Linking the Peptidoglycan Synthesis Protein Complex with Asymmetric Cell Division during Bacillus subtilis Sporulation
Abstract
1. Introduction
2. Results and Discussion
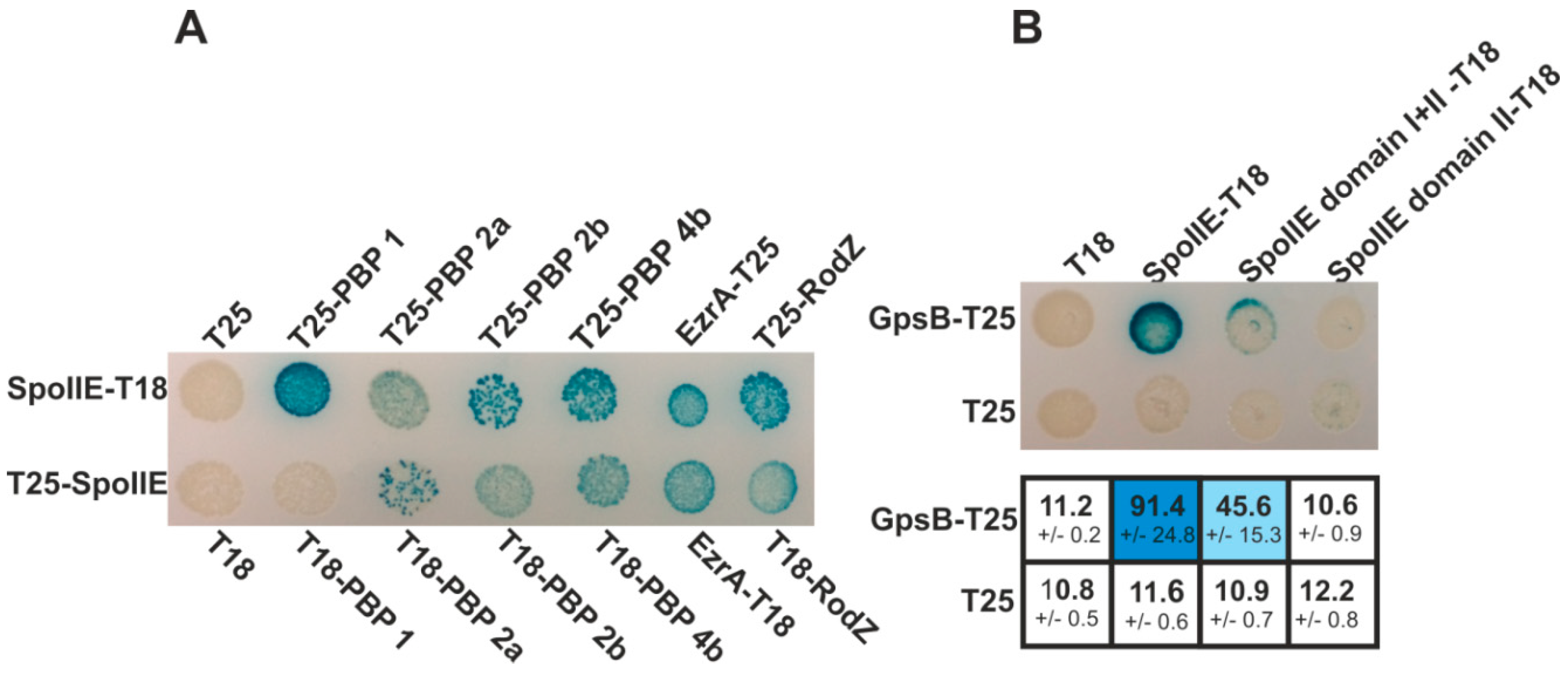
2.1. GpsB Interacts with SpoIIE
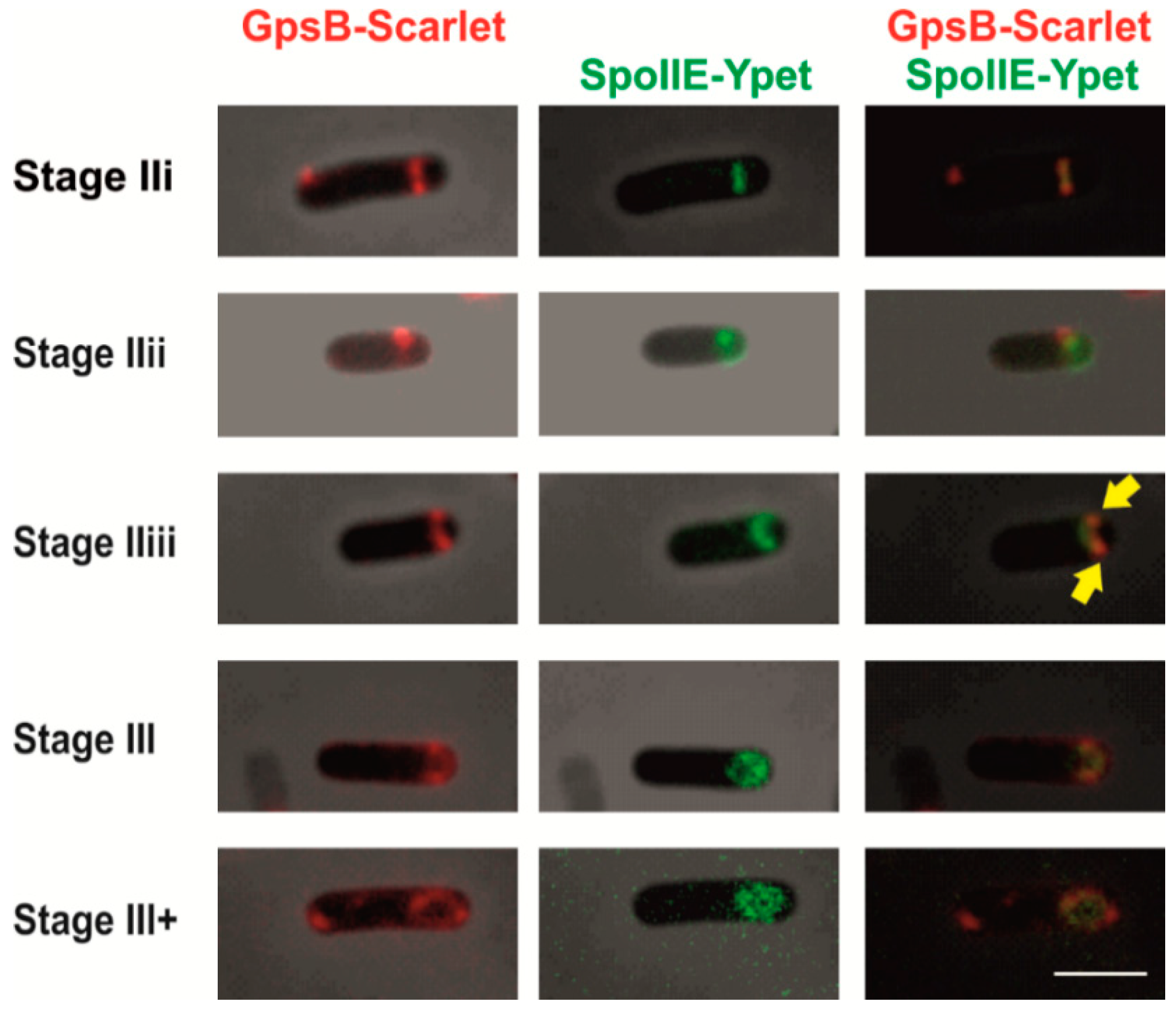
2.2. GpsB Co-Localizes with SpoIIE during Sporulation
2.3. Localization of SpoIIE and GpsB Is Mutually Independent
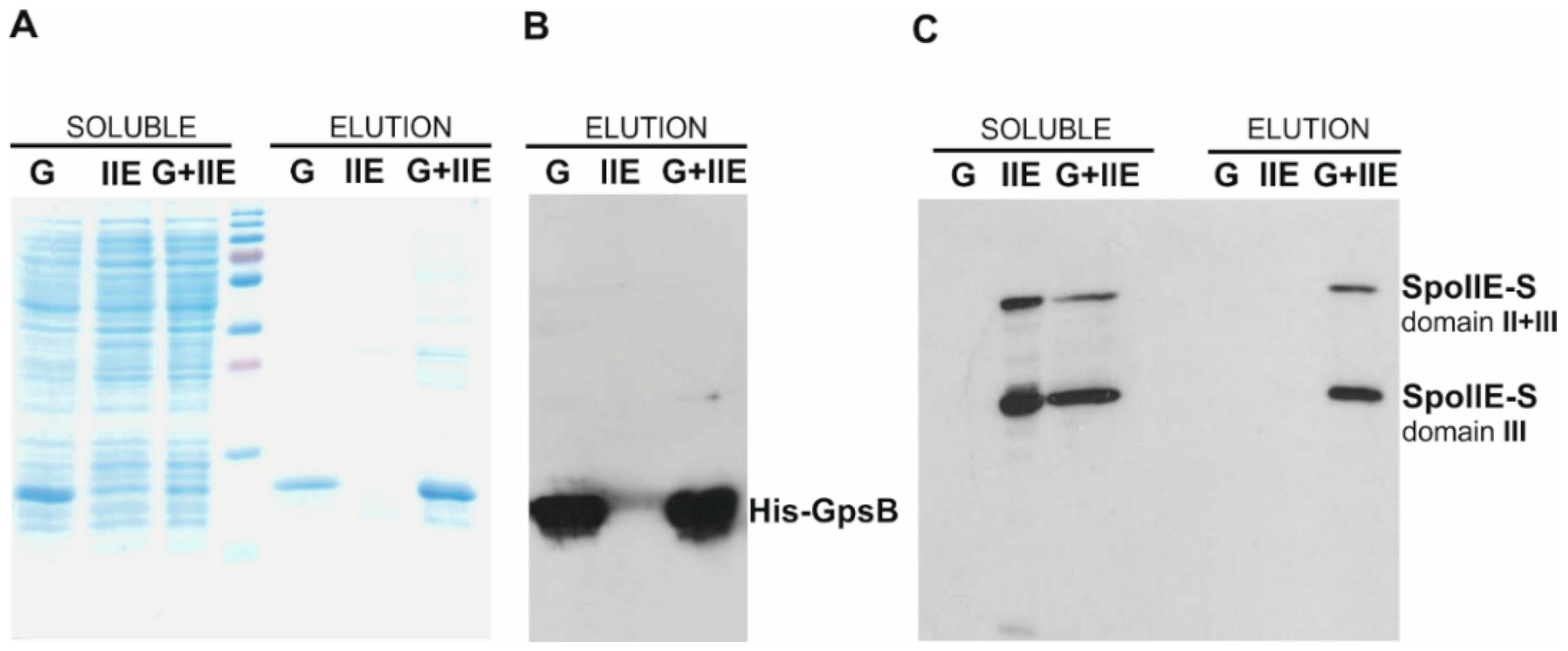
2.4. SpoIIE and GpsB Are Components of a Multi-Protein Complex
3. Materials and Methods
3.1. Media and General Methods
3.2. Bacterial Strains and Plasmids
3.3. Bacterial Two-Hybrid System
3.4. Protein Isolation and Purification
3.5. Fluorescence Microscopy and Image Acquisition
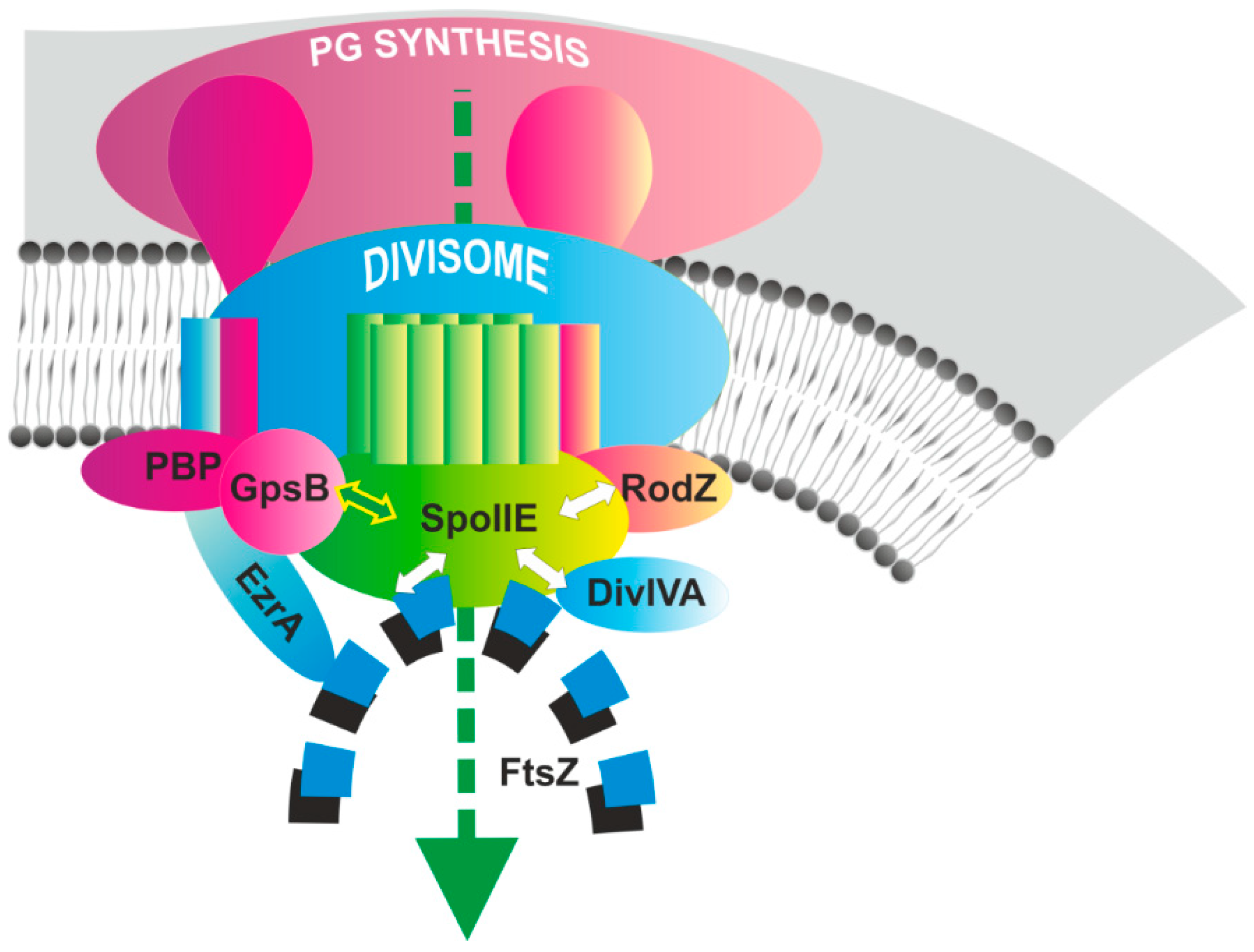
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Higgins, D.; Dworkin, J. Recent progress in Bacillus subtilis sporulation. FEMS Microbiol. Rev. 2012, 36, 131–148. [Google Scholar] [CrossRef] [PubMed]
- Tan, I.S.; Ramamurthi, K.S. Spore formation in Bacillus subtilis. Environ. Microbiol. Rep. 2014, 6, 212–225. [Google Scholar] [CrossRef] [PubMed]
- Levin, P.A.; Losick, R. Transcription factor SpoOA switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes Dev. 1996, 10, 478–488. [Google Scholar] [CrossRef] [PubMed]
- Ben-Yehuda, S.; Losick, R. Asymmetric cell division in B. subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell 2002, 109, 257–266. [Google Scholar] [PubMed]
- Eswaramoorthy, P.; Winter, P.W.; Wawrzusin, P.; York, A.G.; Shroff, H.; Ramamurthi, K.S. Asymmetric Division and Differential Gene Expression during a Bacterial Developmental Program Requires DivIVA. PLoS Genet. 2014, 10, e1004526. [Google Scholar] [CrossRef] [PubMed]
