon, H.; Illingworth, S.; Kueberuwa, G.; Tedcastle, A.; Jakeman, P.; Chia, S.L.; Brown, A.; Silva, M.A.; et al. Oncolytic group B adenovirus enadenotucirev mediates non-apoptotic cell death with membrane disruption and release of inflammatory mediators. Mol. Oncolytics 2016, 4, 18–30. [Google Scholar] [CrossRef]
- Ko, E.-J.; Hait, S.H.; Enyindah-Asonye, G.; Rahman, M.A.; Hoang, T.; Robert-Guroff, M. Replicating adenovirus-SIV immunization of rhesus macaques induces mucosal dendritic cell activation and function leading to rectal immune responses. Front. Immunol. 2019, 10, 779. [Google Scholar] [CrossRef] [PubMed]
- Quinn, K.M.; Zak, D.E.; Costa, A.; Yamamoto, A.; Kastenmüller, K.; Hill, B.J.; Lynn, G.M.; Darrah, P.A.; Lindsay, R.W.; Wang, L.; et al. Antigen expression determines adenoviral vaccine potency independent of IFN and STING signaling. J. Clin. Investig. 2015, 125, 1129–1146. [Google Scholar] [CrossRef]
- Cheng, C.; Wang, L.; Ko, S.-Y.; Kong, W.-P.; Schmidt, S.D.; Gall, J.G.; Colloca, S.; Seder, R.A.; Mascola, J.R.; Nabel, G.J. Combination recombinant simian or chimpanzee adenoviral vectors for vaccine development. Vaccine 2015, 33, 7344–7351. [Google Scholar] [CrossRef]
- Colloca, S.; Barnes, E.; Folgori, A.; Ammendola, V.; Capone, S.; Cirillo, A.; Siani, L.; Naddeo, M.; Grazioli, F.; Esposito, M.L.; et al. Vaccine vectors derived from a large collection of simian adenoviruses induce potent cellular immunity across multiple species. Sci. Transl. Med. 2012, 4, 115ra2. [Google Scholar] [CrossRef]
- Limbach, K.; Stefaniak, M.; Chen, P.; Patterson, N.B.; Liao, G.; Weng, S.; Krepkiy, S.; Ekberg, G.; Torano, H.; Ettyreddy, D.; et al. New gorilla adenovirus vaccine vectors induce potent immune responses and protection in a mouse malaria model. Malar. J. 2017, 16, 263. [Google Scholar] [CrossRef]
- Herath, S.; Le Heron, A.; Colloca, S.; Patterson, S.; Tatoud, R.; Weber, J.; Dickson, G. Strain-dependent and distinctive T-cell responses to HIV antigens following immunisation of mice with differing chimpanzee adenovirus vaccine vectors. Vaccine 2016, 34, 4378–4385. [Google Scholar] [CrossRef][Green Version]
- Paris, R.; Kuschner, R.A.; Binn, L.; Thomas, S.J.; Colloca, S.; Nicosia, A.; Cortese, R.; Bailer, R.T.; Sullivan, N.; Koup, R.A. Adenovirus type 4 and 7 vaccination or adenovirus type 4 respiratory infection elicits minimal cross-reactive antibody responses to nonhuman adenovirus vaccine vectors. Clin. Vaccine Immunol. 2014, 21, 783–786. [Google Scholar] [CrossRef]
