Microorganisms and Their Metabolic Capabilities in the Context of the Biogeochemical Nitrogen Cycle at Extreme Environments
Abstract
1. Introduction
2. Classification of Extreme Microorganisms
3. Extreme Microorganisms in the Context of Biogeochemical Nitrogen Cycle
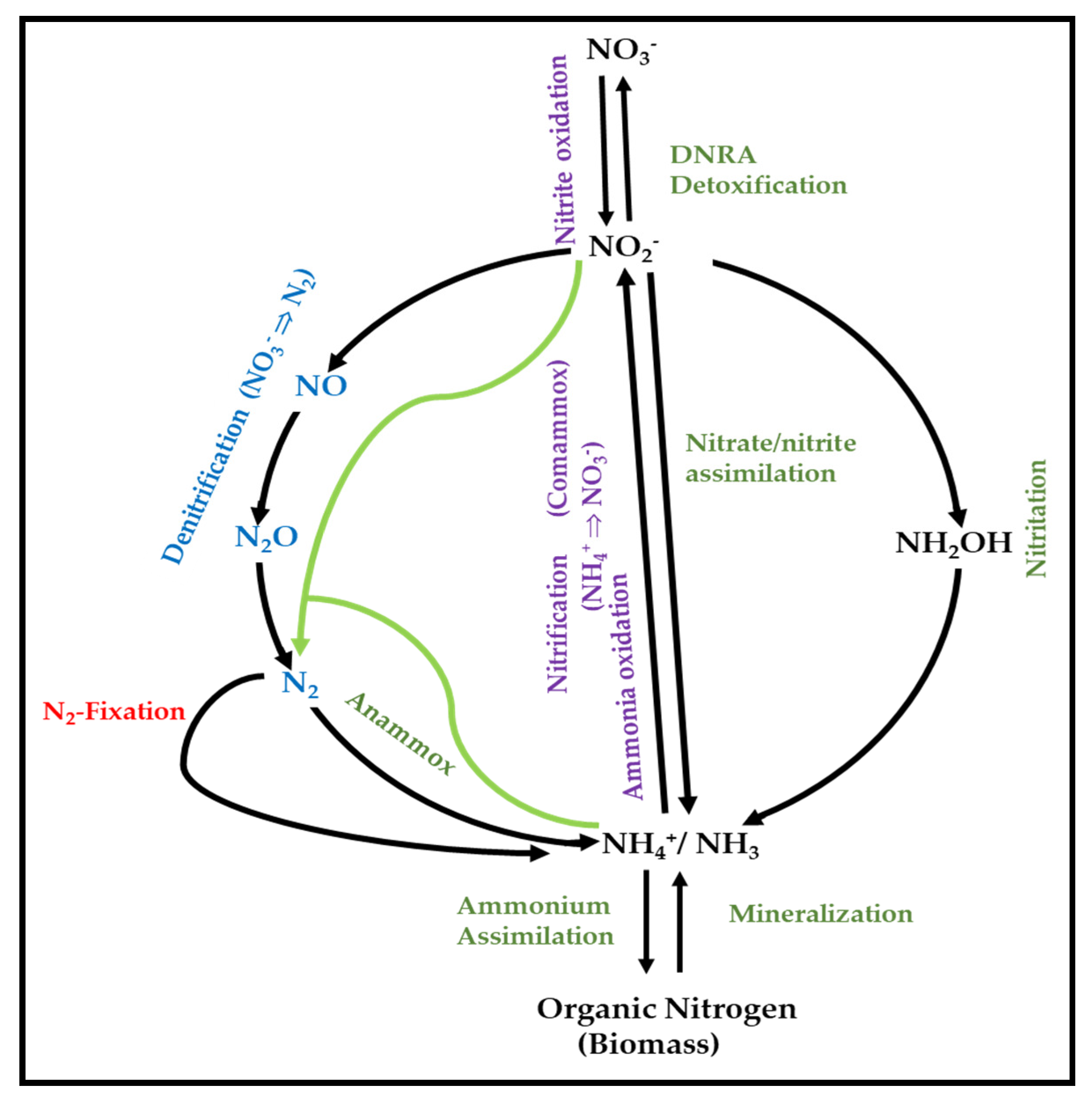
3.1. General Overview of the Role of Microorganisms in N-Cycle
3.2. Metabolic Pathways of N-Cycle Carried out by Extremophiles
4. Potential Applications of N-Cycle Pathways Driven by Extremophiles in Biotechnology and in Studies on Climate Change and Environmental Global Warming
4.1. Wastewater Treatments and Bioremediation
4.2. Environmental Studies
4.3. N-Cycle Enzymes
5. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Rothschild, L.J.; Mancinelli, R.L. Life in extreme environments. Nature 2001, 409, 1092–1101. [Google Scholar] [CrossRef] [PubMed]
- Rampelotto, P.H. Extremophiles and extreme environments. Life 2013, 3, 482–485. [Google Scholar] [CrossRef] [PubMed]
- Maccario, L.; Sanguino, L.; Vogel, T.M.; Larose, C. Snow and ice ecosystems: Not so extreme. Res. Microbiol. 2015, 166, 782–795. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Cen, Z.; Zhao, J. The survival mechanisms of thermophiles at high temperatures: An angle of omics. Physiology 2015, 30, 97–106. [Google Scholar] [CrossRef]
- Feller, G. Protein folding at extreme temperatures: Current issues. Semin. Cell. Dev. Biol. 2018, 84, 129–137. [Google Scholar] [CrossRef]
- Gunde-Cimerman, N.; Plemenitaš, A.; Oren, A. Strategies of adaptation of microorganisms of the three domains of life to high salt concentrations. FEMS Microbiol. Rev. 2018, 42, 353–375. [Google Scholar] [CrossRef]
- Altinisik, K.F.E.; Avci, F.G.; Sayar, N.A.; Kazan, D.; Sayar, A.A.; Akbulut, S.B. What are the multi-omics mechanisms for adaptation by microorganisms to high alkalinity? A transcriptomic and proteomic study of a Bacillus strain with industrial potential. Omics 2018, 22, 717–732. [Google Scholar] [CrossRef]
- Adam, P.S.; Borrel, G.; Brochier-Armanet, C.; Gribaldo, S. The growing tree of Archaea: New perspectives on their diversity, evolution and ecology. ISME J. 2017, 11, 2407–2425. [Google Scholar] [CrossRef]
- Sayed, A.M.; Hassan, M.H.A.; Alhadrami, H.A.; Hassan, H.M.; Goodfellow, M.; Rateb, M.E. Extreme environments: Microbiology leading to specialized metabolites. J. Appl. Microbiol. 2020, 128, 630–657. [Google Scholar] [CrossRef]
- Gribaldo, S.; Brochier-Armanet, C. The origin and evolution of Archaea: A state of the art. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2006, 361, 1007–1022. [Google Scholar] [CrossRef] [PubMed]
- Camprubi, E.; Jordan, S.F.; Vasiliadou, R.; Lane, N. Iron catalysis at the origin of life. IUBMB Life 2017, 69, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Goldford, J.E.; Hartman, H.; Marsland, R.; Segrè, D. Environmental boundary conditions for the origin of life converge to an organo-sulfur metabolism. Nat. Ecol. Evol. 2019, 3, 1715–1724. [Google Scholar] [CrossRef] [PubMed]
- Martin, W.; Russell, M.J. On the origin of biochemistry at an alkaline hydrothermal vent. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2007, 362, 1887–1925. [Google Scholar] [CrossRef] [PubMed]
- Sousa, F.L.; Thiergart, T.; Landan, G.; Nelson-Sathi, S.; Pereira, I.A.; Allen, J.F.; Lane, N.; Martin, W.F. Early bioenergetic evolution. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2013, 368, 20130088. [Google Scholar] [CrossRef]
- Pikuta, E.V.; Hoover, R.B.; Tang, J. Microbial extremophiles at the limits of life. Crit. Rev. Microbiol. 2007, 33, 183–209. [Google Scholar] [CrossRef]
- Williams, J.P.; Hallsworth, J.E. Limits of life in hostile environments: No barriers to biosphere function? Environ. Microbiol. 2009, 11, 3292–3308. [Google Scholar] [CrossRef]
- Canganella, F.; Wiegel, J. Extremophiles: From abyssal to terrestrial ecosystems and possibly beyond. Naturwissenschaften 2011, 98, 253–279. [Google Scholar] [CrossRef]
- Schulze-Makuch, D.; Airo, A.; Schirmack, J. The Adaptability of life on earth and the diversity of planetary habitats. Front. Microbiol. 2017, 8, 2011. [Google Scholar] [CrossRef]
- Sorokin, D.Y.; Berben, T.; Melton, E.D.; Overmars, L.; Vavourakis, C.D.; Muyzer, G. Microbial diversity and biogeochemical cycling in soda lakes. Extremophiles 2014, 18, 791–809. [Google Scholar] [CrossRef]
- Spang, A.; Caceres, E.F.; Ettema, T.J.G. Genomic exploration of the diversity, ecology, and evolution of the archaeal domain of life. Science 2017, 357, eaaf3883. [Google Scholar] [CrossRef]
- He, H.; Fu, L.; Liu, Q.; Fu, L.; Bi, N.; Yang, Z.; Zhen, Y. Community structure, abundance and potential functions of bacteria and Archaea in the Sansha Yongle Blue Hole, Xisha, south China sea. Front. Microbiol. 2019, 10, 2404. [Google Scholar] [CrossRef] [PubMed]
- Santoro, A.E.; Casciotti, K.L.; Francis, C.A. Activity, abundance and diversity of nitrifying Archaea and bacteria in the central California current. Environ. Microbiol. 2010, 12, 1989–2006. [Google Scholar] [CrossRef] [PubMed]
- Edwards, H.G. Will-o’-the-Wisp: An ancient mystery with extremophile origins? Philos. Trans. A. Math. Phys. Eng. Sci. 2014, 372, 20140206. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Coker, J.A. Recent advances in understanding extremophiles. F1000 Res. 2019, 8. [Google Scholar] [CrossRef]
- Venkateswaran, K.; La Duc, M.T.; Horneck, G. Microbial existence in controlled habitats and their resistance to space conditions. Microbes Environ. 2014, 29, 243–249. [Google Scholar] [CrossRef]
