Comparative Transcriptome Analysis of Two Contrasting Soybean Varieties in Response to Aluminum Toxicity
Abstract
1. Introduction
2. Results
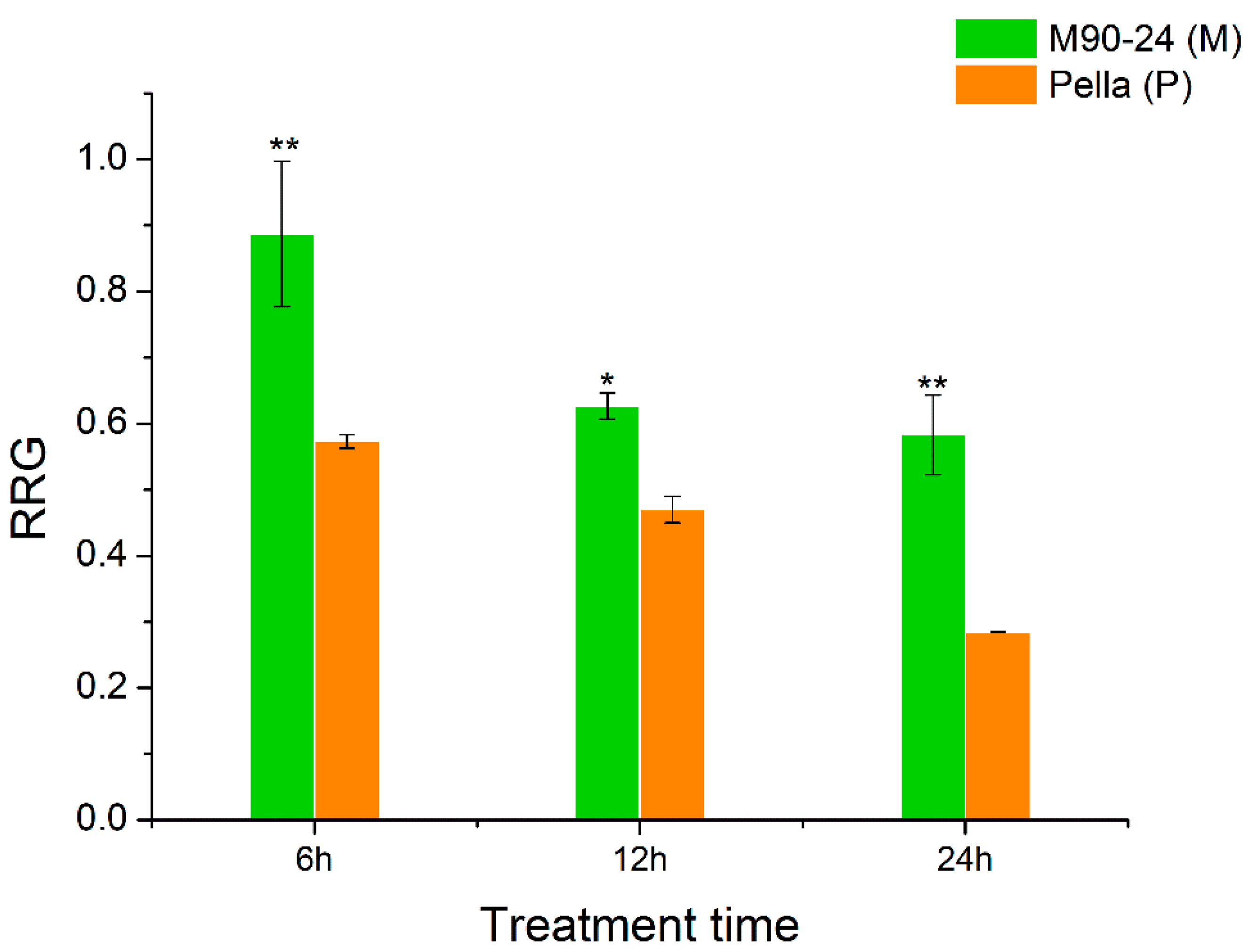
2.1. Relative Root Growth of Two Soybean Varieties under Al Stress
2.2. RNA-seq Data Quality, Assembly and Annotation
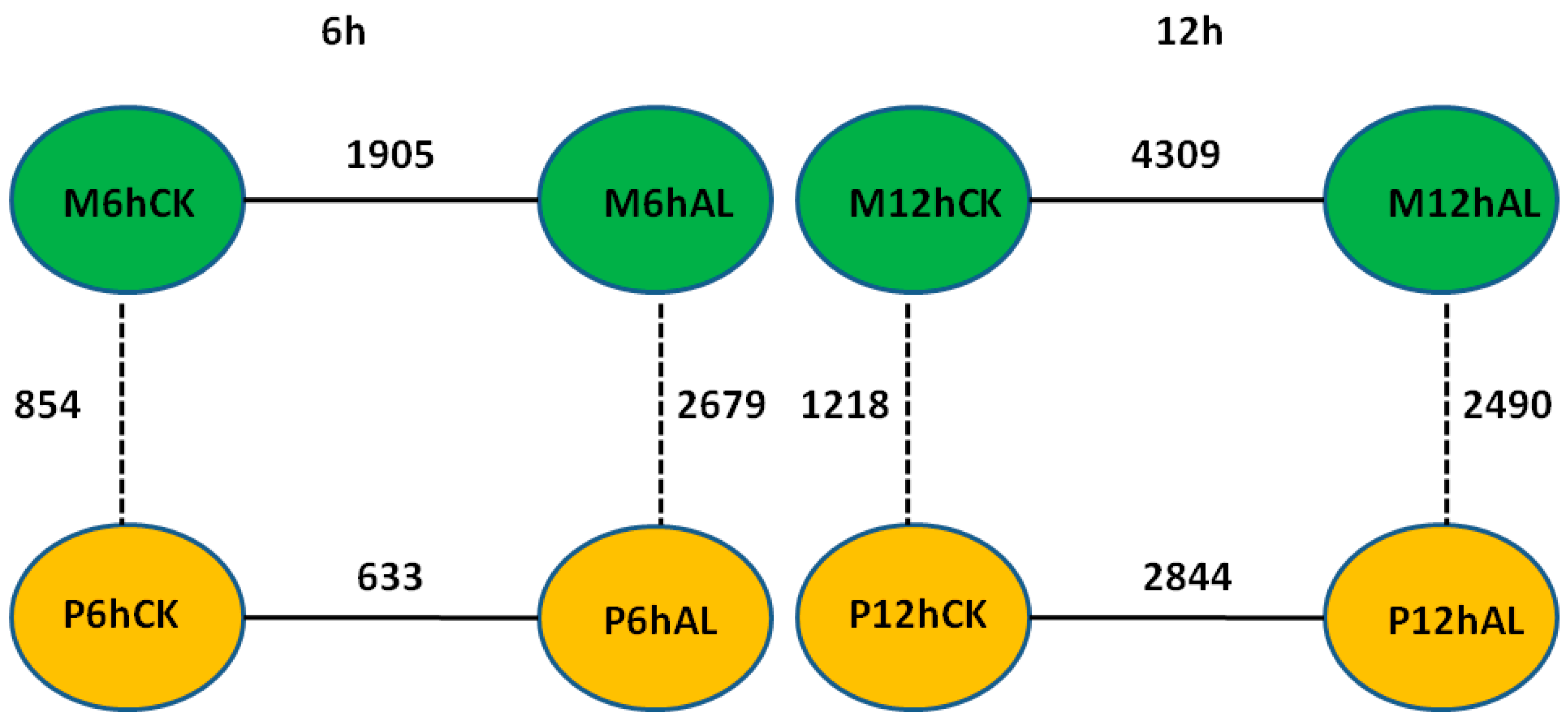
2.3. Differentially Expressed Genes (DEGs) in Soybean Roots in Response to Al Stress
2.4. DEGs between Two Soybean Varieties
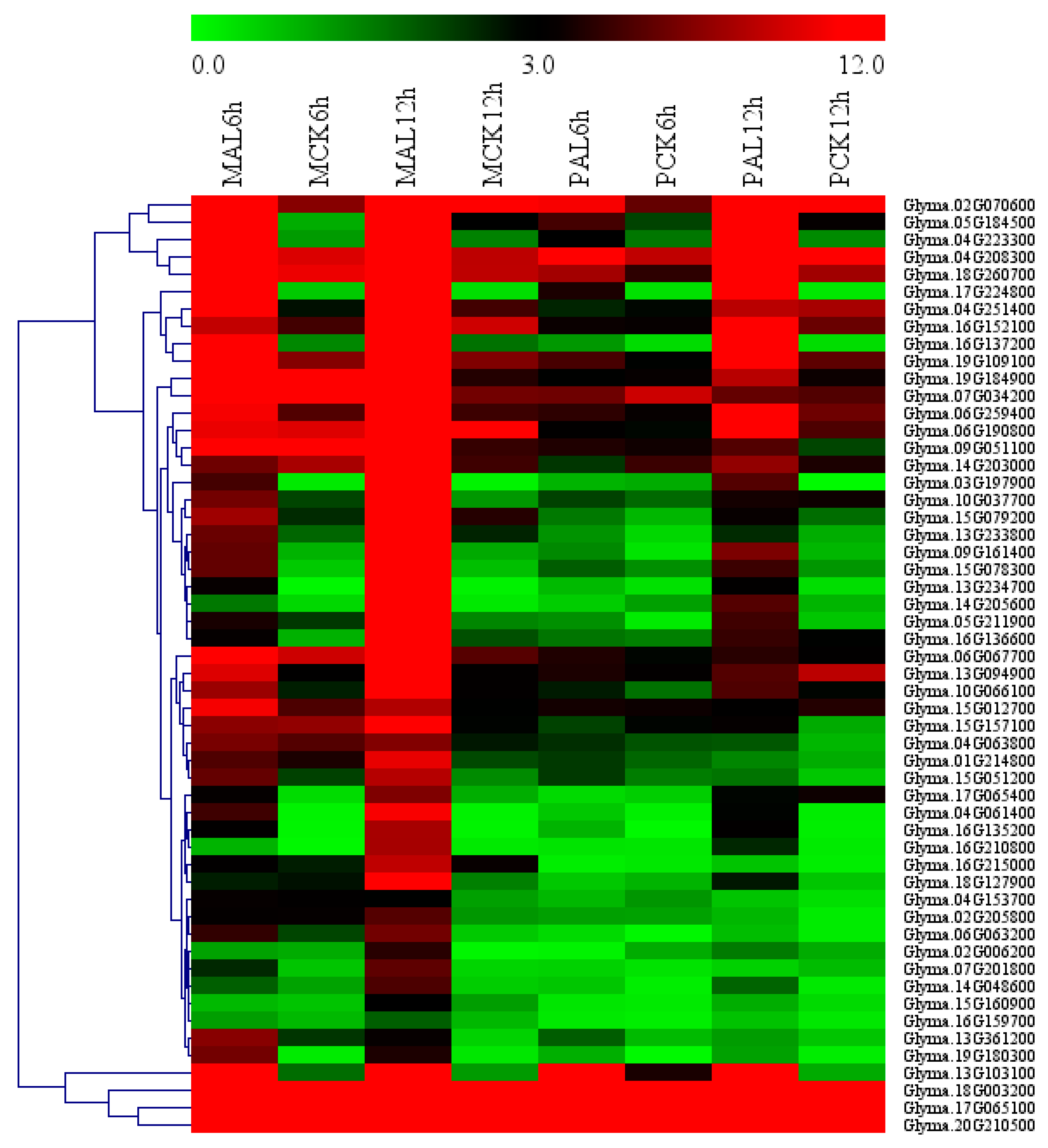
2.5. Identification of Candidate Genes Related to Al Tolerance in Soybean
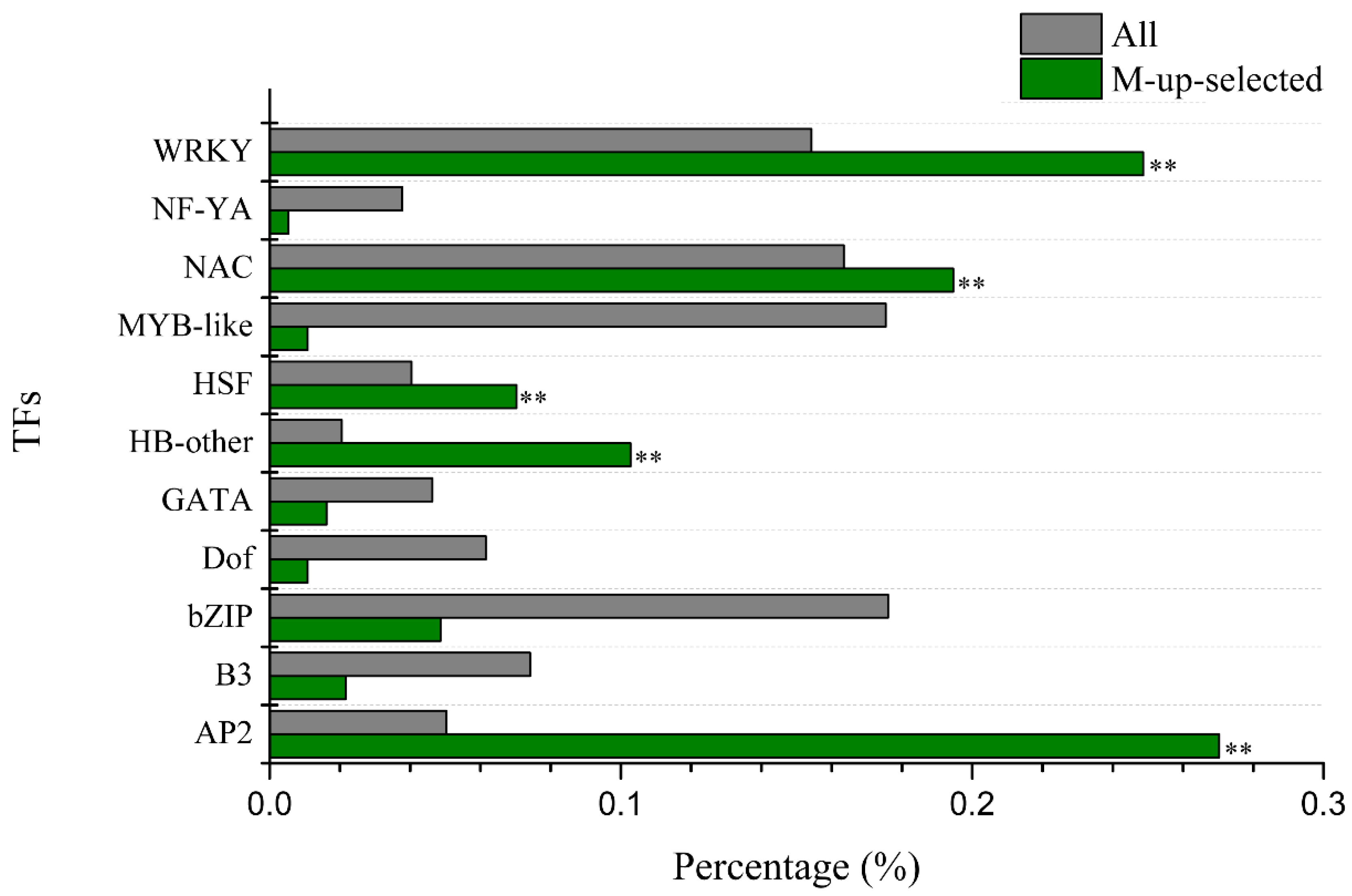
2.6. Gene Ontology (GO) Enrichment of Candidate Al Tolerance Genes in Soybean
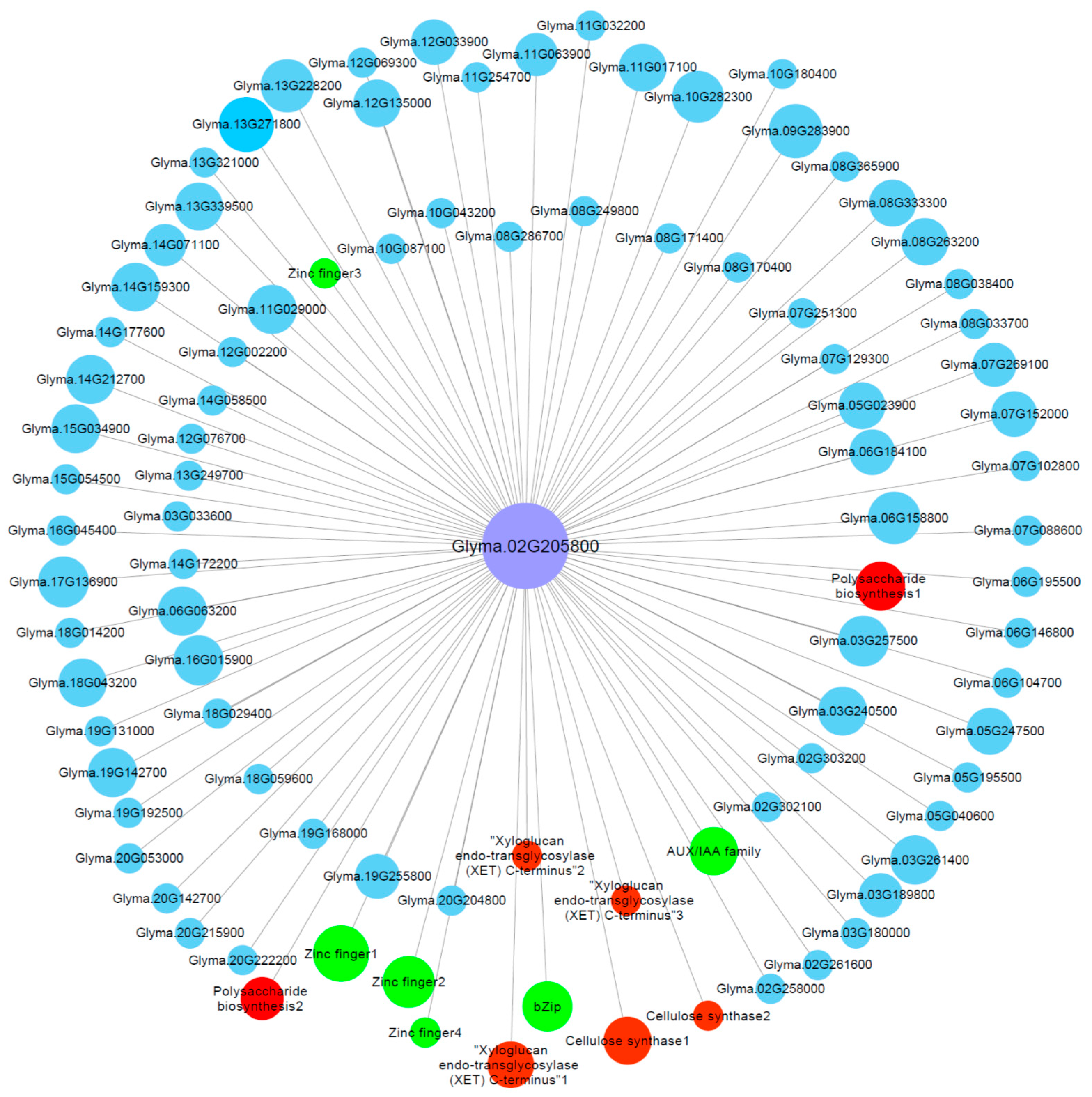
2.7. Identification of Hub Genes through Co-Expression Network
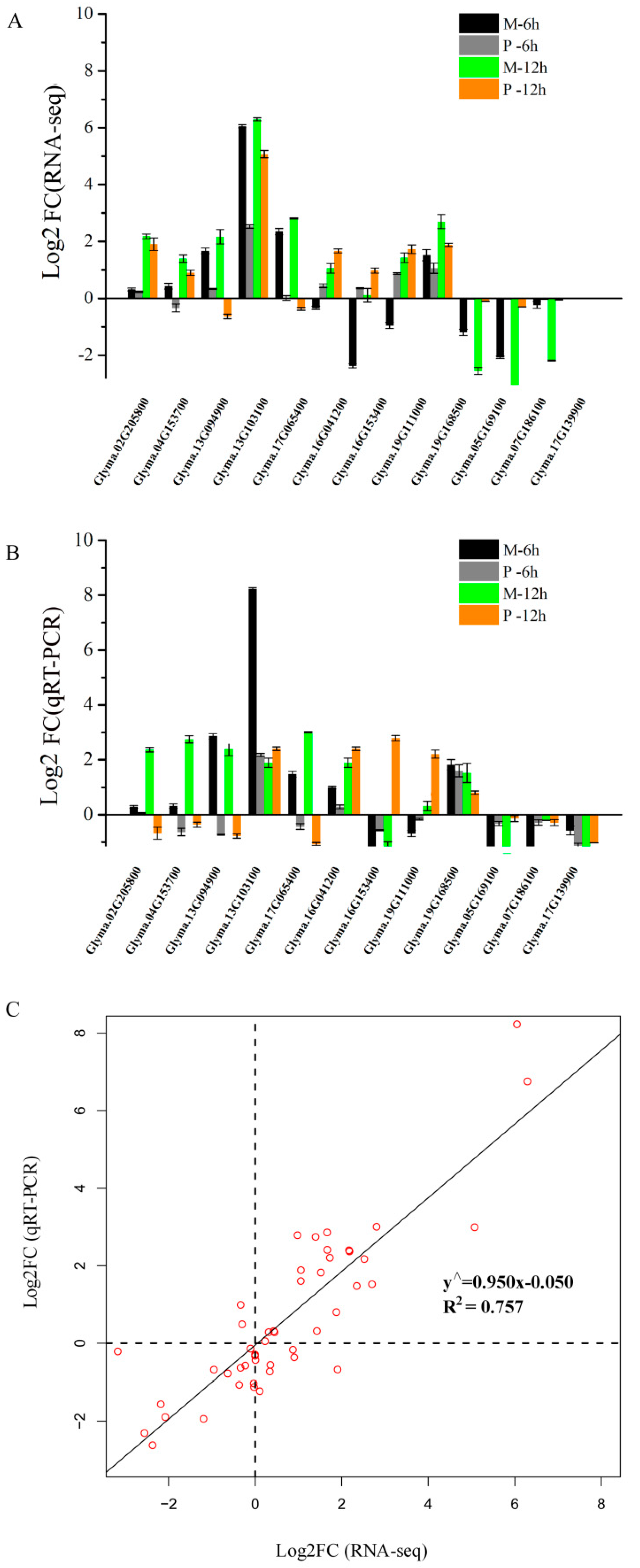
2.8. Validation of RNA-seq Data by Quantitative Real Time PCR (qRT-PCR) Analysis
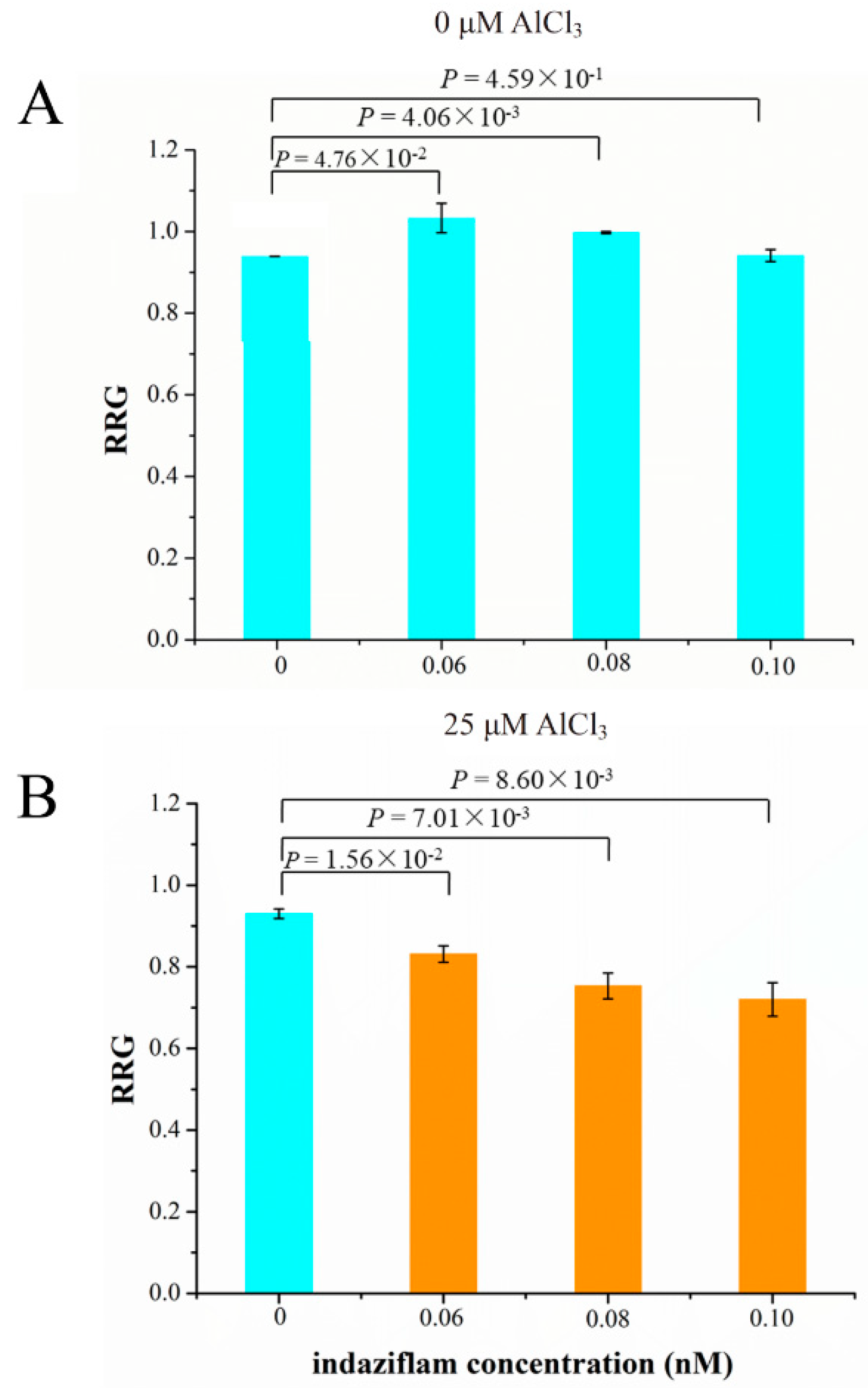
2.9. Role of Cellulose Synthase Genes in Soybean Tolerance to Al Toxicity
3. Discussion
4. Materials and Methods
4.1. Plant Growth Conditions and Stress Treatment
4.2. Root Length Measurement and Relative Root Growth
4.3. Sample Collection
4.4. RNA Isolation and RNA-seq Library Preparation
4.5. RNA-seq Analysis
4.6. Gene Annotation, Classification and Enrichment Analysis
4.7. Gene Co-Expression Network Analysis
4.8. Quantitative Real-Time PCR
4.9. Indaziflam Treatment
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ABA | abscisic acid |
| ALMT | aluminum-activated malate transporter |
| CBI | cellulose biosynthesis inhibitor |
| DEG | differently expressed gene |
| FDR | false discovery rate |
| FPKM | fragments per kilobase per million |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MATE | multidrug and toxin extrusion |
| RG | root growth |
| RRG | relative root growth |
| RNA-Seq | RNA sequencing |
| SA | salicylic acid |
| SEA | singular enrichment analysis |
| TF | transcription factor |
| XET | xyloglucan endo-transglycosylase |
References
- Uexküll, H.R.V.; Mutert, E. Global extent, development and economic impact of acid soils. Plant Soil 1995, 171, 1–15. [Google Scholar] [CrossRef]
- Kochian, L.V.; Pineros, M.A.; Liu, J.; Magalhaes, J.V. Plant adaptation to acid soils: The molecular basis for crop aluminum resistance. Annu. Rev. Plant Biol. 2015, 66, 571–598. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.Q.; Bai, J.H.; Xue, X.Q. Advances in studies on aluminum toxicity and tolerance in plants. Chin. Bull. Bot. 2001, 18, 385–395. [Google Scholar]