- Cappuccini, F.; Stribbling, S.; Pollock, E.; Hill, A.V.S.; Redchenko, I. Immunogenicity and efficacy of the novel cancer vaccine based on simian adenovirus and MVA vectors alone and in combination with PD-1 mAb in a mouse model of prostate cancer. Cancer Immunol. Immunother. 2016, 65, 701–713. [Google Scholar] [CrossRef]
- Tan, W.G.; Jin, H.-T.; West, E.E.; Penaloza-MacMaster, P.; Wieland, A.; Zilliox, M.J.; McElrath, M.J.; Barouch, D.H.; Ahmed, R. Comparative analysis of simian immunodeficiency virus gag-specific effector and memory CD8+T cells induced by different adenovirus vectors. J. Virol. 2012, 87, 1359–1372. [Google Scholar] [CrossRef]
- Reading, J.L.; Galvez-Cancino, F.; Swanton, C.; Lladser, A.; Peggs, K.S.; Quezada, S.A. The function and dysfunction of memory CD8+T cells in tumor immunity. Immunol. Rev. 2018, 283, 194–212. [Google Scholar] [CrossRef]
- Swadling, L.; Capone, S.; Antrobus, R.D.; Brown, A.; Richardson, R.; Newell, E.W.; Halliday, J.; Kelly, C.; Bowen, D.; Fergusson, J.; et al. A human vaccine strategy based on chimpanzee adenoviral and MVA vectors that primes, boosts, and sustains functional HCV-specific T cell memory. Sci. Transl. Med. 2014, 6, 261ra153. [Google Scholar] [CrossRef]
- Penaloza-MacMaster, P.; Provine, N.M.; Ra, J.; Borducchi, E.N.; McNally, A.; Simmons, N.; Iampietro, M.J.; Barouch, D.H. Alternative serotype adenovirus vaccine vectors elicit memory T cells with enhanced anamnestic capacity compared to Ad5 vectors. J. Virol. 2012, 87, 1373–1384. [Google Scholar] [CrossRef][Green Version]
- Rollier, C.S.; Verschoor, E.J.; Verstrepen, B.E.; Drexhage, J.A.R.; Paranhos-Baccala, G.; Liljeström, P.; Sutter, G.; Arribillaga, L.; Lasarte, J.J.; Bartosch, B.; et al. T- and B-cell responses to multivalent prime-boost DNA and viral vectored vaccine combinations against hepatitis C virus in non-human primates. Gene Ther. 2016, 23, 753–759. [Google Scholar] [CrossRef]
- Li, H.; Liu, J.; Carville, A.; Mansfield, K.G.; Lynch, D.; Barouch, D.H. Durable mucosal simian immunodeficiency virus-specific effector memory T lymphocyte responses elicited by recombinant adenovirus vectors in rhesus monkeys. J. Virol. 2011, 85, 11007–11015. [Google Scholar] [CrossRef][Green Version]
