Association between P2X7 Polymorphisms and Post-Transplant Outcomes in Allogeneic Haematopoietic Stem Cell Transplantation
Abstract
1. Introduction
2. Results
2.1. Allelic Frequency
2.2. Paired Cohort Patients
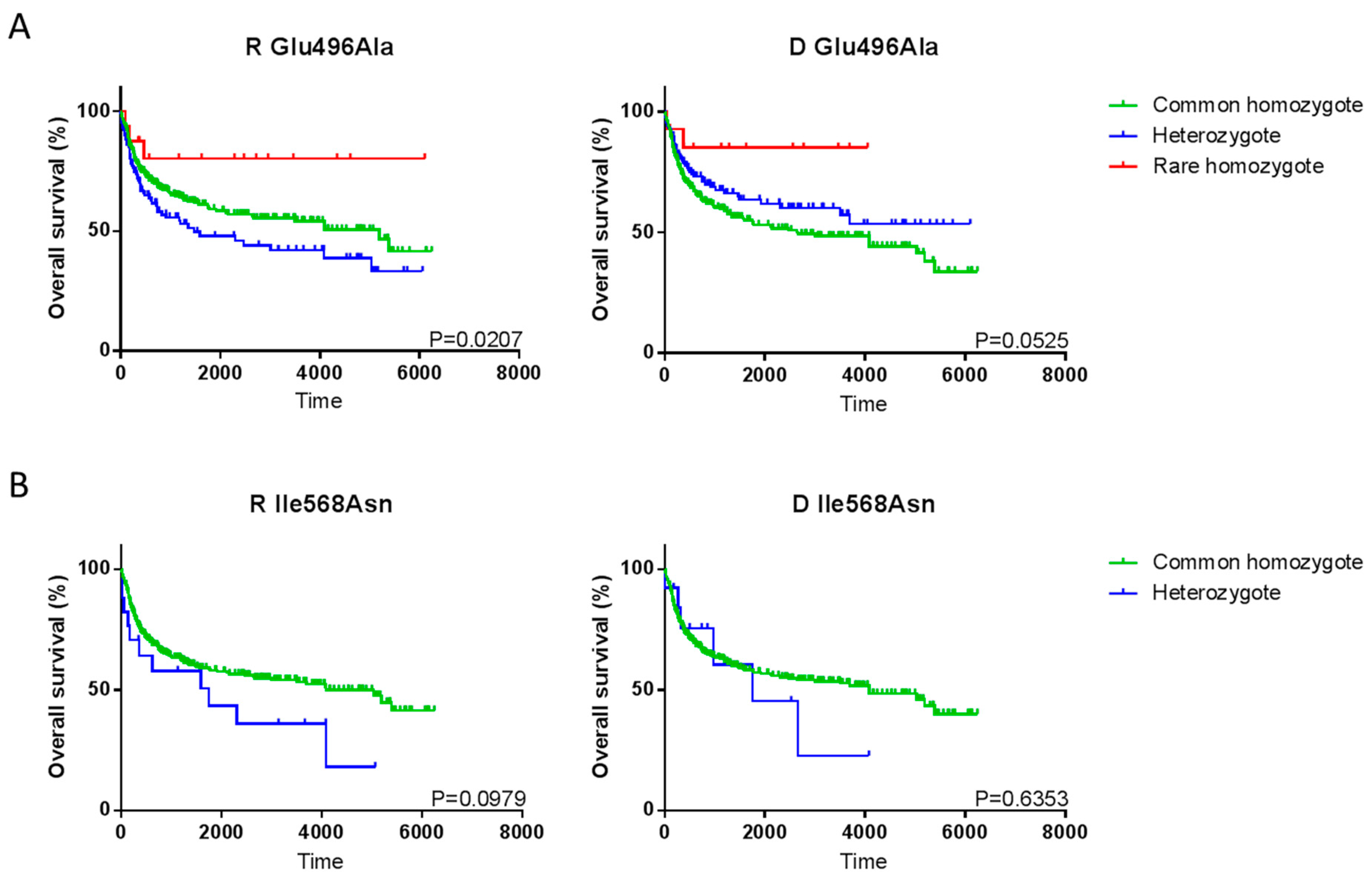
2.3. Single SNP Associations with Transplant Outcome
2.4. P2X7 Haplotype Associations with Transplant Outcome
3. Discussion
4. Materials and Methods
4.1. Patient Samples
4.2. P2X7 Polymorphism Analysis
4.3. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Copelan, E.A.; Chojecki, A.; Lazarus, H.M.; Avalos, B.R. Allogeneic hematopoietic cell transplantation; the current renaissance. Blood Rev. 2019, 34, 34–44. [Google Scholar] [CrossRef]
- Stikvoort, A.; Gaballa, A.; Solders, M.; Nederlof, I.; Onfelt, B.; Sundberg, B.; Remberger, M.; Sundin, M.; Mattsson, J.; Uhlin, M. Risk Factors for Severe Acute Graft-versus-Host Disease in Donor Graft Composition. Biol. Blood Marrow Transplant. 2018, 24, 467–477. [Google Scholar] [CrossRef]
- Hossain, M.S.; Kunter, G.M.; El-Najjar, V.F.; Jaye, D.L.; Al-Kadhimi, Z.; Taofeek, O.K.; Li, J.M.; Waller, E.K. PD-1 and CTLA-4 up regulation on donor T cells is insufficient to prevent GvHD in allo-HSCT recipients. PLoS ONE 2017, 12, e0184254. [Google Scholar] [CrossRef] [PubMed]
- Nair, A.P.; Walker, P.; Kalff, A.; Bergin, K.; Hocking, J.; Avery, S.; Curtis, D.J.; Patil, S.; Das, T.; Klarica, D.; et al. Adverse impact of high donor CD3+ cell dose on outcome following tandem auto-NMA allogeneic transplantation for high-risk myeloma. Bone Marrow Transplant. 2017, 52, 839–845. [Google Scholar] [CrossRef] [PubMed]
- Sairafi, D.; Stikvoort, A.; Gertow, J.; Mattsson, J.; Uhlin, M. Donor Cell Composition and Reactivity Predict Risk of Acute Graft-versus-Host Disease after Allogeneic Hematopoietic Stem Cell Transplantation. J. Immunol. Res. 2016, 2016, 5601204. [Google Scholar] [CrossRef] [PubMed]
- Hartwell, M.J.; Ozbek, U.; Holler, E.; Renteria, A.S.; Major-Monfried, H.; Reddy, P.; Aziz, M.; Hogan, W.J.; Ayuk, F.; Efebera, Y.A.; et al. An early-biomarker algorithm predicts lethal graft-versus-host disease and survival. JCI Insight 2017, 2, e89798. [Google Scholar] [CrossRef]
- Major-Monfried, H.; Renteria, A.S.; Pawarode, A.; Reddy, P.; Ayuk, F.; Holler, E.; Efebera, Y.A.; Hogan, W.J.; Wolfl, M.; Qayed, M.; et al. MAGIC biomarkers predict long-term outcomes for steroid-resistant acute GVHD. Blood 2018, 131, 2846–2855. [Google Scholar] [CrossRef]
