Diagnostic and Therapeutic Value of Aptamers in Envenomation Cases
Abstract
1. Introduction
2. Envenomation is still a Major Public Health Problem
3. Major Threats to Antivenom Production and Usage
4. Emerging Technological Alternatives to the Production and Use of Polyclonal Antibody-Based Antivenoms
4.1. Toxin Activity-Based Antivenoms
4.2. Bioinformatic-Assisted Rationale Snake Antivenom Design
4.3. Monoclonal Antibodies
4.4. Other Technological and Chemical Initiatives
5. Comparative Advantages in Using Aptamers over Antibodies
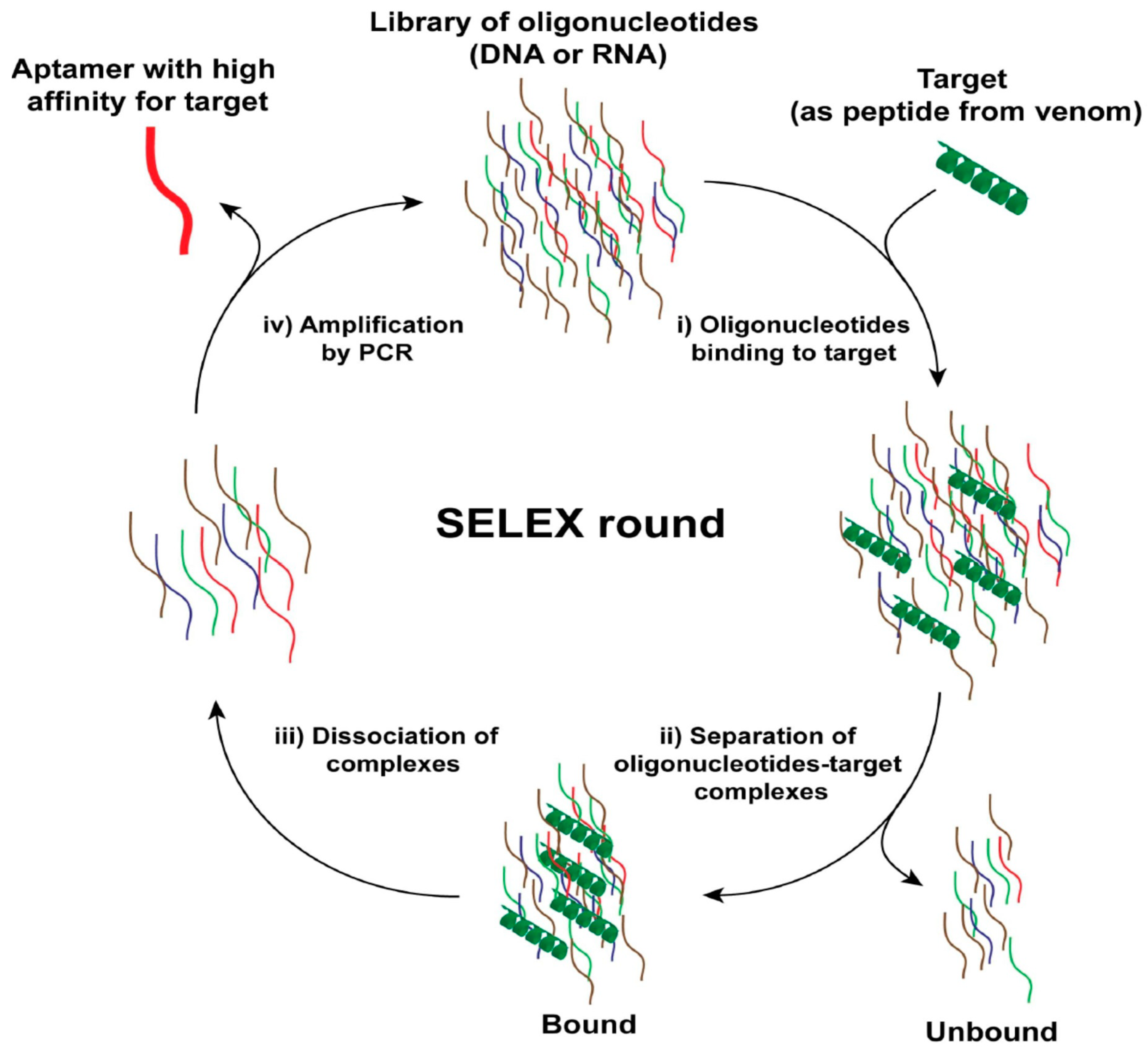
6. Aptamer Selection Procedures and Property Improvements
7. Use of Aptamers as Antitoxins
7.1. DNA Aptamers Directed against Bungarotoxin from Venom of the Elapid Bungarus Multicinctus
7.2. Engineering of DNA Aptamers as an Approach to Identify New Ligands from the Indian Bungarus Caeruleus (Krait) Snake Venom
7.3. DNA Aptamers against Cardiotoxin from Naja Atra Snake Venom
7.4. Neutralization of a Lethal Venom Toxin, αC-Conotoxin PrXA, In Vivo by a DNA Aptamer
7.5. First RNA Aptamers Targeted against Loxosceles Laeta Spider Toxins
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Calmette, A. The treatment of animals poisoned with snake venom by the injection of antivenomous serum. BMJ 1896, 2, 399–400. [Google Scholar] [CrossRef] [PubMed]
- Hawgood, B.J. Doctor Albert Calmette 1863-1933: founder of antivenomous serotherapy and of antituberculous BCG vaccination. Toxicon. 1999, 37, 1241–1258. [Google Scholar] [CrossRef]
- Calmette, A. Le venin des serpents. Physiologie de l’envenimation. Traitement des morsures venimeuses par le sérum des animaux vaccinés; Société d’Editions Scientifiques: Paris, France, 1896. [Google Scholar]
- Santiesteban, B.G. Realidades del Alacran; Impresiones Graficas: Herfa, Durango, 1996. [Google Scholar]
- Snake serum received at city hospital to guard miamians. Miami Daily News and Metropolis, 18 May 1927.
- Dohme, M.S. Lyovac Achievement Report, US Pharmaceutical Industry, 1936-1966; Merck Sharop & Dohme, Biologicals: Kenilworth, NJ, USA, 1966. [Google Scholar]
- Parfentjev, I.A. Method for purification of antitoxins and the like. U.S. Patent No 2,065,196, 22 December 1936. [Google Scholar]
- Parfentjev, I.A. Purification of antibody compositions. U.S. Patent No 2,175,090, 3 October 1939. [Google Scholar]
- Hansen, A. Reinigung und konzentrierung von diphtherie-antitoxin durch adsorption nach de autolyse mit pepsin. Biochemistry 1938, 299, 377. [Google Scholar]
- Pope, C. The action of proteolytic enzymes on the antitoxins and proteins in immune sera. I. True digestion of proteins. Br. J. Exp. Pathol. 1939, 20, 132. [Google Scholar]
- Sullivan, J.B., Jr. In search of a better snake trap. Wilderness Environ. Med. 1999, 10, 140–141. [Google Scholar] [CrossRef]
- Olvera, A.; Ramos-Cerrillo, B.; Estevez, J.; Clement, H.; de Roodt, A.; Paniagua-Solis, J.; Vazquez, H.; Zavaleta, A.; Arruz, M.S.; Stock, R.P.; et al. North and South American Loxosceles spiders: development of a polyvalent antivenom with recombinant sphingomyelinases D as antigens. Toxicon 2006, 48, 64–74. [Google Scholar] [CrossRef]
- Becerril, B.; Riano, L.; Possani, L. Development of novel scorpion anti-venoms in Mexico. Toxicon 2012, 60, 190. [Google Scholar] [CrossRef]
- Pucca, M.B.; Cerni, F.A.; Janke, R.; Bermudez-Mendez, E.; Ledsgaard, L.; Barbosa, J.E.; Laustsen, A.H. History of envenoming therapy and current perspectives. Front. Immunol. 2019, 10, 1598. [Google Scholar] [CrossRef]
- Boyer, L. History of scorpion antivenom: one Arizonan’s view. Toxicon 2013, 69, 14–20. [Google Scholar] [CrossRef]
- Bochner, R. Paths to the discovery of antivenom serotherapy in France. J. Venom. Anim. Toxins Incl. Trop. Dis. 2016, 22, 20. [Google Scholar] [CrossRef]
- World Health Organization. Snakebite envenoming. Available online: https://www.who.int/snakebites/disease/en/ (accessed on 17 May 2020).
