Abstract
Aluminum (Al) stress is a major limiting factor for plant growth and crop production in acid soils. At present, only a few transcription factors involved in the regulation of Al resistance have been characterized. Here, we used reversed genetic approach through phenotype analysis of overexpressors and mutants to demonstrate that AtHB7 and AtHB12, two HD-Zip I transcription factors, participate in Al resistance. In response to Al stress, AtHB7 and AtHB12 displayed different dynamic expression patterns. Although both AtHB7 and AtHB12 positively regulate root growth in the absence of Al stress, our results showed that AtHB7 antagonizes with AtHB12 to control root growth in response to Al stress. The athb7/12 double mutant displayed a wild-type phenotype under Al stress. Consistently, our physiological analysis showed that AtHB7 and AtHB12 oppositely regulate the capacity of cell wall to bind Al. Yeast two hybrid assays showed that AtHB7 and AtHB12 could form homo-dimers and hetero-dimers in vitro, suggesting the interaction between AtHB7 and AtHB12 in the regulation of root growth. The conclusion was that AtHB7 and AtHB12 oppositely regulate Al resistance by affecting Al accumulation in root cell wall.
1. Introduction
A major factor constraint crop production on acidic soils worldwide is aluminum (Al) toxicity [1]. Excess Al3+ in acid soils is toxic to plants. It has been reported that the rapid inhibition of root elongation is the earliest and most dramatic symptom of Al toxicity, resulting in a reduced and damaged root system that limits mineral nutrient and water uptake, and eventually leads to reduced production [2,3,4,5].
The root transition zone (TZ), located between the apical meristem and basal elongation zone (EZ) in root apex, is the critical site of perception of Al toxicity, which has been reported in the model plant Arabidopsis (Arabidopsis thaliana), wheat (Triticum aestivum), maize (Zea mays), sorghum (Sorghum bicolor), and common bean (Phaseolus vulgaris) [6,7,8,9,10,11]. The root TZ has also been thought to be the active site for phytohormone cross-talks. Al stress induced local auxin biosynthesis through the up-regulation of TAA1 and YUCCAs. The local accumulated auxin in root TZ further increased local cytokinin biosynthesis and enhanced cytokinin responses and eventually inhibited root growth, a process which was dependent on ethylene signaling [7,12,13]. However, in maize, external supply of auxin to the root EZ is able to partly overcome the inhibition of root under Al stress, indicating the repressed root growth might be result from low auxin accumulation in root tips [14]. This hypothesis was further confirmed by characterization of the zmpgp1 mutants which are deficient in the maize auxin efflux carrier P-glycoprotein (ZmPGP1) and displayed higher levels of auxin in root tips and increased Al tolerance [15]. All these investigations suggest that auxin plays an important role in root growth regulation under Al stress.
When plants suffer Al toxicity, cell wall is the main site for Al accumulation. For example, 80%–90% of Al accumulated in the cell wall of barley [16]; in total, 95% of Al deposited in the cell wall of hypocotyl epidermal cells in Abelmoschus esculentus [17]. It has been widely accepted that the large amount of Al deposited in the cell wall alters its property, reduces its ductility and arrests cell expansion in root elongation zone [18], which are the main causes of Al-induced inhibition of root growth. Pectin methylation and modifications of xyloglucan oligosaccharides which are the major components of cell wall hemicellulose in non-poalean monocotyledons and dicotyledons, are recognized as two vital strategies to affect cell wall Al binding capacity. PME46 (PECTIN METHYLESTERASE46), an inhibitor of pectin methylesterase activity, increased methylated pectin and reduced Al binding to cell wall and thus conferred plants Al resistance [19]. Besides, Al binding capacity of hemicelluloses could be positively regulated by xyloglucan endotransglucosylase/hydrolases AtXTH17 and AtXTH31, and accordingly, lacking each of the XTH enzymes leads to increased Al-sensitivity [20,21]. Mutants of TRICHOME BIREFRINGENCE-LIKE27 (BL27), encoding a putative O-acetyltransferase for modulation of the O-acetylation level of xyloglucan, showed increased Al sensitivity by enhancing Al-binding in the hemicellulose [21]. In rice (Oryza sativa), Arabidopsis (Arabidopsis thaliana) and buckwheat (Fagopyrum esculentum), STAR1 interact with STAR2/ALS3 to form an ABC transporter to regulate Al resistance possibly via transporting UDP-glucose for cell wall modification [22,23,24,25,26]. Knockout of OsEXPA10 in rice affected Al binding to cell wall, but did not affect Al-induced root-growth inhibition [27].
Besides modifying cell wall, plants have also employed other strategies to encounter with Al toxicity, such as organic acid exudation and internal detoxification like vacuolar compartment [28]. Normally, few malic acid and citric acid were secreted from the roots. However, in response to Al stress, both AtALMT1 (Al-activated Malate Transporter1) and AtMATE (Multidrug and Toxic Compound Extrusion) were significantly up-regulated at transcription levels, which caused the secretion of malic acid and cirtic acid into rhizosphere for Al chelation, and thus alleviated Al-induced inhibition of root elongation [29,30]. Furthermore, ALS1 (ALUMINUM SENSITIVE1), which located in tonoplast, seemed to participate in transporting Al to vacuole for segregation [31].
In Arabidopsis thaliana, SENSITIVE TO PROTON RHIZOTOXICITY1 (STOP1), a type of C2H2 zinc finger transcription factor, was identified as a key regulator in response to Al stress through the transcriptional regulation of AtALMT1, AtMATE, and ALS3, which encoded organic acids transporters and a half-type ABC transporter, respectively [32,33]. Subsequently, orthologous genes have been identified in rice and sweet sorghum [34,35,36]. STOP2 is a homolog of STOP1, having a shorter C-terminus compared to STOP1. AtSTOP2 activates the expression of some genes involved in Al- and low pH-tolerance that are also regulated by AtSTOP1 such as AtMATE and AtALS3 [37]. Recently, an F-box protein encoded by the gene regulating AtALMT1 expression 1 (RAE1) was shown to regulate STOP1 protein abundance. The RAE1 protein interacts with STOP1 and hence promotes STOP1 ubiquitination and degradation [38]. AtWRKY46 was reported as a negative regulator of AtALMT1 transcription to restrain malate secretion under Al stress [39]. AtWRKY47 directly regulated ELP (EXTENSIN-LIKE PROTEIN) and XTH17 which are responsible for cell wall modification under Al stress [40]. As a STOP1 ortholog in rice, ART1 functions as a central modulator of Al response, regulating expression of a broad of crucial Al resistance genes, including OsSTAR1/2, OsNrat1, OsALS1, OsFRDL4, OsMGT1 and OsCDT3 [34]. ART2, a homology of ART1, plays a supplementary role in Al tolerance in rice. [35]. OsWRKY22 was involved in regulation of Al-activated citrate acid secretion by regulating expression of OsFRDL4 which encodes a citrate transporter in rice [41]. ABSCISIC ACID, STRESS AND RIPENING (ASR) transcription factor ASR5 collaboratively acts with ASR1 in Al-stress response [42,43].
HD-ZIP transcription factors which are unique in plants are characterized as containing a homeodomain (HD) for binding DNA and a leucine zipper (Zip) motif for protein dimerization [44]. HD-ZIP I proteins from different species were reported to participate in various abiotic adversity responses. In rice, OsHOX22 acted as a negative regulator of drought and salt tolerance through an ABA-dependent pathway [45]. MtHB1 from Medicago truncatula controlled lateral root emergence under abiotic stresses [46]. ZmHDZ10 positively regulated drought and salt stress dependent on ABA signal pathway [47]. In Arabidopsis, AtHB6 was reported to be involved in ABA response by interacting with the PP2C phosphatase ABI1 and mediated drought stress responses acting downstream of ABI1 and ABI2 [48,49]. AtHB13 regulated response to freezing temperature via inducing genes expression which involved in cell membrane stabilization and inhibition of ice growth [50]. In addition, AtHB13 positively regulates a NAC transcription factor gene JUB1 expression to confer Arabidopsis drought tolerance [51]. AtHB7 and AtHB12 negatively module ABA signaling pathway by activating PP2C genes and repressing ABA receptors PYL5 and PYL8 [52]. Olsson et al. showed that AtHB7 and AtHB12 are negative developmental regulators in response to drought [53]. Our previous data showed that AtHB7 and AtHB12 were highly up-regulated among the differential expression genes in Col-0 wild type treated with or without Al [7].
Since AtHB7 and AtHB12 as two members of HD-ZIP I subfamily play important roles in regulation of tolerance to abiotic stress and were significantly induced by Al, we speculated that these two transcription factors might relate to regulation of Al resistance. Here, we showed that AtHB7 and AtHB12 acted in a cooperative manner under normal condition but in an antagonistic manner to regulate primary root growth under Al treatment.
