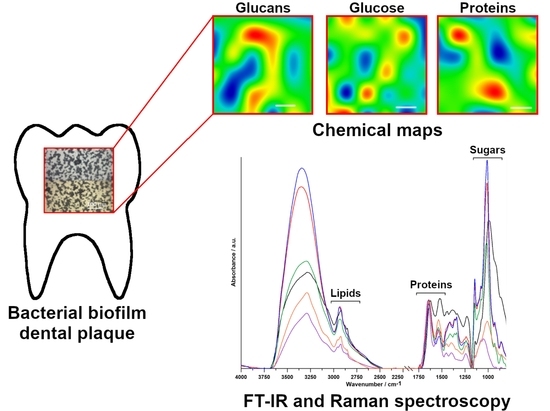
The FT-IR and Raman Spectroscopies as Tools for Biofilm Characterization Created by Cariogenic Streptococci
Abstract
1. Introduction
2. Results
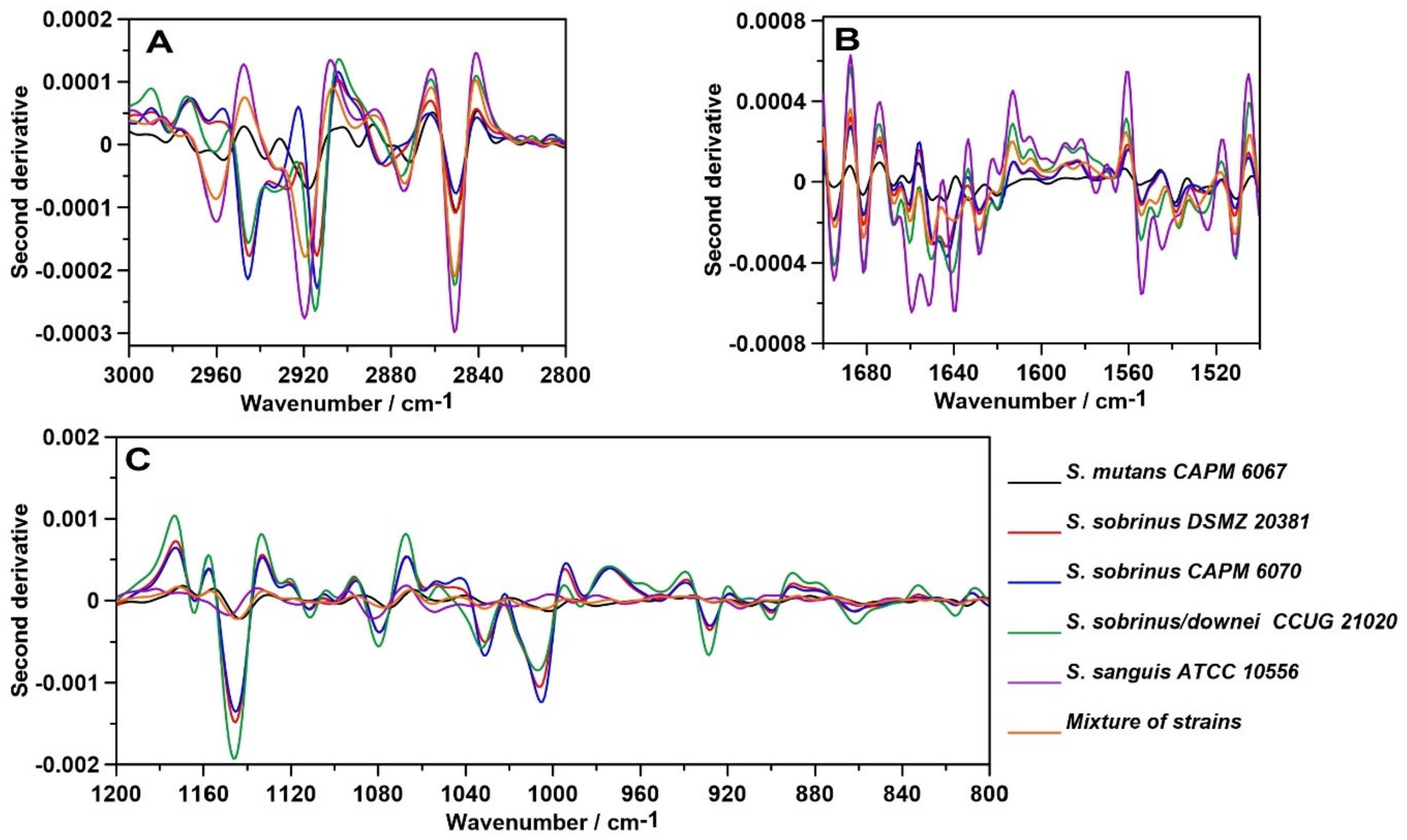
2.1. FT-IR Spectroscopy
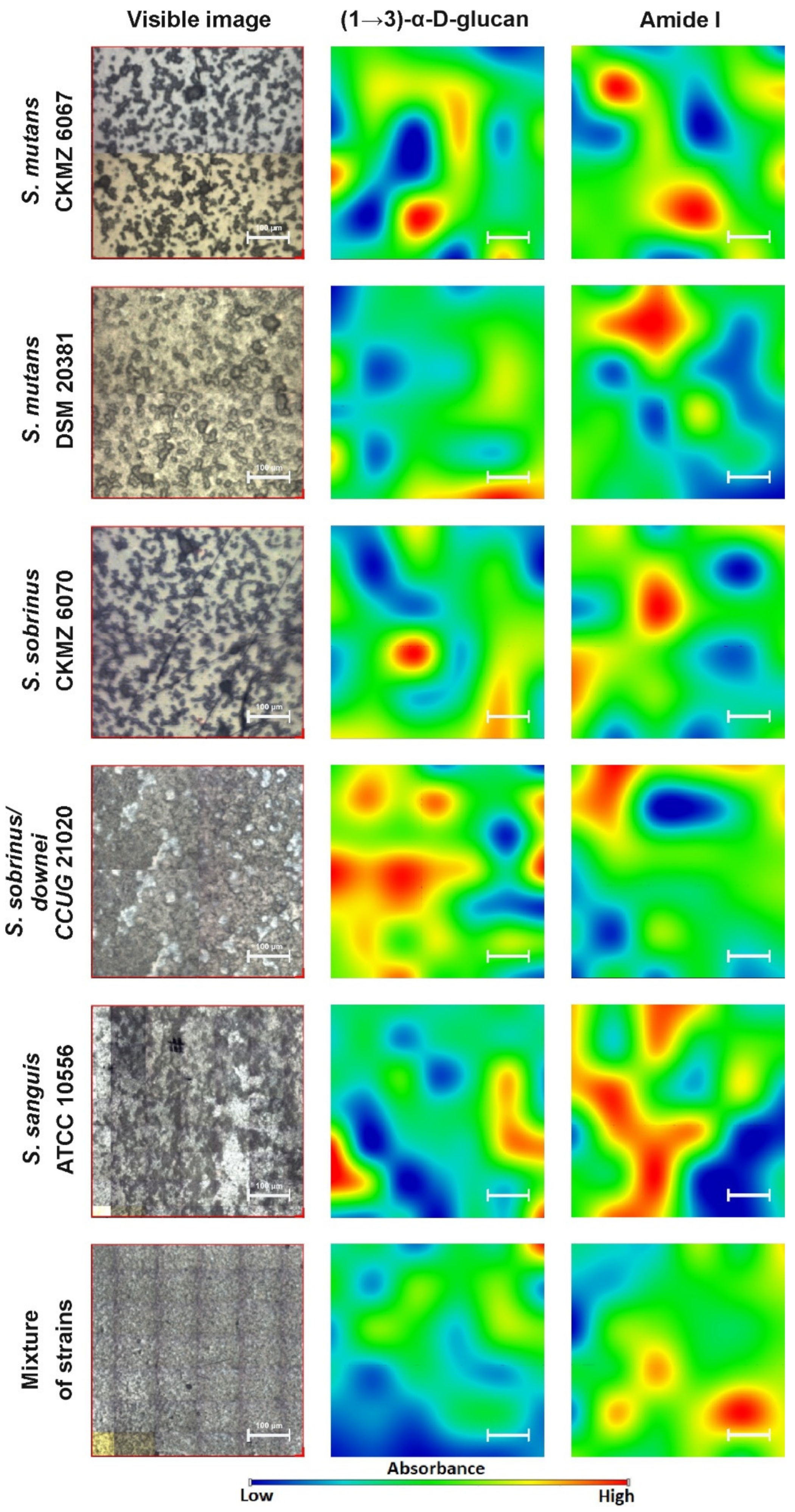
2.2. FT-IR Imaging
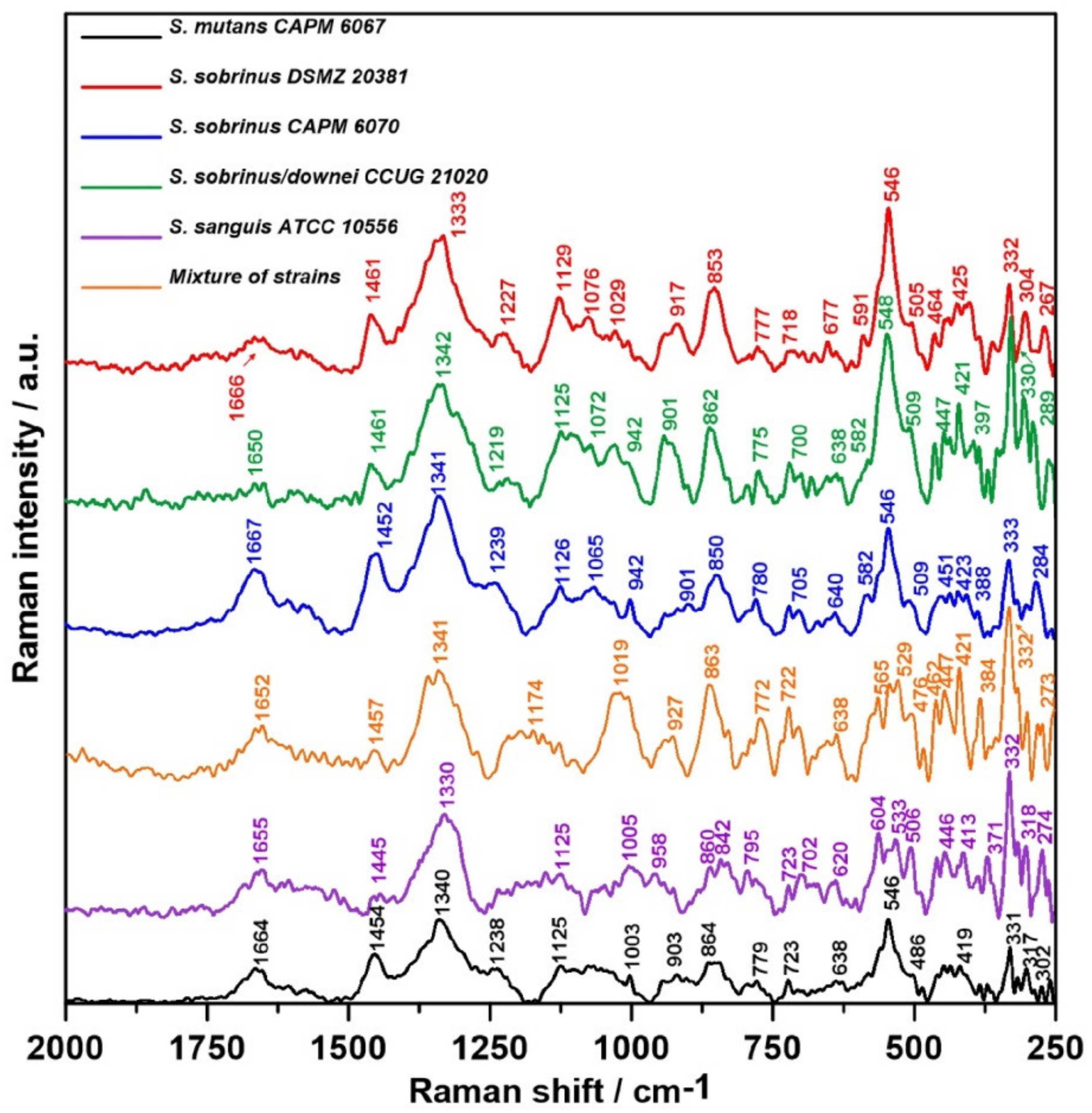
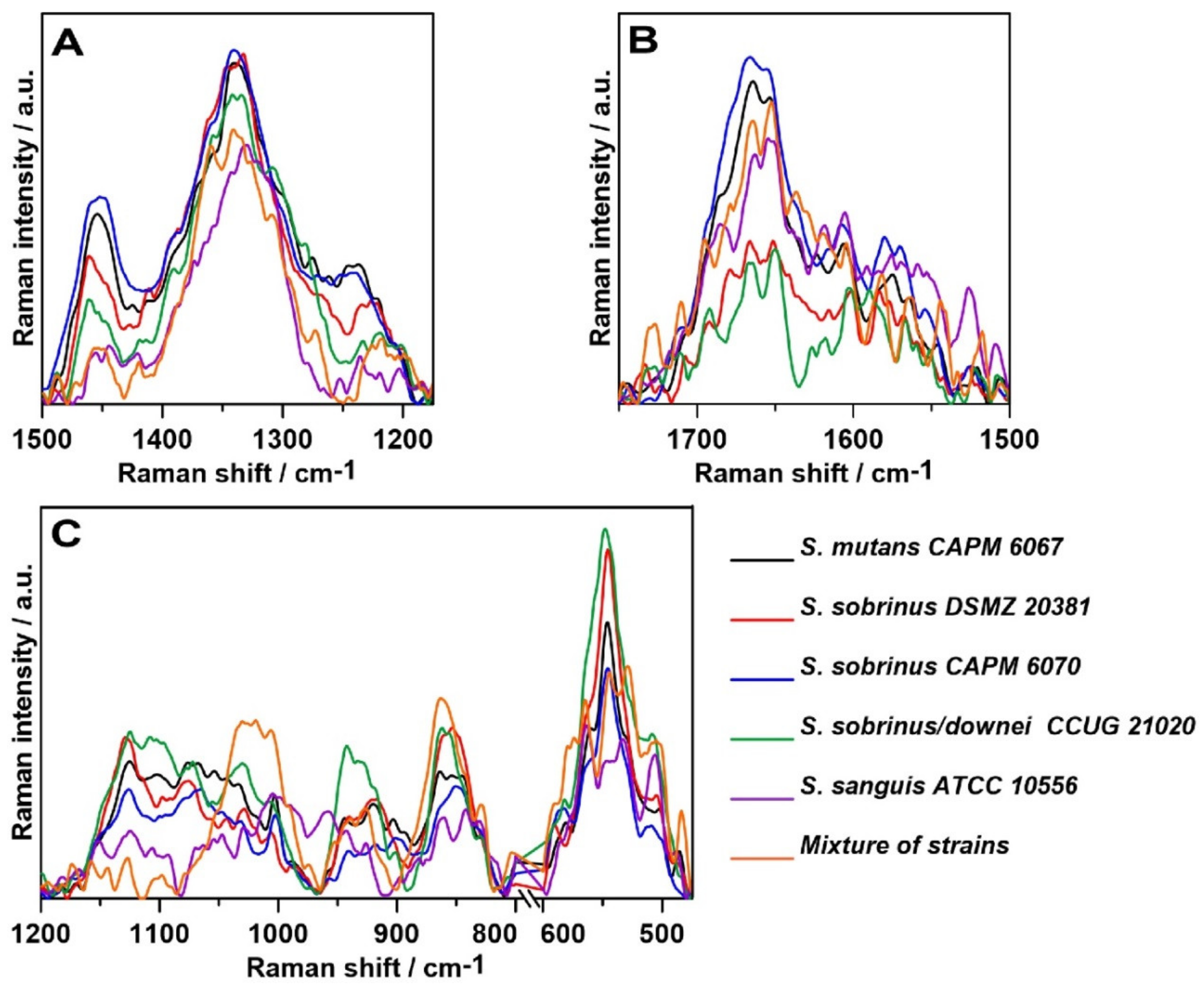
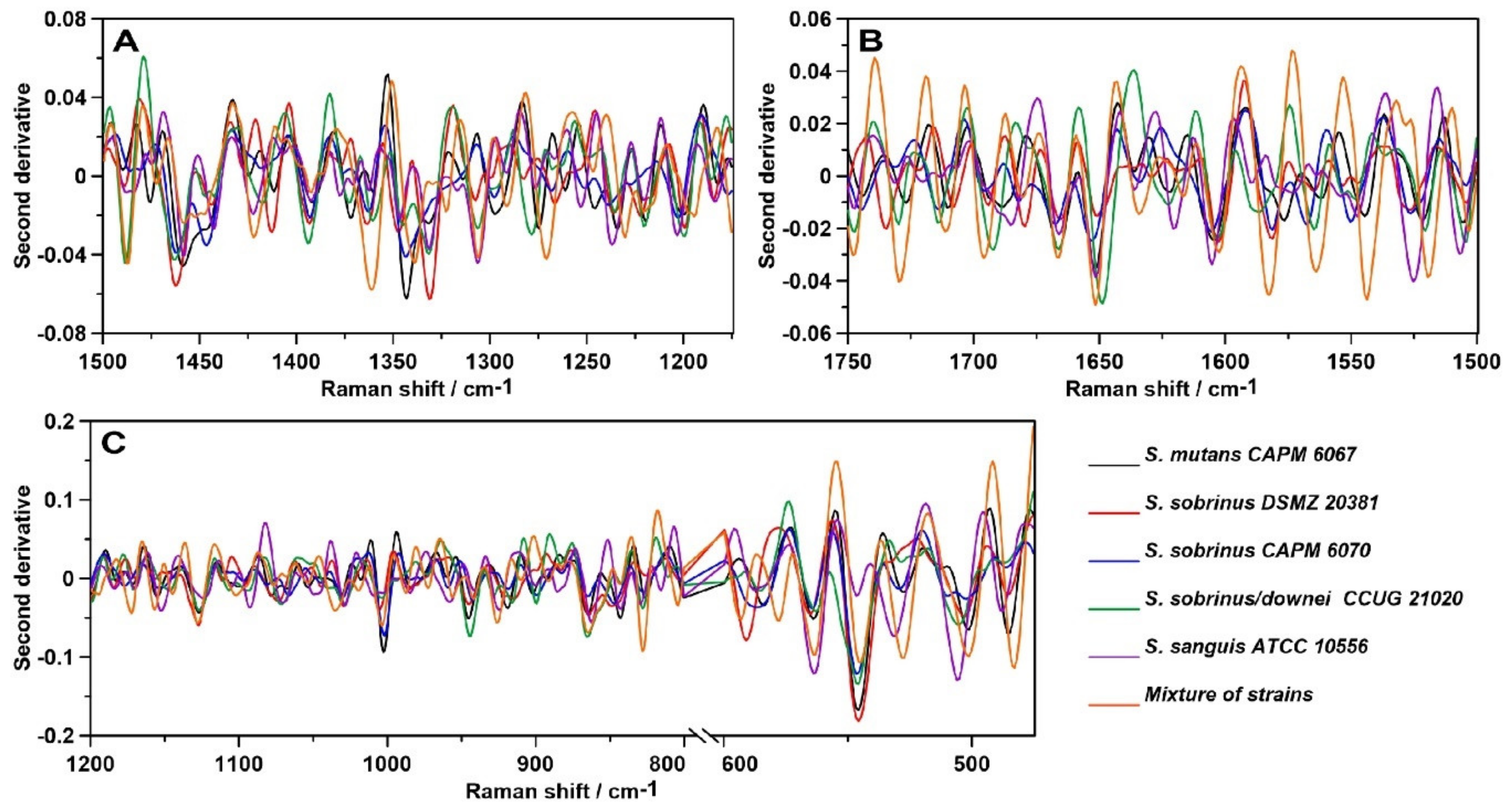
2.3. Raman Spectroscopy
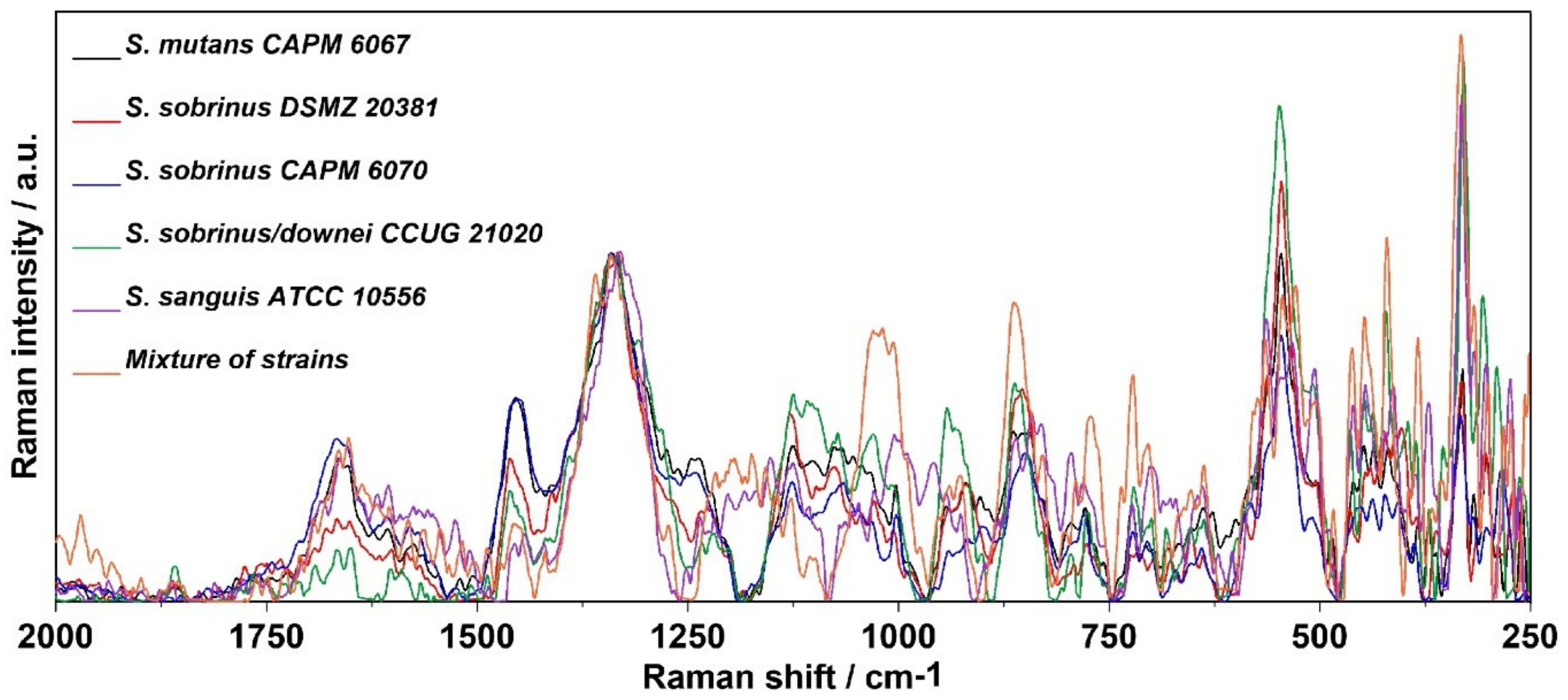
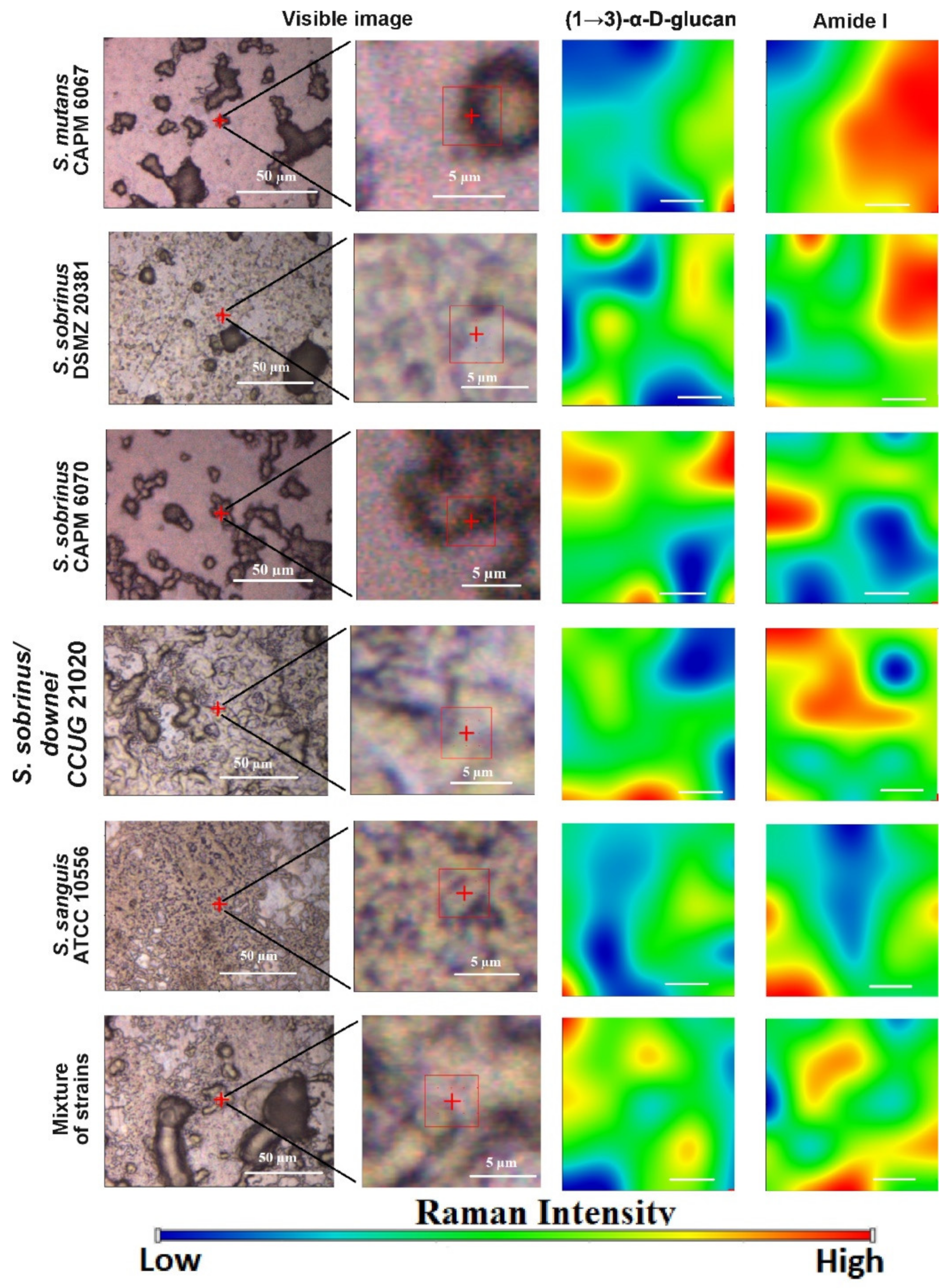
2.4. Raman Imaging
3. Discussion
4. Materials and Methods
4.1. Microorganisms
4.2. Streptococcal Biofilm Formation
4.3. FT-IR Microspectroscopy
4.4. Raman Microspectroscopy
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| FT-IR | Fourier transform infrared spectroscopy |
| Gbps | Glucan-binding proteins |
| Gtfs | Glucosyltransferases |
| PAc | Cell surface protein antigen c |
| MS | Mutans streptococci |
References
- Marsh, P.D. Dental diseases—Are these examples of ecological catastrophes? Int. J. Dent. Hyg. 2006, 4 (Suppl. 1), 3–10. [Google Scholar] [CrossRef]
- Pleszczyńska, M.; Wiater, A.; Janczarek, M.; Szczodrak, J. (1→3)-α-d-Glucan hydrolases in dental biofilm prevention and control: A review. Int. J. Biol. Macromol. 2015, 79, 761–778. [Google Scholar] [CrossRef] [PubMed]
- Loesche, W.J. Role of Streptococcus mutans in human dental decay. Microbiol. Rev. 1986, 50, 353–380. [Google Scholar] [CrossRef] [PubMed]
- Aoki, H.; Shiroza, T.; Hayakawa, M.; Sato, S.; Kuramitsu, H.K. Cloning of a streptococcus mutans glucosyltransferase gene coding for insoluble glucan synthesis. Infect. Immun. 1986, 53, 587–594. [Google Scholar] [CrossRef] [PubMed]
- Hanada, N.; Kuramitsu, H.K. Isolation and characterization of the Streptococcus mutans gtfD gene, coding for primer-dependent soluble glucan synthesis. Infect. Immun. 1989, 57, 2079–2085. [Google Scholar] [CrossRef] [PubMed]
- Hanada, N.; Kuramitsu, H.K. Isolation and characterization of the streptococcus mutans gtfc gene, coding for synthesis of both soluble and insoluble glucans. Infect. Immun. 2005, 56, 1999–2005. [Google Scholar] [CrossRef]
- Matsumoto-nakano, M. Role of Streptococcus mutans surface proteins for biofilm formation. Jpn. Dent. Sci. Rev. 2018, 54, 22–29. [Google Scholar] [CrossRef]
- Nanbu, A.; Hayakawa, M.; Takada, K.; Shinozaki, N. Production, characterization, and application of monoclonal antibodies which distinguish four glucosyltransferases from Streptococcus sobrinus. FEMS Immunol. Med. Microbiol. 2000, 27, 9–15. [Google Scholar] [CrossRef]
- Lynch, D.J.; Fountain, T.L.; Mazurkiewicz, J.E.; Banas, J.A. Glucan-binding proteins are essential for shaping Streptococcus mutans biofilm architecture. FEMS Microbiol. Lett. 2007, 268, 158–165. [Google Scholar] [CrossRef]
