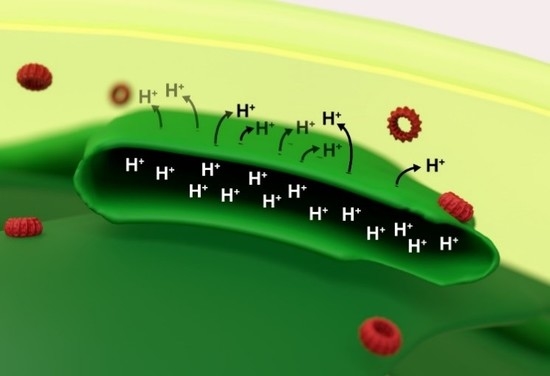
Proton Leakage Is Sensed by IM30 and Activates IM30-Triggered Membrane Fusion
Abstract
1. Introduction
2. Results
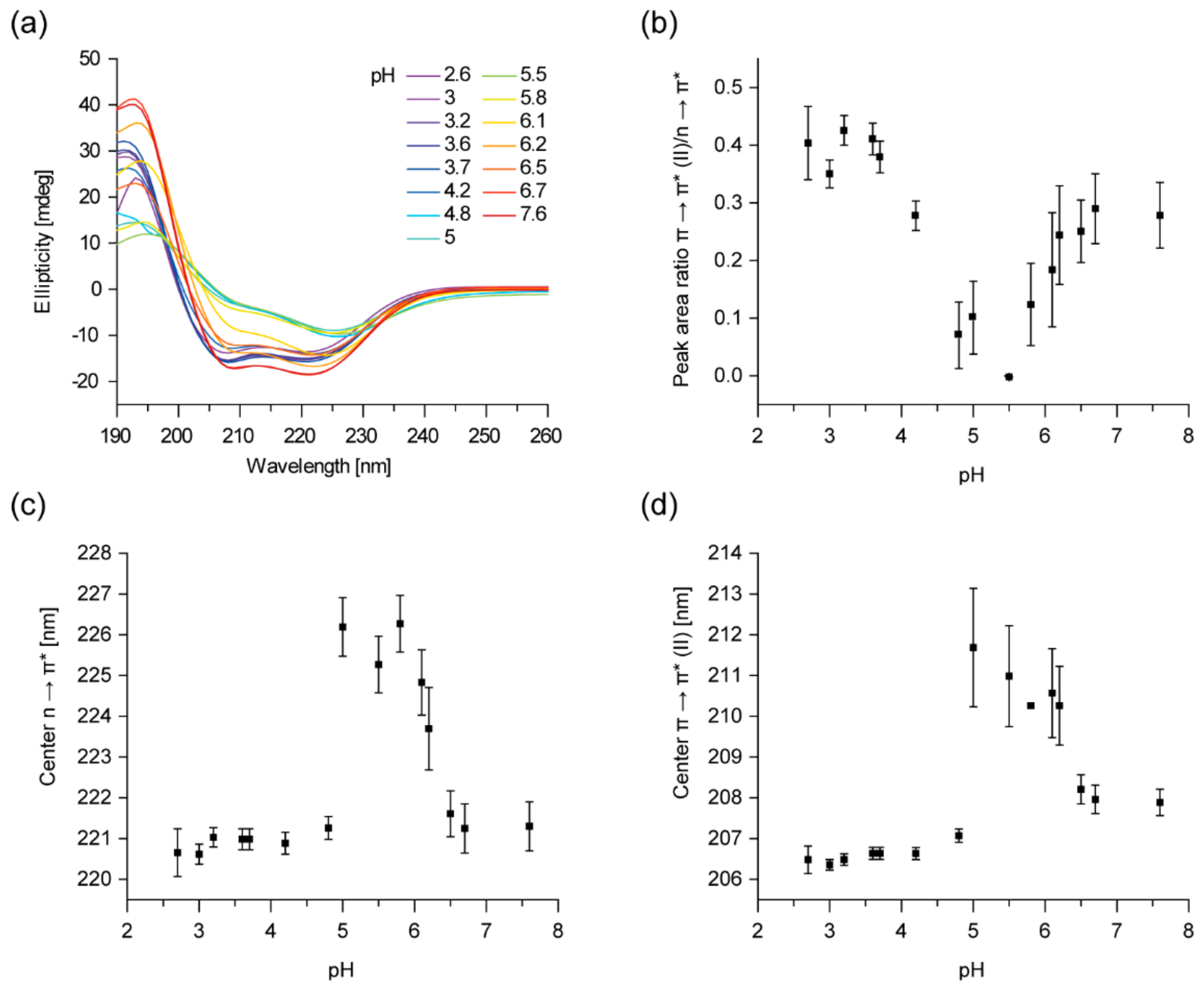
2.1. IM30 Has an Unconventional Secondary Structure at pH 5.5
2.2. The Tertiary Structure of IM30 Changes upon Acidification
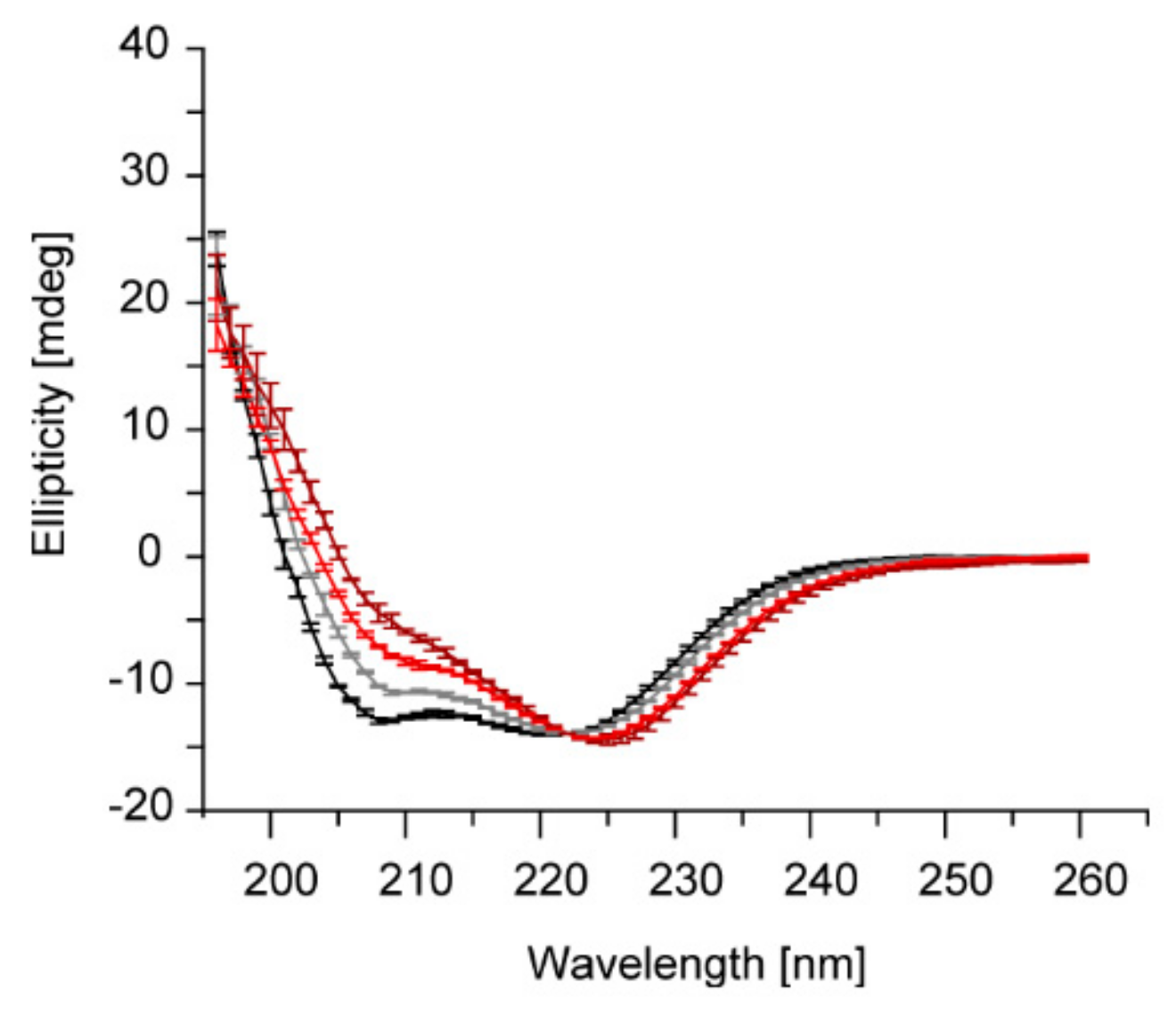
2.3. Acidic pH and Mg2+ Have Similar Effects on the IM30 Structure
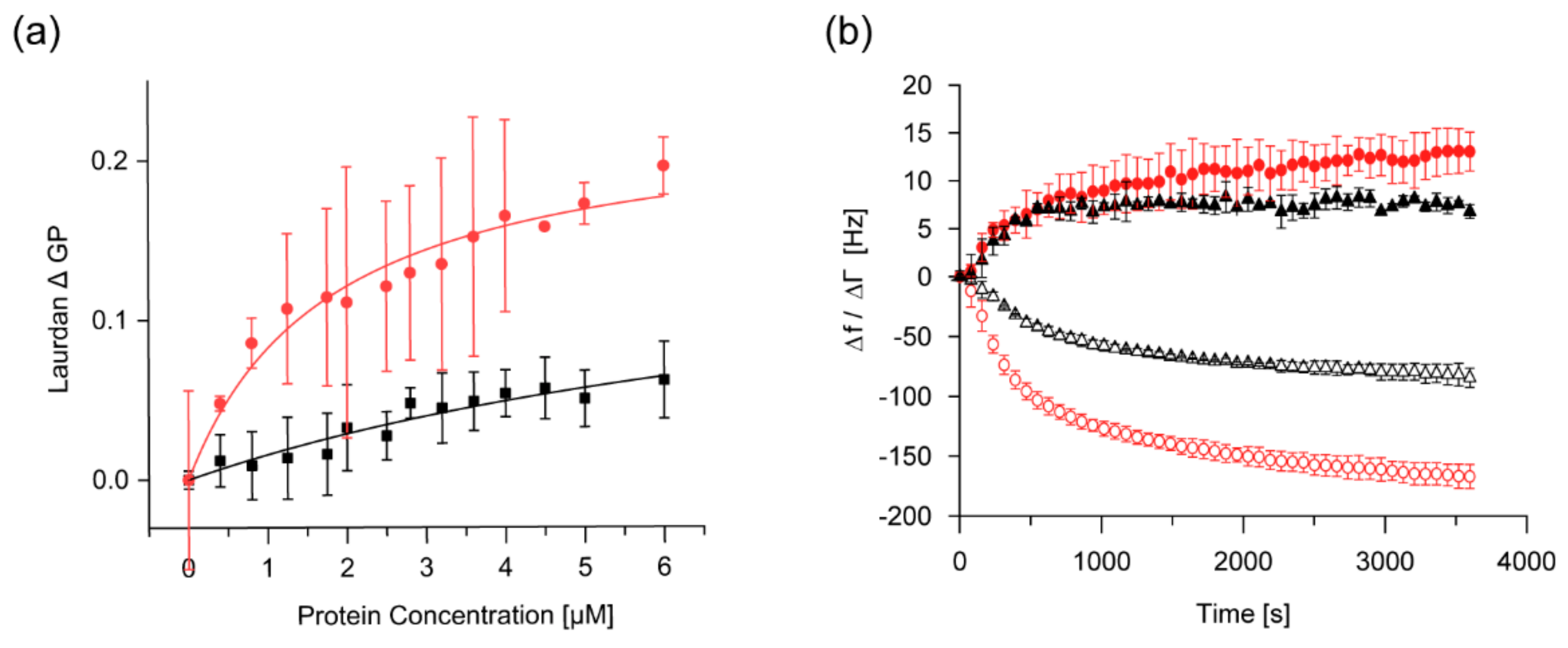
2.4. The Membrane-Binding Affinity of IM30 is Increased at Low pH
2.5. IM30-Mediated Membrane Fusion is Enhanced at Low pH
3. Discussion
3.1. The Structure and Membrane Interaction of IM30 is Modulated by a More Acidic pH
3.2. Physiological Implications of a pH-Responsive IM30 Structure and Function
4. Materials and Methods
4.1. Cloning, Expression, and Purification of IM30
4.2. Circular Dichroism Spectroscopy
4.3. ANS Fluorescence Measurements
4.4. Trp Fluorescence Measurements
4.5. Liposome Preparation
4.6. Laurdan Fluorescence Measurements
4.7. Membrane Fusion Assay
4.8. Quartz Crystal Microbalance
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| IM30 | Inner-membrane associated protein of 30 kDa |
| PspA | Phage shock protein A |
| PMF | Proton motive force |
| TM | Thylakoid Membrane |
| CD | Circular Dichroism |
| SD | Standard Deviation |
| QCM | Quartz Crystal Microbalance |
References
- Vothknecht, U.C.; Otters, S.; Hennig, R.; Schneider, D. Vipp1: A very important protein in plastids. J. Exp. Bot. 2012, 63, 1699–1712. [Google Scholar] [CrossRef] [PubMed]
- Westphal, S.; Heins, L.; Soll, J.; Vothknecht, U.C. Vipp1 deletion mutant of synechocystis: A connection between bacterial phage shock and thylakoid biogenesis? Proc. Natl. Acad. Sci. USA 2001, 98, 4243–4248. [Google Scholar] [CrossRef] [PubMed]
- Joly, N.; Engl, C.; Jovanovic, G.; Huvet, M.; Toni, T.; Sheng, X.; Stumpf, M.P.H.; Buck, M. Managing membrane stress: The phage shock protein (Psp) response, from molecular mechanisms to physiology. FEMS Microbiol. Rev. 2010, 34, 797–827. [Google Scholar] [CrossRef] [PubMed]
- Bultema, J.B.; Fuhrmann, E.; Boekema, E.J.; Schneider, D. Vipp1 and PspA. Commun. Integr. Biol. 2010, 3, 162–165. [Google Scholar] [CrossRef] [PubMed]
