Involvement of POLA2 in Double Strand Break Repair and Genotoxic Stress
Abstract
1. Introduction
2. Results
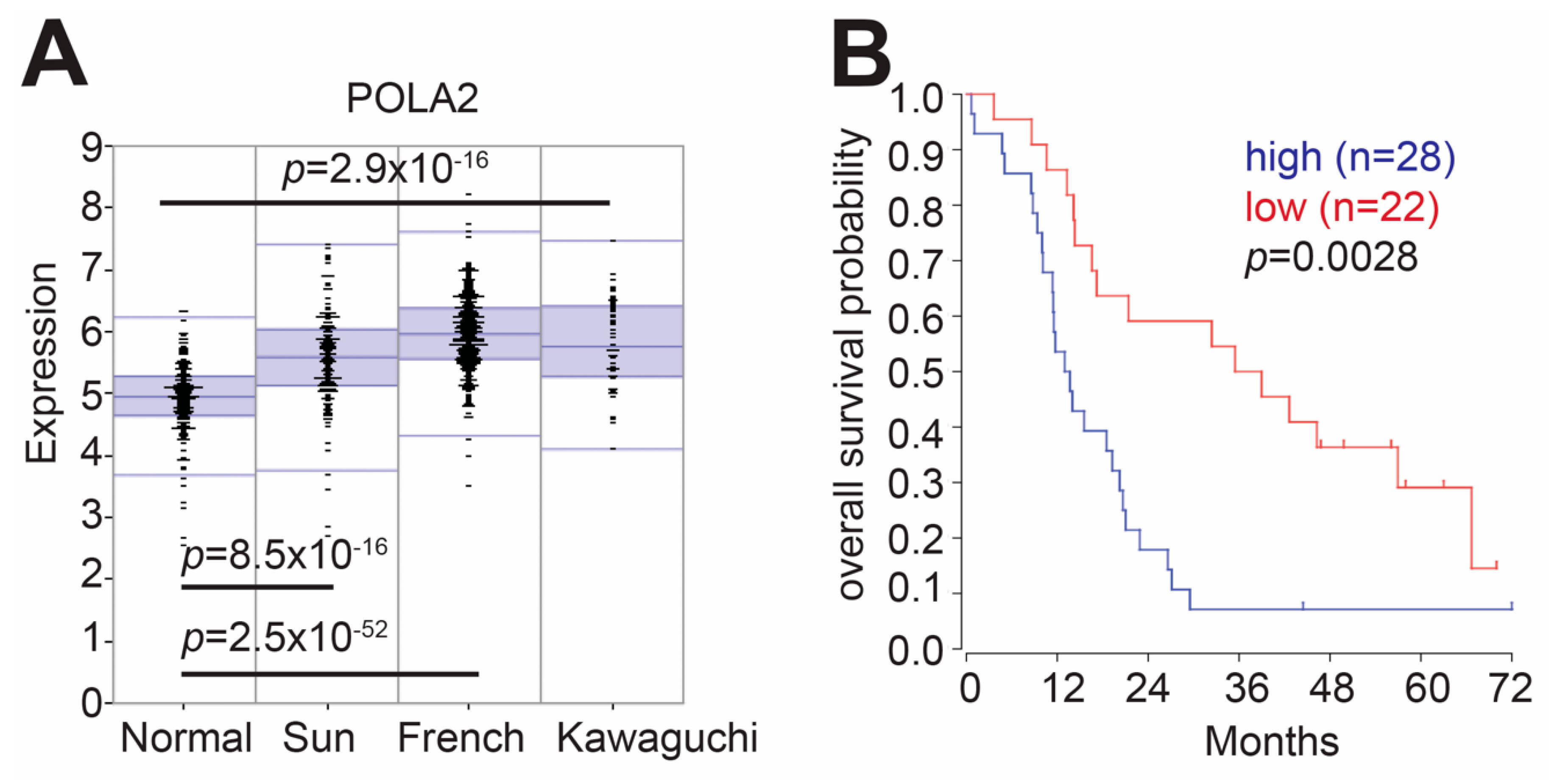
2.1. Elevated POLA2 Expression Is Detrimental to GBM Patients
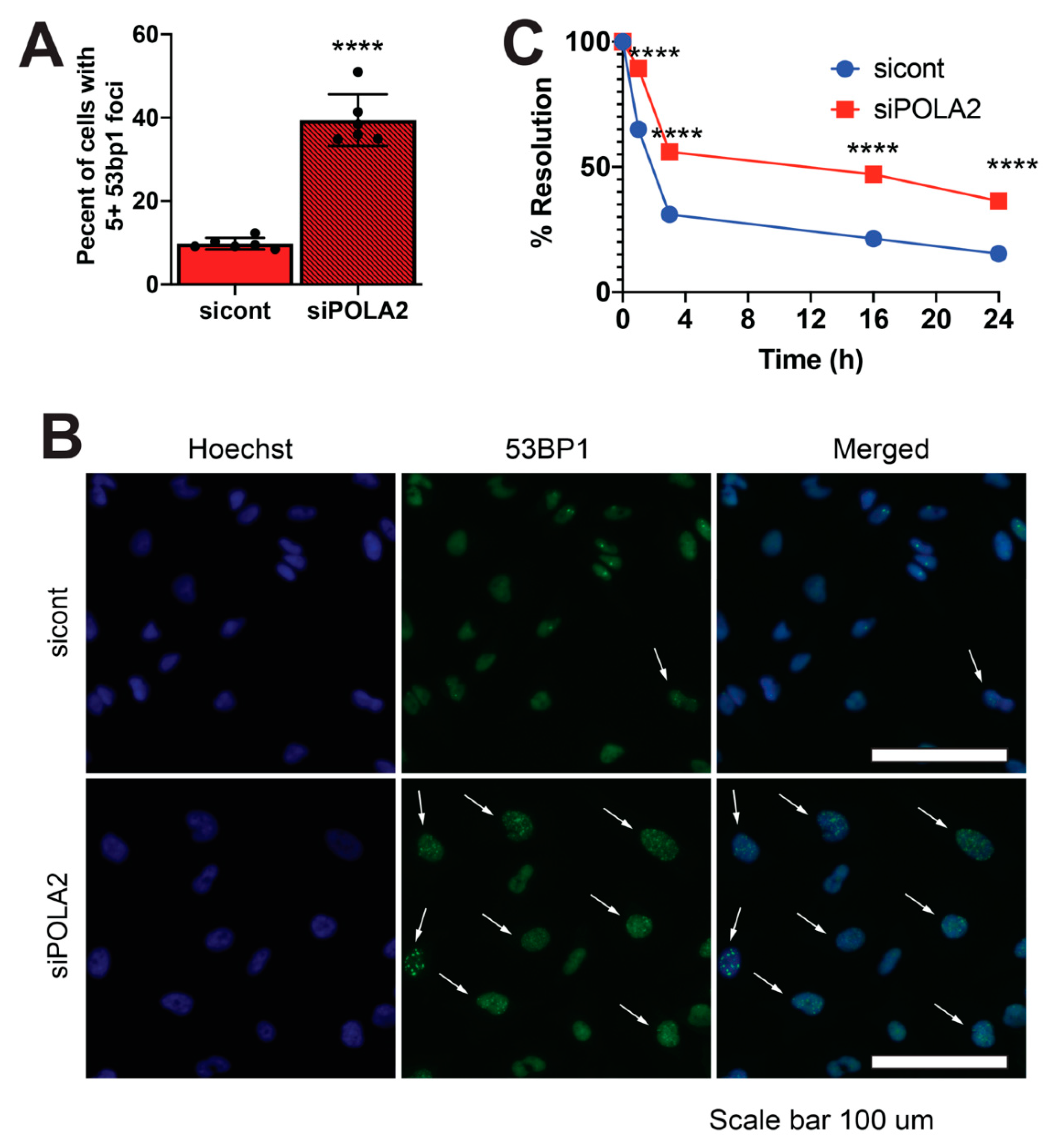
2.2. Loss of POLA2 Leads to Increased Spontaneous DSB Formation and Impairs DSB Repair Rates
2.3. POLA2 Loss Sensitizes Cells to Genomic Insult
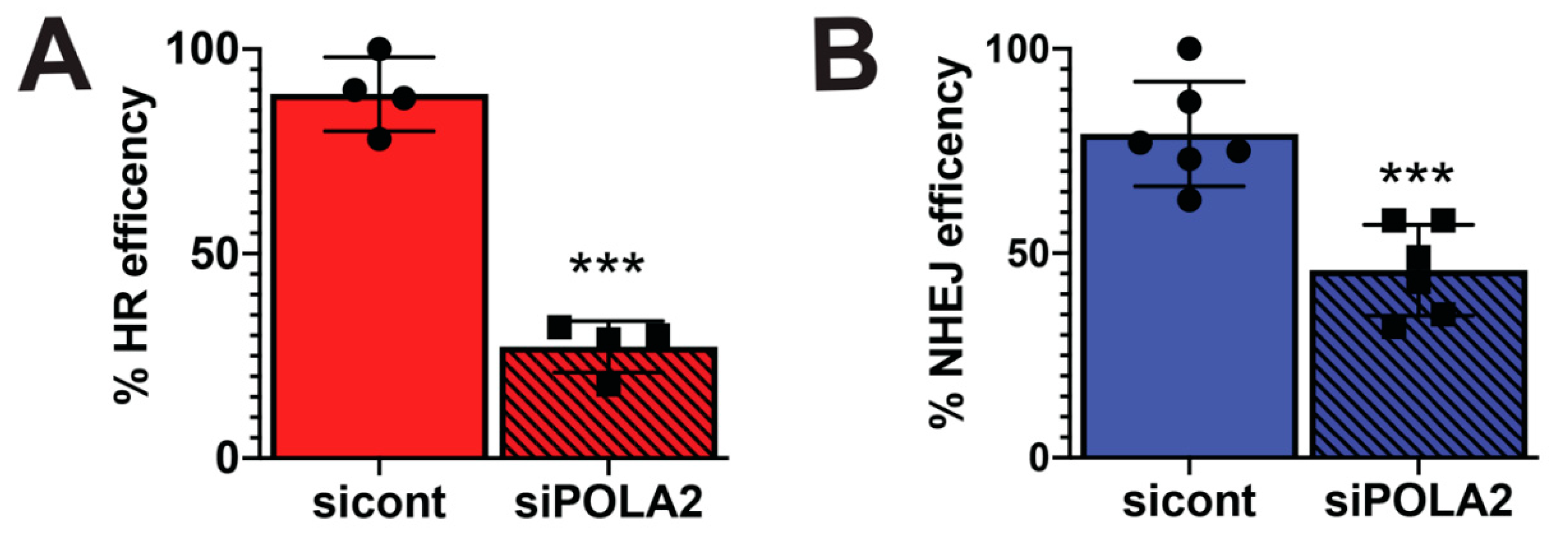
2.4. Loss of POLA2 Inhibits HR and NHEJ Repair
3. Discussion
3.1. Connecting POLA2 and the DNA Damage Response
3.2. POLA2 as a Therapeutic Target
4. Materials and Methods
- siPOLA2, Sigma Aldrich (St. Louis, MO, USA),
- SASI_Hs02_00334255, CUAUGAGUCGUUCUAUGUU
- siPOLA2, Sigma Aldrich (St. Louis, MO, USA),
- SASI_Hs01_00177968, CAUUCAAGAGCUAAUUGAA
- siPOLA2, Sigma Aldrich (St. Louis, MO, USA),
- SASI_Hs01_00177970, CUGAACAACAAGUCAGUGA
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Fragkos, M.; Ganier, O.; Coulombe, P.; Méchali, M. DNA replication origin activation in space and time. Nat. Rev. Mol. Cell Biol. 2015, 16, 360–374. [Google Scholar] [CrossRef] [PubMed]
- Núñez-Ramírez, R.; Klinge, S.; Sauguet, L.; Melero, R.; Recuero-Checa, M.A.; Kilkenny, M.; Perera, R.L.; García-Alvarez, B.; Hall, R.J.; Nogales, E.; et al. Flexible tethering of primase and DNA Pol α in the eukaryotic primosome. Nucleic Acids Res. 2011, 39, 8187–8199. [Google Scholar] [CrossRef] [PubMed]
- Baranovskiy, A.G.; Duong, V.N.; Babayeva, N.D.; Zhang, Y.; Pavlov, Y.I.; Anderson, K.S.; Tahirov, T.H. Activity and fidelity of human DNA polymerase α depend on primer structure. J. Biol. Chem. 2018, 293, 6824–6843. [Google Scholar] [CrossRef] [PubMed]