2. Results
2.1. AtHB7 and AtHB12 Induced by Al Stress, Which Was Independent of STOP1
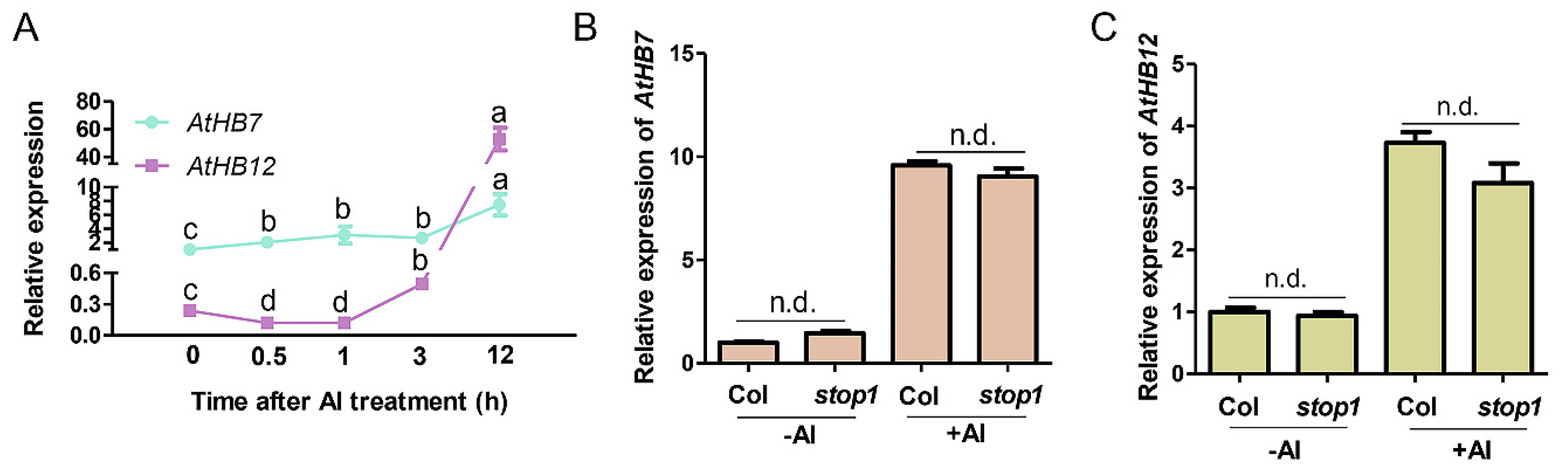
AtHB7 and AtHB12, the most closed members in HD-ZIP I subfamily, were up-regulated 13 folds and 206 folds respectively by Al treatment [7]. To characterize the potential roles of AtHB7 and AtHB12 in root growth response to Al stress, we further analyzed the expression patterns of AtHB7 and AtHB12 under Al stress through a time-course analysis. Real-time qPCR results showed that AtHB7 mRNA level was dominant in early response stage and increased with Al treatment, whereas AtHB12 expression level appeared to be down-regulated initially, and then up-regulated at 6 h after Al exposure and surpassed AtHB7 accumulation level at 12 h (Figure 1A). The different dynamic expression of AtHB7 and AtHB12 in response to Al stimuli suggests that AtHB7 and AtHB12 might play different roles in response to Al stress. To clarify the molecular mechanism of transcriptional regulation of AtHB7 and AtHB12 in response to Al stress, we firstly examined if STOP1, which is a key transcription factor during Al stress response and regulates multiple Al resistance genes expression, was involved in this process. The results showed that the expression patterns of AtHB7 and AtHB12 were not changed in stop1 mutant with or without Al treatment, indicating that Al-regulated AtHB7 and AtHB12 expression is independent of STOP1 (Figure 1B,C).
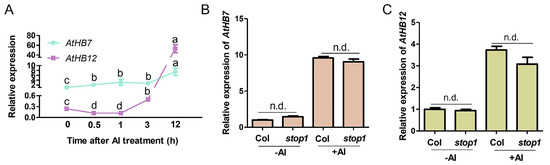
Figure 1.
Two HD-Zip I transcription factors AtHB7 and AtHB12 response to Al stress independent of STOP1 regulation. (A) Time-course analysis of AtHB7 and AtHB12 relative expression in roots in response to 20 μM Al exposure. The relative expression was normalized to ACTIN2 expression and AtHB7 expression untreated with Al was set as 1. Relative expression of AtHB7 (B) and AtHB12 (C) in roots of wild type and stop1 mutant in response to 20 μM Al exposure. Data are means ± SD (n = 3). Different letters indicate significant differences by Tukey’s test.
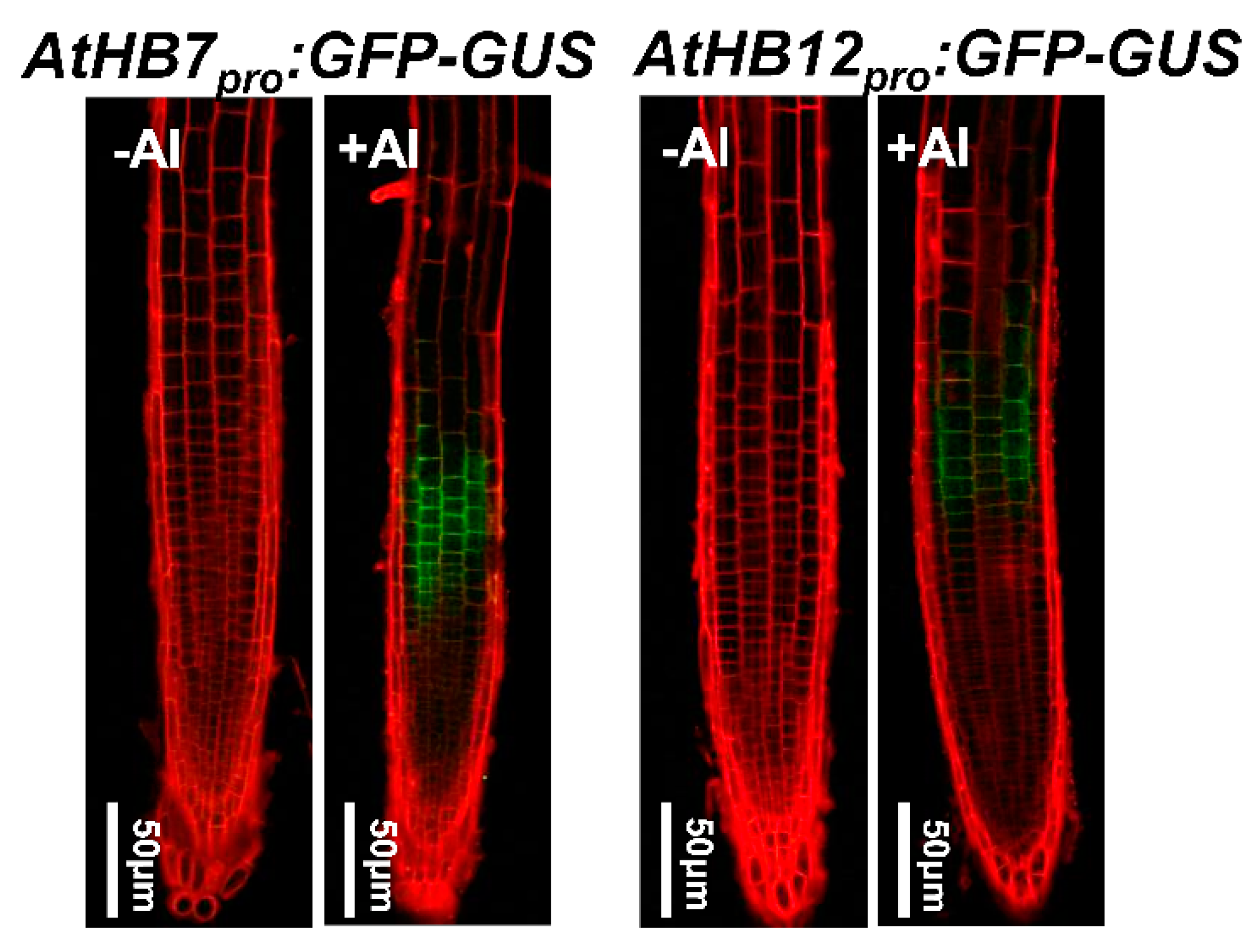
To address the expression patterns of AtHB7 and AtHB12 in the presence of Al stress, the effect of exposure to Al stress on the spatial expression of AtHB7 and AtHB12 was analyzed by monitoring the expression of the AtHB7pro:eGFP-GUS and AtHB12pro:eGFP-GUS transgenes. After a 6 h exposure to 25 µM Al, GFP signals were highly detected in the root apex TZ, while there is no GFP signal in the root tips under non-stressed conditions (Figure 2).
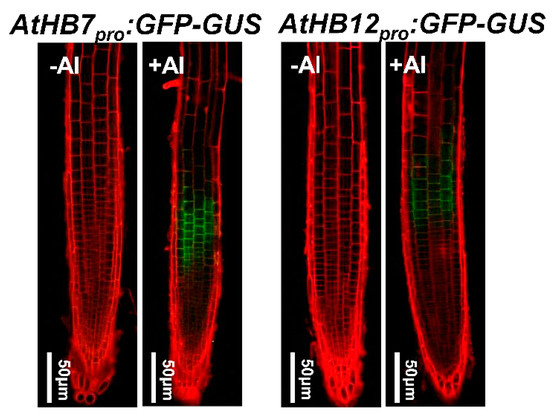
Figure 2.
Both AtHB7 and AtHB12 were induced by Al toxicity in transition zone. Expression of AtHB7pro:GFP-GUS and AtHB12pro:GFP-GUS in root tips of seedlings exposed without or with 20 μM Al for 6 h were analyzed by observation using confocal laser scanning microscopy.