- Arigoni, F.; Guérout-Fleury, A.M.; Barák, I.; Stragier, P. The SpoIIE phosphatase, the sporulation septum and the establishment of forespore-specific transcription in Bacillus subtilis: A reassessment. Mol. Microbiol. 1999, 31, 1407–1415. [Google Scholar] [CrossRef]
- Rawlings, A.E.; Levdikov, V.M.; Blagova, E.; Colledge, V.L.; Mas, P.J.; Tunaley, J.; Vavrova, L.; Wilson, K.S.; Barak, I.; Hart, D.J.; et al. Expression of soluble, active fragments of the morphogenetic protein SpoIIE from Bacillus subtilis using a library-based construct screen. Protein Eng. Des. Sel. 2010, 23, 817–825. [Google Scholar] [CrossRef]
- Barák, I.; Youngman, P. SpoIIE mutants of Bacillus subtilis comprise two distinct phenotypic classes consistent with a dual functional role for the SpoIIE protein. J. Bacteriol. 1996, 178, 4984–4989. [Google Scholar] [CrossRef]
- Feucht, A.; Magnin, T.; Yudkin, M.D.; Errington, J. Bifunctional protein requried for asymmetric cell division and cell-specific transcription in Bacillus subtilis. Genes Dev. 1996, 10, 794–803. [Google Scholar] [CrossRef]
- Duncan, L.; Alper, S.; Arigoni, F.; Losick, R.; Stragier, P. Activation of cell-specific transcription by a serine phosphatase at the site of asymmetric division. Science 1995, 270, 641–644. [Google Scholar] [CrossRef]
- Arigoni, F.; Duncan, L.; Alper, S.; Losick, R.; Stragier, P. SpoIIE governs the phosphorylation state of a protein regulating transcription factor sigma F during sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 1996, 93, 3238–3242. [Google Scholar] [CrossRef] [PubMed]
- Iber, D.; Clarkson, J.; Yudkin, M.D.; Campbell, I.D. The mechanism of cell differentiation in Bacillus subtilis. Nature 2006, 441, 371–374. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, N.; Losick, R. Asymmetric division triggers cell-specific gene expression through coupled capture and stabilization of a phosphatase. Elife 2015, 4, e08145. [Google Scholar] [CrossRef] [PubMed]