- Reijns, M.A.M.; Kemp, H.; Ding, J.; Marion de Procé, S.; Jackson, A.P.; Taylor, M.S. Lagging-strand replication shapes the mutational landscape of the genome. Nature 2015, 518, 502–506. [Google Scholar] [CrossRef] [PubMed]
- Coloma, J.; Johnson, R.E.; Prakash, L.; Prakash, S.; Aggarwal, A.K. Human DNA polymerase α in binary complex with a DNA: DNA template-primer. Sci. Rep. 2016, 6, 23784. [Google Scholar] [CrossRef]
- Macheret, M.; Halazonetis, T.D. DNA Replication Stress as a Hallmark of Cancer. Annu. Rev. of Pathol. Mech. Dis. 2015, 10, 425–448. [Google Scholar] [CrossRef] [PubMed]
- Blank, A.; Kim, B.; Loeb, L.A. DNA polymerase delta is required for base excision repair of DNA methylation damage in Saccharomyces cerevisiae. Proc. Natl. Acad Sci. USA 1994, 91, 9047–9051. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.R.; Jiang, Y.; Zhang, S.J.; Hao, H.; Lee, M.Y. DNA polymerase delta is involved in the cellular response to UV damage in human cells. J. Biol. Chem. 1994, 269, 13748–13751. [Google Scholar]
- Moiseeva, T.N.; Gamper, A.M.; Hood, B.L.; Conrads, T.P.; Bakkenist, C.J. Human DNA polymerase ε is phosphorylated at serine-1940 after DNA damage and interacts with the iron-sulfur complex chaperones CIAO1 and MMS19. DNA Repair 2016, 43, 9–17. [Google Scholar] [CrossRef]
- Feng, W.; D’Urso, G. Schizosaccharomyces pombe cells lacking the amino-terminal catalytic domains of DNA polymerase epsilon are viable but require the DNA damage checkpoint control. Mol. Cell. Biol. 2001, 21, 4495–4504. [Google Scholar] [CrossRef]
- Navas, T.A.; Sanchez, Y.; Elledge, S.J. RAD9 and DNA polymerase epsilon form parallel sensory branches for transducing the DNA damage checkpoint signal in Saccharomyces cerevisiae. Genes Dev. 1996, 10, 2632–2643. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Chao, H.H.; Wang, X.; Zhang, Z.; Lee, E.Y.C.; Lee, M.Y.W.T. Loss of the p12 subunit of DNA polymerase delta leads to a defect in HR and sensitization to PARP inhibitors. DNA Repair 2019, 73, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.A.; Kwon, Y.; Xu, Y.; Chung, W.-H.; Chi, P.; Niu, H.; Mayle, R.; Chen, X.; Malkova, A.; Sung, P.; et al. Pif1 helicase and Polδ promote recombination-coupled DNA synthesis via bubble migration. Nature 2013, 502, 393–396. [Google Scholar] [CrossRef] [PubMed]
- Rogers, R.F.; Walton, M.I.; Cherry, D.L.; Collins, I.; Clarke, P.A.; Garrett, M.D.; Workman, P. CHK1 inhibition is synthetically lethal with loss of B-family DNA polymerase function in human lung and colorectal cancer cells. Cancer Res. 2020, 80, 1735–1747. [Google Scholar] [CrossRef] [PubMed]
- Røe, O.D.; Szulkin, A.; Anderssen, E.; Flatberg, A.; Sandeck, H.; Amundsen, T.; Erlandsen, S.E.; Dobra, K.; Sundstrøm, S.H. Molecular Resistance Fingerprint of Pemetrexed and Platinum in a Long-Term Survivor of Mesothelioma. PLoS ONE 2012, 7, e40521. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, A.; Yajima, N.; Tsuchiya, N.; Homma, J.; Sano, M.; Natsumeda, M.; Takahashi, H.; Fujii, Y.; Kakuma, T.; Yamanaka, R. Gene expression signature-based prognostic risk score in patients with glioblastoma. Cancer Sci. 2013, 104, 1205–1210. [Google Scholar] [CrossRef]
- Sun, L.; Hui, A.-M.; Su, Q.; Vortmeyer, A.; Kotliarov, Y.; Pastorino, S.; Passaniti, A.; Menon, J.; Walling, J.; Bailey, R.; et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell 2006, 9, 287–300. [Google Scholar] [CrossRef]
- Gravendeel, L.A.M.; Kouwenhoven, M.C.M.; Gevaert, O.; de Rooi, J.J.; Stubbs, A.P.; Duijm, J.E.; Daemen, A.; Bleeker, F.E.; Bralten, L.B.C.; Kloosterhof, N.K.; et al. Intrinsic Gene Expression Profiles of Gliomas Are a Better Predictor of Survival than Histology. Cancer Res. 2009, 69, 9065. [Google Scholar] [CrossRef]