- Merino, N.; Aronson, H.S.; Bojanova, D.P.; Feyhl-Buska, J.; Wong, M.L.; Zhang, S.; Giovannelli, D. Living at the extremes: Extremophiles and the limits of life in a planetary context. Front. Microbiol. 2019, 10, 780. [Google Scholar] [CrossRef]
- Moissl-Eichinger, C.; Cockell, C.; Rettberg, P. Venturing into new realms? Microorganisms in space. FEMS Microbiol. Rev. 2016, 40, 722–737. [Google Scholar] [CrossRef]
- Das Sarma, S.; DasSarma, P.; Laye, V.J.; Schwieterman, E.W. Extremophilic models for astrobiology: Haloarchaeal survival strategies and pigments for remote sensing. Extremophiles 2020, 24, 31–41. [Google Scholar] [CrossRef]
- Cavicchioli, R. Extremophiles and the search for extraterrestrial life. Astrobiology 2002, 2, 281–292. [Google Scholar] [CrossRef]
- Javaux, E.J. Extreme life on earth-past, present and possibly beyond. Res. Microbiol. 2006, 157, 37–48. [Google Scholar] [CrossRef]
- McKay, C.P. Requirements and limits for life in the context of exoplanets. Proc. Natl. Acad. Sci. USA 2014, 111, 12628–12633. [Google Scholar] [CrossRef] [PubMed]
- Dodsworth, J.A.; Hungate, B.; de la Torre, J.R.; Jiang, H.; Hedlund, B.P. Measuring nitrification, denitrification, and related biomarkers in terrestrial geothermal ecosystems. Methods Enzymol. 2011, 486, 171–203. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, D.E.; Mancinelli, R.L. Bio-markers and the search for extinct life on Mars. Adv. Space Res. 1989, 9, 155–158. [Google Scholar] [CrossRef]
- Seckbach, J.; Chela-Flores, J. Extremophiles and chemotrophs as contributors to astrobiological signatures on Europa: A review of biomarkers of sulfate-reducers and other microorganisms. In Proceedings of the Proceedings Instruments, Methods, and Missions for Astrobiology X, San Diego, CA, USA, 26–30 August 2007; Volume 6694. [Google Scholar] [CrossRef]
- Rüttimann, C.; Cotorás, M.; Zaldívar, J.; Vicuña, R. DNA polymerases from the extremely thermophilic bacterium Thermus thermophilus HB-8. Eur. J. Biochem. 1985, 149, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, A.; Saha, J.; Haldar, S.; Bhowmic, A.; Mukhopadhyay, U.K.; Mukherjee, J. Production of poly-3-(hydroxybutyrate-co-hydroxyvalerate) by Haloferax mediterranei using rice-based ethanol stillage with simultaneous recovery and re-use of medium salts. Extremophiles 2014, 18, 463–470. [Google Scholar] [CrossRef]
- Raddadi, N.; Cherif, A.; Daffonchio, D.; Neifar, M.; Fava, F. Biotechnological applications of extremophiles, extremozymes and extremolytes. Appl. Microbiol. Biotechnol. 2015, 99, 7907–7913. [Google Scholar] [CrossRef]
- Rodrigo-Baños, M.; Garbayo, I.; Vílchez, C.; Bonete, M.J.; Martínez-Espinosa, R.M. Carotenoids from Haloarchaea and their potential in biotechnology. Mar. Drugs. 2015, 13, 5508–5532. [Google Scholar] [CrossRef]
- Torregrosa-Crespo, J.; Martínez-Espinosa, R.M.; Esclapez, J.; Bautista, V.; Pire, C.; Camacho, M.; Richardson, D.J.; Bonete, M.J. Anaerobic metabolism in Haloferax genus: Denitrification as case of study. Adv. Microb. Physiol. 2016, 68, 41–85. [Google Scholar] [CrossRef]
- Aracil-Gisbert, S.; Torregrosa-Crespo, J.; Martínez-Espinosa, R.M. Recent trend on bioremediation of polluted salty soils and waters using Haloarchaea. In Advances in Bioremediation and Phytoremediation; Shiomi, N., Ed.; Intech: London, UK, 2018; pp. 63–77. ISBN 978-953-51-3958-4. [Google Scholar]
- Colman, D.R.; Poudel, S.; Stamps, B.W.; Boyd, E.S.; Spear, J.R. The deep, hot biosphere: Twenty-five years of retrospection. Proc. Natl. Acad. Sci. USA 2017, 114, 6895–6903. [Google Scholar] [CrossRef]
- Huang, J.M.; Baker, B.J.; Li, J.T.; Wang, Y. New microbial lineages capable of carbon fixation and nutrient cycling in deep-sea sediments of the northern south China sea. Appl. Environ. Microbiol. 2019, 85, e005419–e005523. [Google Scholar] [CrossRef]
- Tkacz, A.; Hortala, M.; Poole, P.S. Absolute quantitation of microbiota abundance in environmental samples. Microbiome 2018, 6, 110. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Espinosa, R.M.; Cole, J.A.; Richardson, D.J.; Watmough, N.J. Enzymology and ecology of the nitrogen cycle. Biochem. Soc. Trans. 2011, 39, 175–178. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Wu, L.; Clark, I.M.; Xue, K.; Yang, Y.; Van Nostrand, J.D.; Deng, Y.; He, Z.; McGrath, S.; Storkey, J.; et al. Over 150 years of long-term fertilization alters spatial scaling of microbial biodiversity. mBio 2015, 6, e00240–e002415. [Google Scholar] [CrossRef] [PubMed]
- Zerkle, A.L.; Mikhail, S. The geobiological nitrogen cycle: From microbes to the mantle. Geobiology 2017, 15, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Niu, S.; Classen, A.T.; Dukes, J.S.; Kardol, P.; Liu, L.; Luo, Y.; Rustad, L.; Sun, J.; Tang, J.; Templer, P.H.; et al. Global patterns and substrate-based mechanisms of the terrestrial nitrogen cycle. Ecol. Lett. 2016, 19, 697–709. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Bakshi, B.R. Accounting for the biogeochemical cycle of nitrogen in input-output life cycle assessment. Environ. Sci. Technol. 2013, 47, 9388–9396. [Google Scholar] [CrossRef]
- Bonete, M.J.; Martínez-Espinosa, R.M.; Pire, C.; Zafrilla, B.; Richardson, D.J. Nitrogen metabolism in haloarchaea. Saline Syst. 2008, 4, 9. [Google Scholar] [CrossRef]
- Sorokin, D.Y.; Banciu, H.L.; Muyzer, G. Functional microbiology of soda lakes. Curr. Opin. Microbiol. 2015, 25, 88–96. [Google Scholar] [CrossRef]
- Torregrosa-Crespo, J.; Bergaust, L.; Pire, C.; Martínez-Espinosa, R.M. Denitrifying haloarchaea: Sources and sinks of nitrogenous gases. FEMS Microbiol. Lett. 2018, 365. [Google Scholar] [CrossRef]