- Bojórquez-Quintal, E.; Escalante-Magaña, C.; Echevarría-Machado, I.; Martínez-Estévez, M. Aluminum, a friend or foe of higher plants in acid soils. Front. Plant. Sci. 2017, 8, 1767. [Google Scholar] [CrossRef] [PubMed]
- Ryan, P.R.; Tyerman, S.D.; Sasaki, T.; Furuichi, T.; Yamamoto, Y.; Zhang, W.H. The identification of aluminium-resistance genes provides opportunities for enhancing crop production on acid soils. J. Exp. Bot. 2011, 62, 9–20. [Google Scholar] [CrossRef]
- Claudio, I.; Braulio, S.; Pilar, U.; Felipe, A.; Marjorie, R. Resistance mechanisms of Aluminum (Al3+) phytotoxicity in cereals: Physiological, genetic and molecular basis. J. Soil Sci. Plant Nutr. 2008, 8, 57–71. [Google Scholar]
- Ligaba-Osena, A.; Fei, Z.; Liu, J.; Xu, Y.; Shaff, J.; Lee, S.C. Loss-of-function mutation of the calcium sensor CBL1 increases aluminum sensitivity in Arabidopsis. New Phytol. 2017, 214, 830–841. [Google Scholar] [CrossRef]
- Ikka, T.; Kobayashi, Y.; Iuchi, S.; Sakurai, N.; Shibata, D.; Kobayashi, M.; Koyama, H. Natural variation of Arabidopsis thaliana reveals that aluminum resistance and proton resistance are controlled by different genetic factors. Theor. Appl. Genet. 2007, 115, 709–719. [Google Scholar] [CrossRef]
- Marschner, H. Mechanisms of adaptation of plants to acid soil. Plant Soil 1991, 134, 1–20. [Google Scholar] [CrossRef]
- Kochian, L.V.; Hoekenga, O.A.; Pineros, M.A. How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu. Rev. Plant Biol. 2004, 55, 459–493. [Google Scholar] [CrossRef]
- Kochian, L.V. Cellular mechanisms of aluminum toxicity and resistance in plants. Annu. Rev. Plant Biol. Mol. Biol. 1995, 46, 237–260. [Google Scholar] [CrossRef]
- Ma, J.F.; Chen, Z.C.; Shen, R.F. Molecular mechanisms of Al tolerance in gramineous plants. Plant Soil 2014, 381, 1–12. [Google Scholar] [CrossRef]
- Ma, J.F.; Yamamoto, R.; Nevins, D.J.; Matsumoto, H.; Brown, P.H. Al binding in the epidermis cell wall inhibits cell elongation of okra hypocotyl. Plant Cell Physiol. 1999, 40, 549–556. [Google Scholar] [CrossRef]
- Wang, Y.; Stass, A.; Horst, W.J. Apoplastic binding of aluminum is involved in silicon-induced amelioration of aluminum toxicity in maize. Plant Physiol. 2004, 136, 3762–3770. [Google Scholar] [CrossRef]
- Chang, Y.C.; Yamamoto, Y.; Matsumoto, H. Accumulation of aluminium in the cell wall pectin in cultured tobacco (Nicotiana tabacum L.) cells treated with a combination of aluminium and iron. Plant Cell Environ. 1999, 22, 1009–1017. [Google Scholar] [CrossRef]