- Carter, E.A.; Tam, K.K.; Armstrong, R.S.; Lay, P.A. Vibrational spectroscopic mapping and imaging of tissues and cells. Biophys. Rev. 2009, 1, 95–103. [Google Scholar] [CrossRef][Green Version]
- Song, C.L.; Kazarian, S.G. Three-dimensional depth profiling of prostate tissue by micro ATR-FTIR spectroscopic imaging with variable angles of incidence. Analyst 2019, 144, 2954–2964. [Google Scholar] [CrossRef] [PubMed]
- Song, C.L.; Vardaki, M.Z.; Goldin, R.D.; Kazarian, S.G. Fourier transform infrared spectroscopic imaging of colon tissues: Evaluating the significance of amide I and C–H stretching bands in diagnostic applications with machine learning. Anal. Bioanal. Chem. 2019, 411, 6969–6981. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Li, M.; Cheng, W. FT-IR and Raman vibrational microspectroscopies used for spectral biodiagnosis of human tissues. Spectroscopy 2007, 21, 1–30. [Google Scholar] [CrossRef]
- Kazarian, S.G.; Chan, K.L.A. Applications of ATR-FTIR spectroscopic imaging to biomedical samples. Biochim. Biophys. Acta 2006, 1758, 858–867. [Google Scholar] [CrossRef] [PubMed]
- Prince, R.C.; Potma, E.O. Going visible: High-resolution coherent Raman imaging of cells and tissues. Light Sci. Appl. 2019, 8, 8–9. [Google Scholar] [CrossRef]
- Henry, V.A.; Jessop, J.L.P.; Peeples, T.L. Differentiating Pseudomonas sp. strain ADP cells in suspensions and biofilms using Raman spectroscopy and scanning electron microscopy. Anal. Bioanal. Chem. 2017, 409, 1441–1449. [Google Scholar] [CrossRef]
- Ricciardelli, A.; Casillo, A.; Vergara, A.; Balasco, N.; Corsaro, M.M.; Tutino, M.L.; Parrilli, E. Environmental conditions shape the biofilm of the Antarctic bacterium Pseudoalteromonas haloplanktis TAC125. Microbiol. Res. 2019, 218, 66–75. [Google Scholar] [CrossRef]
- Imbert, L.; Gourion-Arsiquaud, S.; Villarreal-Ramirez, E.; Spevak, L.; Taleb, H.; van der Meulen, M.C.; Boskey, A.L. Dynamic structure and composition of bone investigated by nanoscale infrared spectroscopy. PLoS ONE 2018, 13, e0202833. [Google Scholar] [CrossRef]
- Fu, B.; Sun, X.; Qian, W.; Shen, Y.; Chen, R.; Hannig, M. Evidence of chemical bonding to hydroxyapatite by phosphoric acid esters. Biomaterials 2005, 26, 5104–5110. [Google Scholar] [CrossRef]
- Tsuda, H.; Ruben, J.; Arends, J. Raman spectra of human dentin mineral. Eur. J. Oral. Sci. 1996, 104, 123–131. [Google Scholar] [CrossRef]
- Bista, R.K.; Bruch, R.F. Near-infrared spectroscopic studies of self-forming lipids and nanovesicles. In Nanoscale Imaging, Sensing, and Actuation for Biomedical Applications VI; SPIE—International Society for Optics and Photonics: San Francisco, CA, USA, 2009; p. 718809. [Google Scholar]
- Liu, Y.; Yao, X.; Liu, Y.W.; Wang, Y. A Fourier transform infrared spectroscopy analysis of carious dentin from transparent zone to normal zone. Caries Res. 2014, 48, 320–329. [Google Scholar] [CrossRef] [PubMed]