- Schiraldi, C.; De Rosa, M. Mesophilic organisms. In Encyclopedia of Membranes; Drioli, E., Giorno, L., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 1–2. [Google Scholar] [CrossRef]
- Gonzalez, O.; Oberwinkler, T.; Mansueto, L.; Pfeiffer, F.; Mendoza, E.; Zimmer, R.; Oesterhelt, D. Characterization of growth and metabolism of the haloalkaliphile Natronomonas pharaonis. PLoS Comput. Biol. 2010, 6, e1000799. [Google Scholar] [CrossRef]
- Lee, B.D.; Apel, W.A.; DeVeaux, L.C.; Sheridan, P.P. Concurrent metabolism of pentose and hexose sugars by the polyextremophile Alicyclobacillus acidocaldarius. J. Ind. Microbiol. Biotechnol. 2017, 44, 1443–1458. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo, F.D.; Palmigiano, A.; Paciello, I.; Pallach, M.; Garozzo, D.; Bernardini, M.L.; Cono, V.; Yakimov, M.M.; Molinaro, A.; Silipo, A. The Deep-sea polyextremophile Halobacteroides lacunaris TB21 rough-type LPS: Structure and inhibitory activity towards toxic LPS. Mar. Drugs. 2017, 15, 201. [Google Scholar] [CrossRef] [PubMed]
- Colman, D.R.; Poudel, S.; Hamilton, T.L.; Having, J.R.; Selensky, M.J.; Shock, E.L.; Boyd, E.S. Geobiological feedbacks and the evolution of thermoacidophiles. ISME J. 2018, 12, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Monballiu, A.; Cardon, N.; Nguyen, T.M.; Cornelly, C.; Meesschaert, B.; Chiang, Y.W. Tolerance of chemoorganotrophic bioleaching microorganisms to heavy metal and alkaline stresses. Bioinorg. Chem. Appl. 2015, 2015, 861874. [Google Scholar] [CrossRef] [PubMed]
- Casas-Flores, S.; Gómez-Rodríguez, E.Y.; García-Meza, J.V. Community of thermoacidophilic and arsenic resistant microorganisms isolated from a deep profile of mine heaps. AMB Express 2015, 5, 132. [Google Scholar] [CrossRef]
- Lima, M.A.; Urbieta, M.S.; Donati, E. Arsenic-tolerant microbial consortia from sediments of Copahue geothermal system with potential applications in bioremediation. J. Basic. Microbiol. 2019. [Google Scholar] [CrossRef]
- Li, J.; Liu, Y.R.; Zhang, L.M.; He, J.Z. Sorption mechanism and distribution of cadmium by different microbial species. J. Environ. Manage. 2019, 237, 552–559. [Google Scholar] [CrossRef]
- Ramos-Zúñiga, J.; Gallardo, S.; Martínez-Bussenius, C.; Norambuena, R.; Navarro, C.A.; Paradela, A.; Jerez, C.A. Response of the biomining Acidithiobacillus ferrooxidans to high cadmium concentrations. J. Proteom. 2019, 198, 132–144. [Google Scholar] [CrossRef]
- Mangold, S.; Potrykus, J.; Björn, E.; Lövgren, L.; Dopson, M. Extreme zinc tolerance in acidophilic microorganisms from the bacterial and archaeal domains. Extremophiles 2013, 17, 75–85. [Google Scholar] [CrossRef]
- Teng, Y.; Du, X.; Wang, T.; Mi, C.; Yu, H.; Zou, L. Isolation of a fungus Pencicillium sp. with zinc tolerance and its mechanism of resistance. Arch. Microbiol. 2018, 200, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Glendinning, K.J.; Macaskie, L.E.; Brown, N.L. Mercury tolerance of thermophilic Bacillus sp. and Ureibacillus sp. Biotechnol. Lett. 2005, 27, 1657–1662. [Google Scholar] [CrossRef] [PubMed]
- Geesey, G.G.; Barkay, T.; King, S. Microbes in mercury-enriched geothermal springs in western north America. Sci. Total. Environ. 2016, 569–570, 321–331. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ma, K.; Zirngibl, C.; Linder, D.; Stetter, K.O.; Thauer, R.K. N5, N10-methylenetetrahydromethanopterin dehydrogenase (H2-forming) from the extreme thermophile Methanopyrus kandleri. Arch. Microbiol. 1991, 156, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Ciaramella, M.; Napoli, A.; Rossi, M. Another extreme genome: How to live at pH 0. Trends Microbiol. 2005, 13, 49–51. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.S.; Naushad, S.; Baker, S. Phylogenomic analyses and molecular signatures for the class Halobacteria and its two major clades: A proposal for division of the class Halobacteria into an emended order Halobacteriales and two new orders, Haloferacales ord. nov. and Natrialbales ord. nov., containing the novel families Haloferacaceae fam. nov. and Natrialbaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2015, 65, 1050–1069. [Google Scholar] [CrossRef] [PubMed]
- Cava, F.; Hidalgo, A.; Berenguer, J. Thermus thermophilus as biological model. Extremophiles 2009, 13, 213–231. [Google Scholar] [CrossRef] [PubMed]
- Oren, A. Salinibacter: An extremely halophilic bacterium with archaeal properties. FEMS Microbiol. Lett. 2013, 342, 1–9. [Google Scholar] [CrossRef]
- Rasuk, M.C.; Kurth, D.; Flores, M.R.; Contreras, M.; Novoa, F.; Poire, D.; Farias, M.E. Microbial characterization of microbial ecosystems associated to evaporites domes of gypsum in Salar de Llamara in Atacama desert. Microb. Ecol. 2014, 68, 483–494. [Google Scholar] [CrossRef]
- Santiago, I.F.; Gonçalves, V.N.; Gómez-Silva, B.; Galetovic, A.; Rosa, L.H. Fungal diversity in the Atacama Desert. Antonie Van Leeuwenhoek. 2018, 111, 1345–1360. [Google Scholar] [CrossRef]
- Banerjee, M.; Everroad, R.C.; Castenholz, R.W. An unusual cyanobacterium from saline thermal waters with relatives from unexpected habitats. Extremophiles 2009, 13, 707–716. [Google Scholar] [CrossRef]
- Stierle, A.A.; Stierle, D.B.; Girtsman, T.; Mou, T.C.; Antczak, C.; Djaballah, H. Azaphilones from an acid mine extremophile strain of a Pleurostomophora sp. J. Nat. Prod. 2015, 78, 2917–2923. [Google Scholar] [CrossRef] [PubMed]