- Yang, J.L.; Li, Y.Y.; Zhang, Y.J.; Zhang, S.S.; Wu, Y.R.; Wu, P.; Zheng, S.J. Cell wall polysaccharides are specifically involved in the exclusion of aluminum from the rice root apex. Plant Physiol. 2008, 146, 602–611. [Google Scholar] [CrossRef]
- Yang, J.L.; Zhu, X.F.; Peng, Y.X.; Zheng, C.; Li, G.X.; Liu, Y.; Shi, Y.Z.; Zheng, S.J. Cell wall hemicellulose contributes significantly to aluminum adsorption and root growth in Arabidopsis. Plant Physiol. 2011, 155, 1885–1892. [Google Scholar] [CrossRef]
- Van, H.L.; Kuraishi, S.; Sakurai, N. Aluminum-induced rapid root inhibition and changes in cell-wall components of squash seedlings. Plant Physiol. 1994, 106, 971–976. [Google Scholar] [CrossRef]
- Ellis, C.; Karafyllidis, I.; Wasternack, C.; Turner, J.G. The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell. 2002, 14, 1557–1566. [Google Scholar] [CrossRef]
- Cano-Delgado, A.; Penfield, S.; Smith, C.; Catley, M.; Bevan, M. Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. Plant Mol. Biol. 2003, 34, 351–362. [Google Scholar] [CrossRef]
- Horst, W.J.; Wang, Y.; Eticha, D. The role of the root apoplast in aluminium-induced inhibition of root elongation and in aluminium resistance of plants: A review. Ann. Bot. 2010, 106, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.F.; Hiradate, S.; Matsumoto, H. High aluminum resistance in buckwheat II. Oxalic acid detoxifies aluminum internally. Plant Physiol. 1998, 117, 753–759. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.F.; Hiradate, S. Form of aluminium for uptake and translocation in buckwheat (Fagopyrum esculentum, Moench). Planta 2000, 211, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Shen, R.; Ma, J.F.; Kyo, M.; Iwashita, T. Compartmentation of aluminium in leaves of an Al-accumulator, Fagopyrum esculentum Moench. Planta 2002, 215, 394–398. [Google Scholar] [CrossRef] [PubMed]
- Shen, R.; Iwashita, T.; Ma, J.F. Form of Al changes with Al concentration in leaves of buckwheat. J. Exp. Bot. 2004, 55, 131–136. [Google Scholar] [CrossRef]
- Lei, G.J.; Yokosho, K.; Yamaji, N.; Fujiikashino, M.; Ma, J.F. Functional characterization of two half-size ABC transporter genes in aluminium-accumulating buckwheat. New Phytol. 2017, 215, 1080–1089. [Google Scholar] [CrossRef]
- Ma, J.F.; Ryan, P.R.; Delhaize, E. Aluminium tolerance in plants and the complexing role of organic acids. Trends Plant Sci. 2001, 6, 273–278. [Google Scholar] [CrossRef]
- Maron, L.G.; Piñeros, M.A.; Guimarães, C.T.; Magalhaes, J.V.; Pleiman, J.K.; Mao, C.Z. Two functionally distinct members of the MATE (multi-drug and toxic compound extrusion) family of transporters potentially underlie two major aluminum tolerance QTLs in maize. Plant Mol. Biol. 2010, 61, 728–740. [Google Scholar] [CrossRef]