- Wu, X.; Xie, Y.; Wang, C.; Han, Y.; Bao, X.; Ma, S.; Yilmaz, A.; Yang, B.; Ji, Y.; Xu, J.; et al. Prediction of acute GVHD and relapse by metabolic biomarkers after allogeneic hematopoietic stem cell transplantation. JCI Insight 2018, 3. [Google Scholar] [CrossRef]
- MacMillan, M.L.; DeFor, T.E.; Holtan, S.G.; Rashidi, A.; Blazar, B.R.; Weisdorf, D.J. Validation of Minnesota acute graft-versus-host disease Risk Score. Haematologica 2020, 105, 519–524. [Google Scholar] [CrossRef]
- Armand, P.; Gibson, C.J.; Cutler, C.; Ho, V.T.; Koreth, J.; Alyea, E.P.; Ritz, J.; Sorror, M.L.; Lee, S.J.; Deeg, H.J.; et al. A disease risk index for patients undergoing allogeneic stem cell transplantation. Blood 2012, 120, 905–913. [Google Scholar] [CrossRef]
- Zeiser, R.; Socie, G.; Blazar, B.R. Pathogenesis of acute graft-versus-host disease: From intestinal microbiota alterations to donor T cell activation. Br. J. Haematol. 2016, 175, 191–207. [Google Scholar] [CrossRef]
- DeWolf, S.; Sykes, M. Alloimmune T cells in transplantation. J. Clin. Investig. 2017, 127, 2473–2481. [Google Scholar] [CrossRef]
- Wilhelm, K.; Ganesan, J.; Muller, T.; Durr, C.; Grimm, M.; Beilhack, A.; Krempl, C.D.; Sorichter, S.; Gerlach, U.V.; Juttner, E.; et al. Graft-versus-host disease is enhanced by extracellular ATP activating P2X7R. Nat. Med. 2010, 16, 1434–1438. [Google Scholar] [CrossRef]
- Wiley, J.S.; Sluyter, R.; Gu, B.J.; Stokes, L.; Fuller, S.J. The human P2X7 receptor and its role in innate immunity. Tissue Antigens 2011, 78, 321–332. [Google Scholar] [CrossRef]
- Mariathasan, S.; Weiss, D.S.; Newton, K.; McBride, J.; O’Rourke, K.; Roose-Girma, M.; Lee, W.P.; Weinrauch, Y.; Monack, D.M.; Dixit, V.M. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature 2006, 440, 228–232. [Google Scholar] [CrossRef]
- Qu, Y.; Franchi, L.; Nunez, G.; Dubyak, G.R. Nonclassical IL-1 beta secretion stimulated by P2X7 receptors is dependent on inflammasome activation and correlated with exosome release in murine macrophages. J. Immunol. 2007, 179, 1913–1925. [Google Scholar] [CrossRef]
- Sluyter, R.; Stokes, L. Significance of P2X7 receptor variants to human health and disease. Recent Pat. DNA Gene Seq. 2011, 5, 41–54. [Google Scholar] [CrossRef]