- Morales, J.C.; Richard, P.; Rommel, A.; Fattah, F.J.; Motea, E.A.; Patidar, P.L.; Xiao, L.; Leskov, K.; Wu, S.-Y.; Hittelman, W.N.; et al. Kub5-Hera, the human Rtt103 homolog, plays dual functional roles in transcription termination and DNA repair. Nucleic Acids Res. 2014, 42, 4996–5006. [Google Scholar] [CrossRef]
- Morales, J.C.; Richard, P.; Patidar, P.L.; Motea, E.A.; Dang, T.T.; Manley, J.L.; Boothman, D.A. XRN2 Links Transcription Termination to DNA Damage and Replication Stress. PLoS Genet. 2016, 12, e1006107. [Google Scholar] [CrossRef]
- Gil del Alcazar, C.R.; Todorova, P.K.; Habib, A.A.; Mukherjee, B.; Burma, S. Augmented HR Repair Mediates Acquired Temozolomide Resistance in Glioblastoma. Mol. Cancer Res. 2016, 14, 928. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.-X.; Weaver, Z.; Xu, X.; Li, C.; Weinstein, M.; Chen, L.; Guan, X.-Y.; Ried, T.; Deng, C.-X. A targeted disruption of the murine Brca1 gene causes γ-irradiation hypersensitivity and genetic instability. Oncogene 1998, 17, 3115–3124. [Google Scholar] [CrossRef] [PubMed]
- Ziv, Y.; Bar-Shira, A.; Pecker, I.; Russell, P.; Jorgensen, T.J.; Tsarfati, I.; Shiloh, Y. Recombinant ATM protein complements the cellular A-T phenotype. Oncogene 1997, 15, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Morales, J.C.; Xia, Z.; Lu, T.; Aldrich, M.B.; Wang, B.; Rosales, C.; Kellems, R.E.; Hittelman, W.N.; Elledge, S.J.; Carpenter, P.B. Role for the BRCA1 C-terminal Repeats (BRCT) Protein 53BP1 in Maintaining Genomic Stability. J. Biol. Chem. 2003, 278, 14971–14977. [Google Scholar] [CrossRef] [PubMed]