- Yokosho, K.; Yamaji, N.; Jian, F.M. Isolation and characterisation of two MATE genes in rye. Funct. Plant Biol. 2010, 37, 296–303. [Google Scholar] [CrossRef]
- Sasaki, T.; Yamamoto, Y.; Ezaki, B.; Katsuhara, M.; Ahn, S.J.; Ryan, P.R.; Delhaize, E.; Matsumoto, H. A wheat gene encoding an aluminum-activated malate transporter. Plant J. 2004, 37, 645–653. [Google Scholar] [CrossRef]
- Ligaba, A.; Matsumoto, H. The BnALMT1 and BnALMT2 genes from rape encode aluminum-activated malate transporters that enhance the aluminum resistance of plant cells. Plant Physiol. 2006, 142, 1294–1303. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Man, Z.; Shen, S.; Fu, Y.; Sasaki, T.; Yamamoto, Y. Al-induced secretion of organic acid, gene expression and root elongation in soybean roots. Acta Physiol. Plant. 2013, 35, 223–232. [Google Scholar] [CrossRef]
- Liang, C.; Pineros, M.A.; Tian, J.; Yao, Z.; Sun, L.; Liu, J.; Shaff, J.; Coluccio, A.; Kochian, L.V.; Liao, H. Low pH, aluminum, and phosphorus coordinately regulate malate exudation through GmALMT1 to improve soybean adaptation to acid soils. Plant Physiol. 2013, 161, 1347–1361. [Google Scholar] [CrossRef] [PubMed]
- Ligaba, A.; Dreyer, I.; Margaryan, A.; Schneider, D.J.; Kochian, L.; Pineros, M. Functional, structural and phylogenetic analysis of domains underlying the Al sensitivity of the aluminum-activated malate/anion transporter, TaALMT1. Plant J. 2013, 76, 766–780. [Google Scholar] [CrossRef]
- Magalhaes, J.V.; Liu, J.; Guimarães, C.T.; Lana, U.G.; Alves, V.M.; Wang, Y.H. A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nat. Genet. 2007, 39, 1156–1161. [Google Scholar] [CrossRef]
- Liu, J.P.; Magalhaes, J.V.; Shaff, J.; Kochian, L.V. Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. Plant J. 2009, 57, 389–399. [Google Scholar] [CrossRef]
- Yokosho, K.; Yamaji, N.; Ma, J.F. An Al-inducible MATE gene is involved in external detoxification of Al in rice. Plant Mol. Biol. 2011, 68, 1061–1069. [Google Scholar] [CrossRef]
- Liu, K.S. Soybeans: Chemistry, Technology and Utilization; Springer: Berlin, Germany, 1997. [Google Scholar]
- Ying, X.F.; Liu, P.; Xu, G.; Lu, Q.; Zhu, S. Screening of soybean genotypes with tolerance to aluminum toxicity and study of the screening indices. Chin. J. Oil Crop Sci. 2005, 27, 46–51. [Google Scholar]
- Bianchihall, C.M.; Carter, T.E.; Bailey, M.A.; Mian, M.a.R.; Rufty, T.W.; Ashley, D.A.; Boerma, H.R.; Arellano, C.; Hussey, R.S.; Parrott, W.A. Aluminum tolerance associated with quantitative trait loci derived from soybean PI 416937 in hydroponics. Crop Sci. 2000, 40, 538–545. [Google Scholar] [CrossRef]