- Simpson, I.D.; Blaylock, R.S. The anti snake venom crisis in Africa: a suggested manufacturers product guide. Wilderness Environ. Med. 2009, 20, 275–282. [Google Scholar] [CrossRef]
- Wilde, H.; Thipkong, P.; Sitprija, V.; Chaiyabutr, N. Heterologous antisera and antivenins are essential biologicals: perspectives on a worldwide crisis. Ann. Intern. Med. 1996, 125, 233–236. [Google Scholar] [CrossRef]
- Theakston, R.D.; Warrell, D.A. Crisis in snake antivenom supply for Africa. Lancet 2000, 356, 2104. [Google Scholar] [CrossRef]
- Habib, A.G.; Brown, N.I. The snakebite problem and antivenom crisis from a health-economic perspective. Toxicon 2018, 150, 115–123. [Google Scholar] [CrossRef]
- Brown, N.I. Consequences of neglect: analysis of the sub-Saharan African snake antivenom market and the global context. PLoS Negl. Trop. Dis. 2012, 6, e1670. [Google Scholar] [CrossRef]
- de Silva, H.A.; Ryan, N.M.; de Silva, H.J. Adverse reactions to snake antivenom, and their prevention and treatment. Br. J. Clin. Pharmacol. 2016, 81, 446–452. [Google Scholar] [CrossRef]
- Varga, Z.; Gurrola-Briones, G.; Papp, F.; Rodriguez de la Vega, R.C.; Pedraza-Alva, G.; Tajhya, R.B.; Gaspar, R.; Cardenas, L.; Rosenstein, Y.; Beeton, C.; et al. Vm24, a natural immunosuppressive peptide, potently and selectively blocks Kv1.3 potassium channels of human T cells. Mol. Pharmacol. 2012, 82, 372–382. [Google Scholar] [CrossRef]
- Potet, J.; Smith, J.; McIver, L. Reviewing evidence of the clinical effectiveness of commercially available antivenoms in sub-Saharan Africa identifies the need for a multi-centre, multi-antivenom clinical trial. PLoS Negl. Trop. Dis. 2019, 13, e0007551. [Google Scholar] [CrossRef]
- Williams, D.J.; Gutierrez, J.M.; Calvete, J.J.; Wuster, W.; Ratanabanangkoon, K.; Paiva, O.; Brown, N.I.; Casewell, N.R.; Harrison, R.A.; Rowley, P.D.; et al. Ending the drought: new strategies for improving the flow of affordable, effective antivenoms in Asia and Africa. J. Proteomics 2011, 74, 1735–1767. [Google Scholar] [CrossRef]
- Chippaux, J.P. WHO Guidelines for the production, control and regulation of snake antivenom immunoglobulins. Biol. Aujourdhui 2010, 204, 87–91. [Google Scholar] [CrossRef]
- Williams, D.J.; Faiz, M.A.; Abela-Ridder, B.; Ainsworth, S.; Bulfone, T.C.; Nickerson, A.D.; Habib, A.G.; Junghanss, T.; Fan, H.W.; Turner, M.; et al. Strategy for a globally coordinated response to a priority neglected tropical disease: Snakebite envenoming. PLoS Negl. Trop. Dis. 2019, 13, e0007059. [Google Scholar] [CrossRef] [PubMed]
- Ainsworth, S.; Slagboom, J.; Alomran, N.; Pla, D.; Alhamdi, Y.; King, S.I.; Bolton, F.M.S.; Gutierrez, J.M.; Vonk, F.J.; Toh, C.H.; et al. The paraspecific neutralisation of snake venom induced coagulopathy by antivenoms. Commun. Biol. 2018, 1, 34. [Google Scholar] [CrossRef] [PubMed]
- Wagstaff, S.C.; Laing, G.D.; Theakston, R.D.; Papaspyridis, C.; Harrison, R.A. Bioinformatics and multiepitope DNA immunization to design rational snake antivenom. PLoS Med. 2006, 3, e184. [Google Scholar] [CrossRef] [PubMed]
- Bahraoui, E.; Pichon, J.; Muller, J.M.; Darbon, H.; Elayeb, M.; Granier, C.; Marvaldi, J.; Rochat, H. Monoclonal antibodies to scorpion toxins. Characterization and molecular mechanisms of neutralization. J. Immunol. 1988, 141, 214–220. [Google Scholar]
- Fernandes, I.; Assumpcao, G.G.; Silveira, C.R.; Faquim-Mauro, E.L.; Tanjoni, I.; Carmona, A.K.; Alves, M.F.; Takehara, H.A.; Rucavado, A.; Ramos, O.H.; et al. Immunochemical and biological characterization of monoclonal antibodies against BaP1, a metalloproteinase from Bothrops asper snake venom. Toxicon. 2010, 56, 1059–1065. [Google Scholar] [CrossRef]
- Morine, N.; Matsuda, S.; Terada, K.; Eto, A.; Ishida, I.; Oku, H. Neutralization of hemorrhagic snake venom metalloproteinase HR1a from Protobothrops flavoviridis by human monoclonal antibody. Toxicon 2008, 51, 345–352. [Google Scholar] [CrossRef]
- Pucca, M.B.; Zoccal, K.F.; Roncolato, E.C.; Bertolini, T.B.; Campos, L.B.; Cologna, C.T.; Faccioli, L.H.; Arantes, E.C.; Barbosa, J.E. Serrumab: a human monoclonal antibody that counters the biochemical and immunological effects of Tityus serrulatus venom. J. Immunotoxicol. 2012, 9, 173–183. [Google Scholar] [CrossRef]
- Bugli, F.; Graffeo, R.; Paroni Sterbini, F.; Torelli, R.; Masucci, L.; Sali, M.; Grasso, A.; Rufini, S.; Ricci, E.; Fadda, G.; et al. Monoclonal antibody fragment from combinatorial phage display library neutralizes alpha-latrotoxin activity and abolishes black widow spider venom lethality, in mice. Toxicon 2008, 51, 547–554. [Google Scholar] [CrossRef]
- Meng, J.; John, T.R.; Kaiser, I.I. Specificity and binding affinity of an anti-crotoxin combinatorial antibody selected from a phage-displayed library. Biochem. Pharmacol. 1995, 50, 1969–1977. [Google Scholar]
- Lafaye, P.; Choumet, V.; Demangel, C.; Bon, C.; Mazie, J.C. Biologically active human anti-crotoxin scFv isolated from a semi-synthetic phage library. Immunotechnology 1997, 3, 117–125. [Google Scholar] [CrossRef]
- Laustsen, A.H. Toxin-centric development approach for next-generation antivenoms. Toxicon 2018, 150, 195–197. [Google Scholar] [CrossRef] [PubMed]
- Laustsen, A.H.; Karatt-Vellatt, A.; Masters, E.W.; Arias, A.S.; Pus, U.; Knudsen, C.; Oscoz, S.; Slavny, P.; Griffiths, D.T.; Luther, A.M.; et al. In vivo neutralization of dendrotoxin-mediated neurotoxicity of black mamba venom by oligoclonal human IgG antibodies. Nat. Commun. 2018, 9, 3928. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, T.P.; Fryer, T.; Dehli, R.I.; Jurgensen, J.A.; Fuglsang-Madsen, A.; Fons, S.; Laustsen, A.H. Toxin neutralization using alternative binding proteins. Toxins 2019, 11, 53. [Google Scholar] [CrossRef] [PubMed]
- Karain, B.D.; Lee, M.K.H.; Hayes, W.K. C60 fullerenes as a novel treatment for poisoning and envenomation: a proof-of-concept study for snakebite. J. Nanosci. Nanotechnol. 2016, 16, 7764–7771. [Google Scholar] [CrossRef]
- Bryan-Quiros, W.; Fernandez, J.; Gutierrez, J.M.; Lewin, M.R.; Lomonte, B. Neutralizing properties of LY315920 toward snake venom group I and II myotoxic phospholipases A2. Toxicon 2019, 157, 1–7. [Google Scholar] [CrossRef]