- Hȩdzelek, W.; Wachowiak, R.; Marcinkowska, A.; Domka, L. Infrared spectroscopic identification of chosen dental materials and natural teeth. Acta Phys. Pol. A 2008, 114, 471–484. [Google Scholar] [CrossRef]
- El-Sharkawyi, Y.H. Detection and characterization of human teeth caries using 2D correlation raman spectroscopy. J. Biomed. Phys. Eng. 2019, 9, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Buchwald, T.; Buchwald, Z. Assessment of the Raman spectroscopy effectiveness in determining the early changes in human enamel caused by artificial caries. Analyst 2019, 144, 1409–1419. [Google Scholar] [CrossRef] [PubMed]
- Baiz, C.R.; Reppert, M.; Tokmakoff, A. Amide I two-dimensional infrared spectroscopy: Methods for visualizing the vibrational structure of large proteins. J. Phys. Chem. A 2013, 117, 5955–5961. [Google Scholar] [CrossRef] [PubMed]
- Zarnowiec, P. Fourier Transform Infrared Spectroscopy (FTIR) as a tool for the identification and differentiation of pathogenic bacteria. Curr. Med. Chem. 2015, 22, 1710–1718. [Google Scholar] [CrossRef]
- Lin, H.; Deng, K.; Zhang, J.; Wang, L.; Zhang, Z.; Luo, Y.; Huang, P. Biochemical detection of fatal hypothermia and hyperthermia in affected rat hypothalamus tissues by Fourier transform infrared spectroscopy. Biosci. Rep. 2019, 39, BSR20181633. [Google Scholar] [CrossRef]
- Hernadez, B.; Pfluger, F.; Adenier, A.; Kurglik, S.G.; Ghomi, M. Vibrational analysis of amino acids and short peptides in hydrated media. IV. Amino acids with hydrophobic side chains: l-Alanine, l-Valine, and l-Isoleucine. J. Phys. Chem. 2009, 114, 15319–15330. [Google Scholar]
- Gao, Y.; Huo, X.; Dong, L.I.U.; Sun, X.; Sai, H.E.; Wei, G.; Wu, J. Fourier transform infrared microspectroscopy monitoring of 5-fluorouracil-induced apoptosis in SW620 colon cancer cells. Mol. Med. Rep. 2015, 11, 2585–2591. [Google Scholar] [CrossRef]
- Choo-Smith, L.P.; Maquelin, K.; Van Vreeswijk, T.; Bruining, H.A.; Puppels, G.J.; Thi, N.N.; Orsini, F. Investigating Microbial (Micro) colony heterogeneity by vibrational spectroscopy. Appl. Environ. Microbiol. 2001, 67, 1461–1469. [Google Scholar] [CrossRef]
- Gieroba, B.; Arczewska, M.; Sławińska-Brych, A.; Rzeski, W.; Stepulak, A.; Gagoś, M. Prostate and breast cancer cells death induced by xanthohumol investigated with Fourier transform infrared spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 231, 118112. [Google Scholar] [CrossRef] [PubMed]
- Júnior, Z.S.S.; Botta, S.B.; Ana, P.A.; França, C.M.; Fernandes, K.P.S.; Mesquita-Ferrari, R.A.; Bussadori, S.K. Effect of papain-based gel on type I collagen—Spectroscopy applied for microstructural analysis. Sci. Rep. 2015, 5, 11448. [Google Scholar] [CrossRef] [PubMed]
- Synytsya, A.; Novák, M. Structural diversity of fungal glucans. Carbohydr. Polym. 2013, 92, 792–809. [Google Scholar] [CrossRef] [PubMed]
