Identification of Novel microRNA Profiles Dysregulated in Plasma and Tissue of Abdominal Aortic Aneurysm Patients
Abstract
1. Introduction
2. Results
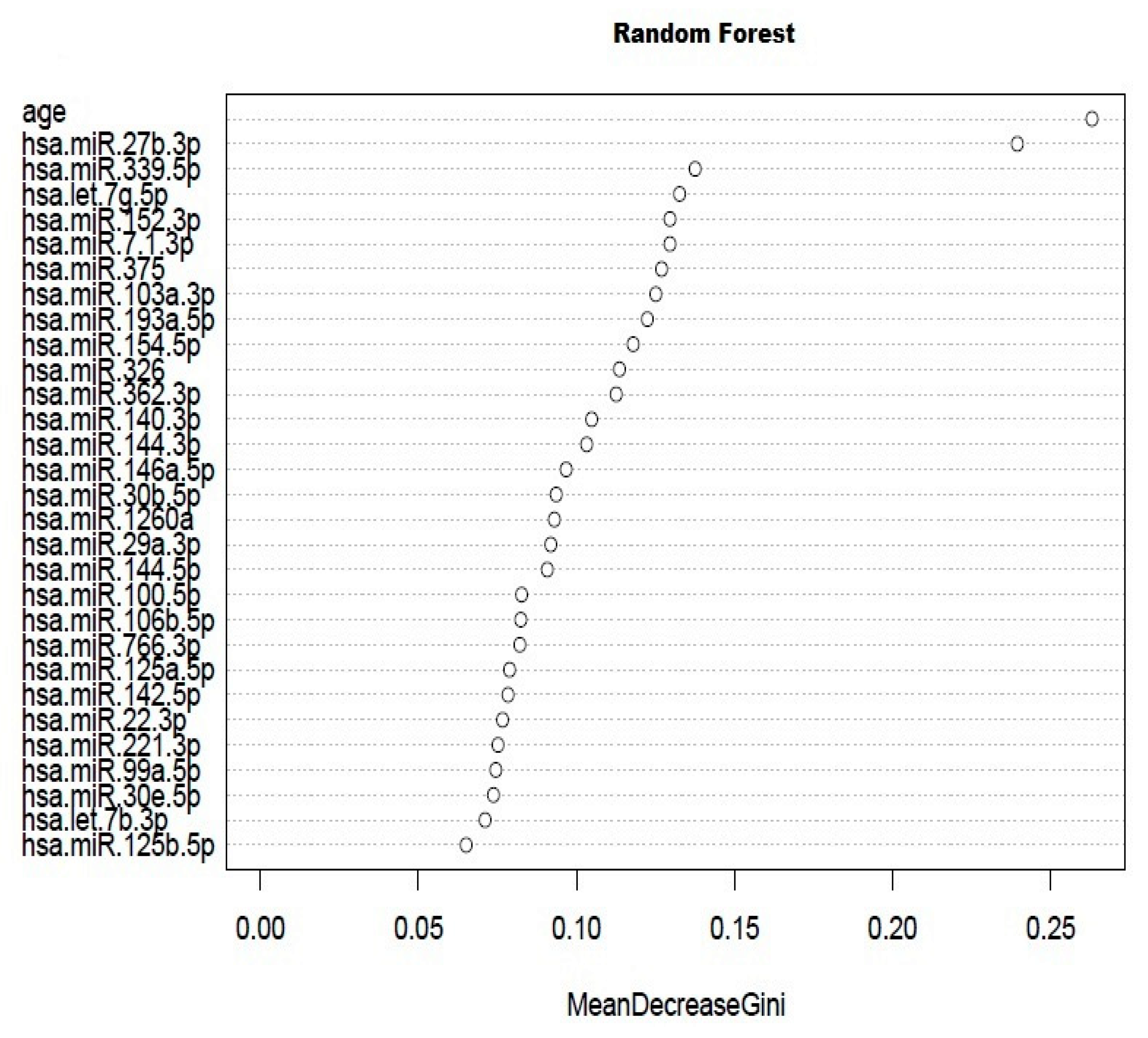
2.1. Screening Stage
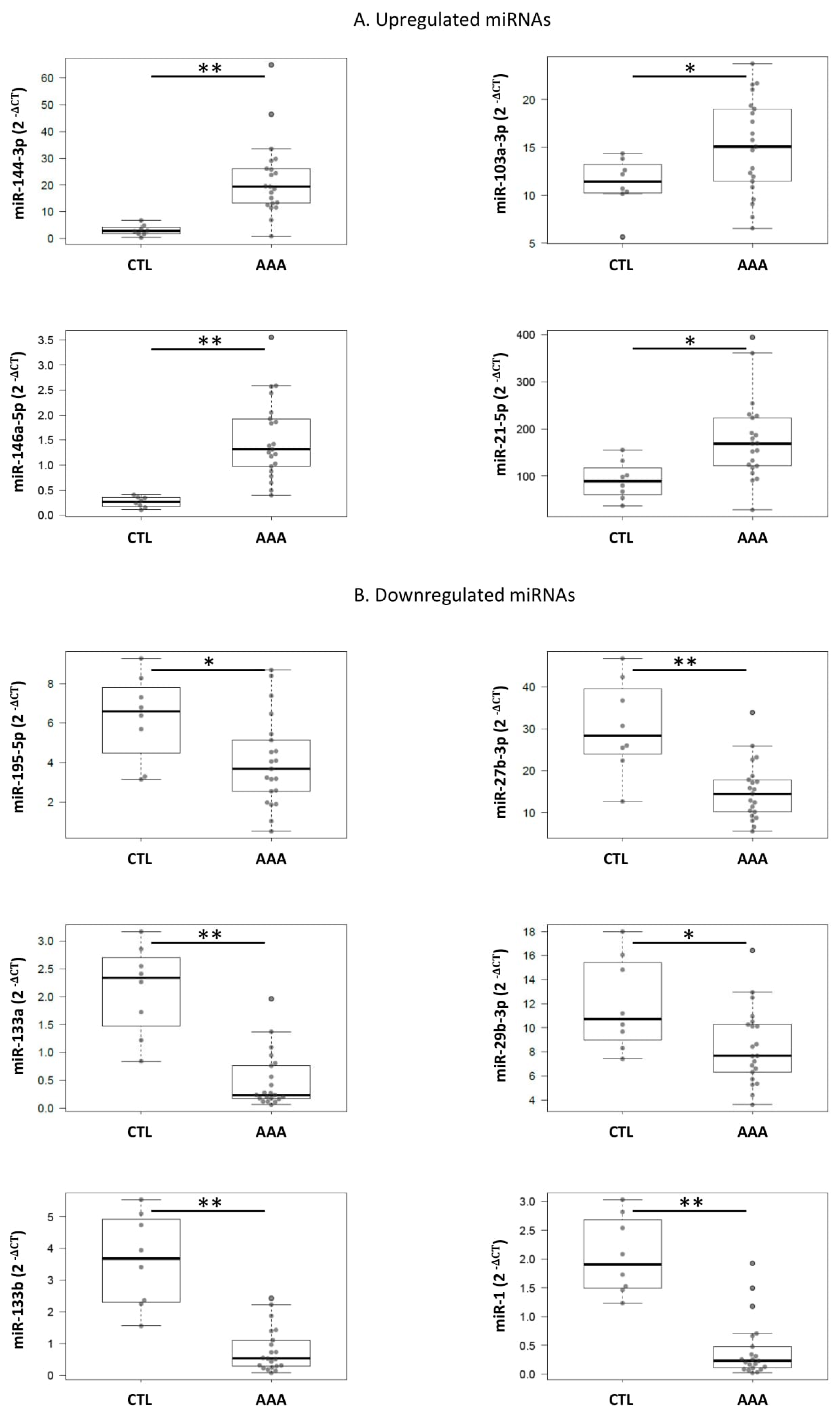
2.2. Quantification of Selected miRNA in Plasma Samples
2.3. Quantification of Selected miRNA in Tissue Samples
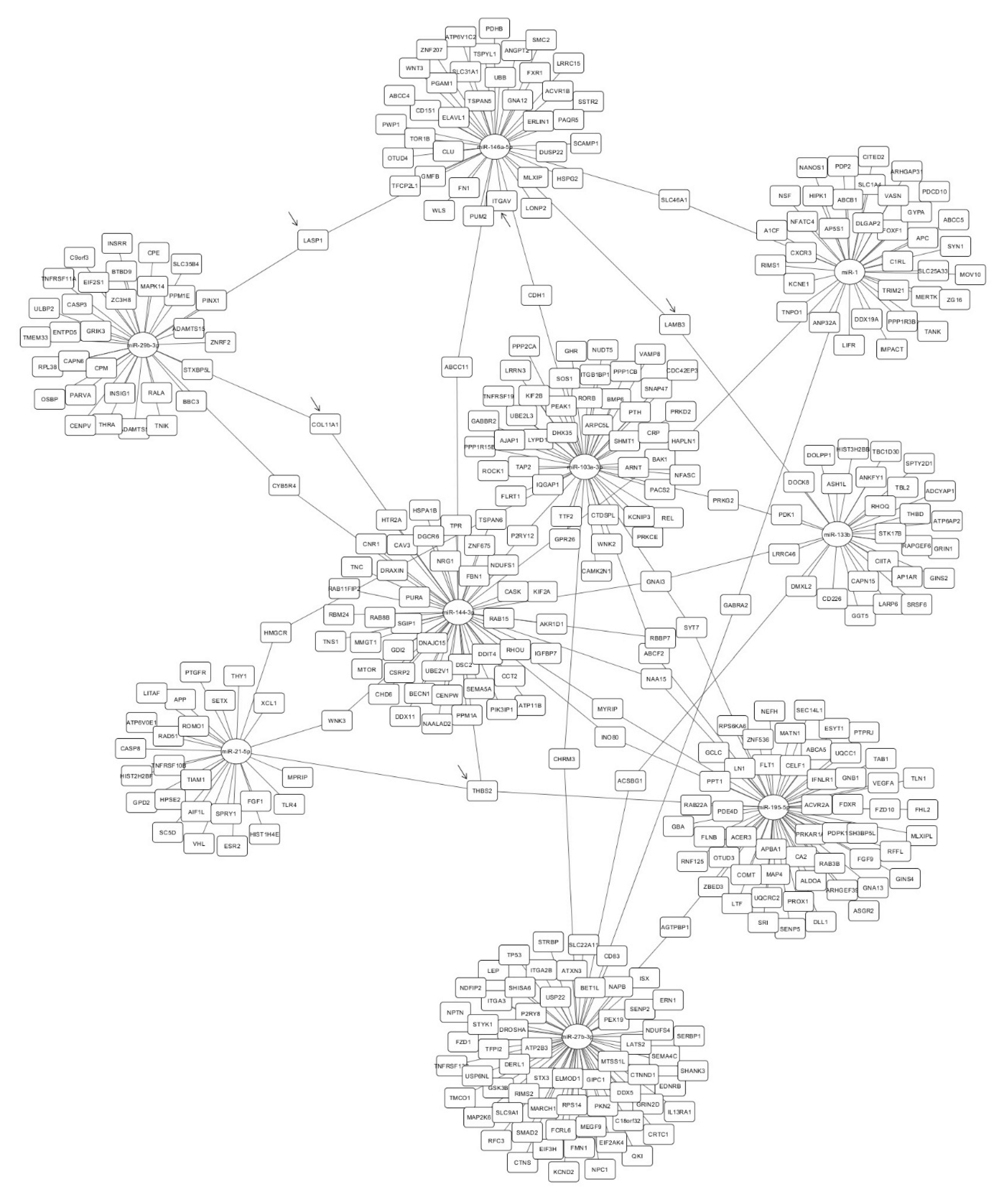
2.4. Identification of miRNA Targets
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Sample Collection
4.3. RNA Isolation and cDNA Synthesis from Plasma and Tissue Samples
4.4. miRNA Quantification
4.5. Identification of miRNA Targets
4.6. Western Blot Analysis of Target Proteins
4.7. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AAA | Abdominal aortic aneurysm |
| AUC | Area under curve |
| ASA | Acetyl salicylic acid |
| BP | Biological processes |
| CE | Carotid endarterectomy |
| CC | Cellular components |
| COL11A1A | Collagen XI α1 |
| CT | Cycle threshold |
| CTL | Control |
| DM | Diabetes mellitus |
| DL | Dyslipidemia |
| ECM | Extracellular matrix |
| GO | Gene Ontology |
| GSEA | Gene set enrichment analysis |
| HTN | Hypertension |
| ITGAV | Integrin α5 |
| LAMB3 | Laminin 3β subunit |
| LASP1 | LIM and SH3 domain protein 1 |
| miRNA | microRNA |
| MF | Molecular function |
| MMP | Matrix Metalloproteinases |
| THBS2 | Thrombospondin 2 |
| VSMC | Vascular smooth muscle cells |
References
- Cornuz, J.; Sidoti Pinto, C.; Tevaearai, H.; Egger, M. Risk factors for asymptomatic abdominal aortic aneurysm: Systematic review and meta-analysis of population-based screening studies. Eur. J. Public Health 2004, 14, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Sakalihasan, N.; Limet, R.; Defawe, O.D. Abdominal aortic aneurysm. Lancet 2005, 365, 1577–1589. [Google Scholar] [CrossRef]
- Kumar, S.; Boon, R.A.; Maegdefessel, L.; Dimmeler, S.; Jo, H. Role of Noncoding RNAs in the Pathogenesis of Abdominal Aortic Aneurysm. Circ. Res. 2019, 124, 619–630. [Google Scholar] [CrossRef]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Zorio, E.; Medina, P.; Rueda, J.; Millan, J.M.; Arnau, M.A.; Beneyto, M.; Marin, F.; Gimeno, J.R.; Osca, J.; Salvador, A.; et al. Insights into the role of microRNAs in cardiac diseases: From biological signalling to therapeutic targets. Cardiovasc. Hematol. Agents Med. Chem. 2009, 7, 82–90. [Google Scholar] [CrossRef]
- Faruq, O.; Vecchione, A. microRNA: Diagnostic Perspective. Front. Med. 2015, 2, 51. [Google Scholar] [CrossRef] [PubMed]
- Raffort, J.; Lareyre, F.; Clement, M.; Mallat, Z. Micro-RNAs in abdominal aortic aneurysms: Insights from animal models and relevance to human disease. Cardiovasc. Res. 2016, 110, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Biros, E.; Moran, C.S.; Wang, Y.; Walker, P.J.; Cardinal, J.; Golledge, J. microRNA profiling in patients with abdominal aortic aneurysms: The significance of miR-155. Clin. Sci. (Lond.) 2014, 126, 795–803. [Google Scholar] [CrossRef]