- Lewin, M.; Samuel, S.; Merkel, J.; Bickler, P. Varespladib (LY315920) appears to be a potent, broad-spectrum, inhibitor of snake venom phospholipase A2 and a possible pre-referral treatment for envenomation. Toxins 2016, 8, 248. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, J.; Zhang, D.; Xiao, H.; Xiong, S.; Huang, C. Exploration of the inhibitory potential of varespladib for snakebite envenomation. Molecules 2018, 23, 391. [Google Scholar] [CrossRef]
- Gomez-Betancur, I.; Gogineni, V.; Salazar-Ospina, A.; Leon, F. Perspective on the therapeutics of anti-snake venom. Molecules 2019, 24, 3276. [Google Scholar] [CrossRef]
- Houghton, P.J.; Osibogun, I.M. Flowering plants used against snakebite. J. Ethnopharmacol. 1993, 39, 1–29. [Google Scholar] [CrossRef]
- Giovannini, P.; Howes, M.-J.R. Medicinal plants used to treat snakebites in Central America: review and assessment of scientific evidence. J. Ethnopharmacol. 2017, 199, 240–256. [Google Scholar] [CrossRef]
- Amui, S.F.; Puga, R.D.; Soares, A.M.; Giuliatti, S. Plant-antivenom: database of anti-venom medicinal plants. Electronic J. Biotechnol. 2010, 14, 6–7. [Google Scholar] [CrossRef]
- Vale, L.H.; Mendes, M.M.; Hamaguchi, A.; Soares, A.M.; Rodrigues, V.M.; Homsi-Brandeburgo, M.I. Neutralization of pharmacological and toxic activities of bothrops snake venoms by Schizolobium parahyba (Fabaceae) aqueous extract and its fractions. Basic Clin. Pharmacol. Toxicol. 2008, 103, 104–107. [Google Scholar] [CrossRef]
- Gomez-Betancur, I.; Pereanez, J.A.; Patino, A.C.; Benjumea, D. Inhibitory effect of pinostrobin from Renealmia alpinia, on the enzymatic and biological activities of a PLA2. Int. J. Biol. Macromol. 2016, 89, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Jabeen, T.; Pal, A.; Sharma, S.; Perbandt, M.; Betzel, C.; Singh, T.P. Crystal structures of the complexes of a group IIA phospholipase A2 with two natural anti-inflammatory agents, anisic acid, and atropine reveal a similar mode of binding. Proteins 2006, 64, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Mors, W.B.; Nascimento, M.C.; Pereira, B.M.; Pereira, N.A. Plant natural products active against snake bite--the molecular approach. Phytochemistry 2000, 55, 627–642. [Google Scholar] [CrossRef]
- Toyama, D.O.; Marangoni, S.; Diz-Filho, E.B.; Oliveira, S.C.; Toyama, M.H. Effect of umbelliferone (7-hydroxycoumarin, 7-HOC) on the enzymatic, edematogenic and necrotic activities of secretory phospholipase A2 (sPLA2) isolated from Crotalus durissus collilineatus venom. Toxicon 2009, 53, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Leanpolchareanchai, J.; Pithayanukul, P.; Bavovada, R.; Saparpakorn, P. Molecular docking studies and anti-enzymatic activities of Thai mango seed kernel extract against snake venoms. Molecules 2009, 14, 1404–1422. [Google Scholar] [CrossRef]
- Soares, A.M.; Ticli, F.K.; Marcussi, S.; Lourenco, M.V.; Januario, A.H.; Sampaio, S.V.; Giglio, J.R.; Lomonte, B.; Pereira, P.S. Medicinal plants with inhibitory properties against snake venoms. Curr. Med. Chem. 2005, 12, 2625–2641. [Google Scholar] [CrossRef]
- Jimenez-Estrada, M.; Velazquez-Contreras, C.; Garibay-Escobar, A.; Sierras-Canchola, D.; Lapizco-Vazquez, R.; Ortiz-Sandoval, C.; Burgos-Hernandez, A.; Robles-Zepeda, R.E. In vitro antioxidant and antiproliferative activities of plants of the ethnopharmacopeia from northwest of Mexico. BMC Complement. Altern. Med. 2013, 13, 12. [Google Scholar] [CrossRef]
- Chen, Y.J.; Tsai, C.Y.; Hu, W.P.; Chang, L.S. DNA aptamers against Taiwan banded Krait alpha-bungarotoxin recognize Taiwan cobra cardiotoxins. Toxins 2016, 8, 66. [Google Scholar]
- Dhiman, A.; Anand, A.; Malhotra, A.; Khan, E.; Santra, V.; Kumar, A.; Sharma, T.K. Rational truncation of aptamer for cross-species application to detect krait envenomation. Sci. Rep. 2018, 8, 17795. [Google Scholar] [CrossRef] [PubMed]
- El-Aziz, T.M.A.; Ravelet, C.; Molgo, J.; Fiore, E.; Pale, S.; Amar, M.; Al-Khoury, S.; Dejeu, J.; Fadl, M.; Ronjat, M.; et al. Efficient functional neutralization of lethal peptide toxins in vivo by oligonucleotides. Sci. Rep. 2017, 7, 7202. [Google Scholar] [CrossRef] [PubMed]
- Taiwe, G.S.; Montnach, J.; Nicolas, S.; De Waard, S.; Fiore, E.; Peyrin, E.; El-Aziz, T.M.A.; Amar, M.; Molgo, J.; Ronjat, M.; et al. Aptamer efficacies for in vitro and in vivo modulation of alphaC-conotoxin PrXA pharmacology. Molecules 2019, 24, 229. [Google Scholar]
- Lauridsen, L.H.; Shamaileh, H.A.; Edwards, S.L.; Taran, E.; Veedu, R.N. Rapid one-step selection method for generating nucleic acid aptamers: development of a DNA aptamer against alpha-bungarotoxin. PLoS One 2012, 7, e41702. [Google Scholar] [CrossRef]
- Lauridsen, L.H.; Veedu, R.N. Nucleic acid aptamers against biotoxins: a new paradigm toward the treatment and diagnostic approach. Nucleic Acid Ther. 2012, 22, 371–379. [Google Scholar] [CrossRef]
- Laustsen, A.H.; Sola, M.; Jappe, E.C.; Oscoz, S.; Lauridsen, L.P.; Engmark, M. Biotechnological trends in spider and scorpion antivenom development. Toxins 2016, 8, 226. [Google Scholar] [CrossRef]
- Sapag, A.; Salinas-Luypaert, C.; Constenla-Munoz, C. First report of in vitro selection of RNA aptamers targeted to recombinant Loxosceles laeta spider toxins. Biol. Res. 2014, 47, 2. [Google Scholar] [CrossRef]
- Ye, F.; Zheng, Y.; Wang, X.; Tan, X.; Zhang, T.; Xin, W.; Wang, J.; Huang, Y.; Fan, Q.; Wang, J. Recognition of Bungarus multicinctus venom by a DNA aptamer against beta-bungarotoxin. PLoS One 2014, 9, e105404. [Google Scholar] [CrossRef]
- Laustsen, A.H.; Johansen, K.H.; Engmark, M.; Andersen, M.R. Recombinant snakebite antivenoms: A cost-competitive solution to a neglected tropical disease? PLoS Negl. Trop. Dis. 2017, 11, e0005361. [Google Scholar] [CrossRef]
- Kini, R.M.; Sidhu, S.S.; Laustsen, A.H. Biosynthetic Oligoclonal Antivenom (BOA) for snakebite and next-generation treatments for snakebite victims. Toxins 2018, 10, 534. [Google Scholar] [CrossRef]