- Campo, N.; Marquis, K.A.; Rudner, D.Z. SpoIIQ anchors membrane proteins on both sides of the sporulation septum in Bacillus subtilis. J. Biol. Chem. 2008, 283, 4975–4982. [Google Scholar] [CrossRef] [PubMed]
- Muchová, K.; Chromiková, Z.; Bradshaw, N.; Wilkinson, A.J.; Barák, I. Morphogenic Protein RodZ Interacts with Sporulation Specific SpoIIE in Bacillus subtilis. PLoS ONE 2016, 11, e0159076. [Google Scholar] [CrossRef]
- Barák, I.; Muchová, K. The positioning of the asymmetric septum during sporulation in Bacillus subtilis. PLoS ONE 2018, 13, e0201979. [Google Scholar] [CrossRef]
- Claessen, D.; Emmins, R.; Hamoen, L.W.; Daniel, R.A.; Errington, J.; Edwards, D.H. Control of the cell elongation-division cycle by shuttling of PBP1 protein in Bacillus subtilis. Mol. Microbiol. 2008, 68, 1029–1046. [Google Scholar] [CrossRef]
- Fleurie, A.; Manuse, S.; Zhao, C.; Campo, N.; Cluzel, C.; Lavergne, J.P.; Freton, C.; Combet, C.; Guiral, S.; Soufi, B.; et al. Interplay of the Serine/Threonine-Kinase StkP and the Paralogs DivIVA and GpsB in Pneumococcal Cell Elongation and Division. PLoS Genet. 2014, 10, e1004275. [Google Scholar] [CrossRef]
- Rismondo, J.; Cleverley, R.M.; Lane, H.V.; Großhennig, S.; Steglich, A.; Möller, L.; Mannala, G.K.; Hain, T.; Lewis, R.J.; Halbedel, S. Structure of the bacterial cell division determinant GpsB and its interaction with penicillin-binding proteins. Mol. Microbiol. 2016, 99, 978–998. [Google Scholar] [CrossRef]
- Tavares, J.R.; De Souza, R.F.; Meira, G.L.S.; Gueiros-Filho, F.J. Cytological characterization of YpsB, a novel component of the Bacillus subtilis divisome. J. Bacteriol. 2008, 190, 7096–7107. [Google Scholar] [CrossRef]