- Hankamer, B.D.; Elderkin, S.L.; Buck, M.; Nield, J. Organization of the AAA+ Adaptor Protein PspA Is an Oligomeric Ring. J. Biol. Chem. 2004, 279, 8862–8866. [Google Scholar] [CrossRef]
- Otters, S.; Braun, P.; Hubner, J.; Wanner, G.; Vothknecht, U.C.; Chigri, F. The first α-helical domain of the vesicle-inducing protein in plastids 1 promotes oligomerization and lipid binding. Planta 2013, 237, 529–540. [Google Scholar] [CrossRef]
- Brissette, J.L.; Russel, M.; Weiner, L.; Model, P. Phage shock protein, a stress protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 1990, 87, 862–866. [Google Scholar] [CrossRef]
- Osadnik, H.; Schöpfel, M.; Heidrich, E.; Mehner, D.; Lilie, H.; Parthier, C.; Risselada, H.J.; Grubmüller, H.; Stubbs, M.T.; Brüser, T. PspF-binding domain PspA1-144and the PspA·F complex: New insights into the coiled-coil-dependent regulation of AAA+ proteins. Mol. Microbiol. 2015, 98, 743–759. [Google Scholar] [CrossRef]
- Aseeva, E.; Ossenbühl, F.; Eichacker, L.A.; Wanner, G.; Soll, J.; Vothknecht, U.C. Complex formation of Vipp1 depends on its α-helical PspA-like domain. J. Biol. Chem. 2004, 279, 35535–35541. [Google Scholar] [CrossRef]
- Liu, C.; Willmund, F.; Whitelegge, J.P.; Hawat, S.; Knapp, B.; Lodha, M.; Schroda, M. J-domain protein CDJ2 and HSP70B are a plastidic chaperone pair that interacts with vesicle-inducing protein in plastids 1. Mol. Biol. Cell 2005, 16, 1165–1177. [Google Scholar] [CrossRef]
- Fuhrmann, E.; Bultema, J.B.; Kahmann, U.; Rupprecht, E.; Boekema, E.J.; Schneider, D. The vesicle-inducing protein 1 from Synechocystis sp. PCC 6803 organizes into diverse higher-ordered ring structures. Mol. Biol. Cell 2009, 20, 4620–4628. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Wang, W.; Zhang, W.; Liu, C. α-Helical Domains Affecting the Oligomerization of Vipp1 and Its Interaction with Hsp70/DnaK in Chlamydomonas. Biochemistry 2015, 54, 4877–4889. [Google Scholar] [CrossRef] [PubMed]
- Heidrich, J.; Wulf, V.; Hennig, R.; Saur, M.; Markl, J.; Sönnichsen, C.; Schneider, D. Organization into higher ordered ring structures counteracts membrane binding of IM30, a protein associated with inner membranes in chloroplasts and cyanobacteria. J. Biol. Chem. 2016, 291, 14954–14962. [Google Scholar] [CrossRef] [PubMed]