- Sahu, R.K.; Salman, A.; Mordechai, S. Tracing overlapping biological signals in mid-infrared using colonic tissues as a model system. World J. Gastroenterol. 2017, 23, 286–296. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, S.; Miyazaki, M.; Sakai, K.; Takeshita, M.; Yuasa, S.; Sato, A.; Okuyama, H. Fourier transform infrared spectroscopic analysis of rat brain microsomal membranes modified by dietary fatty acids: Possible correlation with altered learning behavior. Biospectroscopy 1997, 3, 281–290. [Google Scholar] [CrossRef]
- Baeva, E.; Bleha, R.; Lavrova, E.; Sushytskyi, L.; Čopíková, J.; Jablonsky, I.; Synytsya, A. Polysaccharides from basidiocarps of cultivating mushroom pleurotus ostreatus: Isolation and structural characterization. Molecules 2019, 24, 2740. [Google Scholar] [CrossRef]
- Nauman, D.; Helm, D.; Labischinski, H. Microbiological characterizations by FT-IR spectroscopy. Nature 1991, 351, 81–82. [Google Scholar] [CrossRef]
- Derenne, A.; Vandersleyen, O.; Goormaghtigh, E. Lipid quantification method using FTIR spectroscopy applied on cancer cell extracts. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2014, 1841, 1200–1209. [Google Scholar] [CrossRef]
- Shapaval, V.; Brandenburg, J.; Blomqvist, J.; Tafintseva, V.; Passoth, V. Biotechnology for Biofuels Biochemical profiling, prediction of total lipid content and fatty acid profile in oleaginous yeasts by FTIR spectroscopy. Biotechnol. Biofuels 2019, 12, 140. [Google Scholar] [CrossRef]
- Barth, A. Infrared spectroscopy of proteins. Biochim. Biophys. Acta Bioenerg. 2007, 1767, 1073–1101. [Google Scholar] [CrossRef]
- Sabbatini, S.; Conti, C.; Orilisi, G.; Giorgini, E. Infrared spectroscopy as a new tool for studying single living cells: Is there a niche? Biomed. Spectrosc. Imaging 2017, 6, 85–99. [Google Scholar] [CrossRef]
- Synytsya, A.; Novak, M. Structural analysis of glucans. Ann. Transl. Med. 2014, 2, 17. [Google Scholar] [PubMed]
- Tanaka, S.; Kojić, D.; Tsenkova, R.; Yasui, M. Quantification of anomeric structural changes of glucose solutions using near-infrared spectra. Carbohydr. Res. 2018, 463, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Shakeel, F.; Baboota, S.; Ahuja, A.; Ali, J.; Shafiq, S. Skin permeation mechanism and bioavailability enhancement of celecoxib from transdermally applied nanoemulsion. J. Nanobiotechnol. 2008, 6, 1–11. [Google Scholar] [CrossRef]
- Kong, J.; Yu, S. Fourier transform infrared spectroscopic analysis of protein secondary structures protein FTIR data analysis and band assign. Acta Biochim. Biophys. Sin. 2007, 39, 549–559. [Google Scholar] [CrossRef]
- Huang, H.; Shi, H.; Feng, S.; Chen, W.; Yu, Y.; Lin, D.; Chen, R. Confocal Raman spectroscopic analysis of the cytotoxic response to cisplatin in nasopharyngeal carcinoma cells. Anal. Methods 2013, 5, 260–266. [Google Scholar] [CrossRef]
- Czamara, K.; Majzner, K.; Pacia, M.Z.; Kochan, K.; Kaczor, A.; Baranska, M. Raman spectroscopy of lipids: A review. J. Raman Spectrosc. 2015, 46, 4–20. [Google Scholar] [CrossRef]