- Cleverley, R.M.; Rutter, Z.J.; Rismondo, J.; Corona, F.; Tsui, H.C.T.; Alatawi, F.A.; Daniel, R.A.; Halbedel, S.; Massidda, O.; Winkler, M.E.; et al. The cell cycle regulator GpsB functions as cytosolic adaptor for multiple cell wall enzymes. Nat. Commun. 2019, 10, 261. [Google Scholar] [CrossRef] [PubMed]
- Bhavsar, A.P.; Brown, E.D. Cell wall assembly in Bacillus subtilis: How spirals and spaces challenge paradigms. Mol. Microbiol. 2006, 60, 1077–1090. [Google Scholar] [CrossRef] [PubMed]
- Barák, I.; Muchová, K.; Labajová, N. Asymmetric cell division during Bacillus subtilis sporulation. Future Microbiol. 2019, 14, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Levin, P.A.; Kurtser, I.G.; Grossman, A.D. Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 1999, 96, 9642–9647. [Google Scholar] [CrossRef] [PubMed]
- Scheffers, D.; Errington, J. PBP1 Is a Component of the Bacillus subtilis cell division machinery. J Bacteriol 2004, 186, 5153–5156. [Google Scholar] [CrossRef]
- Daniel, R.A.; Harry, E.J.; Errington, J. Role of penicillin-binding protein PBP 2B in assembly and functioning of the division machinery of Bacillus subtilis. Mol. Microbiol. 2000, 35, 299–311. [Google Scholar] [CrossRef]
- Scheffers, D.J. Dynamic localization of penicillin-binding proteins during spore development in Bacillus subtilis. Microbiology 2005, 151, 999–1012. [Google Scholar] [CrossRef]
- Wei, Y.; McPherson, D.C.; Popham, D.L. A Mother Cell-Specific Class B Penicillin-Binding Protein, PBP4b, in Bacillus subtilis. J. Bacteriol. 2004, 186, 258–261. [Google Scholar] [CrossRef][Green Version]