- Laustsen, A.H.; Gutierrez, J.M.; Rasmussen, A.R.; Engmark, M.; Gravlund, P.; Sanders, K.L.; Lohse, B.; Lomonte, B. Danger in the reef: Proteome, toxicity, and neutralization of the venom of the olive sea snake, Aipysurus laevis. Toxicon 2015, 107, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Calvete, J.J.; Lomonte, B. A bright future for integrative venomics. Toxicon 2015, 107, 159–162. [Google Scholar] [CrossRef] [PubMed]
- Frauches, T.S.; Petretski, J.H.; Arnholdt, A.C.; Lasunskaia, E.B.; de Carvalho, E.C.; Kipnis, T.L.; da Silva, W.D.; Kanashiro, M.M. Bothropic antivenom based on monoclonal antibodies, is it possible? Toxicon 2013, 71, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Stumpp, M.T.; Binz, H.K.; Amstutz, P. DARPins: a new generation of protein therapeutics. Drug Discov. Today 2008, 13, 695–701. [Google Scholar] [CrossRef]
- Santos-Filho, N.A.; Boldrini-Franca, J.; Santos-Silva, L.K.; Menaldo, D.L.; Henrique-Silva, F.; Sousa, T.S.; Cintra, A.C.; Mamede, C.C.; Oliveira, F.; Arantes, E.C.; et al. Heterologous expression and biochemical and functional characterization of a recombinant alpha-type myotoxin inhibitor from Bothrops alternatus snake. Biochimie 2014, 105, 119–128. [Google Scholar] [CrossRef]
- Chijiwa, T.; So, S.; Hattori, S.; Yoshida, A.; Oda-Ueda, N.; Ohno, M. Suppression of severe lesions, myonecrosis and hemorrhage, caused by Protobothrops flavoviridis venom with its serum proteins. Toxicon 2013, 76, 197–205. [Google Scholar] [CrossRef]
- Shi, Y.; Ji, M.K.; Xu, J.W.; Lin, X.; Lin, J.Y. High-level expression, purification, characterization and structural prediction of a snake venom metalloproteinase inhibitor in Pichia pastoris. Protein J. 2012, 31, 212–221. [Google Scholar] [CrossRef]
- Shirai, R.; Toriba, M.; Hayashi, K.; Ikeda, K.; Inoue, S. Identification and characterization of phospholipase A2 inhibitors from the serum of the Japanese rat snake, Elaphe climacophora. Toxicon 2009, 53, 685–692. [Google Scholar] [CrossRef]
- Scire, A.; Tanfani, F.; Bertoli, E.; Furlani, E.; Nadozie, H.O.; Cerutti, H.; Cortelazzo, A.; Bini, L.; Guerranti, R. The belonging of gpMuc, a glycoprotein from Mucuna pruriens seeds, to the Kunitz-type trypsin inhibitor family explains its direct anti-snake venom activity. Phytomedicine 2011, 18, 887–895. [Google Scholar] [CrossRef]
- Quiros, S.; Alape-Giron, A.; Angulo, Y.; Lomonte, B. Isolation, characterization and molecular cloning of AnMIP, a new alpha-type phospholipase A2 myotoxin inhibitor from the plasma of the snake Atropoides nummifer (Viperidae: Crotalinae). Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 2007, 146, 60–68. [Google Scholar] [CrossRef]
- Jurgilas, P.B.; Neves-Ferreira, A.G.; Domont, G.B.; Perales, J. PO41, a snake venom metalloproteinase inhibitor isolated from Philander opossum serum. Toxicon 2003, 42, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Neves-Ferreira, A.G.; Perales, J.; Fox, J.W.; Shannon, J.D.; Makino, D.L.; Garratt, R.C.; Domont, G.B. Structural and functional analyses of DM43, a snake venom metalloproteinase inhibitor from Didelphis marsupialis serum. J. Biol. Chem. 2002, 277, 13129–13137. [Google Scholar] [CrossRef] [PubMed]
- Lewin, M.R.; Gutierrez, J.M.; Samuel, S.P.; Herrera, M.; Bryan-Quiros, W.; Lomonte, B.; Bickler, P.E.; Bulfone, T.C.; Williams, D.J. Delayed oral LY333013 rescues mice from highly neurotoxic, lethal doses of papuan taipan (Oxyuranus scutellatus) venom. Toxins 2018, 10, 380. [Google Scholar] [CrossRef] [PubMed]
- Arias, A.S.; Rucavado, A.; Gutierrez, J.M. Peptidomimetic hydroxamate metalloproteinase inhibitors abrogate local and systemic toxicity induced by Echis ocellatus (saw-scaled) snake venom. Toxicon 2017, 132, 40–49. [Google Scholar] [CrossRef]
- Laustsen, A.H.; Engmark, M.; Milbo, C.; Johannesen, J.; Lomonte, B.; Gutierrez, J.M.; Lohse, B. From fangs to pharmacology: The future of snakebite envenoming therapy. Curr. Pharm. Des. 2016, 22, 5270–5293. [Google Scholar] [CrossRef]
- Knudsen, C.; Laustsen, A.H. Recent advances in next generation snakebite antivenoms. Trop. Med. Infect. Dis. 2018, 3, 42. [Google Scholar]
- Santhosh, M.S.; Hemshekhar, M.; Sunitha, K.; Thushara, R.M.; Jnaneshwari, S.; Kemparaju, K.; Girish, K.S. Snake venom induced local toxicities: plant secondary metabolites as an auxiliary therapy. Mini Rev. Med. Chem. 2013, 13, 106–123. [Google Scholar] [CrossRef]
- Ali, M.H.; Elsherbiny, M.E.; Emara, M. Updates on aptamer research. Int. J. Mol. Sci. 2019, 20, 2511. [Google Scholar] [CrossRef]
- Bagalkot, V.; Zhang, L.; Levy-Nissenbaum, E.; Jon, S.; Kantoff, P.W.; Langer, R.; Farokhzad, O.C. Quantum dot-aptamer conjugates for synchronous cancer imaging, therapy, and sensing of drug delivery based on bi-fluorescence resonance energy transfer. Nano. Lett. 2007, 7, 3065–3070. [Google Scholar] [CrossRef]
- Champanhac, C.; Teng, I.T.; Cansiz, S.; Zhang, L.; Wu, X.; Zhoa, Z.; Fu, T.; Tan, W. Development of a panel of DNA aptamers with high affinity for pancreatic ductal adenocarcinoma. Sci. Rep. 2015, 5, 16788. [Google Scholar] [CrossRef]
- Cox, J.C.; Rudolph, P.; Ellington, A.D. Automated RNA selection. Biotechnol. Prog. 1998, 14, 845–850. [Google Scholar] [CrossRef]
- Dausse, E.; Da Rocha Gomes, S.; Toulme, J.J. Aptamers: a new class of oligonucleotides in the drug discovery pipeline? Curr. Opin. Pharmacol. 2009, 9, 602–607. [Google Scholar] [CrossRef] [PubMed]
- Drabovich, A.P.; Berezovski, M.; Okhonin, V.; Krylov, S.N. Selection of smart aptamers by methods of kinetic capillary electrophoresis. Anal. Chem. 2006, 78, 3171–3178. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Keefe, A.D.; Pai, S.; Ellington, A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010, 9, 537–550. [Google Scholar] [CrossRef]
- Hicke, B.J.; Stephens, A.W. Escort aptamers: a delivery service for diagnosis and therapy. J. Clin. Invest. 2000, 106, 923–928. [Google Scholar] [CrossRef] [PubMed]
- Hori, S.I.; Herrera, A.; Rossi, J.J.; Zhou, J. Current advances in aptamers for cancer diagnosis and therapy. Cancers 2018, 10, 9. [Google Scholar] [CrossRef]
- Fu, Z.; Xiang, J. Aptamers, the nucleic acid antibodies, in cancer therapy. Int. J. Mol. Sci. 2020, 21, 2793. [Google Scholar] [CrossRef]
- Lokesh, G.L.; Wang, H.; Lam, C.H.; Thiviyanathan, V.; Ward, N.; Gorenstein, D.G.; Volk, D.E. X-aptamer selection and validation. Methods Mol. Biol. 2017, 1632, 151–174. [Google Scholar]