- Rohleder, D.; Kiefer, W.; Petrich, W. Quantitative analysis of serum and serum ultrafiltrate by means of Raman spectroscopy. Analyst 2004, 129, 906–911. [Google Scholar] [CrossRef]
- Wagner, M.; Ivleva, N.P.; Haisch, C.; Niessner, R.; Horn, H. Combined use of confocal laser scanning microscopy ( CLSM ) and Raman microscopy (RM): Investigations on EPS—Matrix. Water Res. 2009, 43, 63–76. [Google Scholar] [CrossRef]
- Keleştemur, S.; Avci, E.; Çulha, M. Raman and surface-enhanced Raman scattering for biofilm characterization. Chemosensors 2018, 6, 5. [Google Scholar] [CrossRef]
- Ivleva, N.P.; Wagner, M.; Horn, H.; Niessner, R.; Haisch, C. Raman microscopy and Surface-Enhanced Raman Scattering (SERS) for in situ analysis of biofilms. J. Biophotonics. 2010, 3, 548–556. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Cai, W.; Du, B.; Qian, M.; Sun, Z. Raman spectroscopic investigation on the interaction of malignant hepatocytes with doxorubicin. Biophys. Chem. 2009, 140, 57–61. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.P.; Wallach, D.F.H. Raman spectra of some saturated, unsaturated and deuterated C18 fatty acids in the hch-deformation and ch-stretching regions. Biochim. Biophys. Acta 1977, 486, 217–227. [Google Scholar] [CrossRef]
- Pearman, W.F.; Lawrence-Snyder, M.; Angel, S.M.; Decho, A.W. Surface-enhanced raman spectroscopy for in situ measurements of signaling molecules (autoinducers) relevant to bacteria quorum sensing. Appl. Spectrosc. 2007, 61, 1295–1300. [Google Scholar] [CrossRef]
- Schulz, H.; Baranska, M. Identification and quantification of valuable plant substances by IR and Raman spectroscopy. Vib. Spectrosc. 2007, 43, 13–25. [Google Scholar] [CrossRef]
- Szymańska-Chargot, M.; Chylińska, M.; Pieczywek, P.M.; Rösch, P.; Schmitt, M.; Popp, J.; Zdunek, A. Raman imaging of changes in the polysaccharides distribution in the cell wall during apple fruit development and senescence. Planta 2016, 243, 935–945. [Google Scholar] [CrossRef]
- Agarwal, U.P. Raman imaging to investigate ultrastructure and composition of plant cell walls: Distribution of lignin and cellulose in black spruce wood (Picea mariana). Planta 2006, 224, 1141–1153. [Google Scholar] [CrossRef]
- Synytsya, A. Spectroscopic estimation of feruloyl groups in sugar beet pulp and pectin. Int. Sugar J. 2003, 105, 481–488. [Google Scholar]
- Ramirez-Mora, T.; Dávila-Pérez, C.; Torres-Méndez, F.; Valle-Bourrouet, G. Raman spectroscopic characterization of endodontic biofilm matrices. J. Spectrosc. 2019, 2019, 1307397. [Google Scholar] [CrossRef]
- Shao, J.; Lin, M.; Li, Y.; Li, X.; Liu, J.; Liang, J.; Yao, H. In vivo blood glucose quantification using raman spectroscopy. PLoS ONE 2012, 7, e48127. [Google Scholar] [CrossRef]