- Pompeo, F.; Foulquier, E.; Serrano, B.; Grangeasse, C.; Galinier, A. Phosphorylation of the cell division protein GpsB regulates PrkC kinase activity through a negative feedback loop in Bacillus subtilis. Mol. Microbiol. 2015, 97, 139–150. [Google Scholar] [CrossRef]
- Halbedel, S.; Lewis, R.J. Structural basis for interaction of DivIVA/GpsB proteins with their ligands. Mol. Microbiol. 2019, 111, 1404–1415. [Google Scholar] [CrossRef]
- Reyes-Lamothe, R.; Sherratt, D.J.; Leake, M.C. Stoichiometry and architecture of active DNA replication machinery in Escherichia coli. Science 2010, 328, 498–501. [Google Scholar] [CrossRef] [PubMed]
- Illing, N.; Errington, J. Genetic Regulation of Morphogenesis in Bacillus subtilis: Roles of σE and σF in Prespore Engulfment. J. Bacteriol. 1991, 173, 3159–3169. [Google Scholar] [CrossRef] [PubMed]
- Levin, P.A.; Losick, R.; Stragier, P.; Arigoni, F. Localization of the sporulation protein SpoIIE in Bacillus subtilis is dependent upon the cell division protein FtsZ. Mol. Microbiol. 1997, 25, 839–846. [Google Scholar] [CrossRef] [PubMed]
- Gamba, P.; Veening, J.W.; Saunders, N.J.; Hamoen, L.W.; Daniel, R.A. Two-step assembly dynamics of the Bacillus subtilis divisome. J. Bacteriol. 2009, 191, 4186–4194. [Google Scholar] [CrossRef] [PubMed]
- Lucet, I.; Feucht, A.; Yudkin, M.D.; Errington, J. Direct interaction between the cell division protein FtsZ and the cell differentiation protein SpoIIE. EMBO J. 2000, 19, 1467–1475. [Google Scholar] [CrossRef]
- Król, E.; De Sousa Borges, A.; Kopacz, M.; Scheffers, D.J. Metal-dependent SpoIIE oligomerization stabilizes FtsZ during asymmetric division in Bacillus subtilis. PLoS ONE 2017, 12, e0174713. [Google Scholar] [CrossRef]
- Wu, L.J.; Errington, J. Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell 2004, 117, 915–925. [Google Scholar] [CrossRef]