- Saur, M.; Hennig, R.; Young, P.; Rusitzka, K.; Hellmann, N.; Heidrich, J.; Morgner, N.; Markl, J.; Schneider, D. A Janus-Faced IM30 Ring Involved in Thylakoid Membrane Fusion Is Assembled from IM30 Tetramers. Structure 2017, 25, 1380–1390.e5. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Willmund, F.; Golecki, J.R.; Cacace, S.; Heß, B.; Markert, C.; Schroda, M. The chloroplast HSP70B-CDJ2-CGE1 chaperones catalyse assembly and disassembly of VIPP1 oligomers in Chlamydomonas. Plant. J. 2007, 50, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Wolf, D.; Kalamorz, F.; Wecke, T.; Juszczak, A.; Mäder, U.; Homuth, G.; Jordan, S.; Kirstein, J.; Hoppert, M.; Voigt, B.; et al. In-depth profiling of the LiaR response of bacillus subtilis. J. Bacteriol. 2010, 192, 4680–4693. [Google Scholar] [CrossRef]
- Heidrich, J.; Thurotte, A.; Schneider, D. Specific interaction of IM30/Vipp1 with cyanobacterial and chloroplast membranes results in membrane remodeling and eventually in membrane fusion. Biochim. Biophys. Acta - Biomembr. 2017, 1859, 537–549. [Google Scholar] [CrossRef]
- Siebenaller, C.; Junglas, B.; Schneider, D. Functional Implications of Multiple IM30 Oligomeric States. Front. Plant. Sci. 2019, 10, 1–11. [Google Scholar] [CrossRef]
- Kobayashi, R.; Suzuki, T.; Yoshida, M. Escherichia coli phage-shock protein A (PspA) binds to membrane phospholipids and repairs proton leakage of the damaged membranes. Mol. Microbiol. 2007, 66, 100–109. [Google Scholar] [CrossRef]
- Kleerebezem, M.; Crielaard, W.; Tommassen, J. Involvement of stress protein PspA (phage shock protein A) of Escherichia coli in maintenance of the protonmotive force under stress conditions. Embo J. 1996, 15, 162–171. [Google Scholar] [CrossRef]
- Hennig, R.; Heidrich, J.; Saur, M.; Schmüser, L.; Roeters, S.J.; Hellmann, N.; Woutersen, S.; Bonn, M.; Weidner, T.; Markl, J.; et al. IM30 triggers membrane fusion in cyanobacteria and chloroplasts. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- McDonald, C.; Jovanovic, G.; Ces, O.; Buck, M. Membrane stored curvature elastic stress modulates recruitment of maintenance proteins pspa and vipp1. MBio 2015, 6, e01188-15. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Kato, Y.; Otters, S.; Vothknecht, U.C.; Sakamoto, W. Essential role of VIPP1 in chloroplast envelope maintenance in Arabidopsis. Plant. Cell 2012, 24, 3695–3707. [Google Scholar] [CrossRef] [PubMed]