- Maier, K.E.; Levy, M. From selection hits to clinical leads: progress in aptamer discovery. Mol. Ther. Methods Clin. Dev. 2016, 5, 16014. [Google Scholar] [CrossRef]
- Moutsiopoulou, A.; Broyles, D.; Dikici, E.; Daunert, S.; Deo, S.K. Molecular aptamer beacons and their applications in sensing, imaging, and diagnostics. Small 2019, 15, e1902248. [Google Scholar] [CrossRef]
- Nimjee, S.M.; White, R.R.; Becker, R.C.; Sullenger, B.A. Aptamers as therapeutics. Annu. Rev. Pharmacol. Toxicol. 2017, 57, 61–79. [Google Scholar] [CrossRef]
- Parashar, A. Aptamers in therapeutics. J. Clin. Diagn. Res. 2016, 10, BE01–BE06. [Google Scholar] [CrossRef] [PubMed]
- Song, K.M.; Lee, S.; Ban, C. Aptamers and their biological applications. Sensors 2012, 12, 612–631. [Google Scholar] [CrossRef]
- Kedzierski, S.; Khoshnejad, M.; Caltagirone, T.G. Synthetic antibodies: The emerging field of aptamers. BioProcess J. 2012, 11, 46–49. [Google Scholar] [CrossRef]
- Thiel, K. Oligo oligarchy-the surprisingly small world of aptamers. Nat. Biotechnol. 2004, 22, 649–651. [Google Scholar] [CrossRef] [PubMed]
- Studnicka, J.; Rihova, B.; Rencova, E.; Rozsival, P.; Dubska, Z.; Chrapek, O.; Kolar, P.; Kandrnal, V.; Demlova, R.; Pitrova, S.; et al. Cost and effectiveness of therapy for wet age-related macular degeneration in routine clinical practice. Ophthalmologica 2013, 230, 34–42. [Google Scholar] [CrossRef]
- Odeh, F.; Nsairat, H.; Alshaer, W.; Ismail, M.A.; Esawi, E.; Qaqish, B.; Bawab, A.A.; Ismail, S.I. Aptamers chemistry: Chemical modifications and conjugation strategies. Molecules 2019, 25, 3. [Google Scholar] [CrossRef]
- Krasheninina, O.A.; Novopashina, D.S.; Apartsin, E.K.; Venyaminova, A.G. Recent advances in nucleic acid targeting probes and supramolecular constructs based on pyrene-modified oligonucleotides. Molecules 2017, 22, 2108. [Google Scholar] [CrossRef]
- Zhang, H.; Ma, Y.; Xie, Y.; An, Y.; Huang, Y.; Zhu, Z.; Yang, C.J. A controllable aptamer-based self-assembled DNA dendrimer for high affinity targeting, bioimaging and drug delivery. Sci. Rep. 2015, 5, 10099. [Google Scholar] [CrossRef]
- Lee, C.H.; Rajendran, R.; Jeong, M.-S.; Ko, H.Y.; Joo, J.Y.; Cho, S.; Chang, Y.W.; Kim, S. Bioimaging of targeting cancers using aptamer-conjugated carbon nanodots. ChemComm. 2013, 49, 6543–6545. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; He, X.; Wang, K.; Wu, X.; Ye, X.; Guo, Q.; Tan, W.; Qing, Z.; Yang, X.; Zhou, B. Activatable aptamer probe for contrast-enhanced in vivo cancer imaging based on cell membrane protein-triggered conformation alteration. Proc. Natl. Acad. Sci. USA 2011, 108, 3900–3905. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Z.; Hu, D.; Lin, C.T.; Li, J.; Lin, Y. Aptamer/graphene oxide nanocomplex for in situ molecular probing in living cells. J. Am. Chem. Soc. 2010, 132, 9274–9276. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Z.; Weber, T.J.; Hu, D.; Lin, C.T.; Li, J.; Lin, Y. In situ live cell sensing of multiple nucleotides exploiting DNA/RNA aptamers and graphene oxide nanosheets. Anal. Chem. 2013, 85, 6775–6782. [Google Scholar] [CrossRef] [PubMed]
- Levy-Nissenbaum, E.; Radovic-Moreno, A.F.; Wang, A.Z.; Langer, R.; Farokhzad, O.C. Nanotechnology and aptamers: applications in drug delivery. Trends Biotechnol. 2008, 26, 442–449. [Google Scholar] [CrossRef]
- Rajabnejad, S.H.; Mokhtarzadeh, A.; Abnous, K.; Taghdisi, S.M.; Ramezani, M.; Razavi, B.M. Targeted delivery of melittin to cancer cells by AS1411 anti-nucleolin aptamer. Drug Dev. Ind. Pharm. 2018, 44, 982–987. [Google Scholar] [CrossRef]
- Ozalp, V.C.; Kavruk, M.; Dilek, O.; Bayrac, A.T. Aptamers: molecular tools for medical diagnosis. Curr. Top. Med. Chem. 2015, 15, 1125–1137. [Google Scholar] [CrossRef]
- Nagarkatti, R.; Bist, V.; Sun, S.; Fortes de Araujo, F.; Nakhasi, H.L.; Debrabant, A. Development of an aptamer-based concentration method for the detection of Trypanosoma cruzi in blood. PLoS One 2012, 7, e43533. [Google Scholar] [CrossRef]
- Tonelli, R.R.; Colli, W.; Alves, M.J. Selection of binding targets in parasites using phage-display and aptamer libraries in vivo and in vitro. Front. Immunol. 2012, 3, 419. [Google Scholar] [CrossRef]
- Sun, H.; Zhu, X.; Lu, P.Y.; Rosato, R.R.; Tan, W.; Zu, Y. Oligonucleotide aptamers: New tools for targeted cancer therapy. Mol. Ther. Nucleic Acids 2014, 3, e182. [Google Scholar] [CrossRef]
- Rosenberg, J.E.; Bambury, R.M.; Van Allen, E.M.; Drabkin, H.A.; Lara, P.N., Jr.; Harzstark, A.L.; Wagle, N.; Figlin, R.A.; Smith, G.W.; Garraway, L.A.; et al. A phase II trial of AS1411 (a novel nucleolin-targeted DNA aptamer) in metastatic renal cell carcinoma. Invest. New Drugs 2014, 32, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Soundararajan, S.; Chen, W.; Spicer, E.K.; Courtenay-Luck, N.; Fernandes, D.J. The nucleolin targeting aptamer AS1411 destabilizes Bcl-2 messenger RNA in human breast cancer cells. Cancer Res. 2008, 68, 2358–2365. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, Y.; Chen, Y.; Hong, S.; Sun, Y.; Sun, N.; Pei, R. In vitro selection of DNA aptamers against renal cell carcinoma using living cell-SELEX. Talanta 2017, 175, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Yuan, B.; Jiang, X.; Chen, Y.; Guo, Q.; Wang, K.; Meng, X.; Huang, Z.; Wen, X. Metastatic cancer cell and tissue-specific fluorescence imaging using a new DNA aptamer developed by Cell-SELEX. Talanta 2017, 170, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Musumeci, D.; Platella, C.; Riccardi, C.; Moccia, F.; Montesarchio, D. Fluorescence sensing using DNA aptamers in cancer research and clinical diagnostics. Cancers 2017, 9, 174. [Google Scholar] [CrossRef]
- Ruckman, J.; Green, L.S.; Beeson, J.; Waugh, S.; Gillette, W.L.; Henninger, D.D.; Claesson-Welsh, L.; Janjic, N. 2’-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J. Biol. Chem. 1998, 273, 20556–20567. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Chen, J.; Wu, M.; Zhao, J.X. Aptamers: active targeting ligands for cancer diagnosis and therapy. Theranostics 2015, 5, 322–344. [Google Scholar] [CrossRef]