- Kin, K.; Miyagawa, S.; Fukushima, S.; Shirakawa, Y.; Torikai, K.; Shimamura, K.; Daimon, T.; Kawahara, Y.; Kuratani, T.; Sawa, Y. Tissue- and plasma-specific MicroRNA signatures for atherosclerotic abdominal aortic aneurysm. J. Am. Heart Assoc. 2012, 1, e000745. [Google Scholar] [CrossRef]
- Maegdefessel, L.; Spin, J.M.; Raaz, U.; Eken, S.M.; Toh, R.; Azuma, J.; Adam, M.; Nakagami, F.; Nagakami, F.; Heymann, H.M.; et al. miR-24 limits aortic vascular inflammation and murine abdominal aneurysm development. Nat. Commun. 2014, 5, 5214. [Google Scholar] [CrossRef]
- Spear, R.; Boytard, L.; Blervaque, R.; Chwastyniak, M.; Hot, D.; Vanhoutte, J.; Staels, B.; Lemoine, Y.; Lamblin, N.; Pruvot, F.R.; et al. Adventitial Tertiary Lymphoid Organs as Potential Source of MicroRNA Biomarkers for Abdominal Aortic Aneurysm. Int. J. Mol. Sci. 2015, 16, 11276–11293. [Google Scholar] [CrossRef]
- Stather, P.W.; Sylvius, N.; Sidloff, D.A.; Dattani, N.; Verissimo, A.; Wild, J.B.; Butt, H.Z.; Choke, E.; Sayers, R.D.; Bown, M.J. Identification of microRNAs associated with abdominal aortic aneurysms and peripheral arterial disease. Br. J. Surg. 2015, 102, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Ramón-Núñez, L.A.; Martos, L.; Fernández-Pardo, Á.; Oto, J.; Medina, P.; España, F.; Navarro, S. Comparison of protocols and RNA carriers for plasma miRNA isolation. Unraveling RNA carrier influence on miRNA isolation. PLoS ONE 2017, 12, e0187005. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant. Mol. Biol. 2012. [Google Scholar] [CrossRef] [PubMed]
- Pahl, M.C.; Derr, K.; Gabel, G.; Hinterseher, I.; Elmore, J.R.; Schworer, C.M.; Peeler, T.C.; Franklin, D.P.; Gray, J.L.; Carey, D.J.; et al. MicroRNA expression signature in human abdominal aortic aneurysms. BMC Med. Genomics 2012, 5, 25. [Google Scholar] [CrossRef] [PubMed]
- Jiao, T.; Yao, Y.; Zhang, B.; Hao, D.C.; Sun, Q.F.; Li, J.B.; Yuan, C.; Jing, B.; Wang, Y.P.; Wang, H.Y. Role of MicroRNA-103a Targeting ADAM10 in Abdominal Aortic Aneurysm. BioMed. Res. Int. 2017, 2017, 9645874. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Song, Y.H.; Li, F.; Yang, T.; Lu, Y.W.; Geng, Y.J. MicroRNA-221 regulates high glucose-induced endothelial dysfunction. Biochem. Biophys. Res. Commun. 2009, 381, 81–83. [Google Scholar] [CrossRef]
- Chen, L.J.; Lim, S.H.; Yeh, Y.T.; Lien, S.C.; Chiu, J.J. Roles of microRNAs in atherosclerosis and restenosis. J. Biomed. Sci. 2012, 19, 79. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Shao, S.; Geng, H.; Yu, Y.; Wang, C.; Liu, Z.; Yu, C.; Jiang, X.; Deng, Y.; Gao, L.; et al. Expression profiles of six circulating microRNAs critical to atherosclerosis in patients with subclinical hypothyroidism: A clinical study. J. Clin. Endocrinol. Metab. 2014, 99, E766–E774. [Google Scholar] [CrossRef]