- Duarte, A.W.F.; Dos Santos, J.A.; Vianna, M.V.; Vieira, J.M.F.; Mallagutti, V.H.; Inforsato, F.J.; Wentzel, L.C.P.; Lario, L.D.; Rodrigues, A.; Pagnocca, F.C.; et al. Cold-adapted enzymes produced by fungi from terrestrial and marine Antarctic environments. Crit. Rev. Biotechnol. 2018, 38, 600–619. [Google Scholar] [CrossRef] [PubMed]
- Møbjerg, N.; Halberg, K.A.; Jørgensen, A.; Persson, D.; Bjørn, M.; Ramløv, H.; Kristensen, R.M. Survival in extreme environments-on the current knowledge of adaptations in tardigrades. Acta Physiol. 2011, 202, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Weronika, E.; Łukasz, K. Tardigrades in space research-past and future. Orig. Life Evol. Biosph. 2017, 47, 545–553. [Google Scholar] [CrossRef] [PubMed]
- Mancinelli, R.L. The nature of nitrogen: An overview. Life Support Biosph. Sci. 1996, 3, 17–24. [Google Scholar] [PubMed]
- Fowler, D.; Coyle, M.; Skiba, U.; Sutton, M.A.; Cape, J.N.; Reis, S.; Sheppard, L.J.; Jenkins, A.; Grizzetti, B.; Galloway, J.N.; et al. The global nitrogen cycle in the twenty-first century. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2013, 368, 20130164. [Google Scholar] [CrossRef] [PubMed]
- Kartal, B.; Keltjens, J.T. Anammox biochemistry: A tale of heme C proteins. Trends Biochem. Sci. 2016, 41, 998–1011. [Google Scholar] [CrossRef]
- Lehtovirta-Morley, L.E. Ammonia oxidation: Ecology, physiology, biochemistry and why they must all come together. FEMS Microbiol. Lett. 2018, 365. [Google Scholar] [CrossRef]
- Peeters, S.H.; van Niftrik, L. Trending topics and open questions in anaerobic ammonium oxidation. Curr. Opin. Chem. Biol. 2019, 49, 45–52. [Google Scholar] [CrossRef]
- Daims, H.; Lebedeva, E.V.; Pjevac, P.; Han, P.; Herbold, C.; Albertsen, M.; Jehmlich, N.; Palatinszky, M.; Vierheilig, J.; Bulaev, A.; et al. Complete nitrification by Nitrospira bacteria. Nature 2015, 528, 504–509. [Google Scholar] [CrossRef]
- Daims, H.; Wagner, M. Nitrospira. Trends Microbiol. 2018, 26, 462–463. [Google Scholar] [CrossRef] [PubMed]
- Sgouridis, F.; Heppell, C.M.; Wharton, G.; Lansdown, K.; Trimmer, M. Denitrification and dissimilatory nitrate reduction to ammonium (DNRA) in a temperate re-connected floodplain. Water Res. 2011, 45, 4909–4922. [Google Scholar] [CrossRef] [PubMed]
- Leigh, J.A. Nitrogen fixation in methanogens: The archaeal perspective. Curr. Issues Mol. Biol. 2000, 2, 125–131. [Google Scholar] [PubMed]
- Barea, J.M.; Pozo, M.J.; Azcón, R.; Azcón-Aguilar, C. Microbial co-operation in the rhizosphere. J. Exp. Bot. 2005, 56, 1761–1778. [Google Scholar] [CrossRef]
- Cheng, Q. Perspectives in biological nitrogen fixation research. J. Integr. Plant Biol. 2008, 50, 786–798. [Google Scholar] [CrossRef]
- Zehr, J.P.; Capone, D.G. Changing perspectives in marine nitrogen fixation. Science 2020, 368, 9514. [Google Scholar] [CrossRef]
- Duc, L.; Neuenschwander, S.; Rehrauer, H.; Zeyer, J. Application of a nifH microarray to assess the impact of environmental factors on free-living diazotrophs in a glacier forefield. Can. J. Microbiol. 2011, 57, 105–114. [Google Scholar] [CrossRef]
- Suleiman, M.K.; Quoreshi, A.M.; Bhat, N.R.; Manuvel, A.J.; Sivadasan, M.T. Divulging diazotrophic bacterial community structure in Kuwait desert ecosystems and their N2-fixation potential. PLoS ONE 2019, 14, e0220679. [Google Scholar] [CrossRef]
- Nishihara, A.; Matsuura, K.; Tank, M.; McGlynn, S.E.; Thiel, V.; Haruta, S. Nitrogenase activity in thermophilic chemolithoautotrophic bacteria in the phylum Aquificae isolated under nitrogen-fixing conditions from Nakabusa hot springs. Microbes Environ. 2018, 33, 394–401. [Google Scholar] [CrossRef]
- Nash, M.V.; Anesio, A.M.; Barker, G.; Tranter, M.; Varliero, G.; Eloe-Fadrosh, E.A.; Nielsen, T.; Turpin-Jelfs, T.; Benning, L.G.; Sánchez-Baracaldo, P. Metagenomic insights into diazotrophic communities across Arctic glacier forefields. FEMS Microbiol. Ecol. 2018, 94. [Google Scholar] [CrossRef]
- Nishihara, A.; Haruta, S.; McGlynn, S.E.; Thiel, V.; Matsuura, K. Nitrogen fixation in thermophilic chemosynthetic microbial communities depending on hydrogen, sulfate, and carbon dioxide. Microbes Environ. 2018, 33, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Nishihara, A.; Thiel, V.; Matsuura, K.; McGlynn, S.E.; Haruta, S. Phylogenetic diversity of nitrogenase reductase genes and possible nitrogen-fixing bacteria in thermophilic chemosynthetic microbial communities in Nakabusa hot springs. Microbes Environ. 2018, 33, 357–365. [Google Scholar] [CrossRef]
- Nishizawa, M.; Miyazaki, J.; Makabe, A.; Koba, K.; Takai, K. Physiological and isotopic characteristics of nitrogen fixation by hyperthermophilic methanogens: Key insights into nitrogen anabolism of the microbial communities in Archean hydrothermal systems. Geochim. Cosmochim. Acta. 2014, 138, 117–135. [Google Scholar] [CrossRef]
- Takai, K. The nitrogen cycle: A large, fast, and mystifying cycle. Microbes Environ. 2019, 34, 223–225. [Google Scholar] [CrossRef] [PubMed]
- Mehta, M.P.; Baross, J.A. Nitrogen fixation at 92 °C by a hydrothermal vent archaeon. Science 2006, 314, 1783–1786. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Espinosa, R.M. Heterologous and homologous expression of proteins from Haloarchaea: Denitrification as case of study. Int. J. Mol. Sci. 2019, 21, 82. [Google Scholar] [CrossRef]
- Ward, B.B.; O’Mullan, G.D. Community level analysis: Genetic and biogeochemical approaches to investigate community composition and function in aerobic ammonia oxidation. Methods Enzymol. 2005, 397, 395–413. [Google Scholar]