- Balandya, E.; Miller, A.D.; Beck, M.; Liu, J.; Li, H.; Borducchi, E.; Smith, K.; Cabral, C.; Stanley, K.; Maxfield, L.F.; et al. Adenovirus serotype 26 and 35 vectors induce simian immunodeficiency virus-specific T lymphocyte responses in foreskin in rhesus monkeys. J. Virol. 2014, 88, 3756–3765. [Google Scholar] [CrossRef][Green Version]
- Maurice, N.; McElrath, M.J.; Andersen-Nissen, E.; Frahm, N.; Prlic, M. CXCR3 enables recruitment and site-specific bystander activation of memory CD8+ T cells. Nat. Commun. 2019, 10, 4987. [Google Scholar] [CrossRef]
- Rosato, P.C.; Wijeyesinghe, S.; Stolley, J.M.; Nelson, C.E.; Davis, R.L.; Manlove, L.S.; Pennell, C.A.; Blazar, B.R.; Chen, C.C.; Geller, M.A.; et al. Virus-specific memory T cells populate tumors and can be repurposed for tumor immunotherapy. Nat. Commun. 2019, 10, 567. [Google Scholar] [CrossRef]
- Sauermann, U.; Radaelli, A.; Stolte-Leeb, N.; Raue, K.; Bissa, M.; Zanotto, C.; Krawczak, M.; Tenbusch, M.; Überla, K.; Keele, B.F.; et al. Vector order determines protection against pathogenic simian immunodeficiency virus infection in a triple-component vaccine by balancing CD4+ and CD8+ T-cell responses. J. Virol. 2017, 91, e01120-17. [Google Scholar] [CrossRef]
- Ahrends, T.; Busselaar, J.; Severson, T.M.; Bąbała, N.; De Vries, E.; Bovens, A.; Wessels, L.; Van Leeuwen, F.; Borst, J. CD4+ T cell help creates memory CD8+ T cells with innate and help-independent recall capacities. Nat. Commun. 2019, 10, 5531. [Google Scholar] [CrossRef]
- Ledgerwood, J.E.; DeZure, A.D.; Stanley, D.A.; Coates, E.E.; Novik, L.; Enama, M.E.; Berkowitz, N.M.; Hu, Z.; Joshi, G.; Ploquin, A.; et al. Chimpanzee adenovirus vector ebola vaccine. N. Engl. J. Med. 2017, 376, 928–938. [Google Scholar] [CrossRef]
- Tuyishime, S.; Haut, L.H.; Kurupati, R.K.; Billingsley, J.M.; Carnathan, D.; Gangahara, S.; Styles, T.M.; Xiang, Z.; Li, Y.; Zopfs, M.; et al. Correlates of protection against SIVmac251 infection in rhesus macaques immunized with chimpanzee-derived adenovirus vectors. EBioMedicine 2018, 31, 25–35. [Google Scholar] [CrossRef]
- Vargas-Inchaustegui, D.A.; Demers, A.; Shaw, J.M.; Kang, G.; Ball, D.; Tuero, I.; Musich, T.; Mohanram, V.; Demberg, T.; Karpova, T.; et al. Vaccine induction of lymph node-resident simian immunodeficiency virus env-specific T follicular helper cells in rhesus macaques. J. Immunol. 2016, 196, 1700–1710. [Google Scholar] [CrossRef]