2.2. AtHB7 and AtHB12 Promote Root Growth by Regulating Cell Number and Cell Length
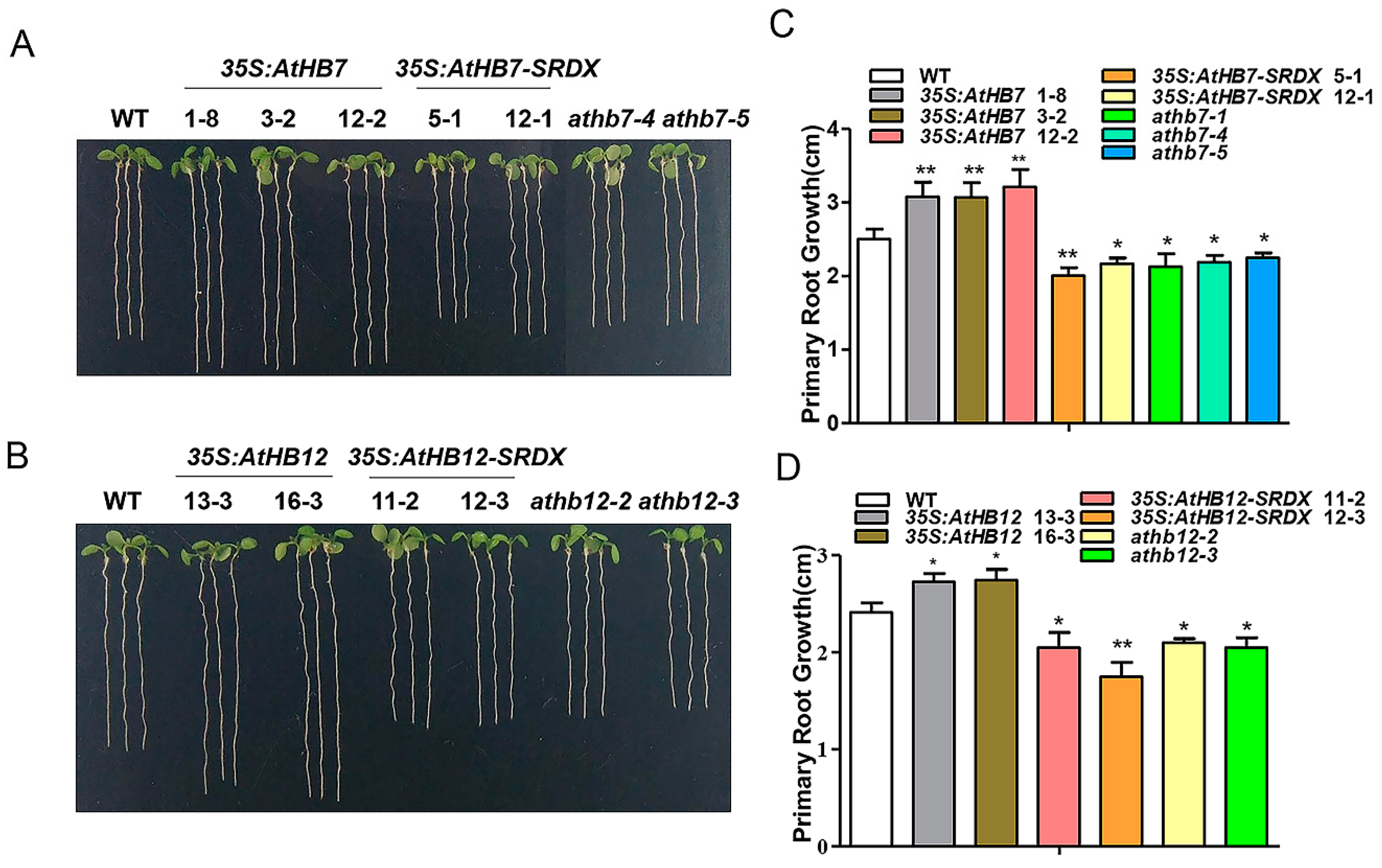
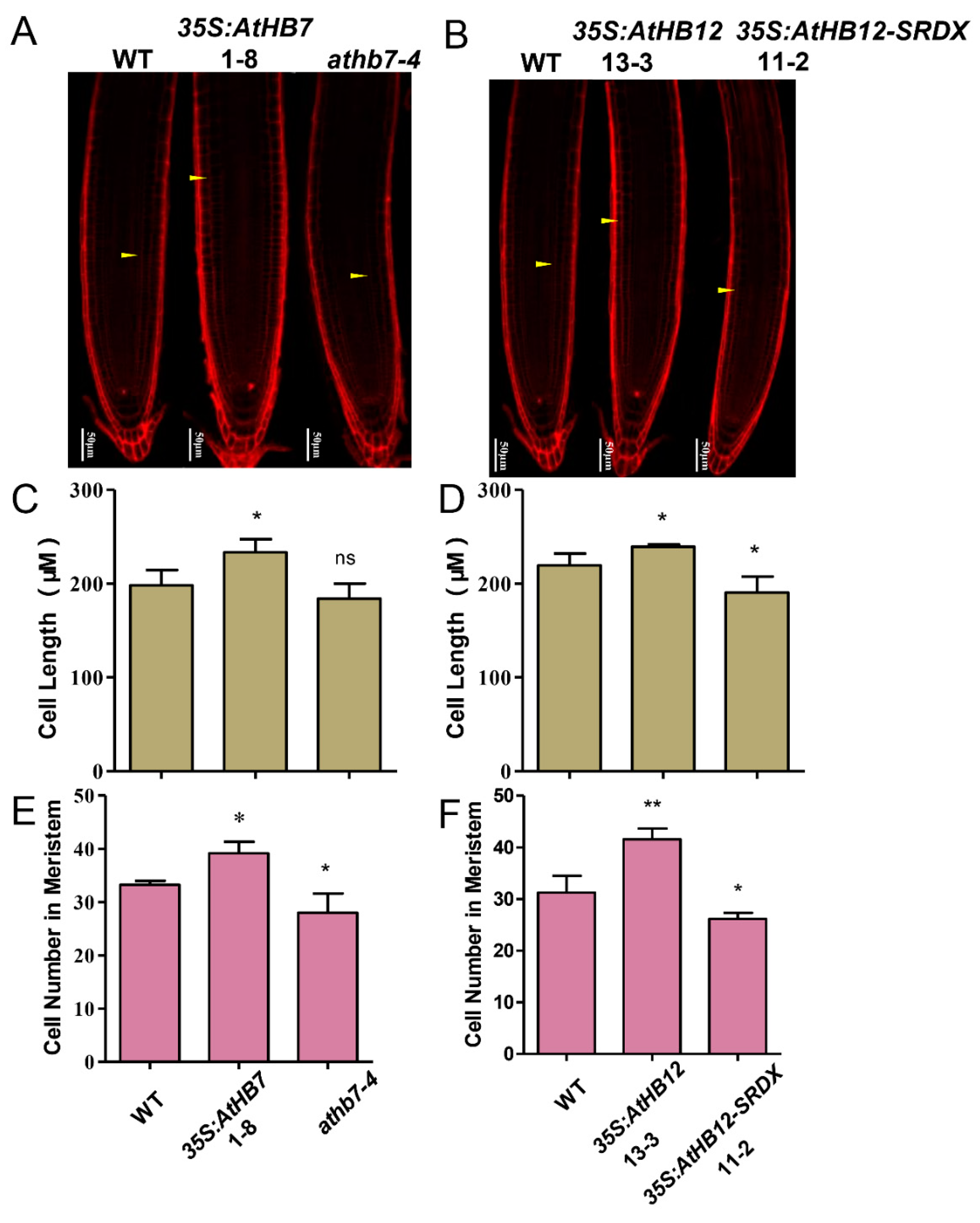
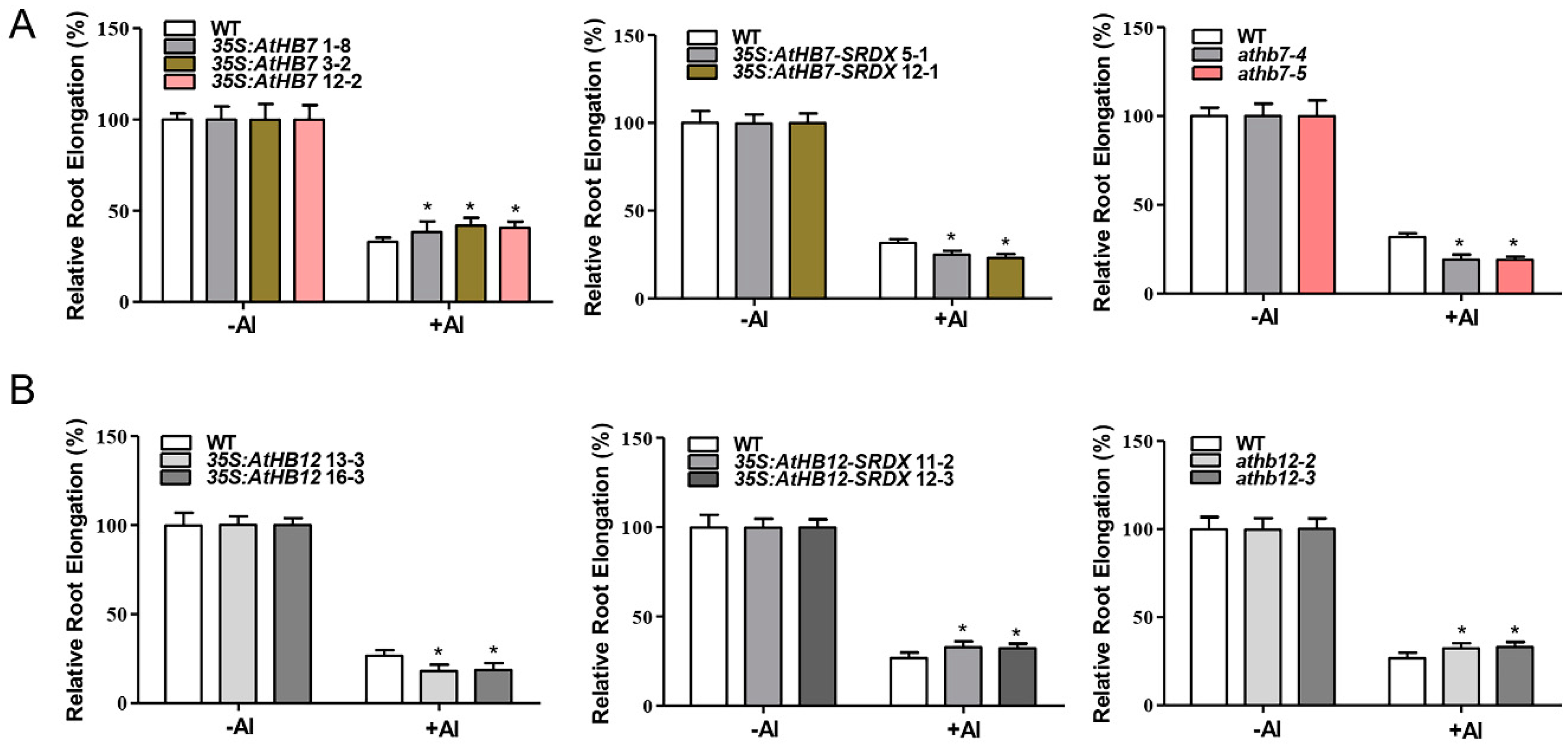
To investigate whether Al responsive AtHB7 and AtHB12 are involved in root growth, we generated 35S:AtHB7/12 overexpression transgenic plants, the dominant-negative constructs 35S:AtHB7/12-SRDX (SUPERMAN repression domain, SRDX) lines and athb7 and athb12 mutants edited by CRISPR CAS9 system (Supplementary Figure S1). Under normal condition, the roots of 35S:AtHB7 and 35S:AtHB12 were longer than those of the wild type, while 35S:AtHB7-SRDX and 35S:AtHB12-SRDX showed short-root phenotypes, which were also confirmed by root length analysis in the athb7 and athb12 mutants (Figure 3). A longitudinal zonation pattern analysis showed that root apical meristem (RAM) of 35S:AtHB7 and 35S:AtHB12 seedlings were significantly longer than that of the WT control (Figure 4A,B). Cell numbers in the meristem zone (MZ) and cell length in the elongation zone (EZ) of 35S:AtHB7 and 35S:AtHB12 seedlings were both increased compared to the WT control (Figure 4C–F). However, the athb7 mutant and the 35S:AtHB12-SRDX transgene showed a reduced size in the root MZ, and cortical cells in the EZ were shorter than those in WT, and cell numbers in the MZ were also strongly reduced (Figure 4C–F). These results suggest that AtHB7 and AtHB12 affect both cell proliferation and cell elongation.