- Gu, B.J.; Sluyter, R.; Skarratt, K.K.; Shemon, A.N.; Dao-Ung, L.P.; Fuller, S.J.; Barden, J.A.; Clarke, A.L.; Petrou, S.; Wiley, J.S. An Arg307 to Gln polymorphism within the ATP-binding site causes loss of function of the human P2X7 receptor. J. Biol. Chem. 2004, 279, 31287–31295. [Google Scholar] [CrossRef]
- Gu, B.J.; Zhang, W.; Worthington, R.A.; Sluyter, R.; Dao-Ung, P.; Petrou, S.; Barden, J.A.; Wiley, J.S. A Glu-496 to Ala polymorphism leads to loss of function of the human P2X7 receptor. J. Biol. Chem. 2001, 276, 11135–11142. [Google Scholar] [CrossRef]
- Stokes, L.; Fuller, S.J.; Sluyter, R.; Skarratt, K.K.; Gu, B.J.; Wiley, J.S. Two haplotypes of the P2X(7) receptor containing the Ala-348 to Thr polymorphism exhibit a gain-of-function effect and enhanced interleukin-1beta secretion. FASEB J. 2010, 24, 2916–2927. [Google Scholar] [CrossRef]
- Stoffels, M.; Zaal, R.; Kok, N.; van der Meer, J.W.; Dinarello, C.A.; Simon, A. ATP-Induced IL-1beta Specific Secretion: True Under Stringent Conditions. Front. Immunol. 2015, 6, 54. [Google Scholar] [CrossRef] [PubMed]
- Sluyter, R.; Dalitz, J.G.; Wiley, J.S. P2X7 receptor polymorphism impairs extracellular adenosine 5′-triphosphate-induced interleukin-18 release from human monocytes. Genes Immun. 2004, 5, 588–591. [Google Scholar] [CrossRef] [PubMed]
- Jankovic, D.; Ganesan, J.; Bscheider, M.; Stickel, N.; Weber, F.C.; Guarda, G.; Follo, M.; Pfeifer, D.; Tardivel, A.; Ludigs, K.; et al. The Nlrp3 inflammasome regulates acute graft-versus-host disease. J. Exp. Med. 2013, 210, 1899–1910. [Google Scholar] [CrossRef] [PubMed]
- Jamieson, S.E.; Peixoto-Rangel, A.L.; Hargrave, A.C.; Roubaix, L.A.; Mui, E.J.; Boulter, N.R.; Miller, E.N.; Fuller, S.J.; Wiley, J.S.; Castellucci, L.; et al. Evidence for associations between the purinergic receptor P2X(7) (P2RX7) and toxoplasmosis. Genes Immun. 2010, 11, 374–383. [Google Scholar] [CrossRef]
- Sharma, S.; Kumar, V.; Khosla, R.; Kajal, N.; Sarin, B.; Sehajpal, P. Association of P2X7 receptor +1513 (A-->C) polymorphism with tuberculosis in a Punjabi population. Int. J. Tuberc. Lung Dis. 2010, 14, 1159–1163. [Google Scholar]
- Jagasia, M.H.; Greinix, H.T.; Arora, M.; Williams, K.M.; Wolff, D.; Cowen, E.W.; Palmer, J.; Weisdorf, D.; Treister, N.S.; Cheng, G.S.; et al. National Institutes of Health Consensus Development Project on Criteria for Clinical Trials in Chronic Graft-versus-Host Disease: I. The 2014 Diagnosis and Staging Working Group report. Biol. Blood Marrow Transplant. 2015, 21, 389–401. [Google Scholar] [CrossRef]