- Ehrenberg, P.K.; Shangguan, S.; Issac, B.; Alter, G.; Geretz, A.; Izumi, T.; Bryant, C.; Eller, M.A.; Wegmann, F.; Apps, R.; et al. A vaccine-induced gene expression signature correlates with protection against SIV and HIV in multiple trials. Sci. Transl. Med. 2019, 11, eaaw4236. [Google Scholar] [CrossRef]
- Appledorn, D.M.; Kiang, A.; McBride, A.; Jiang, H.; Seregin, S.; Scott, J.M.; Stringer, R.; A Kousa, M.Y.; Hoban, M.; Frank, M.M.; et al. Wild-type adenoviruses from groups A–F evoke unique innate immune responses, of which HAd3 and SAd23 are partially complement dependent. Gene Ther. 2008, 15, 885–901. [Google Scholar] [CrossRef]
- Lynch, J.P.; Kajon, A.E. Adenovirus: Epidemiology, global spread of novel serotypes, and advances in treatment and prevention. Semin. Respir. Crit. Care Med. 2016, 37, 586–602. [Google Scholar] [CrossRef]
- Barnadas, C.; Schmidt, D.J.; Fischer, T.K.; Fonager, J. Molecular epidemiology of human adenovirus infections in Denmark, 2011–2016. J. Clin. Virol. 2018, 104, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Sandhu, A.; Medina, A.; Clawson, D.S.; Wilson, J.M. Adenoviruses in fecal samples from asymptomatic rhesus macaques, United States. Emerg. Infect. Dis. 2012, 18, 1081–1088. [Google Scholar] [CrossRef] [PubMed]
- Sukmak, M.; Ostner, J.; Schülke, O.; Wajjwalku, W. A first report of non-invasive adenovirus detection in wild Assamese macaques in Thailand. Primates 2016, 58, 307–313. [Google Scholar] [CrossRef]
- Wasimuddin; Corman, V.M.; Ganzhorn, J.U.; Rakotondranary, J.; Ratovonamana, Y.R.; Drosten, C.; Sommer, S. Adenovirus infection is associated with altered gut microbial communities in a non-human primate. Sci. Rep. 2019, 9, 13410–13412. [Google Scholar] [CrossRef]
- Roy, S.; Vandenberghe, L.H.; Kryazhimskiy, S.; Grant, R.; Calcedo, R.; Yuan, X.; Keough, M.; Sandhu, A.; Wang, Q.; Medina-Jaszek, C.A.; et al. Isolation and characterization of adenoviruses persistently shed from the gastrointestinal tract of non-human primates. PLoS Pathog. 2009, 5, e1000503. [Google Scholar] [CrossRef]
- Borkenhagen, L.K.; Fieldhouse, J.K.; Seto, D.; Gray, G.C. Are adenoviruses zoonotic? A systematic review of the evidence. Emerg. Microbes Infect. 2019, 8, 1679–1687. [Google Scholar] [CrossRef]