- Nussenzweig, A.; Sokol, K.; Burgman, P.; Li, L.; Li, G.C. Hypersensitivity of Ku80-deficient cell lines and mice to DNA damage: The effects of ionizing radiation on growth, survival, and development. Proc. Natl. Acad. Sci. USA 1997, 94, 13588. [Google Scholar] [CrossRef] [PubMed]
- Bryant, H.E.; Schultz, N.; Thomas, H.D.; Parker, K.M.; Flower, D.; Lopez, E.; Kyle, S.; Meuth, M.; Curtin, N.J.; Helleday, T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005, 434, 913–917. [Google Scholar] [CrossRef]
- Jones, P.; Altamura, S.; Boueres, J.; Ferrigno, F.; Fonsi, M.; Giomini, C.; Lamartina, S.; Monteagudo, E.; Ontoria, J.M.; Orsale, M.V.; et al. Discovery of 2-{4-[(3S)-Piperidin-3-yl]phenyl}-2H-indazole-7-carboxamide (MK-4827): A Novel Oral Poly(ADP-ribose)polymerase (PARP) Inhibitor Efficacious in BRCA-1 and -2 Mutant Tumors. J. Med. Chem. 2009, 52, 7170–7185. [Google Scholar] [CrossRef]
- Gunn, A.; Stark, J.M. I-SceI-Based Assays to Examine Distinct Repair Outcomes of Mammalian Chromosomal Double Strand Breaks. In DNA Repair Protocols; Bjergbæk, L., Ed.; Humana Press: Totowa, NJ, USA, 2012; pp. 379–391. [Google Scholar]
- Sollier, J.; Stork, C.T.; García-Rubio, M.L.; Paulsen, R.D.; Aguilera, A.; Cimprich, K.A. Transcription-Coupled Nucleotide Excision Repair Factors Promote R-Loop-Induced Genome Instability. Mol. Cell 2014, 56, 777–785. [Google Scholar] [CrossRef]
- Zhou, B.-B.S.; Elledge, S.J. The DNA damage response: Putting checkpoints in perspective. Nature 2000, 408, 433–439. [Google Scholar] [CrossRef]
- Tykocki, T.; Eltayeb, M. Ten-year survival in glioblastoma. A systematic review. J. Clin. Neurosci. 2018, 54, 7–13. [Google Scholar] [CrossRef]
- Ron, B.; Asna, N.; Pamela, S.; Nicole, F.; Moshe, S. Glioblastoma Multiforme, Diagnosis and Treatment; Recent Literature Review. Curr. Med. Chem. 2017, 24, 3002–3009. [Google Scholar] [CrossRef]