- Jung, M.Y.; Islam, M.A.; Gwak, J.H.; Kim, J.G.; Rhee, S.K. Nitrosarchaeum koreense gen. nov., sp. nov., an aerobic and mesophilic, ammonia-oxidizing archaeon member of the phylum Thaumarchaeota isolated from agricultural soil. Int. J. Syst. Evol. Microbiol. 2018, 68, 3084–3095. [Google Scholar] [CrossRef]
- Mosier, A.C.; Francis, C.A. Relative abundance and diversity of ammonia-oxidizing archaea and bacteria in the San Francisco Bay estuary. Environ. Microbiol. 2008, 10, 3002–3016. [Google Scholar] [CrossRef]
- Schleper, C.; Nicol, G.W. Ammonia-oxidising Archaea-physiology, ecology and evolution. Adv. Microb. Physiol. 2010, 57, 1–41. [Google Scholar] [CrossRef]
- Ragon, M.; Van Driessche, A.E.; García-Ruíz, J.M.; Moreira, D.; López-García, P. Microbial diversity in the deep-subsurface hydrothermal aquifer feeding the giant gypsum crystal-bearing Naica Mine, Mexico. Front. Microbiol. 2013, 4, 37. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.F.; Gu, J.D. Higher diversity of ammonia/ammonium-oxidizing prokaryotes in constructed freshwater wetland than natural coastal marine wetland. Appl. Microbiol. Biotechnol. 2013, 97, 7015–7033. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Jayakumar, A.; Ward, B.B. Community composition of ammonia-oxidizing archaea from surface and anoxic depths of oceanic oxygen minimum zones. Front. Microbiol. 2013, 4, 177. [Google Scholar] [CrossRef] [PubMed]
- Rusch, A.; Gaidos, E. Nitrogen-cycling bacteria and archaea in the carbonate sediment of a coral reef. Geobiology. 2013, 11, 472–484. [Google Scholar] [CrossRef] [PubMed]
- You, J.; Das, A.; Dolan, E.M.; Hu, Z. Ammonia-oxidizing archaea involved in nitrogen removal. Water Res. 2009, 43, 1801–1809. [Google Scholar] [CrossRef]
- Jetten, M.S.; Strous, M.; van de Pas-Schoonen, K.T.; Schalk, J.; van Dongen, U.G.; van de Graaf, A.A.; Logemann, S.; Muyzer, G.; van Loosdrecht, M.C.; Kuenen, J.G. The anaerobic oxidation of ammonium. FEMS Microbiol. Rev. 1998, 22, 421–437. [Google Scholar] [CrossRef]
- Kartal, B.; van Niftrik, L.; Keltjens, J.T.; Op den Camp, H.J.; Jetten, M.S. Anammox-growth physiology, cell biology, and metabolism. Adv. Microb. Physiol. 2012, 60, 211–262. [Google Scholar] [CrossRef]
- Byrne, N.; Strous, M.; Crépeau, V.; Kartal, B.; Birrien, J.L.; Schmid, M.; Lesongeur, F.; Schouten, S.; Jaeschke, A.; Jetten, M.; et al. Presence and activity of anaerobic ammonium-oxidizing bacteria at deep-sea hydrothermal vents. ISME J. 2009, 3, 117–123. [Google Scholar] [CrossRef]
- Jaeschke, A.; Op den Camp, H.J.; Harhangi, H.; Klimiuk, A.; Hopmans, E.C.; Jetten, M.S.; Schouten, S.; Sinninghe Damsté, J.S. 16S rRNA gene and lipid biomarker evidence for anaerobic ammonium-oxidizing bacteria (anammox) in California and Nevada hot springs. FEMS Microbiol. Ecol. 2009, 67, 343–350. [Google Scholar] [CrossRef]
- Zhu, G.; Xia, C.; Shanyun, W.; Zhou, L.; Liu, L.; Zhao, S. Occurrence, activity and contribution of anammox in some freshwater extreme environments. Environ. Microbiol. Rep. 2015, 7, 961–969. [Google Scholar] [CrossRef]
- Francis, C.A.; Beman, J.M.; Kuypers, M.M. New processes and players in the nitrogen cycle: The microbial ecology of anaerobic and archaeal ammonia oxidation. ISME J. 2007, 1, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Torregrosa-Crespo, J.; Pire, C.; Martínez-Espinosa, R.M.; Bergaust, L. Denitrifying haloarchaea within the genus Haloferax display divergent respiratory phenotypes, with implications for their release of nitrogenous gases. Environ. Microbiol. 2019, 21, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Bricio, C.; Alvarez, L.; Gómez, M.J.; Berenguer, J. Partial and complete denitrification in Thermus thermophilus: Lessons from genome drafts. Biochem. Soc. Trans. 2011, 39, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Hedlund, B.P.; McDonald, A.I.; Lam, J.; Dodsworth, J.A.; Brown, J.R.; Hungate, B.A. Potential role of Thermus thermophilus and T. oshimai in high rates of nitrous oxide (N2O) production in ~80 °C hot springs in the US Great Basin. Geobiology 2011, 9, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Zhou, E.M.; Murugapiran, S.K.; Mefferd, C.C.; Liu, L.; Xian, W.D.; Yin, Y.R.; Ming, H.; Yu, T.T.; Huntemann, M.; Clum, A.; et al. High-quality draft genome sequence of the Thermus amyloliquefaciens type strain YIM 77409(T) with an incomplete denitrification pathway. Stand Genomic Sci. 2016, 11, 20. [Google Scholar] [CrossRef] [PubMed]
- Hochstein, L.I.; Lang, F. Purification and properties of a dissimilatory nitrate reductase from Haloferax denitrificans. Arch. Biochem. Biophys. 1991, 288, 380–385. [Google Scholar] [CrossRef]
- Yoshimatsu, K.; Iwasaki, T.; Fujiwara, T. Sequence and electron paramagnetic resonance analyses of nitrate reductase NarGH from a denitrifying halophilic euryarchaeote Haloarcula marismortui. FEBS Lett. 2002, 516, 145–150. [Google Scholar] [CrossRef]
- Hattori, T.; Shiba, H.; Ashiki, K.; Araki, T.; Nagashima, Y.K.; Yoshimatsu, K.; Fujiwara, T. Anaerobic growth of haloarchaeon Haloferax volcanii by denitrification is controlled by the transcription regulator NarO. J. Bacteriol. 2016, 198, 1077–1086. [Google Scholar] [CrossRef]
- Afshar, S.; Kim, C.; Monbouquette, H.G.; Schroder, I.I. Effect of tungstate on nitrate reduction by the hyperthermophilic archaeon Pyrobaculum aerophilum. Appl. Environ. Microbiol. 1998, 64, 3004–3008. [Google Scholar] [CrossRef]
- Cozen, A.E.; Weirauch, M.T.; Pollard, K.S.; Bernick, D.L.; Stuart, J.M.; Lowe, T.M. Transcriptional map of respiratory versatility in the hyperthermophilic crenarchaeon Pyrobaculum aerophilum. J. Bacteriol. 2009, 191, 782–794. [Google Scholar] [CrossRef]