- Song, S.; Wang, L.; Li, J.; Fan, C.; Zhao, J. Aptamer-based biosensors. Trend. Anal. Chem. 2008, 27, 108–117. [Google Scholar] [CrossRef]
- Cash, K.J.; Ricci, F.; Plaxco, K.W. An electrochemical sensor for the detection of protein-small molecule interactions directly in serum and other complex matrices. J. Am. Chem. Soc. 2009, 131, 6955–6957. [Google Scholar] [CrossRef]
- Yuan, H.; Huang, Y.; Yang, J.; Guo, Y.; Zeng, X.; Zhou, S.; Cheng, J.; Zhang, Y. An aptamer-based fluorescence bio-sensor for chiral recognition of arginine enantiomers. Spectrochim. Acta A. Mol. Biomol. Spectrosc. 2018, 200, 330–338. [Google Scholar] [CrossRef]
- Tombelli, S.; Mascini, M. Aptamers biosensors for pharmaceutical compounds. Comb. Chem. High Throughput Screen 2010, 13, 641–649. [Google Scholar] [CrossRef] [PubMed]
- Win, M.N.; Klein, J.S.; Smolke, C.D. Codeine-binding RNA aptamers and rapid determination of their binding constants using a direct coupling surface plasmon resonance assay. Nucleic Acids Res. 2006, 34, 5670–5682. [Google Scholar] [CrossRef]
- Swensen, J.S.; Xiao, Y.; Ferguson, B.S.; Lubin, A.A.; Lai, R.Y.; Heeger, A.J.; Plaxco, K.W.; Soh, H.T. Continuous, real-time monitoring of cocaine in undiluted blood serum via a microfluidic, electrochemical aptamer-based sensor. J. Am. Chem. Soc. 2009, 131, 4262–4266. [Google Scholar] [CrossRef]
- Stojanovic, M.N.; de Prada, P.; Landry, D.W. Aptamer-based folding fluorescent sensor for cocaine. J. Am. Chem. Soc. 2001, 123, 4928–4931. [Google Scholar] [CrossRef] [PubMed]
- Wochner, A.; Menger, M.; Orgel, D.; Cech, B.; Rimmele, M.; Erdmann, V.A.; Glokler, J. A DNA aptamer with high affinity and specificity for therapeutic anthracyclines. Anal. Biochem. 2008, 373, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Hoellenriegel, J.; Zboralski, D.; Maasch, C.; Rosin, N.Y.; Wierda, W.G.; Keating, M.J.; Kruschinski, A.; Burger, J.A. The Spiegelmer NOX-A12, a novel CXCL12 inhibitor, interferes with chronic lymphocytic leukemia cell motility and causes chemosensitization. Blood 2014, 123, 1032–1039. [Google Scholar] [CrossRef]
- Vater, A.; Klussmann, S. Turning mirror-image oligonucleotides into drugs: the evolution of Spiegelmer((R)) therapeutics. Drug Discov. Today 2015, 20, 147–155. [Google Scholar] [CrossRef]
- Boyce, M.; Warrington, S.; Cortezi, B.; Zollner, S.; Vauleon, S.; Swinkels, D.W.; Summo, L.; Schwoebel, F.; Riecke, K. Safety, pharmacokinetics and pharmacodynamics of the anti-hepcidin Spiegelmer lexaptepid pegol in healthy subjects. Br. J. Pharmacol. 2016, 173, 1580–1588. [Google Scholar] [CrossRef]
- Ponce, A.T.; Hong, K.L. A mini-review: Clinical development and potential of aptamers for thrombotic events treatment and monitoring. Biomedicines 2019, 7, 55. [Google Scholar] [CrossRef] [PubMed]
- Steurer, M.; Montillo, M.; Scarfo, L.; Mauro, F.R.; Andel, J.; Wildner, S.; Trentin, L.; Janssens, A.; Burgstaller, S.; Fromming, A.; et al. Olaptesed pegol (NOX-A12) with bendamustine and rituximab: a phase IIa study in patients with relapsed/refractory chronic lymphocytic leukemia. Haematologica 2019, 104, 2053–2060. [Google Scholar] [CrossRef]
- Ni, S.; Yao, H.; Wang, L.; Lu, J.; Jiang, F.; Lu, A.; Zhang, G. Chemical modifications of nucleic acid aptamers for therapeutic purposes. Int. J. Mol. Sci. 2017, 18, 1683. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Berezovski, M.; Musheev, M.; Drabovich, A.; Krylov, S.N. Non-SELEX selection of aptamers. J. Am. Chem. Soc. 2006, 128, 1410–1411. [Google Scholar] [CrossRef] [PubMed]
- Berezovski, M.V.; Musheev, M.U.; Drabovich, A.P.; Jitkova, J.V.; Krylov, S.N. Non-SELEX: selection of aptamers without intermediate amplification of candidate oligonucleotides. Nat. Protoc. 2006, 1, 1359–1369. [Google Scholar] [CrossRef] [PubMed]
- Ahirwar, R.; Nahar, S.; Aggarwal, S.; Ramachandran, S.; Maiti, S.; Nahar, P. In silico selection of an aptamer to estrogen receptor alpha using computational docking employing estrogen response elements as aptamer-alike molecules. Sci. Rep. 2016, 6, 21285. [Google Scholar] [CrossRef]
- Rahimizadeh, K.; AlShamaileh, H.; Fratini, M.; Chakravarthy, M.; Stephen, M.; Shigdar, S.; Veedu, R.N. Development of cell-specific aptamers: Recent advances and insight into the selection procedures. Molecules 2017, 22, 2070. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.; Ren, C.; Wang, L.; Zhu, B.; Jia, W.; Gao, M.; Zeng, F.; Zeng, L.; Xia, X.; Zhang, X.; et al. CD109 is identified as a potential nasopharyngeal carcinoma biomarker using aptamer selected by cell-SELEX. Oncotarget 2016, 7, 55328–55342. [Google Scholar] [CrossRef]
- Yang, X.; Bassett, S.E.; Li, X.; Luxon, B.A.; Herzog, N.K.; Shope, R.E.; Aronson, J.; Prow, T.W.; Leary, J.F.; Kirby, R.; et al. Construction and selection of bead-bound combinatorial oligonucleoside phosphorothioate and phosphorodithioate aptamer libraries designed for rapid PCR-based sequencing. Nucleic Acids Res. 2002, 30, e132. [Google Scholar] [CrossRef]
- Stein, C.A. Exploiting the potential of antisense: beyond phosphorothioate oligodeoxynucleotides. Chem. Biol. 1996, 3, 319–323. [Google Scholar] [CrossRef]
- Yang, X.; Fennewald, S.; Luxon, B.A.; Aronson, J.; Herzog, N.K.; Gorenstein, D.G. Aptamers containing thymidine 3’-O-phosphorodithioates: synthesis and binding to nuclear factor-kappaB. Bioorg. Med. Chem. Lett. 1999, 9, 3357–3362. [Google Scholar] [CrossRef]
- Mou, T.C.; Gray, C.W.; Terwilliger, T.C.; Gray, D.M. Ff gene 5 protein has a high binding affinity for single-stranded phosphorothioate DNA. Biochemistry 2001, 40, 2267–2275. [Google Scholar] [CrossRef] [PubMed]
- Marshall, W.S.; Caruthers, M.H. Phosphorodithioate DNA as a potential therapeutic drug. Science 1993, 259, 1564–1570. [Google Scholar] [CrossRef] [PubMed]
- Klussmann, S.; Nolte, A.; Bald, R.; Erdmann, V.A.; Furste, J.P. Mirror-image RNA that binds D-adenosine. Nat. Biotechnol. 1996, 14, 1112–1115. [Google Scholar] [CrossRef] [PubMed]
- Nolte, A.; Klussmann, S.; Bald, R.; Erdmann, V.A.; Furste, J.P. Mirror-design of L-oligonucleotide ligands binding to L-arginine. Nat. Biotechnol. 1996, 14, 1116–1119. [Google Scholar] [CrossRef] [PubMed]