- Roy, S.; Lahiri, D.; Maji, T.; Biswas, J. Recurrent Glioblastoma: Where we stand. South Asian J. Cancer 2015, 4, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Wen, P.Y.; Kesari, S. Malignant Gliomas in Adults. N. Engl. J. Med. 2008, 359, 492–507. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, Y.P.; Weatherbee, J.L.; Wheelhouse, R.T.; Ross, A.H. Glioblastoma multiforme therapy and mechanisms of resistance. Pharmaceuticals 2013, 6, 1475–1506. [Google Scholar] [CrossRef]
- Erasimus, H.; Gobin, M.; Niclou, S.; Van Dyck, E. DNA repair mechanisms and their clinical impact in glioblastoma. Mutat. Res. Rev. Mutat. Res. 2016, 769, 19–35. [Google Scholar] [CrossRef]
- Pearson, G.W.; Hunter, T. Real-time imaging reveals that noninvasive mammary epithelial acini can contain motile cells. J. Cell Biol. 2007, 179, 1555–1567. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dang, T.T.; Morales, J.C. Involvement of POLA2 in Double Strand Break Repair and Genotoxic Stress. Int. J. Mol. Sci. 2020, 21, 4245. https://doi.org/10.3390/ijms21124245
Dang TT, Morales JC. Involvement of POLA2 in Double Strand Break Repair and Genotoxic Stress. International Journal of Molecular Sciences. 2020; 21(12):4245. https://doi.org/10.3390/ijms21124245
Chicago/Turabian StyleDang, Tuyen T., and Julio C. Morales. 2020. "Involvement of POLA2 in Double Strand Break Repair and Genotoxic Stress" International Journal of Molecular Sciences 21, no. 12: 4245. https://doi.org/10.3390/ijms21124245
APA StyleDang, T. T., & Morales, J. C. (2020). Involvement of POLA2 in Double Strand Break Repair and Genotoxic Stress. International Journal of Molecular Sciences, 21(12), 4245. https://doi.org/10.3390/ijms21124245