- Fernandes, A.T.; Damas, J.M.; Todorovic, S.; Huber, R.; Baratto, M.C.; Pogni, R.; Soares, C.M.; Martins, L.O. The multicopper oxidase from the archaeon Pyrobaculum aerophilum shows nitrous oxide reductase activity. FEBS J. 2010, 277, 3176–3189. [Google Scholar] [CrossRef] [PubMed]
- Kesserü, P.; Kiss, I.; Bihari, Z.; Polyák, B. The effects of NaCl and some heavy metals on the denitrification activity of Ochrobactrum anthropi. J. Basic Microbiol. 2002, 42, 268–276. [Google Scholar] [CrossRef]
- Nakano, M.; Inagaki, T.; Okunishi, S.; Tanaka, R.; Maeda, H. Effect of salinity on denitrification under limited single carbon source by Marinobacter sp. isolated from marine sediment. J. Basic Microbiol. 2010, 50, 285–289. [Google Scholar] [CrossRef] [PubMed]
- Qu, L.; Lai, Q.; Zhu, F.; Hong, X.; Zhang, J.; Shao, Z.; Sun, X. Halomonas daqiaonensis sp. nov., a moderately halophilic, denitrifying bacterium isolated from a littoral saltern. Int. J. Syst. Evol. Microbiol. 2011, 61, 1612–1616. [Google Scholar] [CrossRef]
- Martinez-Espinosa, R.M.; Dridge, E.J.; Bonete, M.J.; Butt, J.N.; Butler, C.S.; Sargent, F.; Richardson, D.J. Look on the positive side! The orientation, identification and bioenergetics of ‘Archaeal’ membrane-bound nitrate reductases. FEMS Microbio. Lett. 2007, 276, 129–139. [Google Scholar] [CrossRef]
- Alvarez, L.; Bricio, C.; Blesa, A.; Hidalgo, A.; Berenguer, J. Transferable denitrification capability of Thermus thermophilus. Appl. Environ. Microbiol. 2014, 80, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Espinosa, R.M.; Marhuenda-Egea, F.C.; Bonete, M.J. Assimilatory nitrate reductase from the haloarchaeon Haloferax mediterranei: Purification and characterisation. FEMS Microbiol. Lett. 2001, 204, 381–385. [Google Scholar] [CrossRef]
- Martínez-Espinosa, R.M.; Marhuenda-Egea, F.C.; Donaire, A.; Bonete, M.J. NMR studies of a ferredoxin from Haloferax mediterranei and its physiological role in nitrate assimilatory pathway. Biochim. Biophys. Acta. 2003, 1623, 47–51. [Google Scholar] [CrossRef]
- Lledó, B.; Marhuenda-Egea, F.C.; Martínez-Espinosa, R.M.; Bonete, M.J. Identification and transcriptional analysis of nitrate assimilation genes in the halophilic archaeon Haloferax mediterranei. Gene 2005, 361, 80–88. [Google Scholar] [CrossRef]
- Martínez-Espinosa, R.M.; Marhuenda-Egea, F.C.; Bonete, M.J. Purification and characterisation of a possible assimilatory nitrite reductase from the halophile archaeon Haloferax mediterranei. FEMS Microbiol. Lett. 2001, 196, 113–118. [Google Scholar] [CrossRef]
- Martínez-Espinosa, R.M.; Esclapez, J.; Bautista, V.; Bonete, M.J. An octameric prokaryotic glutamine synthetase from the haloarchaeon Haloferax mediterranei. FEMS Microbiol. Lett. 2006, 264, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Pire, C.; Martínez-Espinosa, R.M.; Pérez-Pomares, F.; Esclapez, J.; Bonete, M.J. Ferredoxin-dependent glutamate synthase: Involvement in ammonium assimilation in Haloferax mediterranei. Extremophiles 2014, 18, 147–159. [Google Scholar] [CrossRef] [PubMed]
- Hayden, B.M.; Bonete, M.J.; Brown, P.E.; Moir, A.J.; Engel, P.C. Glutamate dehydrogenase of Halobacterium salinarum: Evidence that the gene sequence currently assigned to the NADP+-dependent enzyme is in fact that of the NAD+-dependent glutamate dehydrogenase. FEMS Microbiol. Lett. 2002, 211, 37–41. [Google Scholar] [CrossRef] [PubMed]
- Mary, J.; Révet, B. Isolation and characterization of a protein with high affinity for DNA: The glutamine synthetase of Thermus thermophilus 111. J. Mol. Biol. 1999, 286, 121–134. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, J.L.; Ferrer, J.; Pire, C.; Llorca, F.I.; Bonete, M.J. Denaturation studies by fluorescence and quenching of thermophilic protein NAD+-glutamate dehydrogenase from Thermus thermophilus HB8. J. Protein. Chem. 2003, 22, 295–301. [Google Scholar] [CrossRef]
- Wedler, F.C.; Shreve, D.S.; Kenney, R.M.; Ashour, A.E.; Carfi, J.; Rhee, S.G. Two glutamine synthetases from Bacillus caldolyticus, an extreme thermophile. Isolation, physicochemical and kinetic properties. J. Biol. Chem. 1980, 255, 9507–9516. [Google Scholar]
- Brown, J.R.; Masuchi, Y.; Robb, F.T.; Doolittle, W.F. Evolutionary relationships of bacterial and Archaeal glutamine synthetase genes. J. Mol. Evol. 1994, 38, 566–576. [Google Scholar] [CrossRef]
- Mi, S.; Song, J.; Lin, J.; Che, Y.; Zheng, H.; Lin, J. Complete genome of Leptospirillum ferriphilum ML-04 provides insight into its physiology and environmental adaptation. J. Microbiol. 2011, 49, 890–901. [Google Scholar] [CrossRef]
- Ren, Y.; Hao, N.H.; Guo, W.; Wang, D.; Peng, L.; Ni, B.J.; Wei, W.; Liu, Y. New perspectives on microbial communities and biological nitrogen removal processes in wastewater treatment systems. Bioresour. Technol. 2020, 297, 122491. [Google Scholar] [CrossRef]
- Nájera-Fernández, C.; Zafrilla, B.; Bonete, M.J.; Martínez-Espinosa, R.M. Role of the denitrifying Haloarchaea in the treatment of nitrite-brines. Int. Microbiol. 2012, 15, 111–119. [Google Scholar] [CrossRef]
- Qin, H.; Ji, B.; Zhang, S.; Kong, Z. Study on the bacterial and archaeal community structure and diversity of activated sludge from three wastewater treatment plants. Mar. Pollut. Bull. 2018, 135, 801–807. [Google Scholar] [CrossRef] [PubMed]
- Castro-Barros, C.M.; Ho, L.T.; Winkler, M.K.H.; Volcke, E.I.P. Integration of methane removal in aerobic anammox-based granular sludge reactors. Environ. Technol. 2018, 39, 1615c1625. [Google Scholar] [CrossRef] [PubMed]