- DeLisa, M.P.; Lee, P.; Palmer, T.; Georgiou, G. Phage Shock Protein PspA of Escherichia coli Relieves Saturation of Protein Export via the Tat Pathway. J. Bacteriol. 2004, 186, 366–373. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H. Reactive oxygen species, oxidative signaling and the regulation of photosynthesis. Environ. Exp. Bot. 2018, 154, 134–142. [Google Scholar] [CrossRef]
- Junglas, B.; Schneider, D. What is Vipp1 good for? Mol. Microbiol. 2018, 108, 1–5. [Google Scholar] [CrossRef]
- Cui, H.; Lyman, E.; Voth, G.A. Mechanism of membrane curvature sensing by amphipathic helix containing proteins. Biophys. J. 2011, 100, 1271–1279. [Google Scholar] [CrossRef]
- Gutu, A.; Chang, F.; O’Shea, E.K. Dynamical localization of a thylakoid membrane binding protein is required for acquisition of photosynthetic competency. Mol. Microbiol. 2018, 108, 16–31. [Google Scholar] [CrossRef]
- Thurotte, A.; Brüser, T.; Mascher, T.; Schneider, D. Membrane chaperoning by members of the PspA/IM30 protein family. Commun. Integr. Biol. 2017, 10, e1264546. [Google Scholar] [CrossRef]
- Chuartzman, S.G.; Nevo, R.; Shimoni, E.; Charuvi, D.; Kiss, V.; Ohad, I.; Brumfeld, V.; Reicha, Z. Thylakoid membrane remodeling during state transitions in Arabidopsis. Plant. Cell 2008, 20, 1029–1039. [Google Scholar] [CrossRef]
- Nagy, G.; Posselt, D.; Kovács, L.; Holm, J.K.; Szabó, M.; Ughy, B.; Rosta, L.; Peters, J.; Timmins, P.; Garab, G. Reversible membrane reorganizations during photosynthesis in vivo: Revealed by small-angle neutron scattering. Biochem. J. 2011, 436, 225–230. [Google Scholar] [CrossRef] [PubMed]
- Liberton, M.; Collins, A.M.; Page, L.E.; O’Dell, W.B.; O’Neill, H.; Urban, V.S.; Timlin, J.A.; Pakrasi, H.B. Probing the consequences of antenna modification in cyanobacteria. Photosynth. Res. 2013, 118, 17–24. [Google Scholar] [CrossRef]
- Nestler, H.; Groh, K.J.; Schönenberger, R.; Eggen, R.I.L.; Suter, M.J.F. Linking proteome responses with physiological and biochemical effects in herbicide-exposed Chlamydomonas reinhardtii. J. Proteomics 2012, 75, 5370–5385. [Google Scholar] [CrossRef]