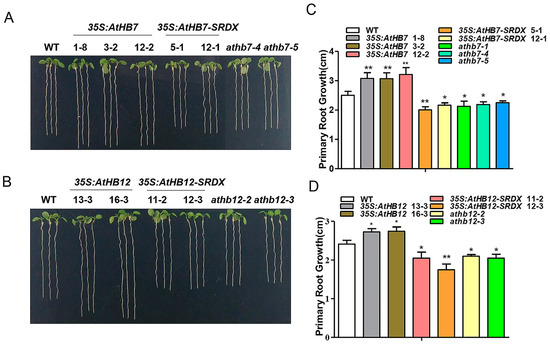
Figure 3.
AtHB7 and AtHB12 contribute to primary root growth during normal development. Phenotype of 6-day-old AtHB7 (A) and AtHB12 (B) overexpression lines, SRDX (SUPERMAN repression domain) transgenic seedlings and mutants under standard growth condition. Primary root length in A (C) and B (D). Data are means ± SD (n = 24). Asterisks indicate significant differences between wild type and transgenic lines at *: p < 0.05 and **: p < 0.01 by Tukey’s test.
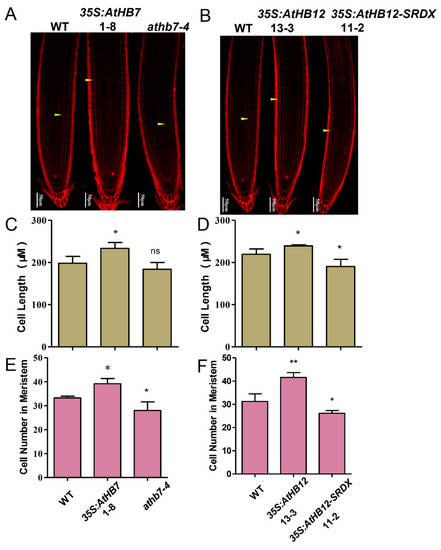
Figure 4.
AtHB7 and AtHB12 module root growth by regulating cell number and cell length. Confocal images of root tips of 6-day-old wild type, AtHB7 (A) and AtHB12 (B) transgenic seedlings including overexpression, SRDX and CRISPR (Clustered regularly interspaced short palindromic repeats) lines stained with PI. White Arrows indicated the first elongated cell in cortex. Cell length of elongation zone in AtHB7 (C) and AtHB12 (D) transgenic plants. Cell number in RAM (root apical meristem) of AtHB7 (E) and AtHB12 (F) transgenic plants. Data are means ± SD (n = 30). Asterisks indicate significant differences between wild type and transgenic seedlings at *: p < 0.05 and **: p < 0.01 by Tukey’s test.
2.3. AtHB7 and AtHB12 Antagonistically Regulate Root Elongation under Al Stress
We further examined the root growth under Al stress. The results showed that 35S:AtHB7 transgenic lines displayed a reduced root-growth inhibition in response to Al stress, while 35S:AtHB7-SRDX transgenic lines and athb7 mutants all showed an enhanced root-growth inhibition after Al treatment (Figure 5A). However, there were no difference between wild type and the 35S:AtHB7, 35S:AtHB7-SRDX transgenic lines and athb7 mutants when plants were exposed to solutions containing different toxic ions including sodium (Na+), copper (Cu2+), lanthanum (La3+) and cadmium (Cd2+), or to various pH solutions (4.5–6.5), indicating that AtHB7 responds to Al stress specifically (Supplementary Figure S2). These results indicate that AtHB7 is a positive regulator of Al resistance. In contrast to AtHB7, 35S:AtHB12 transgenic lines showed an elevated root growth inhibition in response to Al stress, while 35S:AtHB12-SRDX and athb12 mutants showed significantly less inhibition than the WT control (Figure 5B). Similar to AtHB7, no differences in root growth were observed when WT and 35S:AtHB12, 35S:AtHB12-SRDX transgenic lines and athb12 mutant plants were exposed to other metal ions stress and different pH solutions (Supplementary Figure S3). These results indicate that AtHB12 negatively regulates Al resistance.
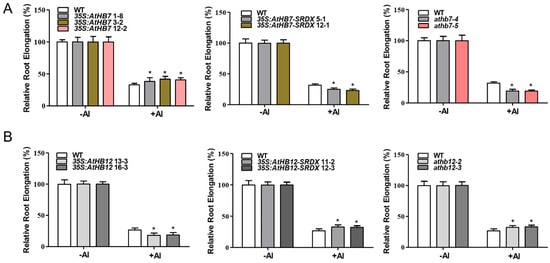
Figure 5.
AtHB7 positively regulates Al resistance (A), whereas AtHB12 negatively regulates Al resistance (B). Relative root elongation of wild type, AtHB7 (A) and AtHB12 (B) overexpression, SRDX and CRISPR lines in response to 6 μM Al for 6 days. Data are means ± SD (n = 30). Asterisks indicate significant differences between wild type and transgenic seedlings at *: p < 0.05 by Tukey’s test.
2.4. AtHB7 and AtHB12 Oppositely Regulate Aluminum Deposition in Cell Wall
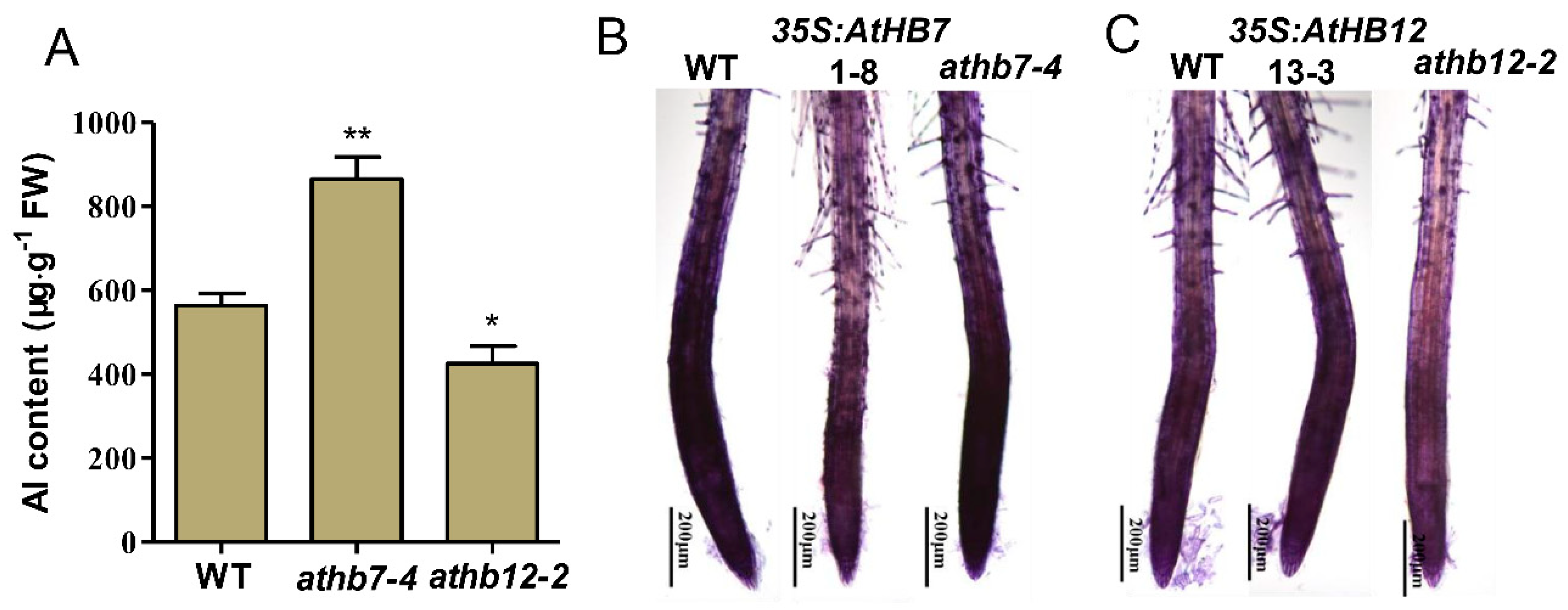
To investigate the underlying mechanisms of how AtHB7/12 regulate Al resistance, Al content in roots of athb7 mutant, athb12 mutant and wild type was detected. Compared with wild type, athb7 mutant accumulated much more Al in roots, but athb12 mutant held less Al in roots (Figure 6A). In depth, hematoxylin staining technique was used to visualize Al accumulation in root cell walls of wild type, AtHB7 or AtHB12 transgenic plants. The staining intensity was stronger in athb7 root tip region and decreased in AtHB7 overexpression plants compared to wild type (Figure 6B). However, stronger intensity in AtHB12 overexpression plants and weaker stain in the athb12 mutant were observed (Figure 6C). Taken together, these results indicate that AtHB7 and AtHB12 differently regulated Al tolerance by affecting Al accumulation in root cell wall.
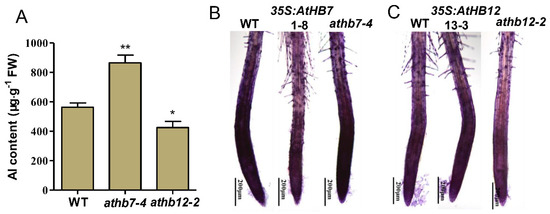
Figure 6.
AtHB7 and AtHB12 oppositely regulate Al binding to cell wall. (A) Analysis of Al content in roots of wild type, athb7 and athb12 mutants. AtHB7 (B) and AtHB12 (C) affect Al accumulations in cell walls of root tips indicated by hematoxylin staining. Asterisks indicate significant differences between wild type and mutants at *: p < 0.05 and **: p < 0.01 by Tukey’s test.