- Nian, H.; Yang, Z.; Huang, H.; Yan, X.; Matsumoto, H. Citrate secretion induced by aluminum stress may not be a key mechanism responsible for differential aluminum tolerance of some soybean genotypes. J. Plant Nutr. 2007, 27, 2047–2066. [Google Scholar] [CrossRef]
- Korir, P.C.; Zhang, J.; Wu, K. Association mapping combined with linkage analysis for aluminum tolerance among soybean cultivars released in Yellow and Changjiang River Valleys in China. Theor. Appl. Genet. 2013, 126, 1659–1675. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Shahla, S.; Baloch, K. Identification and analysis of differentially expressed genes associated with aluminum response in two soybean cultivars by cDNA-RAPD. Pak. J. Bot. 2015, 47, 711–715. [Google Scholar]
- You, J.; Zhang, H.; Liu, N.; Gao, L.; Kong, L.; Yang, Z. Transcriptomic responses to aluminum stress in soybean roots. Genome 2011, 54, 923–933. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.W.; Xu, J.M.; Jin, J.F.; Lou, H.Q.; Fan, W.; Yang, J.L. Genome-wide transcriptome analysis reveals conserved and distinct molecular mechanisms of al resistance in buckwheat (fagopyrum esculentum moench) leaves. Int. J. Mol. Sci. 2017, 18, 1859. [Google Scholar] [CrossRef]
- Tsutsui, T.; Yamaji, N.; Huang, C.F.; Motoyama, R.; Nagamura, Y.; Ma, J.F. Comparative genome-wide transcriptional analysis of Al-responsive genes reveals novel Al tolerance mechanisms in rice. PLoS ONE 2012, 7, e48197. [Google Scholar] [CrossRef]
- Yamaji, N.; Huang, C.F.; Nagao, S. A zinc finger transcription factor ART1 regulates multiple genes implicated in aluminum tolerance in rice. Plant Cell 2009, 21, 3339–3349. [Google Scholar] [CrossRef]
- Yang, Z.B.; Eticha, D.; Rotter, B. Physiological and molecular analysis of polyethylene glycol-induced reduction of aluminium accumulation in the root tips of common bean. J. Exp. Bot. 2012, 63, 3109–3125. [Google Scholar] [CrossRef]
- Li, H.; Wang, B.; Zhang, Q.; Wang, J.; King, G.B. Genome-wide analysis of the auxin / indoleacetic acid, (Aux/IAA) gene family in allotetraploid rapeseed (Brassica napus, L.). BMC Plant Biol. 2017, 17, 204. [Google Scholar] [CrossRef]
- Shani, E.; Salehin, M.; Zhang, Y. Plant stress tolerance requires auxin-sensitive aux/iaa transcriptional repressors. Curr. Biol. 2017, 27, 437–444. [Google Scholar] [CrossRef]
- Brabham, C.; Lei, L.; Gu, Y.; Stork, J.; Barrett, M.; Debolt, S. Indaziflam herbicidal action: A potent cellulose biosynthesis inhibitor. Plant Physiol. 2014, 166, 1177–1185. [Google Scholar] [CrossRef]