- Zampetaki, A.; Attia, R.; Mayr, U.; Gomes, R.S.; Phinikaridou, A.; Yin, X.; Langley, S.R.; Willeit, P.; Lu, R.; Fanshawe, B.; et al. Role of miR-195 in aortic aneurysmal disease. Circ. Res. 2014, 115, 857–866. [Google Scholar] [CrossRef]
- Busch, A.; Busch, M.; Scholz, C.J.; Kellersmann, R.; Otto, C.; Chernogubova, E.; Maegdefessel, L.; Zernecke, A.; Lorenz, U. Aneurysm miRNA Signature Differs, Depending on Disease Localization and Morphology. Int. J. Mol. Sci. 2016, 17, 81. [Google Scholar] [CrossRef]
- Maegdefessel, L.; Azuma, J.; Toh, R.; Deng, A.; Merk, D.R.; Raiesdana, A.; Leeper, N.J.; Raaz, U.; Schoelmerich, A.M.; McConnell, M.V.; et al. MicroRNA-21 blocks abdominal aortic aneurysm development and nicotine-augmented expansion. Sci. Trans. Med. 2012, 4, 122ra122. [Google Scholar] [CrossRef] [PubMed]
- Maegdefessel, L.; Azuma, J.; Toh, R.; Merk, D.R.; Deng, A.; Chin, J.T.; Raaz, U.; Schoelmerich, A.M.; Raiesdana, A.; Leeper, N.J.; et al. Inhibition of microRNA-29b reduces murine abdominal aortic aneurysm development. J. Clin. Investig. 2012, 122, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Iyer, V.; Rowbotham, S.; Biros, E.; Bingley, J.; Golledge, J. A systematic review investigating the association of microRNAs with human abdominal aortic aneurysms. Atherosclerosis 2017, 261, 78–89. [Google Scholar] [CrossRef]
- Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Pereira-da-Silva, T.; Coutinho Cruz, M.; Carrusca, C.; Cruz Ferreira, R.; Napoleão, P.; Mota Carmo, M. Circulating microRNA profiles in different arterial territories of stable atherosclerotic disease: A systematic review. Am. J. Cardiovasc. Dis. 2018, 8, 1–13. [Google Scholar] [PubMed]
- Chen, W.J.; Yin, K.; Zhao, G.J.; Fu, Y.C.; Tang, C.K. The magic and mystery of microRNA-27 in atherosclerosis. Atherosclerosis 2012, 222, 314–323. [Google Scholar] [CrossRef]
- Zhang, W.; Shang, T.; Huang, C.; Yu, T.; Liu, C.; Qiao, T.; Huang, D.; Liu, Z. Plasma microRNAs serve as potential biomarkers for abdominal aortic aneurysm. Clin. Biochem. 2015. [Google Scholar] [CrossRef]
- Spear, R.; Boytard, L.; Blervaque, R.; Chwastyniak, M.; Hot, D.; Vanhoutte, J.; Lamblin, N.; Amouyel, P.; Pinet, F. Let-7f: A New Potential Circulating Biomarker Identified by miRNA Profiling of Cells Isolated from Human Abdominal Aortic Aneurysm. Int. J. Mol. Sci. 2019, 20, 5499. [Google Scholar] [CrossRef] [PubMed]
- Wanhainen, A.; Mani, K.; Vorkapic, E.; De Basso, R.; Bjorck, M.; Lanne, T.; Wagsater, D. Screening of circulating microRNA biomarkers for prevalence of abdominal aortic aneurysm and aneurysm growth. Atherosclerosis 2017, 256, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Schober, A.; Weber, C. Pathogenic arterial remodeling: The good and bad of microRNAs. Am. J. Physiol. Heart Circ. Physiol. 2013, 304, H1050–H1059. [Google Scholar] [CrossRef]