- Herbstová, M.; Tietz, S.; Kinzel, C.; Turkina, M.V.; Kirchhoff, H. Architectural switch in plant photosynthetic membranes induced by light stress. Proc. Natl. Acad. Sci. USA 2012, 109, 20130–20135. [Google Scholar] [CrossRef] [PubMed]
- Findinier, J.; Delevoye, C.; Cohen, M.M. The dynamin-like protein Fzl promotes thylakoid fusion and resistance to light stress in Chlamydomonas reinhardtii. PLOS Genet. 2019, 15, e1008047. [Google Scholar] [CrossRef]
- Thurotte, A.; Schneider, D. The fusion activity of IM30 rings involves controlled unmasking of the fusogenic core. Front. Plant. Sci. 2019, 10, 108. [Google Scholar] [CrossRef]
- Heidrich, J.; Junglas, B.; Grytsyk, N.; Hellmann, N.; Rusitzka, K.; Gebauer, W.; Markl, J.; Hellwig, P.; Schneider, D. Mg2+ binding triggers rearrangement of the IM30 ring structure, resulting in augmented exposure of hydrophobic surfaces competent for membrane binding. J. Biol. Chem. 2018, 293, 8230–8241. [Google Scholar] [CrossRef] [PubMed]
- Pohland, A.C.; Schneider, D. Mg2+ homeostasis and transport in cyanobacteria - at the crossroads of bacterial and chloroplast Mg2+ import. Biol. Chem. 2019, 400, 1289–1301. [Google Scholar] [CrossRef] [PubMed]
- Fuhrmann, E.; Gathmann, S.; Rupprecht, E.; Golecki, J.; Schneider, D. Thylakoid membrane reduction affects the photosystem stoichiometry in the cyanobacterium Synechocystis sp. PCC 6803. Plant. Physiol. 2009, 149, 735–744. [Google Scholar] [CrossRef]
- Micsonai, A.; Wien, F.; Kernya, L.; Lee, Y.-H.; Goto, Y.; Réfrégiers, M.; Kardos, J. Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc. Natl. Acad. Sci. USA 2015, 112, E3095–E3103. [Google Scholar] [CrossRef]
- Urry, D.W.; Ji, T.H. Distortions in circular dichroism patterns of particulate (or membranous) systems. Arch. Biochem. Biophys. 1968, 128, 802–807. [Google Scholar] [CrossRef]
- Duysens, L.N. The flattening of the absorption spectrum of suspensions, as compared to that of solutions. Biochim. Biophys. Acta 1956, 19, 1–12. [Google Scholar] [CrossRef]
- Biswas, S.; Kayastha, A. Spectroscopic Characterization Of Phaseolus Vulgaris Leucoagglutinin. Protein Pept. Lett. 2005, 11, 1–7. [Google Scholar] [CrossRef] [PubMed]