- Singh, S.; Tripathi, D.K.; Singh, S.; Sharma, S.; Dubey, N.K.; Chauhan, D.K. Toxicity of aluminium on various levels of plant cells and organism: A review. Environ. Exp. Bot. 2007, 137, 177–193. [Google Scholar] [CrossRef]
- Teraoka, T.; Kaneko, M.S.; Yoshimura, E. Aluminum rapidly inhibits cellulose synthesis in roots of barley and wheat seedlings. J. Plant Physiol. 2002, 159, 17–23. [Google Scholar] [CrossRef]
- Burn, J.E.; Hurley, U.A.; Birch, R.J.; Arioli, T.; Cork, A.; Williamson, R.E. The cellulose-deficient Arabidopsis mutant rsw3 is defective in a gene encoding a putative glucosidase II, an enzyme processing N-glycans during ER quality control. Plant J. 2002, 32, 949–960. [Google Scholar] [CrossRef] [PubMed]
- Ambroise, V.; Legay, S.; Guerriero, G.; Hausman, J.F.; Cuypers, A.; Sergeant, K. The roots of plant frost hardiness and tolerance. Plant Cell Physiol. 2020, 61, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Arisha, M.H.; Aboelnasr, H.; Ahmad, M.Q.; Liu, Y.; Tang, W.; Gao, R.; Yan, H.; Kou, M.; Wang, X.; Zhang, Y.; et al. Transcriptome sequencing and whole genome expression profiling of hexaploid sweetpotato under salt stress. BMC Genom. 2020, 21, 197. [Google Scholar] [CrossRef]
- Wang, T.; McFarlane, H.E.; Persson, S. The impact of abiotic factors on cellulose synthesis. J. Exp. Bot. 2016, 67, 543–552. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.F.; Shi, Y.Z.; Lei, G.J.; Fry, S.C.; Zhang, B.C.; Zhou, Y.H. XTH31, encoding an in vitro xeh/xet-active enzyme, regulates aluminum sensitivity by modulating in vivo xet action, cell wall xyloglucan content, and aluminum binding capacity in Arabidopsis. Plant Cell 2012, 24, 4731–4747. [Google Scholar] [CrossRef]
- Finkelstein, R.R.; Wang, M.L.; Lynch, T.J.; Rao, S.; Goodman, H.M. The Arabidopsis Abscisic Acid Response Locus ABI4 Encodes an APETALA2 Domain Protein. Plant Cell 1998, 10, 1043–1054. [Google Scholar] [CrossRef]
- Pandey, P.; Srivastava, R.K.; Dubey, R.S. Salicylic acid alleviates aluminum toxicity in rice seedlings better than magnesium and calcium by reducing aluminum uptake, suppressing oxidative damage and increasing antioxidative defense. Ecotoxicology 2013, 22, 656–670. [Google Scholar] [CrossRef]
- Zhang, G.; Chen, M.; Chen, X.; Xu, Z.; Guan, S.; Li, L.C.; Li, A.; Guo, J.; Mao, L.; Ma, Y. Phylogeny, gene structures, and expression patterns of the ERF gene family in soybean (Glycine max L.). J. Exp. Bot. 2008, 59, 4095–4107. [Google Scholar] [CrossRef]