- Maegdefessel, L.; Spin, J.M.; Adam, M.; Raaz, U.; Toh, R.; Nakagami, F.; Tsao, P.S. Micromanaging abdominal aortic aneurysms. Int. J. Mol. Sci. 2013, 14, 14374–14394. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.S.; Wang, H.Y.; Liao, Y.C.; Tsai, P.C.; Chen, K.C.; Cheng, H.Y.; Lin, R.T.; Juo, S.H. MicroRNA-195 regulates vascular smooth muscle cell phenotype and prevents neointimal formation. Cardiovasc. Res. 2012, 95, 517–526. [Google Scholar] [CrossRef]
- Cheng, H.S.; Sivachandran, N.; Lau, A.; Boudreau, E.; Zhao, J.L.; Baltimore, D.; Delgado-Olguin, P.; Cybulsky, M.I.; Fish, J.E. MicroRNA-146 represses endothelial activation by inhibiting pro-inflammatory pathways. EMBO Mol. Med. 2013, 5, 949–966. [Google Scholar] [CrossRef]
- Raitoharju, E.; Lyytikäinen, L.P.; Levula, M.; Oksala, N.; Mennander, A.; Tarkka, M.; Klopp, N.; Illig, T.; Kähönen, M.; Karhunen, P.J.; et al. miR-21, miR-210, miR-34a, and miR-146a/b are up-regulated in human atherosclerotic plaques in the Tampere Vascular Study. Atherosclerosis 2011, 219, 211–217. [Google Scholar] [CrossRef]
- Saba, R.; Sorensen, D.L.; Booth, S.A. MicroRNA-146a: A Dominant, Negative Regulator of the Innate Immune Response. Front. Immunol. 2014, 5, 578. [Google Scholar] [CrossRef]
- Vergadi, E.; Vaporidi, K.; Theodorakis, E.E.; Doxaki, C.; Lagoudaki, E.; Ieronymaki, E.; Alexaki, V.I.; Helms, M.; Kondili, E.; Soennichsen, B.; et al. Akt2 deficiency protects from acute lung injury via alternative macrophage activation and miR-146a induction in mice. J. Immunol. 2014, 192, 394–406. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, H.; Yang, B. miR-146a regulates inflammation and development in patients with abdominal aortic aneurysms by targeting CARD10. Int. Angiol. 2020. [Google Scholar] [CrossRef]
- Venkatesh, P.; Phillippi, J.; Chukkapalli, S.; Rivera-Kweh, M.; Velsko, I.; Gleason, T.; VanRyzin, P.; Aalaei-Andabili, S.H.; Ghanta, R.K.; Beaver, T.; et al. Aneurysm-Specific miR-221 and miR-146a Participates in Human Thoracic and Abdominal Aortic Aneurysms. Int. J. Mol. Sci. 2017, 18, 875. [Google Scholar] [CrossRef]
- Lareyre, F.; Clement, M.; Moratal, C.; Loyer, X.; Jean-Baptiste, E.; Hassen-Khodja, R.; Chinetti, G.; Mallat, Z.; Raffort, J. Differential micro-RNA expression in diabetic patients with abdominal aortic aneurysm. Biochimie 2019, 162, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.W.; Hu, Y.R.; Zhao, J.Y.; Li, S.F.; Ma, X.; Wu, S.G.; Lu, J.B.; Qiu, Y.R.; Sha, Y.H.; Wang, Y.C.; et al. An agomir of miR-144-3p accelerates plaque formation through impairing reverse cholesterol transport and promoting pro-inflammatory cytokine production. PLoS ONE 2014, 9, e94997. [Google Scholar] [CrossRef]
- Araujo, N.N.F.; Lin-Wang, H.T.; Germano, J.F.; Farsky, P.S.; Feldman, A.; Rossi, F.H.; Izukawa, N.M.; Higuchi, M.L.; Savioli Neto, F.; Hirata, M.H.; et al. Dysregulation of microRNAs and target genes networks in human abdominal aortic aneurysm tissues. PLoS ONE 2019, 14, e0222782. [Google Scholar] [CrossRef] [PubMed]