- Podgorski, I.I.; Pantó, L.; Földes, K.; De Winter, I.; Jánoska, M.; Sós, E.; Chenet, B.; Harrach, B.; Benkő, M.; Podgorski, I.I. Adenoviruses of the most ancient primate lineages support the theory on virus−host co-evolution. Acta Veter. Hung. 2018, 66, 474–487. [Google Scholar] [CrossRef]
- Devaux, C.A.; Mediannikov, O.; Medkour, H.; Raoult, D. Infectious disease risk across the growing human-non human primate interface: A review of the evidence. Front. Public Health 2019, 7, 305. [Google Scholar] [CrossRef]
- Folegatti, P.; Bellamy, D.; Roberts, R.; Powlson, J.; Edwards, N.J.; Mair, C.F.; Bowyer, G.; Poulton, I.; Mitton, C.H.; Green, N.; et al. Safety and immunogenicity of a novel recombinant simian adenovirus ChAdOx2 as a vectored vaccine. Vaccines 2019, 7, 40. [Google Scholar] [CrossRef]
- Zhao, H.; Xu, C.; Luo, X.; Wei, F.; Wang, N.; Shi, H.; Ren, X. Seroprevalence of neutralizing antibodies against human adenovirus type-5 and chimpanzee adenovirus type-68 in cancer patients. Front. Immunol. 2018, 9, 335. [Google Scholar] [CrossRef] [PubMed]
- Berkeley, R.A.; Steele, L.P.; Mulder, A.A.; Wollenberg, D.J.M.V.D.; Kottke, T.J.; Thompson, J.; Coffey, M.; Hoeben, R.C.; Vile, R.G.; Melcher, A.A.; et al. Antibody-neutralized reovirus is effective in oncolytic virotherapy. Cancer Immunol. Res. 2018, 6, 1161–1173. [Google Scholar] [CrossRef] [PubMed]
- Zaiss, A.K.; Machado, H.B.; Herschman, H.R. The influence of innate and pre-existing immunity on adenovirus therapy. J. Cell. Biochem. 2009, 108, 778–790. [Google Scholar] [CrossRef] [PubMed]
- Dadáková, E.; Chrudimský, T.; Brožová, K.; Modrý, D.; Celer, V.; Hrazdilová, K. New adenoviruses from new primate hosts—growing diversity reveals taxonomic weak points. Mol. Phylogenet. Evol. 2017, 107, 305–307. [Google Scholar] [CrossRef] [PubMed]
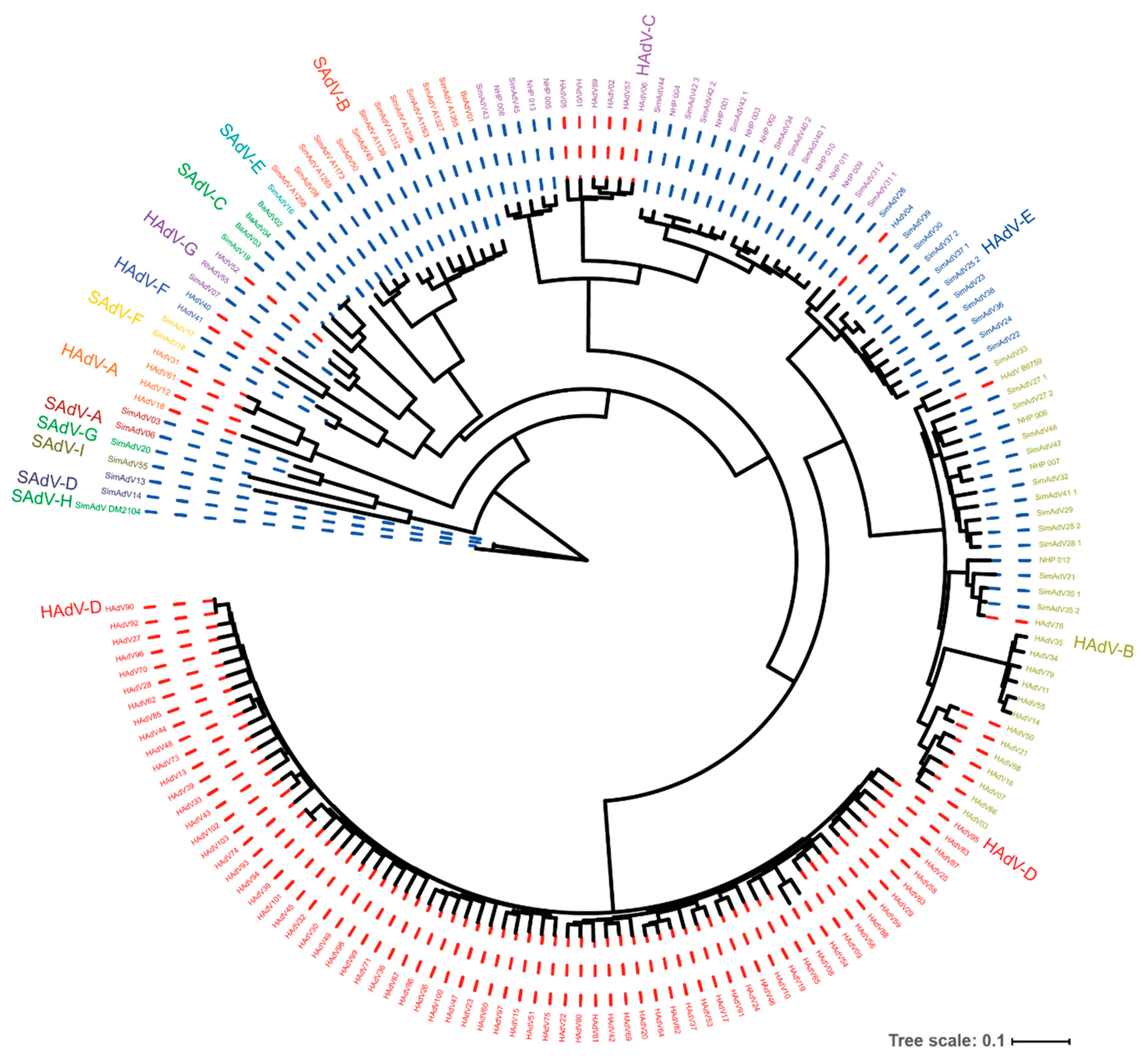
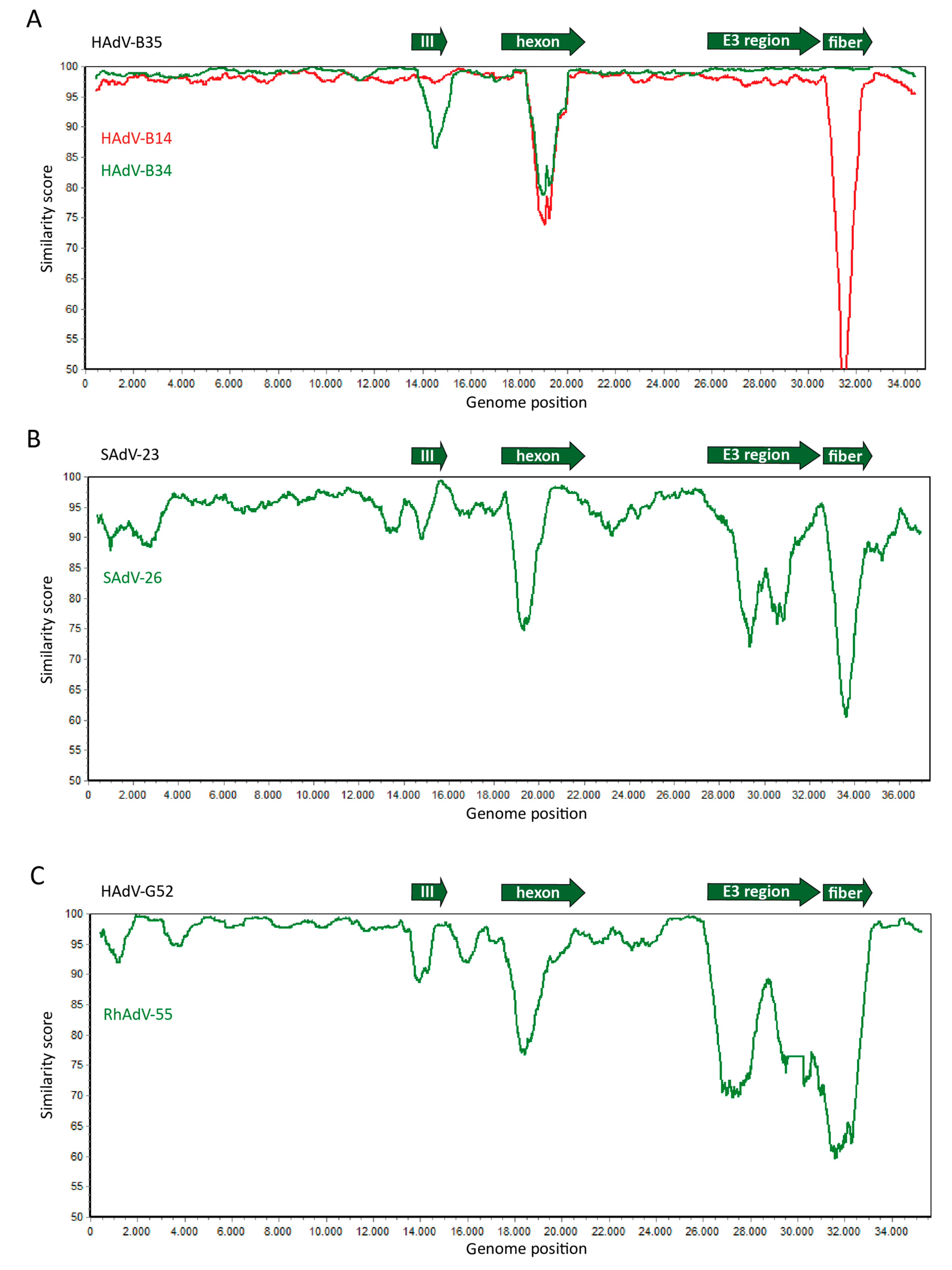
| Nomenclature of Human and Non-Human Primate (NHP)-Derived Adenoviruses |
|---|