- LaPorte, D.C.; Wierman, B.M.; Storm, D.R. Calcium-induced exposure of a hydrophobic surface on calmodulin. Biochemistry 1980, 19, 3814–3819. [Google Scholar] [CrossRef]
- White, A. Effect of pH on fluorescence of tryosine, tryptophan and related compounds. Biochem. J. 1959, 71, 217–220. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Sakamoto, W. Possible function of VIPP1 in thylakoids: Protection but not formation? Plant. Signal. Behav. 2013, 8. [Google Scholar] [CrossRef]
- Zhang, S.; Shen, G.; Li, Z.; Golbeck, J.H.; Bryant, D.A. Vipp1 is essential for the biogenesis of Photosystem I but not thylakoid membranes in Synechococcus sp. PCC 7002. J. Biol. Chem. 2014, 289, 15904–15914. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Sakamoto, W. Possible function of VIPP1 in maintaining chloroplast membranes. Biochim. Biophys. Acta - Bioenerg. 2015, 1847, 831–837. [Google Scholar] [CrossRef]
- Wada, H.; Murata’, N. Synechocystis PCC6803 Mutants Defective in Desaturation of Fatty Acids. Plant Cell Physiol. 1989, 30, 971–978. [Google Scholar]
- Sauerbrey, G. Verwendung von Schwingquarzen zur Wägung dünner Schichten und zur Mikrowägung. Zeitschrift für Phys. 1959, 155, 206–222. [Google Scholar] [CrossRef]
- Reviakine, I.; Johannsmann, D.; Richter, R.P. Hearing What You Cannot See and Visualizing What You Hear: Interpreting Quartz Crystal Microbalance Data from Solvated Interfaces. Anal. Chem. 2011, 83, 8838–8848. [Google Scholar] [CrossRef] [PubMed]
- Voinova, M.V.; Jonson, M.; Kasemo, B. “Missing mass” effect in biosensor’s QCM applications. Biosens. Bioelectron. 2002, 17, 835–841. [Google Scholar] [CrossRef]
- Hennig, R.; West, A.; Debus, M.; Saur, M.; Markl, J.; Sachs, J.N.; Schneider, D. The IM30/Vipp1 C-terminus associates with the lipid bilayer and modulates membrane fusion. Biochim. Biophys. Acta - Bioenerg. 2017, 1858, 126–136. [Google Scholar] [CrossRef] [PubMed]
- McCubbin, W.D.; Oikawa, K.; Kay, C.M. Circular dichroism studies on concanavalin A. Biochem. Biophys. Res. Commun. 1971, 43, 666–674. [Google Scholar] [CrossRef]
- Bañares-Hidalgo, A.; Pérez-Gil, J.; Estrada, P. Acidic pH triggers conformational changes at the NH2-terminal propeptide of the precursor of pulmonary surfactant protein B to form a coiled coil structure. Biochim. Biophys. Acta - Biomembr. 2014, 1838, 1738–1751. [Google Scholar] [CrossRef][Green Version]