- Cerna, V.; Ostasov, P.; Pitule, P.; Molacek, J.; Treska, V.; Pesta, M. The Expression Profile of MicroRNAs in Small and Large Abdominal Aortic Aneurysms. Cardiol. Res. Pract. 2019, 2019, 8645840. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.W.; Kumar, S.; Son, D.J.; Jang, I.H.; Griendling, K.K.; Jo, H. Prevention of abdominal aortic aneurysm by anti-microRNA-712 or anti-microRNA-205 in angiotensin II-infused mice. Arterioscler. Thromb. Vasc. Biol. 2014, 34, 1412–1421. [Google Scholar] [CrossRef]
- Holsti, M.; Wanhainen, A.; Lundin, C.; Björck, M.; Tegler, G.; Svensson, J.; Sund, M. Circulating Vascular Basement Membrane Fragments are Associated with the Diameter of the Abdominal Aorta and Their Expression Pattern is Altered in AAA Tissue. Eur. J. Vasc. Endovasc. Surg. 2018, 56, 110–118. [Google Scholar] [CrossRef]
- Kuivaniemi, H.; Ryer, E.J.; Elmore, J.R.; Tromp, G. Understanding the pathogenesis of abdominal aortic aneurysms. Expert Rev. Cardiovasc. Ther. 2015, 13, 975–987. [Google Scholar] [CrossRef]
- Chen, Q.; Wang, Q.; Zhu, J.; Xiao, Q.; Zhang, L. Reactive oxygen species: Key regulators in vascular health and diseases. Br. J. Pharmacol. 2018, 175, 1279–1292. [Google Scholar] [CrossRef]
- Ghosh, A.; DiMusto, P.D.; Ehrlichman, L.K.; Sadiq, O.; McEvoy, B.; Futchko, J.S.; Henke, P.K.; Eliason, J.L.; Upchurch, G.R. The role of extracellular signal-related kinase during abdominal aortic aneurysm formation. J. Am. Coll. Surg. 2012, 215, 668–680.e661. [Google Scholar] [CrossRef]
- Qureshi, M.I.; Greco, M.; Vorkas, P.A.; Holmes, E.; Davies, A.H. Application of Metabolic Profiling to Abdominal Aortic Aneurysm Research. J. Proteome Res. 2017, 16, 2325–2332. [Google Scholar] [CrossRef] [PubMed]
- Orth, M.F.; Cazes, A.; Butt, E.; Grunewald, T.G. An update on the LIM and SH3 domain protein 1 (LASP1): A versatile structural, signaling, and biomarker protein. Oncotarget 2015, 6, 26–42. [Google Scholar] [CrossRef] [PubMed]
- Choke, E.; Thompson, M.M.; Dawson, J.; Wilson, W.R.; Sayed, S.; Loftus, I.M.; Cockerill, G.W. Abdominal aortic aneurysm rupture is associated with increased medial neovascularization and overexpression of proangiogenic cytokines. Arterioscler. Thromb. Vasc. Biol. 2006, 26, 2077–2082. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.A.; Zavadzkas, J.A.; Chang, E.I.; Sheats, N.; Koval, C.; Stroud, R.E.; Spinale, F.G.; Ikonomidis, J.S. Cellular phenotype transformation occurs during thoracic aortic aneurysm development. J. Thorac. Cardiovasc. Surg. 2010, 140, 653–659. [Google Scholar] [CrossRef]
- Toumpoulis, I.K.; Oxford, J.T.; Cowan, D.B.; Anagnostopoulos, C.E.; Rokkas, C.K.; Chamogeorgakis, T.P.; Angouras, D.C.; Shemin, R.J.; Navab, M.; Ericsson, M.; et al. Differential expression of collagen type V and XI alpha-1 in human ascending thoracic aortic aneurysms. Ann. Thorac. Surg. 2009, 88, 506–513. [Google Scholar] [CrossRef]
- Chistiakov, D.A.; Melnichenko, A.A.; Myasoedova, V.A.; Grechko, A.V.; Orekhov, A.N. Thrombospondins: A Role in Cardiovascular Disease. Int. J. Mol. Sci. 2017, 18, 1540. [Google Scholar] [CrossRef]