- Poillot, C.; Dridi, K.; Bichraoui, H.; Pecher, J.; Alphonse, S.; Douzi, B.; Ronjat, M.; Darbon, H.; De Waard, M. D-Maurocalcine, a pharmacologically inert efficient cell-penetrating peptide analogue. J. Biol. Chem. 2010, 285, 34168–34180. [Google Scholar] [CrossRef]
- di Luccio, E.; Azulay, D.O.; Regaya, I.; Fajloun, Z.; Sandoz, G.; Mansuelle, P.; Kharrat, R.; Fathallah, M.; Carrega, L.; Esteve, E.; et al. Parameters affecting in vitro oxidation/folding of maurotoxin, a four-disulphide-bridged scorpion toxin. Biochem. J. 2001, 358, 681–692. [Google Scholar] [CrossRef]
- Beeton, C.; Smith, B.J.; Sabo, J.K.; Crossley, G.; Nugent, D.; Khaytin, I.; Chi, V.; Chandy, K.G.; Pennington, M.W.; Norton, R.S. The D-diastereomer of ShK toxin selectively blocks voltage-gated K+ channels and inhibits T lymphocyte proliferation. J. Biol. Chem. 2008, 283, 988–997. [Google Scholar] [CrossRef]
- Young, B.E.; Kundu, N.; Sczepanski, J.T. Mirror-image oligonucleotides: History and emerging applications. Chemistry 2019, 25, 7981–7990. [Google Scholar] [CrossRef]
- Gold, L.; Ayers, D.; Bertino, J.; Bock, C.; Bock, A.; Brody, E.N.; Carter, J.; Dalby, A.B.; Eaton, B.E.; Fitzwater, T.; et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One 2010, 5, e15004. [Google Scholar] [CrossRef]
- He, W.; Elizondo-Riojas, M.A.; Li, X.; Lokesh, G.L.; Somasunderam, A.; Thiviyanathan, V.; Volk, D.E.; Durland, R.H.; Englehardt, J.; Cavasotto, C.N.; et al. X-aptamers: a bead-based selection method for random incorporation of druglike moieties onto next-generation aptamers for enhanced binding. Biochemistry 2012, 51, 8321–8323. [Google Scholar] [CrossRef]
- Yang, X.; Li, X.; Prow, T.W.; Reece, L.M.; Bassett, S.E.; Luxon, B.A.; Herzog, N.K.; Aronson, J.; Shope, R.E.; Leary, J.F.; et al. Immunofluorescence assay and flow-cytometry selection of bead-bound aptamers. Nucleic Acids Res. 2003, 31, e54. [Google Scholar] [CrossRef] [PubMed]
- Walss-Bass, C.; Lokesh, G.L.R.; Dyukova, E.; Gorenstein, D.G.; Roberts, D.L.; Velligan, D.; Volk, D.E. X-aptamer technology identifies C4A and ApoB in blood as potential markers for schizophrenia. Mol. Neuropsychiatry 2019, 5, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Lam, C.H.; Li, X.; West, D.L.; Yang, X. Selection of PD1/PD-L1 X-aptamers. Biochimie 2018, 145, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Liang, C.; Lv, Q.; Li, D.; Xu, X.; Liu, B.; Lu, A.; Zhang, G. Molecular selection, modification and development of therapeutic oligonucleotide aptamers. Int. J. Mol. Sci. 2016, 17, 358. [Google Scholar] [CrossRef]
- Yong, K.T.; Wang, Y.; Roy, I.; Rui, H.; Swihart, M.T.; Law, W.C.; Kwak, S.K.; Ye, L.; Liu, J.; Mahajan, S.D.; et al. Preparation of quantum dot/drug nanoparticle formulations for traceable targeted delivery and therapy. Theranostics 2012, 2, 681–694. [Google Scholar] [CrossRef]
- Song, Y.; Shi, Y.; Huang, M.; Wang, W.; Wang, Y.; Cheng, J.; Lei, Z.; Zhu, Z.; Yang, C. Bioinspired engineering of a multivalent aptamer-functionalized nanointerface to enhance the capture and release of circulating tumor cells. Angew. Chem. Int. Ed. Engl. 2019, 58, 2236–2240. [Google Scholar] [CrossRef]
- Vorobyeva, M.; Vorobjev, P.; Venyaminova, A. Multivalent aptamers: Versatile tools for diagnostic and therapeutic applications. Molecules 2016, 21, 1613. [Google Scholar] [CrossRef]
- Davydova, A.S.; Vorobjeva, M.A.; Venyaminova, A.G. Escort aptamers: new tools for the targeted delivery of therapeutics into cells. Acta Naturae 2011, 3, 12–29. [Google Scholar] [CrossRef]
- Muller, C.W.; Rey, F.A.; Sodeoka, M.; Verdine, G.L.; Harrison, S.C. Structure of the NF-kappa B p50 homodimer bound to DNA. Nature 1995, 373, 311–317. [Google Scholar] [CrossRef]
- Chen, F.E.; Huang, D.B.; Chen, Y.Q.; Ghosh, G. Crystal structure of p50/p65 heterodimer of transcription factor NF-kappaB bound to DNA. Nature 1998, 391, 410–413. [Google Scholar] [CrossRef]
- Chen, Y.Q.; Ghosh, S.; Ghosh, G. A novel DNA recognition mode by the NF-kappa B p65 homodimer. Nat. Struct. Biol. 1998, 5, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.C.; Liu, P.Y.; Chiang, L.C.; Liao, S.C.; Su, H.Y.; Hsieh, S.Y.; Yang, C.C. Bungarus multicinctus multicinctus Snakebite in Taiwan. Am. J. Trop. Med. Hyg. 2017, 96, 1497–1504. [Google Scholar] [CrossRef] [PubMed]
- Chu, N.S. Contribution of a snake venom toxin to myasthenia gravis: the discovery of alpha-bungarotoxin in Taiwan. J. Hist. Neurosci. 2005, 14, 138–148. [Google Scholar] [CrossRef] [PubMed]
- Changeux, J.P.; Kasai, M.; Lee, C.Y. Use of a snake venom toxin to characterize the cholinergic receptor protein. Proc. Natl. Acad. Sci. USA 1970, 67, 1241–1247. [Google Scholar] [CrossRef]
- Johnston, C.I.; Ryan, N.M.; Page, C.B.; Buckley, N.A.; Brown, S.G.; O’Leary, M.A.; Isbister, G.K. The Australian Snakebite Project, 2005-2015 (ASP-20). Med. J. Aust. 2017, 207, 119–125. [Google Scholar] [CrossRef]
- White, R.; Rusconi, C.; Scardino, E.; Wolberg, A.; Lawson, J.; Hoffman, M.; Sullenger, B. Generation of species cross-reactive aptamers using “toggle” SELEX. Mol. Ther. 2001, 4, 567–573. [Google Scholar] [CrossRef]
- Levay, A.; Brenneman, R.; Hoinka, J.; Sant, D.; Cardone, M.; Trinchieri, G.; Przytycka, T.M.; Berezhnoy, A. Identifying high-affinity aptamer ligands with defined cross-reactivity using high-throughput guided systematic evolution of ligands by exponential enrichment. Nucleic Acids Res. 2015, 43, e82. [Google Scholar] [CrossRef]
- Anderson, P.D. Bioterrorism: toxins as weapons. J. Pharm. Pract. 2012, 25, 121–129. [Google Scholar] [CrossRef]
- Carstens, B.B.; Clark, R.J.; Daly, N.L.; Harvey, P.J.; Kaas, Q.; Craik, D.J. Engineering of conotoxins for the treatment of pain. Curr. Pharm. Des. 2011, 17, 4242–4253. [Google Scholar] [CrossRef]
- Jimenez, E.C.; Olivera, B.M.; Teichert, R.W. AlphaC-conotoxin PrXA: a new family of nicotinic acetylcholine receptor antagonists. Biochemistry 2007, 46, 8717–8724. [Google Scholar] [CrossRef]
- Abd El Aziz, T.M.; Bourgoin-Voillard, S.; Combemale, S.; Beroud, R.; Fadl, M.; Seve, M.; De Waard, M. Fractionation and proteomic analysis of the Walterinnesia aegyptia snake venom using OFFGEL and MALDI-TOF-MS techniques. Electrophoresis 2015, 36, 2594–2605. [Google Scholar] [CrossRef] [PubMed]