2.5. AtHB7 and AtHB12 Might Act in Homodimer or Heterodimer to Control Root Growth under Al Stress
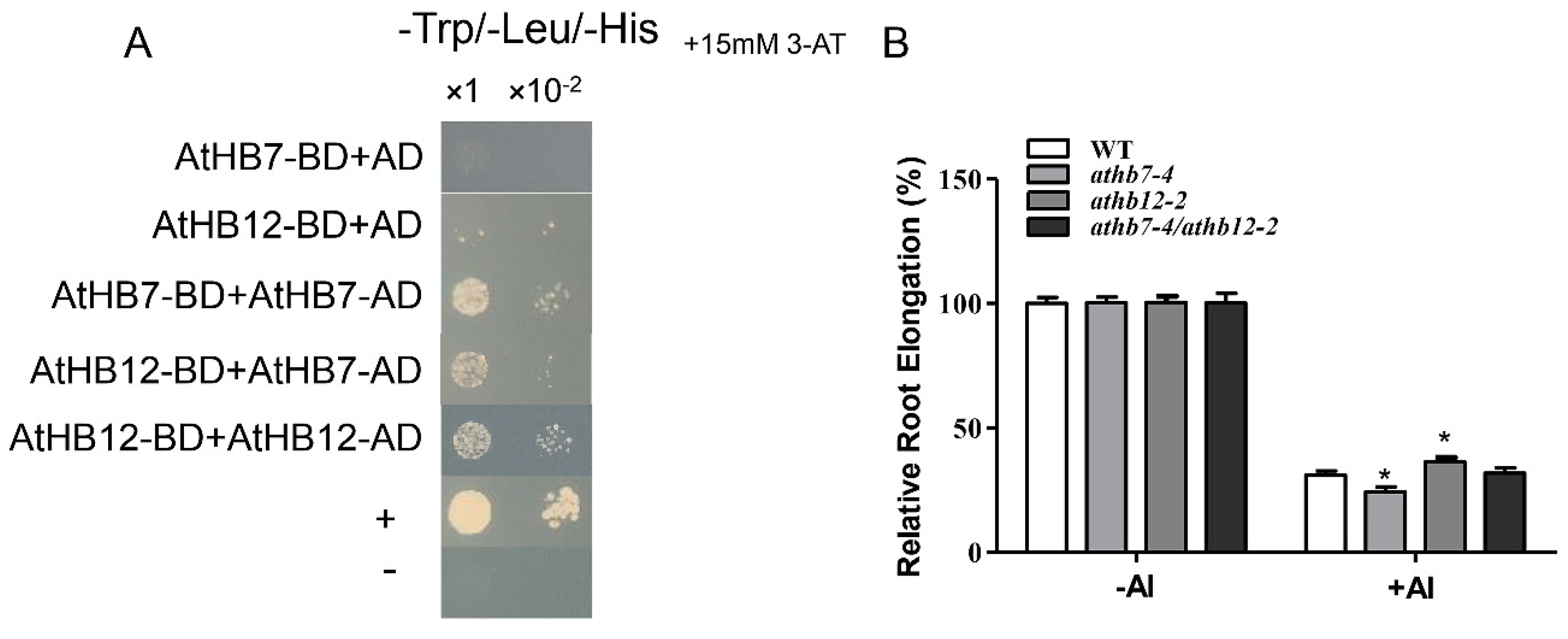
HD ZIP transcription factors are known for containing a homeodomain/leucine zipper domain, the former domain specifically binding to DNA and the latter domain mediating the formation of protein dimers. Based on the high level homology between AtHB7 and AtHB12 and the distinctively opposite function in Al-stress response, we speculated that AtHB7 and AtHB12 may interact to form a heterodimer that affects protein activity or stability to antagonistically regulate Al resistance in Arabidopsis. Yeast two-hybrid assays showed that AtHB7 could not only form homodimer with itself but also form heterodimer with AtHB12 (Figure 7A). Further, the athb7/hb12 double mutant exhibited similar Al resistance with wild type (Figure 7B). These results suggest that AtHB7 and AtHB12 may form a heterodimer to inhibit mutual activities in regulating downstream Al-response genes under Al stress.
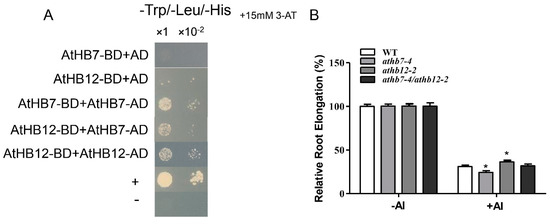
Figure 7.
AtHB7 and AtHB12 might act in homodimer or heterodimer in response to Al stress. (A) AtHB7 and AtHB12 could form homodimer and heterodimer showed by yeast two-hybrid assay. (B) Relative root growth of wild type, athb7, athb12 and athb7/athb12 double mutant in response to Al treatment. Data are means ± SD (n = 30). Asterisks indicate significant differences between wild type and mutants at *: p < 0.05 by Tukey’s test.
3. Discussion
Al stress in one of the most limiting factors for plant growth and crop productivity in acidic soils. Identification of the molecules involved in Al tolerance mechanisms is a current challenge for Al toxicity. Previous studies identified STOP1 as a key transcription factor that regulates primary root growth in response to Al stress. In the present study, we identified two novel regulators of Al resistance. AtHB7 and AtHB12, two HD-Zip I transcription factors, oppositely regulate Al resistance in Arabidopsis. This is directly proved by the fact that AtHB7 overexpression lines and athb12 mutants are tolerant to Al and athb7 mutant, and AtHB12 overexpression lines are sensitive to Al (Figure 5). However, AtHB7 and AtHB12 are not associated with tolerance to other metal such as La3+, Cd2+, Cu2+, Na+ and low pH (Supplementary Figures S2 and S3). Therefore, we conclude that AtHB7 and AtHB12 are specifically involved in Al resistance. The HD-ZIP transcription factor family consists of 47 members in Arabidopsis and several HD-ZIP I subfamily members were found to take part in response to unfavorable conditions such as salt stress, drought stress, ABA and cold stress. In the study, we firstly demonstrated that these two proteins belonging to HD-ZIP I subfamily function in Al resistance mechanisms.
Although AtHB7 and AtHB12 are the most closely related members in HD-Zip I family (Supplementary Figure S4), AtHB7 and AtHB12 showed opposite function in Al resistance. Previous study also showed that AtHB7 was repressed by water loss, while AtHB12 was induced by water loss under water stress conditions; in addition, AtHB7 and AtHB12 play different roles in certain biological process or developmental stage like stomata aperture and senescence [54].
It is interesting to note that AtHB7 and AtHB12 play opposite roles in Al resistance, while both promote root growth during normal development (Figure 3 and Figure 5). The difference of DNA-binding homeodomains between AtHB7 and AtHB12 might lead to distinct target genes in response to Al stress; AtHB7 and AtHB12 also perhaps interact with different partners in Al-response due to the distinct leucine zipper domains between AtHB7 and AtHB12. For instance, some transcription factors interact with transcriptional repressor to negatively regulate target genes. Human transcription factor LEF1 was showed to interact with HDAC1 (Histone Deacetylase1) and its function shift from activation to repression regulated by β-Catenin–Histone Deacetylase [55]. BZR1 acts as either a transcriptional activator or a repressor partly depend on the context of promoters of target genes and trans-factors. BZR1-PIF4 (phytochrome interacting factor 4) heterodimer binds to the E-box motif to activate target expression [56]; while BZR1 interacts with TPL (Groucho/TUP1-like corepressor TOPLESS) through its EAR motif to transcriptionally repress target genes [57]. AtHB7 and AtHB12 were revealed to function as transcription activators in Arabidopsis under normal conditions by transient expression assays [44]. However, recent report showed that AtHB12 interacted with TFIIB through its AHA (aromatic large hydrophobic acidic residues) transactivation motif which locates in carboxy termini, while AtHB7 interacted with both TBP and TFIIB to activate target genes [58].
Our results showed that AtHB7 and AtHB12 were able to form heterodimers (Figure 7A). The leucine zipper domain of HD Zip provides proper structure to allow the formation of dimers [59,60]. For example, AtHB5 was reported to be able to form homodimer and heterodimers with other HD-Zip I members [61]. OsHox1 as a member of HD-Zip Ⅱ family could interact with HAT which is an Arabidopsis HD-Zip I member, whereas the corresponding overexpression transgenic plants show opposite phenotype such as developmental rate [62]. Hetero-dimerization of AtHB7 and AtHB12 seems to inhibit activities of each other under Al stress, and the athb7/hb12 double mutant show similar Al resistance with wild type (Figure 7B).
In this study, we further demonstrated that AtHB7 and AtHB12 differently mediated Al-induced root elongation, which was attributed to distinct capacity of cell wall to bind Al (Figure 6). It is in line with previous findings that AtHB12 regulates expression of cell wall associated genes. AtHB12 overexpression promotes leaf cell expansion by increasing expression of cell wall-related genes such as EXPANSINA10 and DWARF4 [63]. Cell wall is composed of pectin, hemicellulose and cellulose, and pectin and hemicellulose were accepted as the major targets to bind Al [64,65,66]. It is well known that compositional and structural modification of the cell wall, especially pectin methylation and modification of xyloglucan oligosaccharides, exert crucial function in Al resistance. Functional genes associated with cell wall modification were widely characterized in Arabidopsis such as WAK1 (cell wall-associated receptor kinase 1), PME46, LUH (LEUNIG_HOMOLOG), STAR1 (Sensitive to Al rhizotoxicity1), ALS3 (ALUMINUM SENSITIVE 3), and XTH17 and XTH31 [19,20,22,23,24,25,67,68]. Next, we intend to explore whether the above genes mediated AtHB7/12 regulated Al resistance, and discover more novel Al-resistance genes controlled by AtHB7 or AtHB12 based on whole transcriptome sequencing to enrich our knowledge in mechanisms of plants Al resistance.
4. Materials and Methods
4.1. Plant Culture and Treatment
Arabidopsis cultured in Petri dishes with 1/2 MS solid medium or in nutrient solutions or in soils were placed in a plant growth chamber at 22 °C, 150 μmolm−2 s−1 light intensity with a long-day photoperiod (16-h light/8-h dark). For plate culture, seeds were surface disinfected with 10% sodium hypochlorite solution for 5 min, and then washed three times with sterile water [69]. The disinfected seeds were sowed onto 1/2 MS medium and incubated at 4 °C in darkness for 2 days. Then, the plates with seeds were vertically placed in the chamber. For Al and different metal treatment, Arabidopsis seeds soaked in sterile water at 4 °C overnight germinated in 2% MGRL solution (pH 5.0) without or with 6 μM AlCl3, 1 mM LaCl3, 3.5 mM CdCl2, 2 mM CuCl2, 10 mM NaCl for 6 days. For different pH treatment, the soaked Arabidopsis germinated in 2% MGRL solution with pH 6.5, 6.0, 5.5, 5.0, 4.7, 4.5 for 6 days. Treatment solution was renewed every day. Then, the primary root length was analysis by Image J (developed by National Institutes of Health) after treatment.