- Holmes, D.E.; Dang, Y.; Smith, J.A. Nitrogen cycling during wastewater treatment. Adv. Appl. Microbiol. 2019, 106, 113–192. [Google Scholar] [CrossRef] [PubMed]
- De la Torre, J.R.; Walker, C.B.; Ingalls, A.E.; Könneke, M.; Stahl, D.A. Cultivation of a thermophilic ammonia oxidizing Archaeon synthesizing Crenarchaeol. Environ. Microbiol. 2008, 10, 810–818. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, R.; Tao, Y.; Li, G. Archaea in wastewater treatment: Current research and emerging technology. Archaea 2018, 2018, 6973294. [Google Scholar] [CrossRef]
- Oosterkamp, M.J.; Mehboob, F.; Schraa, G.; Plugge, C.M.; Stams, A.J. Nitrate and (per)chlorate reduction pathways in (per)chlorate-reducing bacteria. Biochem. Soc. Trans. 2011, 39, 230–235. [Google Scholar] [CrossRef]
- Martínez-Espinosa, R.M.; Richardson, D.J.; Bonete, M.J. Characterisation of chlorate reduction in the haloarchaeon Haloferax mediterranei. Biochim. Biophys. Acta. 2015, 1850, 587–594. [Google Scholar] [CrossRef]
- Hogue, C. Rocket-fueled river. Chem. Eng. News 2003, 81, 37–46. [Google Scholar] [CrossRef]
- Urbansky, E.T.; Schock, M.R. Issues in managing the risks associated with perchlorate in drinking water. J. Environ. Manag. 1999, 56, 79–95. [Google Scholar] [CrossRef]
- Lehman, S.G.; Badruzzaman, M.; Adham, S.; Roberts, D.J.; Clifford, D.A. Perchlorate and nitrate treatment by ion exchange integrated with biological brine treatment. Water Res. 2008, 42, 969–976. [Google Scholar] [CrossRef]
- Kengen, S.W.M.; Rikken, G.B.; Hagen, W.R.; van Ginkel, C.G.; Stams, A.J.M. Purification and characterization of (per)chlorate reductase from the chlorate-respiring strain GR-1. J. Bacteriol. 1999, 181, 6706–6711. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Espinosa, R.M.; Bonete, M.J. Bioremediation of chlorate and perchlorate salted water using Haloferax mediterranei. J. Biotech. 2007, 2, S227. [Google Scholar] [CrossRef]
- Krzmarzick, M.J.; Taylor, D.K.; Fu, X.; McCutchan, A.L. Diversity and niche of Archaea in bioremediation. Archaea 2018, 2018, 3194108. [Google Scholar] [CrossRef] [PubMed]
- Greaver, T.L.; Clark, C.M.; Compton, J.E.; Vallano, D.; Talhelm, A.F.; Weaver, C.P.; Band, L.E.; Baron, J.S.; Davidson, E.A.; Tague, C.L.; et al. Key ecological responses to nitrogen are altered by climate change. Nat. Clim. Chang. 2016, 6, 836–843. [Google Scholar] [CrossRef]
- Ramos-Barbero, M.D.; Martin-Cuadrado, A.B.; Viver, T.; Santos, F.; Martinez-Garcia, M.; Antón, J. Recovering microbial genomes from metagenomes in hypersaline environments: The good, the bad and the ugly. Syst. Appl. Microbiol. 2019, 42, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Abby, S.S.; Melcher, M.; Kerou, M.; Krupovic, M.; Stieglmeier, M.; Rossel, C.; Pfeifer, K.; Schleper, C. Candidatus Nitrosocaldus cavascurensis, an ammonia oxidizing, extremely thermophilic archaeon with a highly mobile genome. Front. Microbiol. 2018, 9, 28. [Google Scholar] [CrossRef] [PubMed]
- Pitcher, A.; Rychlik, N.; Hopmans, E.C.; Spieck, E.; Rijpstra, W.I.; Ossebaar, J.; Schouten, S.; Wagner, M.; Damsté, J.S. Crenarchaeol dominates the membrane lipids of candidatus Nitrososphaera gargensis, a thermophilic group I.1b Archaeon. ISME J. 2010, 4, 542–552. [Google Scholar] [CrossRef]
- Torregrosa-Crespo, J.; Pire, C.; Bergaust, L.; Martínez-Espinosa, R.M. Haloferax mediterranei, an Archaeal model for denitrification in saline systems, characterized through integrated physiological and transcriptional analyses. Front. Microbiol. 2020, 11, 768. [Google Scholar] [CrossRef]
- Shibata, H.; Branquinho, C.; McDowell, W.H.; Mitchell, M.J.; Monteith, D.T.; Tang, J.; Arvola, L.; Cruz, C.; Cusack, D.F.; Halada, L.; et al. Consequence of altered nitrogen cycles in the coupled human and ecological system under changing climate: The need for long-term and site-based research. Ambio 2015, 44, 178–193. [Google Scholar] [CrossRef]
- Hakeem, K.R.; Sabir, M.; Ozturk, M.; Akhtar, M.S.; Ibrahim, F.H. Nitrate and nitrogen oxides: Sources, health effects and their remediation. Rev. Environ. Contam. Toxicol. 2017, 242, 183–217. [Google Scholar] [CrossRef]
- Cheng, Y.; Wang, J.; Chang, S.X.; Cai, Z.; Müller, C.; Zhang, J. Nitrogen deposition affects both net and gross soil nitrogen transformations in forest ecosystems: A review. Environ. Pollut. 2019, 244, 608–616. [Google Scholar] [CrossRef] [PubMed]
- Samad, M.S.; Biswas, A.; Bakken, L.R.; Clough, T.J.; de Klein, C.A.; Richards, K.G.; Lanigan, G.J.; Morales, S.E. Phylogenetic and functional potential links pH and N2O emissions in pasture soils. Sci. Rep. 2016, 6, 35990. [Google Scholar] [CrossRef]
- Samad, M.S.; Bakken, L.R.; Nadeem, S.; Clough, T.J.; de Klein, C.A.; Richards, K.G.; Lanigan, G.J.; Morales, S.E. High-resolution denitrification kinetics in pasture soils link N2O emissions to pH, and denitrification to C mineralization. PLoS ONE 2016, 11, e0151713. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Mørkved, P.T.; Frostegård, A.; Bakken, L.R. Denitrification gene pools, transcription and kinetics of NO, N2O and N2 production as affected by soil pH. FEMS Microbiol. Ecol. 2010, 72, 407–417. [Google Scholar] [CrossRef] [PubMed]
- Bakken, L.R.; Bergaust, L.; Liu, B.; Frostegård, A. Regulation of denitrification at the cellular level: A clue to the understanding of N2O emissions from soils. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2012, 367, 1226–1234. [Google Scholar] [CrossRef] [PubMed]