- Rygula, A.; Majzner, K.; Marzec, K.M.; Kaczor, A.; Pilarczyk, M.; Baranska, M. Raman spectroscopy of proteins: A review. J. Raman Spectrosc. 2013, 44, 1061–1076. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Iconomidou, V.A.; Chryssikos, G.D.; Gionis, V.; Kumar, K.; Hamodrakas, S.J. International Journal of Biological Macromolecules Secondary structure of chorion proteins of the Lepidoptera Pericallia ricini and Ariadne merione by ATR FT-IR and micro-Raman spectroscopy. Int. J. Biol. Macromol. 2011, 49, 317–322. [Google Scholar] [CrossRef] [PubMed]
- Gelder, J.; Gussem, K.; Vandenabeele, P.; Moens, L. Reference database of Raman spectra of biological molecules. J. Raman Spectrosc. 2007, 1133–1147. [Google Scholar] [CrossRef]
- Khatoon, Z.; Mctiernan, C.D.; Suuronen, E.J.; Mah, T. Bacterial bio fi lm formation on implantable devices and approaches to its treatment and prevention. Heliyon 2018, 4, e01067. [Google Scholar] [CrossRef] [PubMed]
- Jabbouri, S.; Sadovskaya, I. Characteristics of the biofilm matrix and its role as a possible target for the detection and eradication of Staphylococcus epidermidis associated with medical implant infections. FEMS Immunol. Med. Microbiol. 2010, 59, 280–291. [Google Scholar] [CrossRef]
- Wroblewska, M.; Struzycka, I.; Mierzwinska-Nastalska, E. Significance of biofilms in dentistry. Przegl. Epidemiol. 2015, 69, 739–744. [Google Scholar]
- Nadell, C.D.; Xavier, J.B.; Levin, S.A.; Foster, K.R. The evolution of quorum sensing in bacterial biofilms. PLoS Biol. 2008, 6, 0171–0179. [Google Scholar] [CrossRef]
- Mitchell, K.F.; Zarnowski, R.; Sanchez, H.; Edward, J.A.; Reinicke, E.L.; Nett, J.E.; Andes, D.R. Community participation in biofilm matrix assembly and function. Proc. Natl. Acad. Sci. USA 2015, 112, 4092–4097. [Google Scholar] [CrossRef]
- Burmølle, M.; Webb, J.S.; Rao, D.; Hansen, L.H.; Sørensen, S.J.; Kjelleberg, S. Enhanced biofilm formation and increased resistance to antimicrobial agents and bacterial invasion are caused by synergistic interactions in multispecies biofilms. Appl. Environ. Microbiol. 2006, 72, 3916–3923. [Google Scholar] [CrossRef]
- Ren, D.; Madsen, J.S.; Sørensen, S.J.; Burmølle, M. High prevalence of biofilm synergy among bacterial soil isolates in cocultures indicates bacterial interspecific cooperation. ISME J. 2015, 9, 81–89. [Google Scholar] [CrossRef]
- Rice, S.A.; Wuertz, S.; Kjelleberg, S. Next-generation studies of microbial biofilm communities. Microb. Biotechnol. 2016, 9, 677–680. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.C.; Neu, T.R.; Wozniak, D.J. The EPS matrix: The “House of Biofilm Cells”. J. Bacteriol. 2007, 189, 7945–7947. [Google Scholar] [CrossRef] [PubMed]
- Matsuyama, T.; Nakagawa, Y. Surface-active exolipids: Analysis of absolute chemical structures and biological functions. J. Microbiol. Methods 1996, 25, 165–175. [Google Scholar] [CrossRef]
- Ward, O.P. Microbial Biosurfactants and Biodegradation. Adv. Exp. Med. Biol. 2010, 672, 65–74. [Google Scholar] [PubMed]
- Baughn, A.D.; Rhee, K.Y. Biofilm matix proteins. Microbiol. Spectr. 2014, 2, 201–222. [Google Scholar]