4.2. Materials and Transgenic Arabidopsis Construction
athb7 and athb12 mutants were obtained by YAO-promoter driven CRISPR/cas9 gene editing method according to previous reports [70]. Briefly, two sgRNA targets of each gene were designed, then cloned into middle vector in turn and eventually two pYAO:hSpCas9-target1-sgRNA-target2-sgRNA recombinant vectors for editing AtHB7 and AtHB12 respectively were constructed. The amplified AtHB7 and AtHB12 ORF were respectively integrated into pENTR™ TOPO® vector using pENTR™/D-TOPO™ Cloning Kit (Thermo Fisher, catalog number K2400-20), and subsequently transferred to pB7WG2 Gateway destination vector to obtain 35S:AtHB7 and 35S:AtHB12 overexpression vectors. AtHB7 and AtHB12 coding sequences were respectively fused with the EAR-repression domain (SRDX) under the control of the 35S promoter to generate 35S:AtHB7-SRDX and 35S:AtHB12-SRDX recombinant vectors. In total, 2.1 Kb DNA fragment upstream from initiation codon of AtHB7 was cloned to drive repoter genes expression using Gateway cloning technology to construct AtHB7pro:eGFP-GUS. Likewise, approximate 2.2 Kb promoter sequence of AtHB12 was amplified to obtain AtHB12pro:eGFP-GUS plasmid.
The above final expression vectors were separately transformed into Col-0 wild type by floral dip method. Then, three T3 homozygous overexpression lines 35S:AtHB7#1(1-8), #2(3-2), #3(12-2), two AtHB7-SRDX transgenic lines 35S:AtHB7-SRDX#1(5-1), #2 (12-1) and three CRISPR/Cas9-induced athb7 mutant plants athb7-1, -4, -5 were used for research. To explore AtHB12 gene function, two homozygous overexpression lines 35S:AtHB12#1(13-3), #2(16-3), two SRDX lines 35S:AtHB12-SRDX#1(11-2), #2 (12-3) and two athb12 mutants athb12-2, -3 were employed in this study. athb7-4/athb12-2 double mutant was generated by cross athb7-4 (male parent) with athb12-2 (female parent). The homozygous offerings were identified with PCR. All primers used for recombinant constructions and identification of different genotype were listed in Supplementary Table S1.
4.3. Phylogenetic Analysis
The amino acid sequences of HD-ZIP I members were retrieved from TAIR database and then were aligned with Clustal 2.0 software [71]. The phylogenetic tree was constructed using MEGA 7.0 by neighbor-joining (NJ) method with 1000 bootstrap replications [72].
4.4. RNA Isolation and qRT PCR
Total RNA was extracted from intact roots of 6-day-old plants treated without or with 20 μM Al using TaKaRa MiniBEST Plant RNA Extraction Kit (TAKARA, catalog number 9769). At first, genome DNA was removed by gDNase (TIANGEN BIOTECH Co., Ltd., catalog number KR116) (TIANGEN Biotech, Beijing, China). Then, 1 μg RNA was synthesized into the first cDNA strand with FastKing RT Kit according to manufacturers’ protocols (TIANGEN BIOTECH Co., Ltd., catalog number KR116) (TIANGEN Biotech, Beijing, China). The real-time quantitative RT-PCR was performed with SuperReal PreMix Plus (SYBR Green) provided by TIANGEN company using CFX Connect Real-Time System (Bio-Rad, Hercules, CA, USA) [73]. qPCR reaction conditions were 45 cycles at 95 °C for 10 s, 56 °C for 20 s, and 72 °C for 20 s. Expression levels were normalized to the expression level of UBQ10. All qRT PCR experiments were done three repeats from different biological samples.
4.5. Confocal Microscopy
6-day-old AtHB7pro:eGFP-GUS or AtHB12pro:eGFP-GUS seedlings were treated in 2% MGRL solution without or with 20 μM Al at pH 5.0. Then, the roots were immersed in 1 μg/ml propidium iodide (PI) solution and imaged using a confocal laser scanning microscope (Zeiss LSM700). For PI and GFP channels, the 543 nm and the 488 nm wavelengths were used for excitation, respectively. Four-day-old seedlings of wild type, AtHB7 or AtHB12 transgenic plants were stain with PI for growth analysis. Cortical cell numbers in the root apical meristem (RAM) and length of elongation zone cell were determined as previous described [74]. In detail, RAM comprised of the organized cells from QC to the first elongated cell. Elongation zone boundaries were from the first dramatically elongating cell to the cell developing root hairs.
4.6. Al Content Determination
Seeds of wild type, athb7 and athb12 mutants germinated in 2% MGRL solution at pH 5.5 for 6 days and then the seedlings were exposed to 2% MGRL solution with 10 μM Al at pH 5.0. After 24 h, the roots were excised, washed three times with deionized water, dried and digested with 65% HNO3. Al content in roots was measured by GF-AAS (SHIMADZU, Japan).
4.7. Hematoxylin Staining
Arabidopsis seeds were germinated in 2% MGRL solution at pH 5.5 for 6 days and then the seedlings were transferred to 2% MGRL solution supplemented with 20 μM Al at pH 5.0 for 6 h. After treatment, the seedlings were stained with 0.2% hematoxylin containing 0.02% KIO4 for 20 min and washed three times with deionized water before observations. The samples were observed and photographed using an Olympus BX53 microscope equipped with an Olympus DP72 camera system.
4.8. Yeast Two-Hybrid Assay
Analysis of protein–protein interactions in yeast were carried out using the Matchmaker Gold Yeast Two-Hybrid System provided by TAKARA. The coding sequence of AtHB7 and AtHB12 were transferred from pENTRTM to the pGBKT7 (BD) vector by Gateway LR reaction to generate the constructs AtHB7-BD and AtHB12-BD as the preys. Similarly, the amplified coding sequence of AtHB7 and AtHB12 were fused with the PGADT7 (AD), resulting in the baits. The prey and bait were co-transformed into yeast cells. The transformed yeasts were selected on dropout medium without Leu and Trp. Then analysis of interaction was indicated by the growth of yeast colonies on synthetic medium lacking Leu, Trp, His or lacking Leu, Trp, His, and Ade with 3-AT incubated at 30 °C for 3 days.
4.9. Statistical Analysis
Statistical analysis was performed by Tukey’s test among treatments. Three biological replicates for each treatment were conducted for the statistical analysis in this article. Asterisks or different letters in the figures indicated significant differences as follows: * p < 0.05, ** p < 0.01, and *** p < 0.001.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/11/4080/s1, Figure S1: Structure of mutated AtHB7 and AtHB12 genes edited by Crispr/Cas9. Figure S2: AtHB7 specifically regulates Al resistance. Relative root elongation of wild type, AtHB7 transgenic seedlings including overexpression, SRDX and CRISPR lines in response to different metal ions and pH changes. Figure S3: AtHB12 specifically regulates Al resistance. Relative root elongation of wild type, AtHB12 transgenic plants including overexpression, SRDX and CRISPR lines in response to different metal ions and pH changes. Figure S4: Phylogenetic analysis of HD-ZIP I subfamily members. Supplementary Table S1: Primers used in this study.
Author Contributions
Conceptualization, Z.D. and X.K.; methodology, Y.L.; software, Y.L.; validation, Y.L., J.X. and H.T.; formal analysis, J.X.; investigation, Y.L.; resources, Y.L.; data curation, Y.L.; writing—original draft preparation, J.X. and X.K.; writing—review and editing, Z.D., X.K., J.X., H.T., S.G., X.Y., S.Z., S.D., Y.S. and X.L.; visualization, Y.L., J.X.; supervision, Z.D.; project administration, Y.L.; funding acquisition, Z.D. and X.K. All authors have read and agreed to the published version of the manuscript.
Funding
The work was financially supported by Qingdao’s Leading Technology Innovator Project, Youth Interdisciplinary Science and Innovative Research Groups of Shandong University (Grant No. 2020QNQT014), the Natural Science Foundation of Shandong Province (ZR2019PC037), the National Natural Science Foundation of China (No. 31970192), the Young Scholars Program of Shandong University (No. 2018WLJH81), and the Key Research and Development Projects of Shandong Province (No. 2019GSF107011).
Conflicts of Interest
The authors declare that they have no conflict of interest.