- Jisha, V.; Dampanaboina, L.; Vadasser, J.; Mithöfer, A.; Kappara, S.; Ramanan, R. Overexpression of an AP2/ERF Type transcription factor oserebp1 confers biotic and abiotic stress tolerance in rice. PLoS ONE 2015, 10, e0127831. [Google Scholar] [CrossRef]
- Reyna-Llorens, I.; Corrales, I.; Poschenrieder, C.; Barcelo, J.; Cruz-Ortega, R. Both aluminum and ABA induce the expression of an ABC-like transporter gene (FeALS3) in the Al-tolerant species Fagopyrum esculentum. Environ. Exp. Bot. 2015, 111, 74–82. [Google Scholar] [CrossRef]
- Wang, P.; Yu, W.; Zhang, J.; Rengel, Z.; Xu, J.; Han, Q.; Chen, L.; Li, K.; Yu, Y.; Chen, Q. Auxin enhances aluminium-induced citrate exudation through upregulation of GmMATE and activation of the plasma membrane H+-ATPase in soybean roots. Ann. Bot. 2016, 118, 933–940. [Google Scholar] [CrossRef] [PubMed]
- Mattiello, L.; Kirst, M.; Silva, F.R.D.; Jorge, R.A.; Menossi, M. Transcriptional profile of maize roots under acid soil growth. BMC Plant Biol. 2010, 10, 196. [Google Scholar] [CrossRef]
- Goodwin, S.B.; Sutter, T.R. Microarray analysis of Arabidopsis genome response to aluminum stress. Biol. Plant. 2009, 53, 85–99. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, Q.L.; Geng, M.J.; Guo, Z.H.; Zhao, Z.Q. Effect of indole-3-acetic acid on aluminum-induced efflux of malic acid from wheat (Triticum aestivum L). Plant Soil 2011, 346, 215–230. [Google Scholar] [CrossRef]
- Huang, C.F.; Yamaji, N.; Mitani, N.; Yano, M.; Nagamura, Y.; Ma, J.F. A bacterial-type ABC transporter is involved in aluminum tolerance in rice. Plant Cell 2009, 21, 655–667. [Google Scholar] [CrossRef]
- Houde, M.; Diallo, A.O. Identification of genes and pathways associated with aluminum stress and tolerance using transcriptome profiling of wheat near-isogenic lines. BMC Genom. 2008, 9, 400. [Google Scholar] [CrossRef]
- Liu, J.; Li, Y.; Wang, W.; Gai, J.; Li, Y. Genome-wide analysis of MATE transporters and expression patterns of a subgroup of MATE genes in response to aluminum toxicity in soybean. BMC Genom. 2016, 17, 223. [Google Scholar] [CrossRef]
- Wu, W.; Lin, Y.; Chen, Q. Functional conservation and divergence of soybean GmStop1 members in proton and aluminum tolerance. Front Plant Sci. 2018, 9, 570–594. [Google Scholar] [CrossRef]
- Famoso, A.N.; Zhao, K.; Clark, R.T.; Tung, C.W.; Wright, M.H.; Bustamante, C. Genetic architecture of aluminum tolerance in rice (Oryza sativa) determined through genome-wide association analysis and QTL mapping. PLoS Genet. 2011, 7, e1002221. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Cock, P.J.; Fields, C.J.; Goto, N.; Heuer, M.L.; Rice, P.M. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2010, 38, 1767–1771. [Google Scholar] [CrossRef]