- Darwin, A.J. The phage-shock-protein response. Mol. Microbiol. 2005, 57, 621–628. [Google Scholar] [CrossRef]
- Vrancken, K.; Van Mellaert, L.; Anné, J. Characterization of the Streptomyces lividans PspA response. J. Bacteriol. 2008, 190, 3475–3481. [Google Scholar] [CrossRef]
- Schneider, D.; Fuhrmann, E.; Scholz, I.; Hess, W.R.; Graumann, P.L. Fluorescence staining of live cyanobacterial cells suggest non-stringent chromosome segregation and absence of a connection between cytoplasmic and thylakoid membranes. BMC Cell Biol. 2007, 8. [Google Scholar] [CrossRef]
- Belkin, S.; Mehlhorn, R.J.; Packer, L. Proton gradients in intact cyanobacteria. Plant. Physiol. 1987, 84, 25–30. [Google Scholar] [CrossRef]
- Falkner, G.; Horner, F.; Werdan, K.; Heldt, H.W. pH Changes in the Cytoplasm of the Blue-Green Alga Anacystis nidulans Caused by Light-dependent Proton Flux into the Thylakoid Space. Plant. Physiol. 1976, 58, 717–718. [Google Scholar] [CrossRef]
- Gibson, J. Movement of acetate across the cytoplasmic membrane of the unicellular cyanobacteria Synechococcus and Aphanocapsa. Arch. Microbiol. 1981, 130, 175–179. [Google Scholar] [CrossRef]
- Kallas, T.; Dahlquist, F.W. Phosphorus-31 Nuclear Magnetic Resonance Analysis of Internal pH during Photosynthesis in the Cyanobacterium Synechococcus. Biochemistry 1981, 20, 5900–5907. [Google Scholar] [CrossRef] [PubMed]
- Masamoto, K.; Nishimura, M. Estimation of internal pH in cells of blue-green algae in the dark and under illumination. J. Biochem. 1977, 82, 483–487. [Google Scholar] [PubMed]
- Padan, E.; Schuldiner, S. Energy transduction in the photosynthetic membranes of the cyanobacterium (blue-green alga) Plectonema boryanum. J. Biol. Chem. 1978, 253, 3281–3286. [Google Scholar] [PubMed]
- Nitschmann, W.H.; Peschek, G.A. Modes of proton translocation across the cell membrane of respiring cyanobacteria. Arch. Microbiol. 1985, 141, 330–336. [Google Scholar] [CrossRef]
- Scherer, S.; Stürzl, E.; Böger, P. Oxygen-dependent proton efflux in cyanobacteria (blue-green algae). J. Bacteriol. 1984, 158, 609–614. [Google Scholar] [CrossRef] [PubMed]