- Wiley, J.S.; Dao-Ung, L.P.; Li, C.; Shemon, A.N.; Gu, B.J.; Smart, M.L.; Fuller, S.J.; Barden, J.A.; Petrou, S.; Sluyter, R. An Ile-568 to Asn polymorphism prevents normal trafficking and function of the human P2X7 receptor. J. Biol. Chem. 2003, 278, 17108–17113. [Google Scholar] [CrossRef]
- Shemon, A.N.; Sluyter, R.; Fernando, S.L.; Clarke, A.L.; Dao-Ung, L.P.; Skarratt, K.K.; Saunders, B.M.; Tan, K.S.; Gu, B.J.; Fuller, S.J.; et al. A Thr357 to Ser polymorphism in homozygous and compound heterozygous subjects causes absent or reduced P2X7 function and impairs ATP-induced mycobacterial killing by macrophages. J. Biol. Chem. 2006, 281, 2079–2086. [Google Scholar] [CrossRef]
- Aziz, M.D.; Shah, J.; Kapoor, U.; Dimopoulos, C.; Anand, S.; Augustine, A.; Ayuk, F.; Chaudhry, M.; Chen, Y.B.; Choe, H.K.; et al. Disease risk and GVHD biomarkers can stratify patients for risk of relapse and nonrelapse mortality post hematopoietic cell transplant. Leukemia 2020. [Google Scholar] [CrossRef]
- Zhong, X.; Zhu, F.; Qiao, J.; Zhao, K.; Zhu, S.; Zeng, L.; Chen, X.; Xu, K. The impact of P2X7 receptor antagonist, brilliant blue G on graft-versus-host disease in mice after allogeneic hematopoietic stem cell transplantation. Cell Immunol. 2016, 310, 71–77. [Google Scholar] [CrossRef]
- Geraghty, N.J.; Belfiore, L.; Ly, D.; Adhikary, S.R.; Fuller, S.J.; Varikatt, W.; Sanderson-Smith, M.L.; Sluyter, V.; Alexander, S.I.; Sluyter, R.; et al. The P2X7 receptor antagonist Brilliant Blue G reduces serum human interferon-gamma in a humanized mouse model of graft-versus-host disease. Clin. Exp. Immunol. 2017, 190, 79–95. [Google Scholar] [CrossRef] [PubMed]
- Adhikary, S.R.; Geraghty, N.J.; Cuthbertson, P.; Sluyter, R.; Watson, D. Altered donor P2X7 activity in human leukocytes correlates with P2RX7 genotype but does not affect the development of graft-versus-host disease in humanised mice. Purinergic Signal. 2019, 15, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.H.; Park, S.S.; Kim, I.; Kim, J.H.; Ra, E.K.; Yoon, S.S.; Hong, Y.C.; Park, S.; Kim, B.K. P2X7 receptor polymorphism and clinical outcomes in HLA-matched sibling allogeneic hematopoietic stem cell transplantation. Haematologica 2007, 92, 651–657. [Google Scholar] [CrossRef] [PubMed]