- Bornstein, P.; Armstrong, L.C.; Hankenson, K.D.; Kyriakides, T.R.; Yang, Z. Thrombospondin 2, a matricellular protein with diverse functions. Matrix Biol. 2000, 19, 557–568. [Google Scholar] [CrossRef]
- Liu, J.F.; Lee, C.W.; Tsai, M.H.; Tang, C.H.; Chen, P.C.; Lin, L.W.; Lin, C.Y.; Lu, C.H.; Lin, Y.F.; Yang, S.H.; et al. Thrombospondin 2 promotes tumor metastasis by inducing matrix metalloproteinase-13 production in lung cancer cells. Biochem. Pharmacol. 2018, 155, 537–546. [Google Scholar] [CrossRef]
- Helkin, A.; Maier, K.G.; Gahtan, V. Thrombospondin-1, -2 and -5 have differential effects on vascular smooth muscle cell physiology. Biochem. Biophys. Res. Commun. 2015, 464, 1022–1027. [Google Scholar] [CrossRef]
- Golledge, J.; Clancy, P.; Hankey, G.J.; Norman, P.E. Relation between serum thrombospondin-2 and cardiovascular mortality in older men screened for abdominal aortic aneurysm. Am. J. Cardiol. 2013, 111, 1800–1804. [Google Scholar] [CrossRef]
- Kato, K.; Oguri, M.; Kato, N.; Hibino, T.; Yajima, K.; Yoshida, T.; Metoki, N.; Yoshida, H.; Satoh, K.; Watanabe, S.; et al. Assessment of genetic risk factors for thoracic aortic aneurysm in hypertensive patients. Am. J. Hypertens. 2008, 21, 1023–1027. [Google Scholar] [CrossRef] [PubMed]
- Gellert, C.; Schottker, B.; Muller, H.; Holleczek, B.; Brenner, H. Impact of smoking and quitting on cardiovascular outcomes and risk advancement periods among older adults. Eur. J. Epidemiol. 2013, 28, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Liang, B.; Che, J.; Zhao, H.; Zhang, Z.; Shi, G. MiR-195 promotes abdominal aortic aneurysm media remodeling by targeting Smad3. Cardiovasc. Ther. 2017, 35. [Google Scholar] [CrossRef]
- Garcia-Garcia, F.; Panadero, J.; Dopazo, J.; Montaner, D. Integrated gene set analysis for microRNA studies. Bioinformatics 2016, 32, 2809–2816. [Google Scholar] [CrossRef] [PubMed]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstråle, M.; Laurila, E.; et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Oto, J.; Navarro, S.; Larsen, A.C.; Solmoirago, M.J.; Plana, E.; Hervas, D.; Fernandez-Pardo, A.; Espana, F.; Kristensen, S.R.; Thorlacius-Ussing, O.; et al. MicroRNAs and Neutrophil Activation Markers Predict Venous Thrombosis in Pancreatic Ductal Adenocarcinoma and Distal Extrahepatic Cholangiocarcinoma. Int. J. Mol. Sci. 2020, 21, 840. [Google Scholar] [CrossRef]
- Prado, M.S.G.; de Goes, T.C.; de Jesus, M.L.; Mendonca, L.S.O.; Nascimento, J.S.; Kaneto, C.M. Identification of miR-328-3p as an endogenous reference gene for the normalization of miRNA expression data from patients with Diabetic Retinopathy. Sci. Rep. 2019, 9, 19677. [Google Scholar] [CrossRef]
- Breinam, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]



| Variable | CTL | AAA | CE |
|---|---|---|---|
| Participants Included in the Screening Stage of the Plasma Study | |||
| n = 7 | n = 7 | - | |