| Antivenom Class | Target Venoms | Benefits | Disadvantages | References |
|---|---|---|---|---|
| Monovalent | Snake | Demonstrated clinical efficiency over time | Species dependent – Low paraspecificity – Requires unmistakable species determination | [1,2,3,4,5,6,14,15,16] |
| Polyvalent | Snake | Improved paraspecificity | Costly development | [11,14] |
| Based on selected toxins | Spider | Does not require a venom source – Avoids excess needless IgG – Can be polyvalent | Requires i) excellent knowledge of toxic venom components, ii) good toxicovenomic and iii) toxin production capabilities | [12] |
| Bioinformatics-assisted | Snake | Does not require a venom source - Simplifies the production of the antigens – Can be polyvalent | Requires the knowledge of the venom toxic components and good immunogen potential of chosen epitopes | [30] |
| Monoclonal antibodies (IgG) | Snake & scorpion | Polyvalence possible, long half-life and low immunogenicity if human origin, few adverse reactions | Limited tissue distribution, large size, complex structure, costly development | [31,32,33,34,35,36,37,38,39] |
| Fab and F(ab’)2 fragments | Snake & scorpion | Polyvalence possible, enlarged tissue distribution and penetration, fewer adverse reactions than IgG | Higher cost of production | [38] |
| Murine scFv | Snake | Easy to produce, stable, long shelf-life, better tissue distribution and penetration | Shorter half-life in vivo, high immunogenicity | [38] |
| Human scFv | Snake, scorpion and bee | Stable, fewer adverse reactions, large tissue distribution and penetration | Shorter half-life in vivo | [38] |
| Nanobodies | Snake | High affinity and specificity, thermostable, small size (higher tissue penetration), low cost, low immunogenicity | Short half-life (limitation for a longer period time treatment) | [40] |
| Nanoparticles | Snake | Stability, low cost | Pharmacokinetics issues, low solubility | [41] |
| Darpins Affibodies Adnectins Avimers Anticalins | Not yet tested | Small size, high stability and solubility, high affinity, cost-effective production, better tissue penetration, low immunogenicity, polyvalence possible, facilitated chemical conjugation, kidney clearance | Short half-life compared to IgG, efficacy for toxin and venom neutralization remains to be demonstrated | [40] |
| Small molecules | Snake (Varespladib) | High absorbability, low-cost, thermostable, polyvalence | Works on a single class of toxins | [42,43,44] |
| Phytoantivenom | Snake | Viable alternative to modern medicine and pharmacology | Large extent to the global adverse effects at the clinical level | [45,46,47,48,49,50,51,52,53,54,55,56] |
| Aptamers | Snake, scorpion, cone snail | Low cost, high stability, long shelf-life, easier chemical conjugations, polyvalence possible | Demonstration lacking for full venom neutralization | [57,58,59,60,61,62,63,64,65] |
| Name | Method | Target | Target Origin | KD (µM) | Sequence of Random Region (5′ to 3′) | References |
|---|---|---|---|---|---|---|
| Clones 24 & 51* | SELEX | α-bungarotoxin | Bungarus multicinctus | 7.58 | GCGAGGTGTTCGAGAGTTAGGGGCGACATGACCAAACGTT | [61] |
| βB-1 | Plate-SELEX | β-bungarotoxin | Bungarus multicinctus | 0.066 | GTTTTCCCCTTGTCGCTTTTGGTTCGTTCTGCCTCTATCT | [65] |
| βB-20 | Plate-SELEX | β-bungarotoxin | Bungarus multicinctus | 0.084 | ATTAGTCATGTTTGTTTGTCTGGCTTTTTGGGTTTGTGCAGTATTATGAAC | [65] |
| βB-19 | Plate-SELEX | β-bungarotoxin | Bungarus multicinctus | 0.53 | TTTGGTGTGGATCCTGAACATTTATATTCTTTCGTTTTTT | [65] |
| βB-32 | Plate-SELEX | β-bungarotoxin | Bungarus multicinctus | 0.995 | GCAATGCACCTTTGTCTCTTATAGTTTATTTTTTGCCTT | [65] |
| bgt1* | SELEX | α-bungarotoxin | Bungarus multicinctus | 2.21 | GCGAGGTGTTCGAGAGTTAGGGGCGACATGACCAAACGTT | [57] |
| SELEX | CTX1, CTX2, CTX3, CTX4, CTX5, CTXN, CLPB | Naja atra | 2.51, 6.29, 2.25, 8.13, 17.17, 8.85, 7.19 | |||
| bgt2 | SELEX | α-bungarotoxin | Bungarus multicinctus | 0.46 | AGGGCACAGAGAAGAAGTCGTGGATTTGAATGGTTTTGGT | [57] |
| SELEX | CTX3 | Naja atra | 0.26 | |||
| bgt3 | SELEX | α-bungarotoxin | Bungarus multicinctus | 0.14 | ATCATGTCTTTTCGGGATGGGCAAGAAGGGAAATAATGC | [57] |
| SELEX | CTX3 | Naja atra | 1.26 | |||
| bgt4 | SELEX | α-bungarotoxin | Bungarus multicinctus | 0.28 | AGAAACGTAGCGGTAACTGCTAGAATGCGCCGAGAGAGCG | [57] |
| SELEX | CTX3 | Naja atra | 1.17 | |||
| α-Tox-FL* | SELEX | crude venom | Bungarus caeruleus | 0.018 | GCGAGGTGTTCGAGAGTTAGGGGCGACATGACCAAACGTT | [57,58,61] |
| α-Tox-T1 | Bioinformatics tools | crude venom | Bungarus caeruleus | 0.045 | GCGAGGTGTTCGAG | [58] |
| α-Tox-T2 | Bioinformatics tools | crude venom | Bungarus caeruleus | 0.003 | AGTTAGGGGCGACATGACCAAACGTT | [58] |
| D3 | CE-SELEX | αC-conotoxin PrXA | Conus parius | 0.122 | ATCGGTCGTATAGGGTCGATTTGGTCGGCA | [59,60] |
| A5 | CE-SELEX | αC-conotoxin PrXA | Conus parius | 0.184 | GTGCAGGTCTATACAGGACAGTCTTCTGAT | [59,60] |
| D7 | CE-SELEX | αC-conotoxin PrXA | Conus parius | 0.238 | TGCAGCATGGGGGATGTGCTCTTCCGCGTG | [59,60] |
| A4 | CE-SELEX | αC-conotoxin PrXA | Conus parius | 0.246 | AATGCTGTTGTTTGAGTATCAATCAGACCG | [59] |
| B4 | CE-SELEX | αC-conotoxin PrXA | Conus parius | 0.12 | TACGCACATACTGTGTACCTTGAATTTATA | [59] |
| B3 | CE-SELEX | αC-conotoxin PrXA | Conus parius | > 5 | CCGTAGATGCGGGGATGCCAGTCTTGCTTA | [59] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ascoët, S.; De Waard, M. Diagnostic and Therapeutic Value of Aptamers in Envenomation Cases. Int. J. Mol. Sci. 2020, 21, 3565. https://doi.org/10.3390/ijms21103565
Ascoët S, De Waard M. Diagnostic and Therapeutic Value of Aptamers in Envenomation Cases. International Journal of Molecular Sciences. 2020; 21(10):3565. https://doi.org/10.3390/ijms21103565
Chicago/Turabian StyleAscoët, Steven, and Michel De Waard. 2020. "Diagnostic and Therapeutic Value of Aptamers in Envenomation Cases" International Journal of Molecular Sciences 21, no. 10: 3565. https://doi.org/10.3390/ijms21103565
APA StyleAscoët, S., & De Waard, M. (2020). Diagnostic and Therapeutic Value of Aptamers in Envenomation Cases. International Journal of Molecular Sciences, 21(10), 3565. https://doi.org/10.3390/ijms21103565