- Ben-Yehuda, S.; Rudner, D.Z.; Losick, R. RacA, a Bacterial Protein That anchors chromosomes to the cell poles. Science 2003, 299, 532–536. [Google Scholar] [CrossRef]
- Thomaides, H.B.; Freeman, M.; El Karoui, M.; Errington, J. Division site selection protein DivIVA of Bacillus subtilis has a second distinct function in chromosome segregation during sporulation. Genes Dev. 2001, 15, 1662–1673. [Google Scholar] [CrossRef]
- Barák, I.; Behari, J.; Olmedo, G.; Guzmán, P.; Brown, D.P.; Castro, E.; Walker, D.; Westpheling, J.; Youngman, P. Structure and function of the Bacillus SpoIIE protein and its localization to sites of sporulation septum assembly. Mol. Microbiol. 1996, 19, 1047–1060. [Google Scholar] [CrossRef]
- Ausubel, F.M.; Brent, R.; Kingston, R.E.; Moore, D.D.; Seidman, J.G.; Smith, J.A.; Struhl, K. Current Protocols in Molecular Biology; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2001; ISBN 9780471142720. [Google Scholar]
- Harwood, C.R.; Cutting, S.M. Molecular Biological Methods for Bacillus; Wiley: Chichester, UK; New York, NY, USA, 1990; ISBN 9780471923930. [Google Scholar]
- Youngman, P.; Perkins, J.B.; Losick, R. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid 1984, 12, 1–9. [Google Scholar] [CrossRef]
- Backman, K.; Ptashne, M.; Gilbert, W. Construction of plasmids carrying the cI gene of bacteriophage lambda. Proc. Natl. Acad. Sci. USA 1976, 73, 4174–4178. [Google Scholar] [CrossRef] [PubMed]
- Feucht, A.; Lewis, P.J. Improved plasmid vectors for the production of multiple fluorescent protein fusions in Bacillus subtilis. Gene 2001, 264, 289–297. [Google Scholar] [CrossRef]