| Sex (male) | 7 (100.0%) | 7 (100.0%) | - |
| Age (SD) | 62 (3) | 72 (7) | - |
| HTN | 5 (71.4%) | 7 (100.0%) | - |
| DM | 2 (28.6%) | 2 (28.6%) | - |
| DL | 5 (71.4%) | 6 (85.7%) | - |
| Smoking * | 4 (57.1%) | 2 (28.6%) | - |
| Participants included in the complete plasma study | |||
| n = 16 | n = 30 | n = 11 | |
| Sex (male) | 16 (100.0%) | 30 (100.0%) | 11 (100.0%) |
| Age (SD) | 63 (5) | 74 (8) | 68 (6) |
| HTN | 10 (62.5%) | 27 (90.0%) | 7 (63.6%) |
| DM | 2 (12.5%) | 13 (43.3%) | 6 (54.5%) |
| DL | 14 (87.5%) | 20 (66.7%) | 7 (63.6%) |
| Smoking * | 4 (25.0%) | 14 (46.7%) | 8 (72.7%) |
| Open Surgery | - | 10 (33.3%) | - |
| Aneurysm Diameter, mm (SD) | - | 57.3 (10.3) | - |
| Statin use | 4 (25.0%) | 21 (70.0%) | 10 (90.9%) |
| Anti-hypertensives (no beta-blockers) | 10 (62.5%) | 23 (76.7%) | 7 (63.6%) |
| Anti-hypertensives (beta-blockers) | 2 (12.5%) | 8 (26.7%) | 4 (36.4%) |
| ASA | 2 (12.5%) | 11 (36.7%) | 7 (63.6%) |
| Other antiplatelet therapies | 1 (6.3%) | 5 (16.7%) | 3 (27.3%) |
| Anti-coagulants | 0 (0.0%) | 4 (13.3%) | 1 (9.1%) |
| Participants included in the complete tissue study | |||
| n = 8 | n = 21 | - | |
| Sex (male) | - | 20 (95.2%) | - |
| Age (SD) | - | 65.3 (6.5) | - |
| HTN | - | 17 (81.0%) | - |
| DM | - | 10 (47.6%) | - |
| DL | - | 13 (61.9%) | - |
| Smoking * | - | 10 (47.6%) | - |
| Open Surgery | - | 21 (100.0%) | - |
| Aneurysm Diameter, mm (SD) | - | 61.7 (10.9) | - |
| Statin use | - | 14 (66.7%) | - |
| Anti-hypertensives (no beta-blockers) | - | 16 (76.2%) | - |
| Anti-hypertensives (beta-blockers) | - | 6 (28.6%) | - |
| ASA | - | 4 (19.0%) | - |
| Other antiplatelet therapies | - | 3 (14.3%) | - |
| Anti-coagulants | - | 3 (14.3%) | - |
| miRNA Name | Fold-Change (AAA/CTL) |
|---|---|
| miR-27b-3p | 1.39 |
| miR-339-3p | 1.92 |
| let-7g-5p | 1.10 |
| miR-152-3p | 2.52 |
| miR-7-1-3p | −2.27 |
| miR-375 | 2.45 |
| miR-103a-3p | −1.39 |
| miR-193a-5p | 3.44 |
| miR-154-5p | 1.01 |
| miR-326 | 2.59 |
| miR-362-3p | −1.37 |
| miR-140-3p | 1.45 |
| miR-144-3p | −2.27 |
| miR-146a-5p | 1.66 |
| miR-130b-3p | 1.51 |
| miR-1260a | 2.06 |
| miR-29a-3p | 1.34 |
| miR-144-5p | 1.23 |
| miR-100-5p | 1.65 |
| miR-106b-5p | −1.27 |
| miR-766-3p | 1.73 |
| miR-125a-5p | 1.42 |
| miR-142-5p | 1.23 |
| miR-22-3p | 1.96 |
| miR-221-3p | 1.50 |
| miR-99a-5p | 2.71 |
| miR-30e-3p | 1.81 |
| let-7b-3p | 3.24 |
| miR-125b-5p | 1.34 |
| miRNA Name | miRNA Sequence | Fold-Change (AAA vs. CTL) | p-Value | Estimate | Lower 95% | Upper 95% |
|---|---|---|---|---|---|---|
| miR-152-3p | ucagugcaugacagaacuugg | 1.5 | 0.357 | 0.028 | −0.032 | 0.087 |
| miR-144-3p | uacaguauagaugauguacu | 1 | 0.804 | −0.230 | −2.081 | 1.621 |
| miR-27b-3p | uucacaguggcuaaguucugc | 1.6 | 0.043 | 0.427 | 0.014 | 0.841 |
| miR-103a-3p | agcagcauuguacagggcuauga | 1 | 0.284 | 0.528 | −0.452 | 1.509 |
| miR-99a-5p | aacccguagauccgaucuugug | 1.9 | 0.183 | 0.081 | −0.04 | 0.201 |