- Karaesmen, E.; Rizvi, A.A.; Preus, L.M.; McCarthy, P.L.; Pasquini, M.C.; Onel, K.; Zhu, X.; Spellman, S.; Haiman, C.A.; Stram, D.O.; et al. Replication and validation of genetic polymorphisms associated with survival after allogeneic blood or marrow transplant. Blood 2017, 130, 1585–1596. [Google Scholar] [CrossRef]
- Vangsted, A.J.; Klausen, T.W.; Gimsing, P.; Abildgaard, N.; Andersen, N.F.; Gang, A.O.; Holmstrom, M.; Gregersen, H.; Vogel, U.; Schwarz, P.; et al. Genetic variants in the P2RX7 gene are associated with risk of multiple myeloma. Eur. J. Haematol. 2014, 93, 172–174. [Google Scholar] [CrossRef]
- Adinolfi, E.; Melchiorri, L.; Falzoni, S.; Chiozzi, P.; Morelli, A.; Tieghi, A.; Cuneo, A.; Castoldi, G.; Di Virgilio, F.; Baricordi, O.R. P2X7 receptor expression in evolutive and indolent forms of chronic B lymphocytic leukemia. Blood 2002, 99, 706–708. [Google Scholar] [CrossRef]
- Chong, J.H.; Zheng, G.G.; Zhu, X.F.; Guo, Y.; Wang, L.; Ma, C.H.; Liu, S.Y.; Xu, L.L.; Lin, Y.M.; Wu, K.F. Abnormal expression of P2X family receptors in Chinese pediatric acute leukemias. Biochem. Biophys. Res. Commun. 2010, 391, 498–504. [Google Scholar] [CrossRef]
- Zhang, X.J.; Zheng, G.G.; Ma, X.T.; Yang, Y.H.; Li, G.; Rao, Q.; Nie, K.; Wu, K.F. Expression of P2X7 in human hematopoietic cell lines and leukemia patients. Leukemia Res. 2004, 28, 1313–1322. [Google Scholar] [CrossRef]
| dbSNP ID | Amino Acid Change | Effect on Function | Published MAF 1 | Recipient MAF | Donor MAF |
|---|---|---|---|---|---|
| rs28360445 | Arg117Trp | loss | nd | 0.003 | 0.002 |
| rs28360447 | Gly150Arg | loss | 0.018 | 0.016 | 0.017 |
| rs28360451 | Glu186Lys | loss | nd | 0.000 | 0.000 |
| rs28360452 | Leu191Pro | loss | nd | 0.000 | 0.000 |
| rs7958311 | Arg270His | loss | 0.255 | 0.255 | 0.291 |
| rs7958316 | Arg276His | loss | 0.02 | 0.023 | 0.031 |
| rs28360457 | Arg307Gln | loss | 0.013 | 0.012 | 0.018 |
| rs3751143 | Glu496Ala | loss | 0.175 | 0.185 | 0.180 |
| rs1653624 | Ile568Asn | loss | 0.029 | 0.019 | 0.015 |
| rs35933842 | - | loss | 0.008 | 0.008 | 0.007 |
| rs17525809 | Val76Ala | partial loss | 0.062 | 0.069 | 0.089 |
| rs2230911 | Thr357Ser | partial loss | 0.083 | 0.099 | 0.084 |
| rs2230912 | Gln460Arg | partial loss | 0.170 | 0.165 | 0.144 |
| rs2230913 | His521Gln | neutral | 0.02 | 0.001 | 0.001 |