- Cox, M.P.; Peterson, D.A.; Biggs, P.J. SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinform. 2010, 11, 485. [Google Scholar] [CrossRef]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Ghosh, S.; Chan, C.K. Analysis of RNA-Seq data using tophat and cufflinks. Methods Mol. Biol. 2016, 1374, 339–361. [Google Scholar]
- Trapnell, C.; Hendrickson, D.G.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Pachter, L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013, 31, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, A.M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef]
- Jin, J.; Zhang, H.; Kong, L.; Gao, G.; Luo, J. PlantTFDB 3.0: A portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2014, 42, 1182–1187. [Google Scholar] [CrossRef]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Johnson, L.S.; Eddy, S.R.; Portugaly, E. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinform. 2010, 11, 431. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Zhou, X.; Ling, Y.; Zhang, Z.; Su, Z. agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010, 38, W64–W70. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Ernst, J.; Bar-Joseph, Z. STEM: A tool for the analysis of short time series gene expression data. BMC Bioinform. 2006, 7, 191. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, L.; Cui, J.; Cai, Y.; Yang, S.; Liu, J.; Wang, W.; Gai, J.; Hu, Z.; Li, Y. Comparative Transcriptome Analysis of Two Contrasting Soybean Varieties in Response to Aluminum Toxicity. Int. J. Mol. Sci. 2020, 21, 4316. https://doi.org/10.3390/ijms21124316
Zhao L, Cui J, Cai Y, Yang S, Liu J, Wang W, Gai J, Hu Z, Li Y. Comparative Transcriptome Analysis of Two Contrasting Soybean Varieties in Response to Aluminum Toxicity. International Journal of Molecular Sciences. 2020; 21(12):4316. https://doi.org/10.3390/ijms21124316
Chicago/Turabian StyleZhao, Lijuan, Jingjing Cui, Yuanyuan Cai, Songnan Yang, Juge Liu, Wei Wang, Junyi Gai, Zhubing Hu, and Yan Li. 2020. "Comparative Transcriptome Analysis of Two Contrasting Soybean Varieties in Response to Aluminum Toxicity" International Journal of Molecular Sciences 21, no. 12: 4316. https://doi.org/10.3390/ijms21124316
APA StyleZhao, L., Cui, J., Cai, Y., Yang, S., Liu, J., Wang, W., Gai, J., Hu, Z., & Li, Y. (2020). Comparative Transcriptome Analysis of Two Contrasting Soybean Varieties in Response to Aluminum Toxicity. International Journal of Molecular Sciences, 21(12), 4316. https://doi.org/10.3390/ijms21124316