| miR-375 | uuuguucguucggcucgcguga | 2 | 0.139 | 0.075 | −0.025 | 0.174 |
| miR-221-3p | agcuacauugucugcuggguuuc | 1.9 | 0.001 | 0.333 | 0.145 | 0.521 |
| miR-146a-5p | ugagaacugaauuccauggguu | 1.6 | 0.457 | 0.054 | −0.09 | 0.198 |
| miR-1260 | aucccaccucugccacca | 2 | 0.238 | 0.049 | −0.034 | 0.132 |
| miRNA Name | miRNA Sequence | Fold-Change (AAA vs. CTL) | p-Value | Estimate | Lower 95% | Upper 95% |
|---|---|---|---|---|---|---|
| miR-221-3p | agcuacauugucugcuggguuuc | 1.1 | 0.742 | 0.171 | −0.885 | 1.226 |
| miR-27b-3p | uucacaguggcuaaguucugc | −2.0 | <0.001 | −15.244 | −22.319 | −8.169 |
| miR-99a-5p | aacccguagauccgaucuugug | 1.4 | 0.277 | 1.947 | −1.653 | 5.548 |
| miR-103a-3p | agcagcauuguacagggcuauga | 1.3 | 0.050 | 3.856 | 0.007 | 7.705 |
| miR-146a-5p | ugagaacugaauuccauggguu | 5.8 | <0.001 | 1.251 | 0.662 | 1.840 |
| miR-1260 | aucccaccucugccacca | 1.2 | 0.677 | 0.379 | −1.464 | 2.221 |
| miR-144-3p | uacaguauagaugauguacu | 7.2 | 0.001 | 19.002 | 8.656 | 29.348 |
| miR-152-3p | ucagugcaugacagaacuugg | 1.1 | 0.380 | 0.246 | −0.320 | 0.813 |
| miR-195-5p | uagcagcacagaaauauuggc | −1.5 | 0.023 | −2.248 | −4.163 | −0.333 |
| miR-155-5p | uuaaugcuaaucgugauagggguu | 1.9 | 0.082 | 0.316 | −0.043 | 0.676 |
| miR-21-5p | uagcuuaucagacugauguuga | 1.9 | 0.012 | 86.025 | 20.360 | 151.689 |
| miR-133b | uuugguccccuucaaccagcua | −4.6 | <0.001 | −2.82 | −3.637 | −2.002 |
| miR-133a | uuugguccccuucaaccagcug | −4.4 | <0.001 | −1.643 | −2.150 | −1.135 |
| miR-1 | uggaauguaaagaaguauguau | −4.4 | <0.001 | −1.628 | −2.105 | −1.150 |
| miR-29b-3p | uagcaccauuugaaaucaguguu | −1.4 | 0.018 | −3.509 | −6.373 | −0.645 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Plana, E.; Gálvez, L.; Medina, P.; Navarro, S.; Fornés-Ferrer, V.; Panadero, J.; Miralles, M. Identification of Novel microRNA Profiles Dysregulated in Plasma and Tissue of Abdominal Aortic Aneurysm Patients. Int. J. Mol. Sci. 2020, 21, 4600. https://doi.org/10.3390/ijms21134600
Plana E, Gálvez L, Medina P, Navarro S, Fornés-Ferrer V, Panadero J, Miralles M. Identification of Novel microRNA Profiles Dysregulated in Plasma and Tissue of Abdominal Aortic Aneurysm Patients. International Journal of Molecular Sciences. 2020; 21(13):4600. https://doi.org/10.3390/ijms21134600
Chicago/Turabian StylePlana, Emma, Laura Gálvez, Pilar Medina, Silvia Navarro, Victoria Fornés-Ferrer, Joaquín Panadero, and Manuel Miralles. 2020. "Identification of Novel microRNA Profiles Dysregulated in Plasma and Tissue of Abdominal Aortic Aneurysm Patients" International Journal of Molecular Sciences 21, no. 13: 4600. https://doi.org/10.3390/ijms21134600
APA StylePlana, E., Gálvez, L., Medina, P., Navarro, S., Fornés-Ferrer, V., Panadero, J., & Miralles, M. (2020). Identification of Novel microRNA Profiles Dysregulated in Plasma and Tissue of Abdominal Aortic Aneurysm Patients. International Journal of Molecular Sciences, 21(13), 4600. https://doi.org/10.3390/ijms21134600