- Qu, Z.; Wang, J.; Almøy, T.; Bakken, L.R. Excessive use of nitrogen in Chinese agriculture results in high N(2) O/(N(2) O + N(2)) product ratio of denitrification, primarily due to acidification of the soils. Glob. Chang Biol. 2014, 20, 1685–1698. [Google Scholar] [CrossRef]
- Hellsten, S.; Dalgaard, T.; Rankinen, K.; Tørseth, K.; Bakken, L.; Bechmann, M.; Kulmala, A.; Moldan, F.; Olofsson, S.; Piil, K.; et al. Abating N in Nordic agriculture-Policy, measures and way forward. J. Environ. Manage. 2019, 236, 674–686. [Google Scholar] [CrossRef]
- Conthe, M.; Lycus, P.; Arntzen, M.Ø.; Ramos da Silva, A.; Frostegård, Å.; Bakken, L.R.; Kleerebezem, R.; van Loosdrecht, M.C.M. Denitrification as an N2O sink. Water Res. 2019, 151, 381–387. [Google Scholar] [CrossRef]
- Bakken, L.R.; Frostegård, Å. Sources and sinks for N2O, can microbiologist help to mitigate N2O emissions? Environ. Microbiol. 2017, 19, 4801–4805. [Google Scholar] [CrossRef]
- Dumorné, K.; Córdova, D.C.; Astorga-Eló, M.; Renganathan, P. Extremozymes: A potential source for industrial applications. J. Microbiol. Biotechnol. 2017, 27, 649–659. [Google Scholar] [CrossRef]
- Jin, M.; Gai, Y.; Guo, X.; Hou, Y.; Zeng, R. Properties and applications of extremozymes from deep-sea extremophilic microorganisms: A mini review. Mar. Drugs 2019, 17, 656. [Google Scholar] [CrossRef] [PubMed]
- Elleuche, S.; Schröder, C.; Sahm, K.; Antranikian, G. Extremozymes-biocatalysts with unique properties from extremophilic microorganisms. Curr. Opin. Biotechnol. 2014, 29, 116–123. [Google Scholar] [CrossRef] [PubMed]
- Hough, D.W.; Danson, M.J. Extremozymes. Curr. Opin. Chem. Biol. 1999, 3, 39–46. [Google Scholar] [CrossRef]
- Nielsen, M.; Gieseke, A.; de Beer, D.; Revsbech, N.P. Nitrate, nitrite, and nitrous oxide transformations in sediments along a salinity gradient in the Weser Estuary. Aquat. Microb. Ecol. 2009, 55, 39–52. [Google Scholar] [CrossRef]
- Nielsen, M.; Larsen, L.H.; Jetten, M.S.; Revsbech, N.P. Bacterium-based NO2− biosensor for environmental applications. Appl. Environ. Microbiol. 2004, 70, 6551–6558. [Google Scholar] [CrossRef] [PubMed]
- Larsen, L.H.; Revsbech, N.P.; Binnerup, S.J. A microsensor for nitrate based on immobilized denitrifying bacteria. Appl. Environ. Microbiol. 1996, 62, 1248–1251. [Google Scholar] [CrossRef] [PubMed]
- Taylor, C.J.; Bain, L.A.; Richardson, D.J.; Spiro, S.; Russell, D.A. Construction of a whole-cell gene reporter for the fluorescent bioassay of nitrate. Anal. Biochem. 2004, 328, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, T.; Almeida, M.G. Electrochemical enzyme biosensors revisited: Old solutions for new problems. Crit. Rev. Anal. Chem. 2019, 49, 44–66. [Google Scholar] [CrossRef]
- Monteiro, T.; Rodrigues, P.R.; Gonçalves, A.L.; Moura, J.J.; Jubete, E.; Añorga, L.; Piknova, B.; Schechter, A.N.; Silveira, C.M.; Almeida, M.G. Construction of effective disposable biosensors for point of care testing of nitrite. Talanta 2015, 142, 246–251. [Google Scholar] [CrossRef]
- Sohail, M.; Adeloju, S.B. Nitrate biosensors and biological methods for nitrate determination. Talanta 2016, 153, 83–98. [Google Scholar] [CrossRef]
- Aghamiri, Z.S.; Mohsennia, M.; Rafiee-Pour, H.A. Immobilization of cytochrome c and its application as electrochemical biosensors. Talanta 2018, 176, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Revsbech, N.P.; Glud, R.N. Biosensor for laboratory and lander-based analysis of benthic nitrate plus nitrite distribution in marine environments. limnology and oceanography. Methods 2009, 7, 761–770. [Google Scholar]
- Hunter, P. The role of biology in global climate change: Interdisciplinary research in biogeochemistry can help to understand local and global fluxes of carbon and other elements and inform environmental policies. EMBO Rep. 2017, 18, 673–676. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Stein, L.Y.; Klotz, M.G. The nitrogen cycle. Curr. Biol. 2016, 26, R94–R98. [Google Scholar] [CrossRef] [PubMed]
| Term | Factor | Limits |
|---|---|---|
| Acidophile | pH | ≥3 |
| Alkaliphile | pH | ≥9 |
| Halophile | High salt concentration | 1–4 M |
| Hyperthermophile and Thermophile | High temperatures | Hyperthermophile: above 80 °C (176 °F) Thermophile: between 45–122 °C (113–252 °F) |
| Piezophile (also called Barophile) | High pressures | ~1100 bar |
| Psycrophile (also called Cryophile) | Low temperatures | ≤−15 °C (5 °F) |
| Radiophile (also called Radioresistant) | UV radiation, cosmic rays, X-rays | 1500 to 6000 Gy |
| Xerophile | Desiccating conditions | ≤50% relative humidity |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez-Espinosa, R.M. Microorganisms and Their Metabolic Capabilities in the Context of the Biogeochemical Nitrogen Cycle at Extreme Environments. Int. J. Mol. Sci. 2020, 21, 4228. https://doi.org/10.3390/ijms21124228
Martínez-Espinosa RM. Microorganisms and Their Metabolic Capabilities in the Context of the Biogeochemical Nitrogen Cycle at Extreme Environments. International Journal of Molecular Sciences. 2020; 21(12):4228. https://doi.org/10.3390/ijms21124228
Chicago/Turabian StyleMartínez-Espinosa, Rosa María. 2020. "Microorganisms and Their Metabolic Capabilities in the Context of the Biogeochemical Nitrogen Cycle at Extreme Environments" International Journal of Molecular Sciences 21, no. 12: 4228. https://doi.org/10.3390/ijms21124228
APA StyleMartínez-Espinosa, R. M. (2020). Microorganisms and Their Metabolic Capabilities in the Context of the Biogeochemical Nitrogen Cycle at Extreme Environments. International Journal of Molecular Sciences, 21(12), 4228. https://doi.org/10.3390/ijms21124228