- Bryan, S.J.; Burroughs, N.J.; Shevela, D.; Yu, J.; Rupprecht, E.; Liu, L.N.; Mastroianni, G.; Xue, Q.; Llorente-Garcia, I.; Leake, M.C.; et al. Localisation and interactions of the Vipp1 protein in cyanobacteria. Mol. Microbiol. 2014, 94, 1179–1195. [Google Scholar] [CrossRef]
- Pace, C.N.; Grimsley, G.R.; Scholtz, J.M. Protein Ionizable Groups: pK Values and Their Contribution to Protein Stability and Solubility. J. Biol Chem. 2009, 284(20), 13285–13289. [Google Scholar] [CrossRef]
- Sawant, P.; Eissenberger, K.; Karier, L.; Mascher, T.; Bramkamp, M. A dynamin-like protein involved in bacterial cell membrane surveillance under environmental stress. Environ. Microbiol. 2016, 18, 2705–2720. [Google Scholar] [CrossRef]
- Guo, L.; Bramkamp, M. Bacterial dynamin-like protein DynA mediates lipid and content mixing. FASEB J. 2019, 33, 11746–11757. [Google Scholar] [CrossRef]
- Parasassi, T.; Gratton, E. Membrane lipid domains and dynamics as detected by Laurdan fluorescence. J. Fluoresc. 1995, 5, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Meers, P.; Ali, S.; Erukulla, R.; Janoff, A.S. Novel inner monolayer fusion assays reveal differential monolayer mixing associated with cation-dependent membrane fusion. Biochim. Biophys. Acta - Biomembr. 2000, 1467, 227–243. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Siebenaller, C.; Junglas, B.; Lehmann, A.; Hellmann, N.; Schneider, D. Proton Leakage Is Sensed by IM30 and Activates IM30-Triggered Membrane Fusion. Int. J. Mol. Sci. 2020, 21, 4530. https://doi.org/10.3390/ijms21124530
Siebenaller C, Junglas B, Lehmann A, Hellmann N, Schneider D. Proton Leakage Is Sensed by IM30 and Activates IM30-Triggered Membrane Fusion. International Journal of Molecular Sciences. 2020; 21(12):4530. https://doi.org/10.3390/ijms21124530
Chicago/Turabian StyleSiebenaller, Carmen, Benedikt Junglas, Annika Lehmann, Nadja Hellmann, and Dirk Schneider. 2020. "Proton Leakage Is Sensed by IM30 and Activates IM30-Triggered Membrane Fusion" International Journal of Molecular Sciences 21, no. 12: 4530. https://doi.org/10.3390/ijms21124530
APA StyleSiebenaller, C., Junglas, B., Lehmann, A., Hellmann, N., & Schneider, D. (2020). Proton Leakage Is Sensed by IM30 and Activates IM30-Triggered Membrane Fusion. International Journal of Molecular Sciences, 21(12), 4530. https://doi.org/10.3390/ijms21124530