| rs208294 | His155Tyr | gain | 0.439 | 0.470 | 0.447 |
| rs1718119 | Ala348Thr | gain | 0.4 | 0.385 | 0.367 |
| Characteristics (Total n = 453) | n | % |
|---|---|---|
| Recipient Age, median (range), years | 47 | (16–73) |
| Gender | ||
| F | 190 | (41.9%) |
| M | 263 | (58.1%) |
| Stem cell source | ||
| BM | 49 | (10.8%) |
| PB | 404 | (89.2%) |
| Donor | ||
| RD | 247 | (54.5%) |
| MUD | 206 | (45.5%) |
| Conditioning | ||
| MAC | 357 | (78.8%) |
| RIC | 96 | (21.2%) |
| Disease risk index | ||
| Low | 77 | (16.1%) |
| Intermediate | 257 | (56.7%) |
| High | 73 | (16.1%) |
| Very high | 12 | (2.6%) |
| Not determined | 34 | (7.5%) |
| Diagnosis | ||
| AML | 159 | (35.1%) |
| ALL | 63 | (13.9%) |
| MDS | 44 | (9.7%) |
| MM | 28 | (6.2%) |
| FL | 27 | (6.0%) |
| MF | 22 | (4.9%) |
| CLL | 20 | (4.4%) |
| CML | 17 | (3.8%) |
| HL | 15 | (3.3%) |
| Other | 58 | (12.8%) |
| Outcome | Donor or Recipient | SNP | MAF | p Value | RR | 95% CI | |
|---|---|---|---|---|---|---|---|
| Unaffected | Affected | ||||||
| aGVHD (g0 vs. g1–4) | Recipient | Ile568Asn | 0.01 | 0.04 | 0.0173 | 1.027 | 1.003–1.051 |
| aGVHD (g0–1 vs. g2–4) | Recipient | Glu496Ala | 0.19 | 0.13 | 0.0457 | 0.9267 | 0.8683–0.989 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koldej, R.M.; Perera, T.; Collins, J.; Ritchie, D.S. Association between P2X7 Polymorphisms and Post-Transplant Outcomes in Allogeneic Haematopoietic Stem Cell Transplantation. Int. J. Mol. Sci. 2020, 21, 3772. https://doi.org/10.3390/ijms21113772
Koldej RM, Perera T, Collins J, Ritchie DS. Association between P2X7 Polymorphisms and Post-Transplant Outcomes in Allogeneic Haematopoietic Stem Cell Transplantation. International Journal of Molecular Sciences. 2020; 21(11):3772. https://doi.org/10.3390/ijms21113772
Chicago/Turabian StyleKoldej, Rachel M, Travis Perera, Jenny Collins, and David S Ritchie. 2020. "Association between P2X7 Polymorphisms and Post-Transplant Outcomes in Allogeneic Haematopoietic Stem Cell Transplantation" International Journal of Molecular Sciences 21, no. 11: 3772. https://doi.org/10.3390/ijms21113772
APA StyleKoldej, R. M., Perera, T., Collins, J., & Ritchie, D. S. (2020). Association between P2X7 Polymorphisms and Post-Transplant Outcomes in Allogeneic Haematopoietic Stem Cell Transplantation. International Journal of Molecular Sciences, 21(11), 3772. https://doi.org/10.3390/ijms21113772