- Ju, J.; Luo, T.; Haldenwang, W.G. Forespore expression and processing of the SigE transcription factor in wild-type and mutant Bacillus subtilis. J. Bacteriol. 1998, 180, 1673–1681. [Google Scholar] [CrossRef] [PubMed]
- Yanisch-Perron, C.; Vieira, J.; Messing, J. Improved M13 phage cloning vectors and host strains: Nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 1985, 33, 103–119. [Google Scholar] [CrossRef]
- Karimova, G.; Pidoux, J.; Ullmann, A.; Ladant, D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. USA 1998, 95, 5752–5756. [Google Scholar] [CrossRef]
- Miller, J.H. Experiments in Molecular Genetics; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1972; ISBN 0879691069. [Google Scholar]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muchová, K.; Chromiková, Z.; Barák, I. Linking the Peptidoglycan Synthesis Protein Complex with Asymmetric Cell Division during Bacillus subtilis Sporulation. Int. J. Mol. Sci. 2020, 21, 4513. https://doi.org/10.3390/ijms21124513
Muchová K, Chromiková Z, Barák I. Linking the Peptidoglycan Synthesis Protein Complex with Asymmetric Cell Division during Bacillus subtilis Sporulation. International Journal of Molecular Sciences. 2020; 21(12):4513. https://doi.org/10.3390/ijms21124513
Chicago/Turabian StyleMuchová, Katarína, Zuzana Chromiková, and Imrich Barák. 2020. "Linking the Peptidoglycan Synthesis Protein Complex with Asymmetric Cell Division during Bacillus subtilis Sporulation" International Journal of Molecular Sciences 21, no. 12: 4513. https://doi.org/10.3390/ijms21124513
APA StyleMuchová, K., Chromiková, Z., & Barák, I. (2020). Linking the Peptidoglycan Synthesis Protein Complex with Asymmetric Cell Division during Bacillus subtilis Sporulation. International Journal of Molecular Sciences, 21(12), 4513. https://doi.org/10.3390/ijms21124513