| The adenoviruses isolated from humans and NHPs are all classified in the Mastadenovirus genus. Within this genus, the Human Adenoviruses (HAdV) are grouped in “species” (formerly “subgroups”) A through G. Similarly, NHP adenoviruses are grouped in Simian Adenovirus (SAdV) A through I. Within these species the isolates are clustered in “serotypes” or “types”. To date, 103 types are distinguished in adenoviruses isolated from humans. The naming of adenoviruses reflects the host from which the first type of an adenovirus species has been isolated, the adenovirus species, and their type. The Human Adenovirus species C type 5 is indicated as HAdV-C5. |
| Adenovirus 1 | Short Names 2 | Host Origin | Species | Genbank Accession No. |
|---|---|---|---|---|
| Chimpanzee adenovirus Y25 | ChAdOx1 | Chimpanzee | HAdV-E | JN254802 |
| Gorilla beringei adenovirus 7 | GC44 | Gorilla | HAdV-B | KC702813 3 |
| Gorilla beringei adenovirus 8 | GC45 | Gorilla | HAdV-B | KC702814 3 |
| Gorilla beringei adenovirus 9 | GC46 | Gorilla | HAdV-B | KC702815 3 |
| Simian adenovirus 11 | SAdV-11 | Rhesus Macaque | HAdV-G | KP329562 |
| Simian adenovirus 16 | SAdV-16 | Grivet | SAdV-E | KP329564 |
| Simian adenovirus 32 | SAdV-B32 | Chimpanzee | HAdV-B | FJ025911 |
| Simian adenovirus 21 | AdC1 | Chimpanzee | HAdV-B | AC_000010 |
| Simian adenovirus 23 | AdC6 | Chimpanzee | HAdV-E | AY530877 |
| Simian adenovirus 24 | AdC7 | Chimpanzee | HAdV-E | AY530878 |
| Simian adenovirus type 25.2 | ChAd68/ChAdOx2 | Chimpanzee | HAdV-E | FJ025918.1 |
| Pan paniscus adenovirus type 3 | PanAd3 | Bonobo | HAdV-C | N.A. |
| Pan troglodytes adenovirus type 3 | ChAd3 | Chimpanzee | HAdV-C | CS479276 |
| Pan troglodytes adenovirus type 63 | ChAd63 | Chimpanzee | HAdV-E | CS479277 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bots, S.T.F.; Hoeben, R.C. Non-Human Primate-Derived Adenoviruses for Future Use as Oncolytic Agents? Int. J. Mol. Sci. 2020, 21, 4821. https://doi.org/10.3390/ijms21144821
Bots STF, Hoeben RC. Non-Human Primate-Derived Adenoviruses for Future Use as Oncolytic Agents? International Journal of Molecular Sciences. 2020; 21(14):4821. https://doi.org/10.3390/ijms21144821
Chicago/Turabian StyleBots, Selas T.F., and Rob C. Hoeben. 2020. "Non-Human Primate-Derived Adenoviruses for Future Use as Oncolytic Agents?" International Journal of Molecular Sciences 21, no. 14: 4821. https://doi.org/10.3390/ijms21144821
APA StyleBots, S. T. F., & Hoeben, R. C. (2020). Non-Human Primate-Derived Adenoviruses for Future Use as Oncolytic Agents? International Journal of Molecular Sciences, 21(14), 4821. https://doi.org/10.3390/ijms21144821