- Arciola, R.C.; Campoccia, D.; Speziale, P.; Montanaro, L.; William, J. Biomaterials Bio fi lm formation in Staphylococcus implant infections. A review of molecular mechanisms and implications for bio fi lm-resistant materials. Biomaterials 2012, 33, 5967–5982. [Google Scholar] [CrossRef]
- Nishikawara, F.; Nomura, Y.; Imai, S.; Senda, A.; Hanada, N. Evaluation of cariogenic bacteria. Eur. J. Dent. 2007, 01, 031–039. [Google Scholar] [CrossRef]
- Diem, M.; Romeo, M.; Boydston-White, S.; Miljkovic, M.; Matthaus, M. A decade of vibrational micro-spectroscopy of human cells and tissue (1994–2004). Analyst 2004, 129, 880–885. [Google Scholar] [CrossRef]
- Zwielly, A.; Gopas, J.; Brkic, G.; Mordechai, S. Discrimination between drug-resistant and non-resistant human melanoma cell lines by FTIR spectroscopy. Analyst 2009, 134, 294–300. [Google Scholar] [CrossRef]
- Guo, W.; Piao, S.; Yang, T.C.; Guo, J.; Iqbal, K. High-resolution power spectral estimation method using deconvolution. IEEE J. Ocean. Eng. 2020, 45, 489–499. [Google Scholar] [CrossRef]
- Morhac, M.; Matousek, V. High-resolution boosted deconvolution of spectroscopic data. J. Comput. Appl. Math. 2011, 235, 1629–1640. [Google Scholar] [CrossRef]
- Váczi, T. A new, simple approximation for the deconvolution of instrumental broadening in spectroscopic band profiles. Appl. Spectrosc. 2014, 68, 1274–1278. [Google Scholar] [CrossRef]
- Wilson, C.; Lukowicz, R.; Merchant, S.; Valquier-Flynn, H.; Caballero, J.; Sandoval, J.; Clement, B. Quantitative and qualitative assessment methods for biofilm growth: A mini-review. Res. Rev. J. Eng. Technol. 2017, 6, 1–8. [Google Scholar]
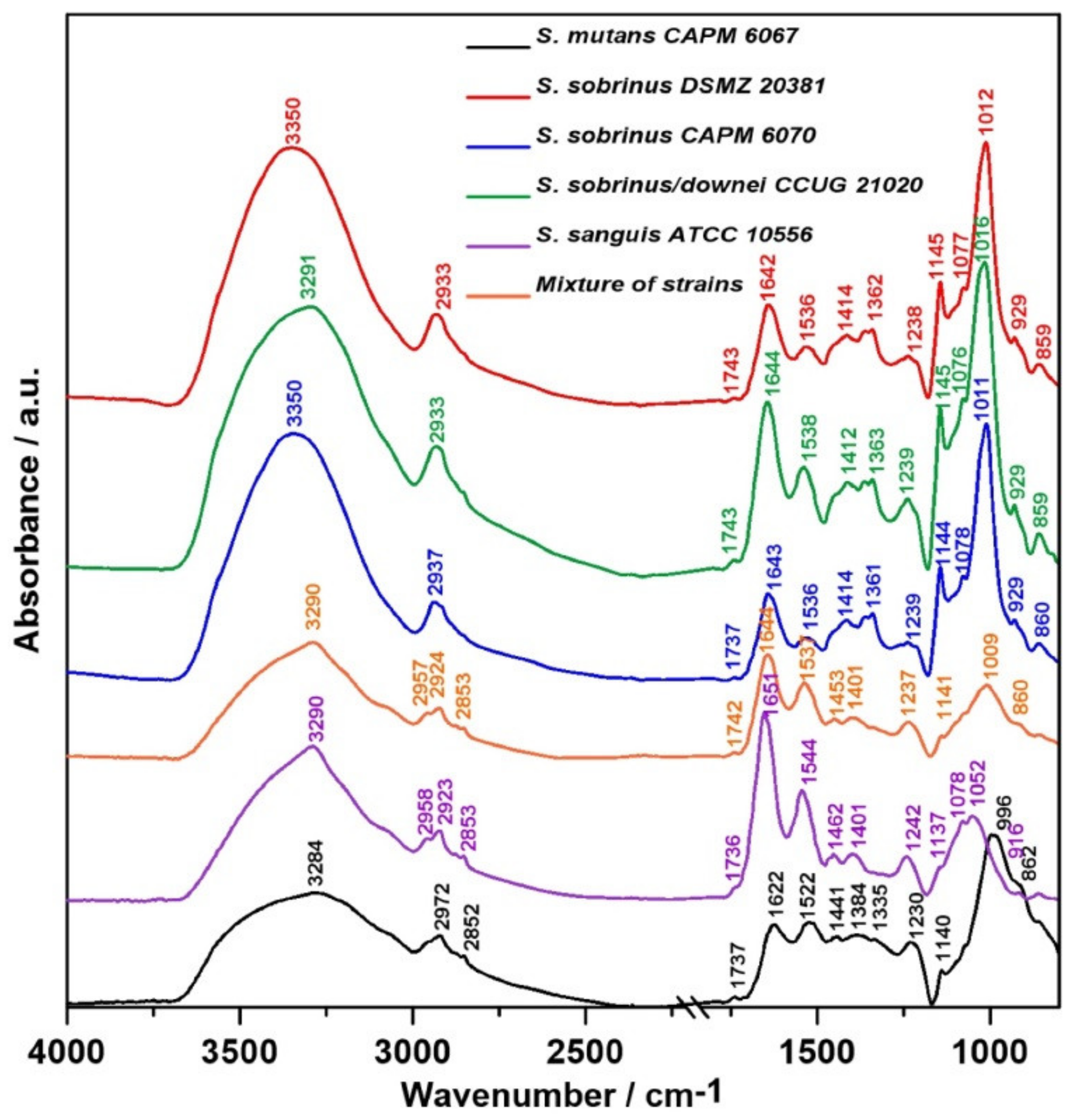
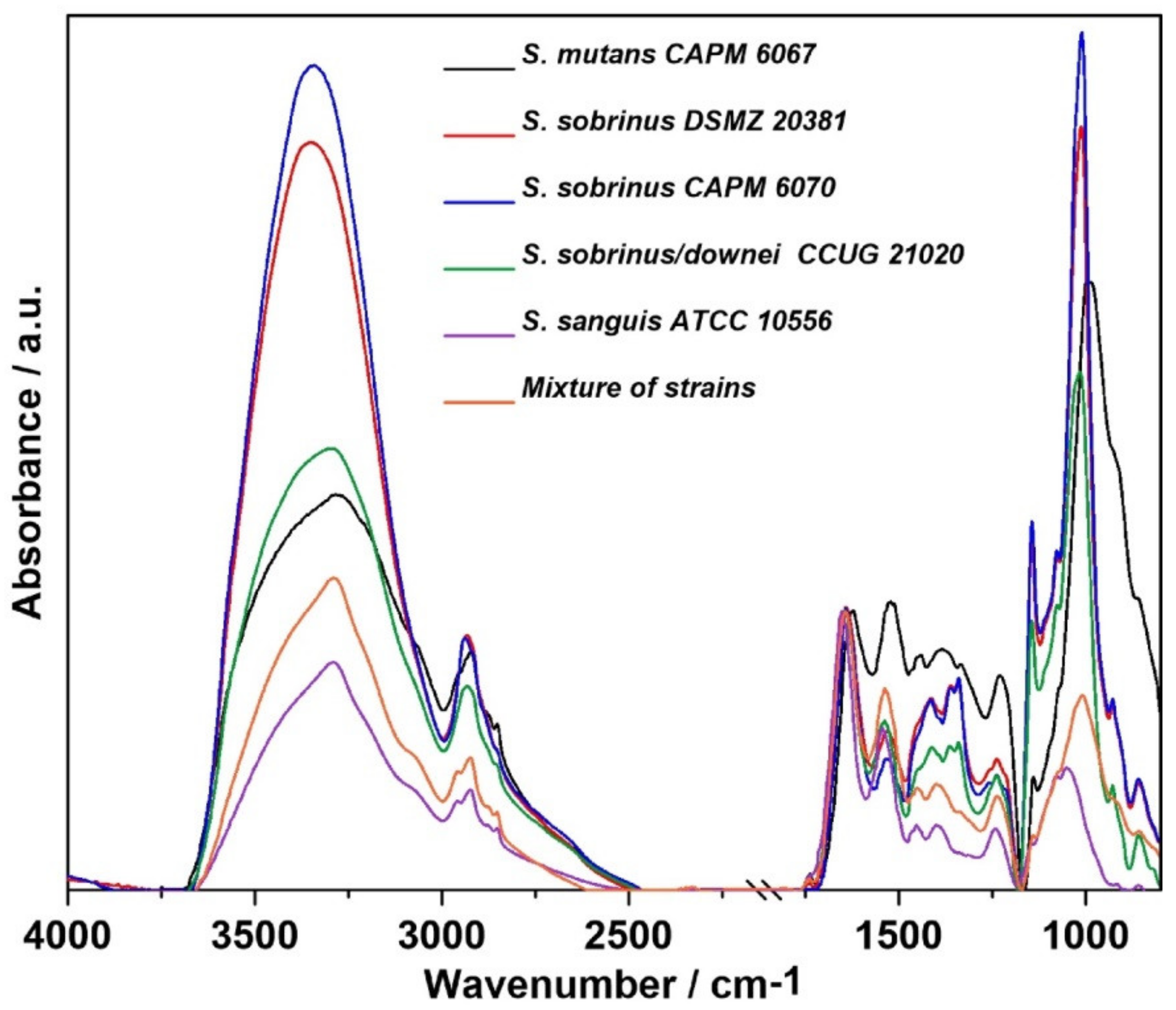
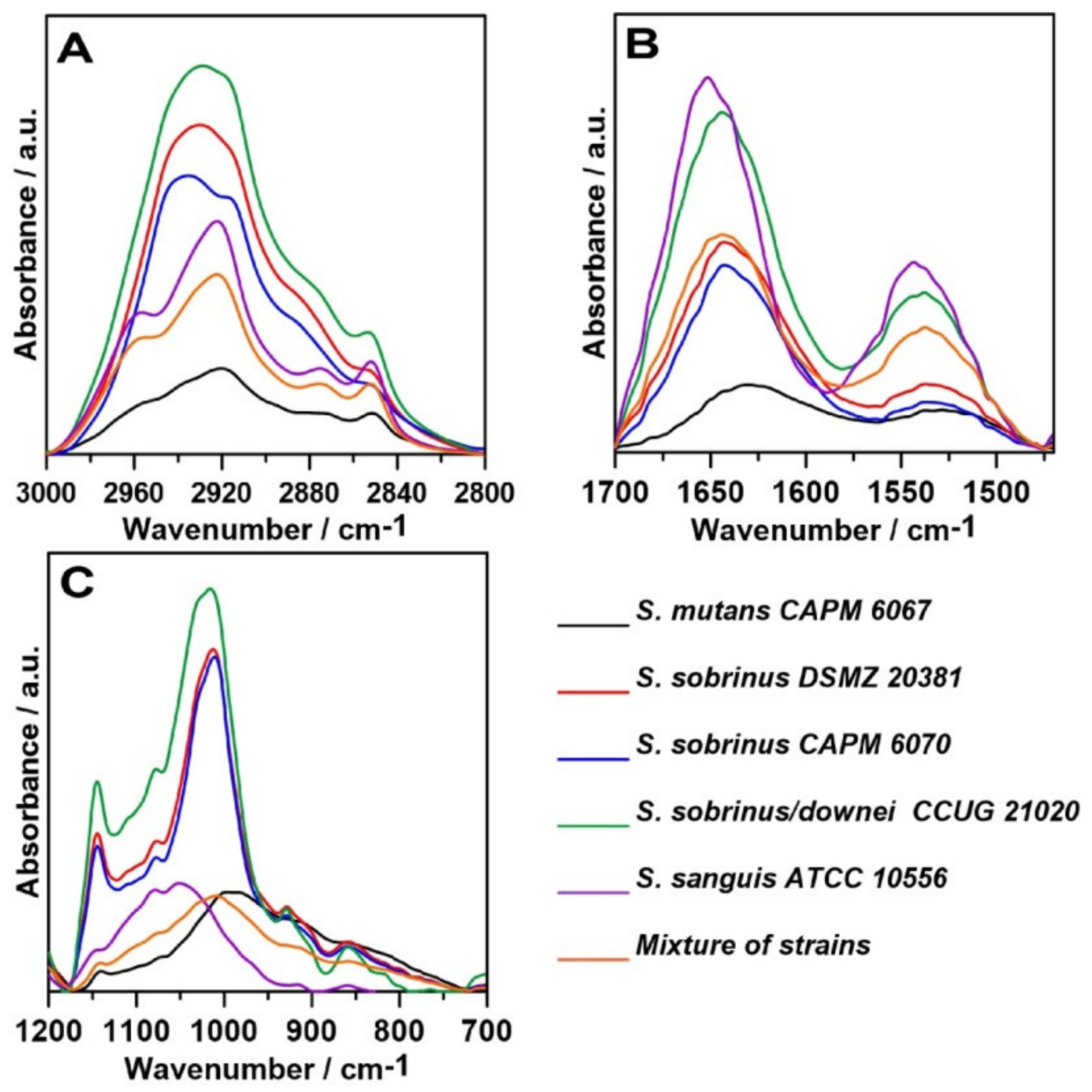
| Wavenumber (cm−1) | Assignment and the Type of Vibration * |
|---|---|
| 3200–3350 | ν (N–H), ν (O-H), Amide A, water |
| 2950–2960 | νas (CH3), lipids |
| 2920–2940 | νas (CH2), lipids |
| 2850–2860 | νs (CH2), lipids |
| 1730–1740 | ν (C=O), phospholipids |
| 1700–1600 | 80% ν (C=O), 20% ν (C-N), τ (HOH), Amide I, water |
| 1600–1500 | 60% τ (N–H), 30% ν (C–N), 10% ν (C–C), Amide II |
| 1441–1462 | pyrrolidine ring vibration of proline and hydroxyproline |
| 1450–1400 | δas (CH3), δas (CH2), proteins, lipids |
| 1400–1350 | δs (CH3), δs (CH2), νs (C=O), proteins, lipids |
| 1350–1200 | τ (N–H), ν (C–N), τ (C=O), ν (C–C), ν (CH3), Amide III, |
| 1242–1230 | νas (PO2–), DNA, RNA, phospholipids, phosphorylated proteins |
| 1144–1137 | Oligosaccharydes |
| ~1086 | νs (PO2–), DNA, RNA, phospholipids, phosphorylated proteins |
| 1080–1070 | ν (C–C), β-glucan bonds |
| 1046–999 | Skeletal vibration connected to anomeric structure of d-glucose |
| 1009–1016 | ν (C–C), RNA, ribose |
| ~972 | ν (C–C), ν (C–O), DNA, deoxirobose |
| 900–700 | anomeric ring vibrations for tryptophan, tyrosine, and phenyloalanine |