Abbreviations
| Kb | 1000 base pair |
| MS | Murashige and Skoog medium |
| GF-AAS | Graphite Furnace Atomic Absorption Spectrometry |
References
- Von Uexküll, H.R.; Mutert, E. Global extent, development and economic impact of acid soils. Plant Soil 1995, 171, 1–15. [Google Scholar] [CrossRef]
- Kochian, L.V. Cellular mechanisms of aluminum toxicity and resistance in plants. Annu. Rev. Plant Biol. 1995, 46, 237–245. [Google Scholar] [CrossRef]
- Ma, J.F. Syndrome of aluminum toxicity and diversity of aluminum resistance in higher plants. Int. Rev. Cytol. 2007, 264, 225–252. [Google Scholar] [PubMed]
- Ryan, P.R.; Tyerman, S.D.; Sasaki, T.; Furuichi, T.; Yamamoto, Y.; Zhang, W.H.; Delhaize, E. The identification of aluminium-resistance genes provides opportunities for enhancing crop production on acid soils. J. Exp. Bot. 2011, 62, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Piñeros, M.A.; Kochian, L.V. The role of aluminum sensing and signaling in plant aluminum resistance. J. Integr. Plant Biol. 2014, 56, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Panda, S.K.; Baluška, F.; Matsumoto, H. Aluminum stress signaling in plants. Plant Signal. Behav. 2009, 4, 592–597. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.B.; Geng, X.Y.; He, C.M.; Zhang, F.; Wang, R.; Horst, W.J.; Ding, Z.J. TAA1-Regulated Local Auxin Biosynthesis in the Root-Apex Transition Zone Mediates the Aluminum-Induced Inhibition of Root Growth inArabidopsis. Plant Cell 2014, 26, 2889–2904. [Google Scholar] [CrossRef]
- Ryan, P.R.; Kochian, L.V. Interaction between aluminum toxicity and calcium uptake at the root apex in near-isogenic lines of wheat (Triticum aestivum L.) differing in aluminum tolerance. Plant Physiol. 1993, 102, 975–982. [Google Scholar] [CrossRef]
- Sivaguru, M.; Horst, W.J. The distal part of the transition zone is the most aluminum-sensitive apical root zone of maize. Plant Physiol. 1998, 116, 155–163. [Google Scholar] [CrossRef]
- Sivaguru, M.; Liu, J.; Kochian, L.V. Targeted expression of SbMATE in the root distal transition zone is responsible for sorghum aluminum resistance. Plant J. 2013, 76, 297–307. [Google Scholar] [CrossRef]
- Rangel, A.F.; Rao, I.M.; Horst, W.J. Spatial aluminium sensitivity of root apices of two common bean (Phaseolus vulgaris L.) genotypes with contrasting aluminium resistance. J. Exp. Bot. 2007, 58, 3895–3904. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Gao, S.; Tian, H.; Wu, W.; Robert, H.S.; Ding, Z. Local Transcriptional Control of YUCCA Regulates Auxin Promoted Root-Growth Inhibition in Response to Aluminium Stress in Arabidopsis. PLoS Genet. 2016, 12, e1006360. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.B.; Liu, G.C.; Liu, J.J.; Zhang, B.; Meng, W.J.; Müller, B.; Hayashi, K.; Zhang, X.S.; Zhao, Z.; De Smet, I.; et al. Synergistic action of auxin and cytokinin mediates aluminum-induced root growth inhibition in Arabidopsis. Embo Rep. 2017, 18, 1213–1230. [Google Scholar] [CrossRef]
- Kollmeier, M.; Felle, H.H.; Horst, W.J. Genotypical differences in aluminum resistance of maize are expressed in the distal part of the transition zone. Is reduced basipetal auxin flow involved in inhibition of root elongation by aluminum? Plant Physiol. 2000, 122, 945–956. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Lu, X.; Li, C.; Zhang, B.; Zhang, C.; Zhang, X.; Ding, Z. Auxin Efflux Carrier ZmPGP1 Mediates Root Growth Inhibition under Aluminum Stress. Plant Physiol. 2018, 177, 819–832. [Google Scholar] [CrossRef]
- Clarkson, D.T. Interactions between aluminium and phosphorus on root surfaces and cell wall material. Plant Soil 1967, 27, 347–356. [Google Scholar] [CrossRef]
- Ma, J.F.; Yamamoto, R.; Nevins, D.J.; Matsumoto, H.; Brown, P.H. Al binding in the epidermis cell wall inhibits cell elongation of okra hypocotyl. Plant Cell Physiol. 1999, 40, 549–556. [Google Scholar] [CrossRef]
- Horst, W.J.; Wang, Y.; Eticha, D. The role of the root apoplast in aluminium-induced inhibition of root elongation and in aluminium resistance of plants: A review. Ann. Bot. 2010, 106, 185–197. [Google Scholar] [CrossRef]
- Geng, X.; Horst, W.J.; Golz, J.F.; Lee, J.E.; Ding, Z.; Yang, Z.B. Leunig_Homolog transcriptional co-repressor mediates aluminium sensitivity through Pectin Methylesterase46-modulated root cell wall pectin methylesterification in Arabidopsis. Plant J. Cell Mol. Biol. 2017, 90, 491–504. [Google Scholar] [CrossRef]
- Zhu, X.F.; Shi, Y.Z.; Lei, G.J.; Fry, S.C.; Zhang, B.C.; Zhou, Y.H.; Braam, J.; Jiang, T.; Xu, X.Y.; Mao, C.Z. XTH31, encoding an in vitro XEH/XET-active enzyme, regulates aluminum sensitivity by modulating in vivo XET action, cell wall xyloglucan content, and aluminum binding capacity in Arabidopsis. Plant Cell 2012, 24, 4731–4747. [Google Scholar] [CrossRef]
- Zhu, X.F.; Sun, Y.; Zhang, B.C.; Mansoori, N.; Wan, J.X.; Liu, Y.; Wang, Z.W.; Shi, Y.Z.; Zhou, Y.H.; Zheng, S.J. Trichome Birefringence-Like27 affects aluminum sensitivity by modulating the O-acetylation of xyloglucan and aluminum-binding capacity in Arabidopsis. Plant Physiol. 2014, 166, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Larsen, P.B.; Geisler, M.J.; Jones, C.A.; Williams, K.M.; Cancel, J.D. ALS3 encodes a phloem-localized ABC transporter-like protein that is required for aluminum tolerance in Arabidopsis. Plant J. Cell Mol. Biol. 2005, 41, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.F.; Yamaji, N.; Ma, J.F. Knockout of a bacterial-type ATP-binding cassette transporter gene, AtSTAR1, results in increased aluminum sensitivity in Arabidopsis. Plant Physiol. 2010, 153, 1669–1677. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.M.; Lou, H.Q.; Jin, J.F.; Chen, W.W.; Wan, J.X.; Fan, W.; Yang, J.L. A half-type ABC transporter FeSTAR1 regulates Al resistance possibly via UDP-glucose-based hemicellulose metabolism and Al binding. Plant Soil 2018, 432, 303–314. [Google Scholar] [CrossRef]
- Xu, J.M.; Wang, Z.Q.; Jin, J.F.; Chen, W.W.; Fan, W.; Zheng, S.J.; Yang, J.L. FeSTAR2 interacted by FeSTAR1 alters its subcellular location and regulates Al tolerance in buckwheat. Plant Soil 2019, 436, 489–501. [Google Scholar] [CrossRef]
- Huang, C.F.; Yamaji, N.; Mitani, N.; Yano, M.; Nagamura, Y.; Ma, J.F. A bacterial-type ABC transporter is involved in aluminum tolerance in rice. Plant Cell 2009, 21, 655–667. [Google Scholar] [CrossRef]
- Che, J.; Yamaji, N.; Shen, R.F.; Ma, J.F. An Al-inducible expansin gene, OsEXPA10 is involved in root cell elongation of rice. Plant J. Cell Mol. Biol. 2016, 88, 132–142. [Google Scholar] [CrossRef]
- Kochian, L.V.; Pineros, M.A.; Liu, J.; Magalhaes, J.V. Plant Adaptation to Acid Soils: The Molecular Basis for Crop Aluminum Resistance. Annu. Rev. Plant Biol. 2015, 66, 571–598. [Google Scholar] [CrossRef]
- Hoekenga, O.A.; Maron, L.G.; Piñeros, M.A.; Cançado, G.M.; Shaff, J.; Kobayashi, Y.; Ryan, P.R.; Dong, B.; Delhaize, E.; Sasaki, T.; et al. AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 9738–9743. [Google Scholar] [CrossRef]
- Liu, J.; Magalhaes, J.V.; Shaff, J.; Kochian, L.V. Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. Plant J. Cell Mol. Biol. 2009, 57, 389–399. [Google Scholar] [CrossRef]
- Larsen, P.B.; Cancel, J.; Rounds, M.; Ochoa, V. Arabidopsis ALS1 encodes a root tip and stele localized half type ABC transporter required for root growth in an aluminum toxic environment. Planta 2007, 225, 1447–1458. [Google Scholar] [CrossRef] [PubMed]
- Iuchi, S.; Koyama, H.; Iuchi, A.; Kobayashi, Y.; Kitabayashi, S.; Kobayashi, Y.; Ikka, T.; Hirayama, T.; Shinozaki, K.; Kobayashi, M. Zinc finger protein STOP1 is critical for proton tolerance in Arabidopsis and coregulates a key gene in aluminum tolerance. Proc. Natl. Acad. Sci. USA 2007, 104, 9900–9905. [Google Scholar] [CrossRef] [PubMed]
- Sawaki, Y.; Iuchi, S.; Kobayashi, Y.; Kobayashi, Y.; Ikka, T.; Sakurai, N.; Fujita, M.; Shinozaki, K.; Shibata, D.; Kobayashi, M.; et al. STOP1 regulates multiple genes that protect arabidopsis from proton and aluminum toxicities. Plant Physiol. 2009, 150, 281–294. [Google Scholar] [CrossRef] [PubMed]
- Yamaji, N.; Huang, C.F.; Nagao, S.; Yano, M.; Sato, Y.; Nagamura, Y.; Ma, J.F. A zinc finger transcription factor ART1 regulates multiple genes implicated in aluminum tolerance in rice. Plant Cell 2009, 21, 3339–3349. [Google Scholar] [CrossRef] [PubMed]
- Che, J.; Tsutsui, T.; Yokosho, K.; Yamaji, N.; Ma, J.F. Functional characterization of an aluminum (Al)-inducible transcription factor, ART2, revealed a different pathway for Al tolerance in rice. New Phytol. 2018, 220, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Gao, J.; You, J.; Liang, Y.; Guan, K.; Yan, S.; Zhan, M.; Yang, Z. Identification of STOP1-like proteins associated with aluminum tolerance in sweet sorghum (Sorghum bicolor L.). Front. Plant Sci. 2018, 9, 258. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Ohyama, Y.; Kobayashi, Y.; Ito, H.; Iuchi, S.; Fujita, M.; Zhao, C.R.; Tanveer, T.; Ganesan, M.; Kobayashi, M. STOP2 activates transcription of several genes for Al- and low pH-tolerance that are regulated by STOP1 in Arabidopsis. Mol. Plant 2014, 7, 311–322. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, J.; Guo, J.; Zhou, F.; Singh, S.; Xu, X.; Xie, Q.; Yang, Z.; Huang, C.-F. F-box protein RAE1 regulates the stability of the aluminum-resistance transcription factor STOP1 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2019, 116, 319–327. [Google Scholar] [CrossRef]