| 929 | (1→3)-α-d-glucan |
| 860–852 | (1→3),(1→6)-α-d-glucan |
| Raman Shift (cm−1) | Assignment and the Type of Vibration * |
|---|---|
| 1700–1600 | ν (C=O), Amide I |
| 1667–1650 | ν (C=C), lipids, proteins |
| 1600–1500 | ν (C–N), δ (N–H), Amide II |
| 1576 | adenine, guanine (DNA bases) |
| 1523 | cytosine (DNA bases) |
| 1500–1400 | in-plane τ and out-of-plane τ (CH2), lipids |
| 1461–1445 | νs (CH2), saturated lipids |
| ~1380 | δ (COH), (HCO), (HCC), νs (COO–), (C–O), polyanionic polysaccharide |
| 1340–1330 | polynucleotide chains, DNA purine bases |
| 1330–1125 | trans ν (C–C), lipids |
| 1300–1250 | in-plane τ and out-of-plane τ (CH3), lipids |
| ~1280 | δ (COH), (HCO), (HCC), νs (COO–), (C–O), polyanionic polysaccharide |
| 1300–1230 | ν (C–N), δ (N–H), Amide III |
| ~1260 (shoulder band) | δ (CH), lipids, proteins |
| 1200–1050 | ν (C–C), lipids |
| 1075, 1055, 980–880 | combination of rhamnose, galactose, and glucose |
| ~1127 | ν (C–N), prolinę |
| 1125 | Glucose |
| ~1120 | νs (COC), glycosidic bonds |
| ~1094 | νas (COC), (1→4)-β-linked glycosidic bonds |
| ~1068 | trans ν (C–C), lipids |
| 1000 | phenyloalanine ring breathing |
| 950–790 | side group δ (COH), (C–CH), (O–CH), carbohydrates |
| ~948 | (1→3)-α-d-glucan |
| 800–640 | out-of-plane τ (N–H), Amide V |
| 852 | (1→6)-α-d-glucan |
| ~783 | ring breathing of cytosine, thymine, uracil; νs (O–P–O), phosphodiester bonds in DNA |
| 770–625 | τ (O=C-N), Amide IV |
| ~757, ~520 | Glucans |
| 600–540 | out-of-plane τ (C=O), Amide VI |
| ~380 | β-d-glucoside |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gieroba, B.; Krysa, M.; Wojtowicz, K.; Wiater, A.; Pleszczyńska, M.; Tomczyk, M.; Sroka-Bartnicka, A. The FT-IR and Raman Spectroscopies as Tools for Biofilm Characterization Created by Cariogenic Streptococci. Int. J. Mol. Sci. 2020, 21, 3811. https://doi.org/10.3390/ijms21113811
Gieroba B, Krysa M, Wojtowicz K, Wiater A, Pleszczyńska M, Tomczyk M, Sroka-Bartnicka A. The FT-IR and Raman Spectroscopies as Tools for Biofilm Characterization Created by Cariogenic Streptococci. International Journal of Molecular Sciences. 2020; 21(11):3811. https://doi.org/10.3390/ijms21113811
Chicago/Turabian StyleGieroba, Barbara, Mikolaj Krysa, Kinga Wojtowicz, Adrian Wiater, Małgorzata Pleszczyńska, Michał Tomczyk, and Anna Sroka-Bartnicka. 2020. "The FT-IR and Raman Spectroscopies as Tools for Biofilm Characterization Created by Cariogenic Streptococci" International Journal of Molecular Sciences 21, no. 11: 3811. https://doi.org/10.3390/ijms21113811
APA StyleGieroba, B., Krysa, M., Wojtowicz, K., Wiater, A., Pleszczyńska, M., Tomczyk, M., & Sroka-Bartnicka, A. (2020). The FT-IR and Raman Spectroscopies as Tools for Biofilm Characterization Created by Cariogenic Streptococci. International Journal of Molecular Sciences, 21(11), 3811. https://doi.org/10.3390/ijms21113811