- Ding, Z.J.; Yan, J.Y.; Xu, X.Y.; Li, G.X.; Zheng, S.J. WRKY46 functions as a transcriptional repressor of ALMT1, regulating aluminum-induced malate secretion in Arabidopsis. Plant J. Cell Mol. Biol. 2013, 76, 825–835. [Google Scholar] [CrossRef]
- Li, C.X.; Yan, J.Y.; Ren, J.Y.; Sun, L.; Xu, C.; Li, G.X.; Ding, Z.J.; Zheng, S.J. A WRKY transcription factor confers aluminum tolerance via regulation of cell wall modifying genes. J. Integr. Plant Biol. 2019. [Google Scholar] [CrossRef]
- Li, G.Z.; Wang, Z.Q.; Yokosho, K.; Ding, B.; Fan, W.; Gong, Q.Q.; Li, G.X.; Wu, Y.R.; Yang, J.L.; Ma, J.F. Transcription factor WRKY 22 promotes aluminum tolerance via activation of Os FRDL 4 expression and enhancement of citrate secretion in rice (Oryza sativa). New Phytol. 2018, 219, 149–162. [Google Scholar] [CrossRef] [PubMed]
- Arenhart, R.A.; Bai, Y.; de Oliveira, L.F.V.; Neto, L.B.; Schunemann, M.; dos Santos Maraschin, F.; Mariath, J.; Silverio, A.; Sachetto-Martins, G.; Margis, R. New insights into aluminum tolerance in rice: The ASR5 protein binds the STAR1 promoter and other aluminum-responsive genes. Mol. Plant 2014, 7, 709–721. [Google Scholar] [CrossRef] [PubMed]
- Arenhart, R.A.; Schunemann, M.; Bucker Neto, L.; Margis, R.; Wang, Z.Y.; Margis-Pinheiro, M. Rice ASR1 and ASR5 are complementary transcription factors regulating aluminium responsive genes. Plant Cell Environ. 2016, 39, 645–651. [Google Scholar]
- Henriksson, E.; Olsson, A.S.; Johannesson, H.; Johansson, H.; Hanson, J.; Engström, P.; Söderman, E. Homeodomain leucine zipper class I genes in Arabidopsis. Expression patterns and phylogenetic relationships. Plant Physiol. 2005, 139, 509–518. [Google Scholar] [CrossRef]
- Zhang, S.; Haider, I.; Kohlen, W.; Jiang, L.; Bouwmeester, H.; Meijer, A.H.; Schluepmann, H.; Liu, C.-M.; Ouwerkerk, P.B.F. Function of the HD-Zip I gene Oshox22 in ABA-mediated drought and salt tolerances in rice. Plant Mol. Biol. 2012, 80, 571–585. [Google Scholar] [CrossRef]
- Ariel, F.; Diet, A.; Verdenaud, M.; Gruber, V.; Frugier, F.; Chan, R.; Crespi, M. Environmental Regulation of Lateral Root Emergence in Medicago truncatula Requires the HD-Zip I Transcription Factor HB1. Plant Cell 2010, 22, 2171–2183. [Google Scholar] [CrossRef]
- Zhao, Y.; Ma, Q.; Jin, X.; Peng, X.; Liu, J.; Deng, L.; Yan, H.; Sheng, L.; Jiang, H.; Cheng, B. A Novel Maize Homeodomain-Leucine Zipper (HD-Zip) I Gene, Zmhdz10, Positively Regulates Drought and Salt Tolerance in Both Rice and Arabidopsis. Plant Cell Physiol. 2014, 55, 1142–1156. [Google Scholar] [CrossRef]
- Himmelbach, A.; Hoffmann, T.; Leube, M.; Höhener, B.; Grill, E. Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. Embo J. 2002, 21, 3029–3038. [Google Scholar] [CrossRef]
- Söderman, E.; Hjellström, M.; Fahleson, J.; Engström, P. The HD-Zip gene ATHB6 in Arabidopsis is expressed in developing leaves, roots and carpels and up-regulated by water deficit conditions. Plant Mol. Biol. 1999, 40, 1073–1083. [Google Scholar] [CrossRef]
- Cabello, J.V.; Arce, A.L.; Chan, R.L. The homologous HD-Zip I transcription factors HaHB1 and AtHB13 confer cold tolerance via the induction of pathogenesis-related and glucanase proteins. Plant J. 2012, 69, 141–153. [Google Scholar] [CrossRef]
- Ebrahimian-Motlagh, S.; Ribone, P.A.; Thirumalaikumar, V.P.; Allu, A.D.; Chan, R.L.; Mueller-Roeber, B.; Balazadeh, S. JUNGBRUNNEN1 confers drought tolerance downstream of the HD-Zip I transcription factor AtHB13. Front. Plant Sci. 2017, 8, 2118. [Google Scholar] [CrossRef] [PubMed]
- Valdés, A.E.; Övernäs, E.; Johansson, H.; Rada-Iglesias, A.; Engström, P. The homeodomain-leucine zipper (HD-Zip) class I transcription factors ATHB7 and ATHB12 modulate abscisic acid signalling by regulating protein phosphatase 2C and abscisic acid receptor gene activities. Plant Mol. Biol. 2012, 80, 405–418. [Google Scholar] [CrossRef] [PubMed]
- Olsson, A.; Engström, P.; Söderman, E. The homeobox genes ATHB12 and ATHB7encode potential regulators of growth in response to water deficit in Arabidopsis. Plant Mol. Biol. 2004, 55, 663–677. [Google Scholar] [CrossRef] [PubMed]
- Ré, D.A.; Capella, M.; Bonaventure, G.; Chan, R.L. Arabidopsis AtHB7 and AtHB12 evolved divergently to fine tune processes associated with growth and responses to water stress. Bmc Plant Biol. 2014, 14, 150. [Google Scholar] [CrossRef] [PubMed]
- Billin, A.N.; Thirlwell, H.; Ayer, D.E. β-Catenin-histone deacetylase interactions regulate the transition of LEF1 from a transcriptional repressor to an activator. Mol. Cell. Biol. 2000, 20, 6882–6890. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Zhu, J.-Y.; Wang, Z.-Y. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 2012, 14, 802–809. [Google Scholar] [CrossRef]
- Oh, E.; Zhu, J.-Y.; Ryu, H.; Hwang, I.; Wang, Z.-Y. TOPLESS mediates brassinosteroid-induced transcriptional repression through interaction with BZR1. Nat. Commun. 2014, 5, 4140. [Google Scholar] [CrossRef]
- Capella, M.; Ré, D.A.; Arce, A.L.; Chan, R.L. Plant homeodomain-leucine zipper I transcription factors exhibit different functional AHA motifs that selectively interact with TBP or/and TFIIB. Plant Cell Rep. 2014, 33, 955–967. [Google Scholar] [CrossRef]
- Sessa, G.; Morelli, G.; Ruberti, I. The Athb-1 and -2 HD-Zip domains homodimerize forming complexes of different DNA binding specificities. Embo J. 1993, 12, 3507–3517. [Google Scholar] [CrossRef]
- Landschulz, W.H.; Johnson, P.F.; McKnight, S.L. The leucine zipper: A hypothetical structure common to a new class of DNA binding proteins. Science 1988, 240, 1759–1764. [Google Scholar] [CrossRef]
- Johannesson, H.; Wang, Y.; Engström, P. DNA-binding and dimerization preferences of Arabidopsis homeodomain-leucine zipper transcription factors in vitro. Plant Mol. Biol. 2001, 45, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Meijer, A.H.; Scarpella, E.; van Dijk, E.L.; Qin, L.; Taal, A.J.; Rueb, S.; Harrington, S.E.; McCouch, S.R.; Schilperoort, R.A.; Hoge, J.H.C. Transcriptional repression by Oshox1, a novel homeodomain leucine zipper protein from rice. Plant J. 1997, 11, 263–276. [Google Scholar] [CrossRef] [PubMed]
- Hur, Y.S.; Um, J.H.; Kim, S.; Kim, K.; Park, H.J.; Lim, J.S.; Kim, W.Y.; Jun, S.E.; Yoon, E.K.; Lim, J. Arabidopsis thaliana homeobox 12 (ATHB12), a homeodomain-leucine zipper protein, regulates leaf growth by promoting cell expansion and endoreduplication. New Phytol. 2015, 205, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Blamey, F.P.C.; Edmeades, D.C.; Wheeler, D.M. Role of root cation-exchange capacity in differential aluminum tolerance of Lotus species. J. Plant Nutr. 1990, 13, 729–744. [Google Scholar] [CrossRef]
- Chang, Y.C.; Yamamoto, Y.; Matsumoto, H. Accumulation of aluminium in the cell wall pectin in cultured tobacco (Nicotiana tabacum L.) cells treated with a combination of aluminium and iron. Plant Cell Environ. 1999, 22, 1009–1017. [Google Scholar] [CrossRef]
- Yang, J.L.; Zhu, X.F.; Peng, Y.X.; Zheng, C.; Li, G.X.; Liu, Y.; Shi, Y.Z.; Zheng, S.J. Cell wall hemicellulose contributes significantly to aluminum adsorption and root growth in Arabidopsis. Plant Physiol. 2011, 155, 1885–1892. [Google Scholar] [CrossRef]
- Sivaguru, M.; Ezaki, B.; He, Z.H.; Tong, H.Y.; Osawa, H.; Baluška, F.; Volkmann, D.; Matsumoto, H. Aluminum-induced gene expression and protein localization of a cell wall-associated receptor kinase in Arabidopsis. Plant Physiol. 2003, 132, 2256–2266. [Google Scholar] [CrossRef]
- Zhu, X.F.; Wan, J.X.; Sun, Y.; Shi, Y.Z.; Braam, J.; Li, G.X.; Zheng, S.J. Xyloglucan Endotransglucosylase-Hydrolase 17 Interacts with Xyloglucan Endotransglucosylase-Hydrolase 31 to Confer Xyloglucan Endotransglucosylase Action and Affect Aluminum Sensitivity in Arabidopsis. Plant Physiol. 2014, 165, 1566–1574. [Google Scholar] [CrossRef]
- Cheng, Z.J.; Zhao, X.Y.; Shao, X.X.; Wang, F.; Zhou, C.; Liu, Y.G.; Zhang, Y.; Zhang, X.S. Abscisic Acid Regulates Early Seed Development in Arabidopsis by ABI5-Mediated Transcription of SHORT HYPOCOTYL UNDER BLUE1. Plant Cell 2014, 26, 1053–1068. [Google Scholar] [CrossRef]
- Yan, L.; Wei, S.; Wu, Y.; Hu, R.; Li, H.; Yang, W.; Xie, Q. High-efficiency genome editing in Arabidopsis using YAO promoter-driven CRISPR/Cas9 system. Mol. Plant 2015, 8, 1820–1823. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Xu, Y.; Zhu, L.; Zhang, L.; Liu, L.; Zhang, D.; Li, D.; Wu, C.; Huang, J.; Yang, G.; et al. The Brassicaceae-specific secreted peptides, STMPs, function in plant growth and pathogen defense. J. Integr. Plant Biol. 2020, 62, 403–420. [Google Scholar] [CrossRef] [PubMed]
- Napsucialy-Mendivil, S.; Alvarez-Venegas, R.; Shishkova, S.; Dubrovsky, J.G. Arabidopsis homolog of trithorax1 (ATX1) is required for cell production, patterning, and morphogenesis in root development. J. Exp. Bot. 2014, 65, 6373–6384. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).