Soil Microbiome Response to Contamination with Bisphenol A, Bisphenol F and Bisphenol S
Abstract
1. Introduction
2. Results
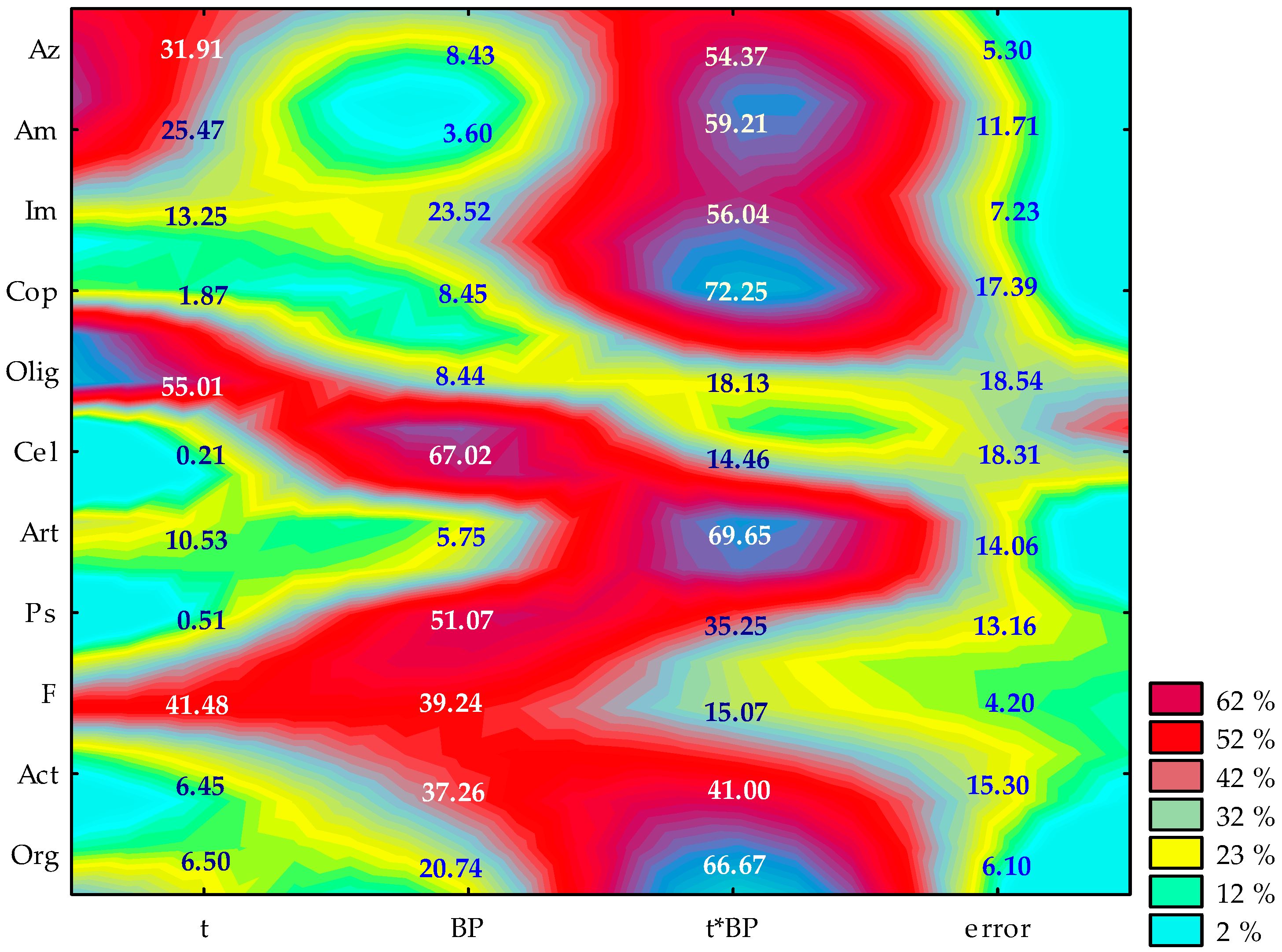
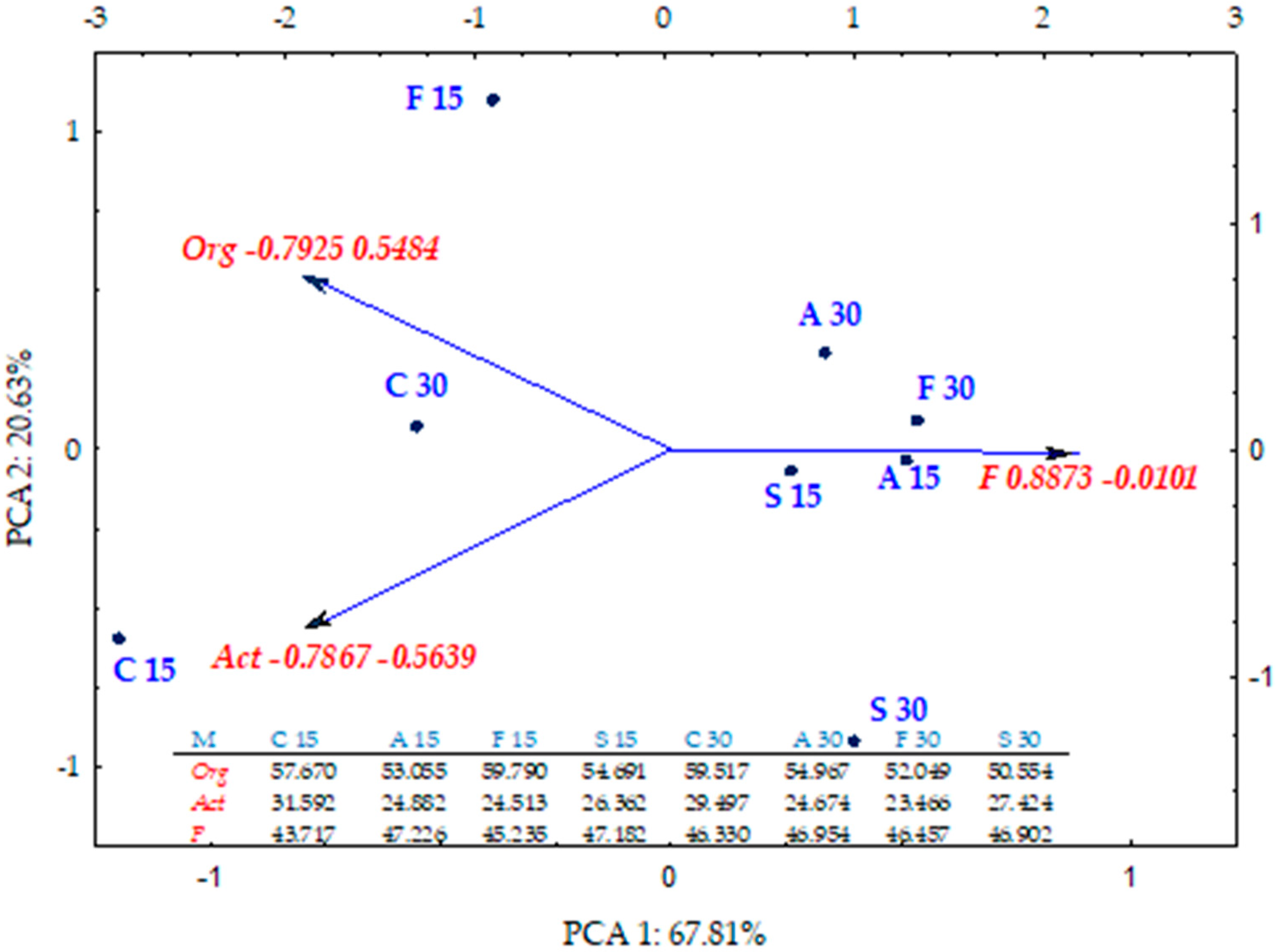
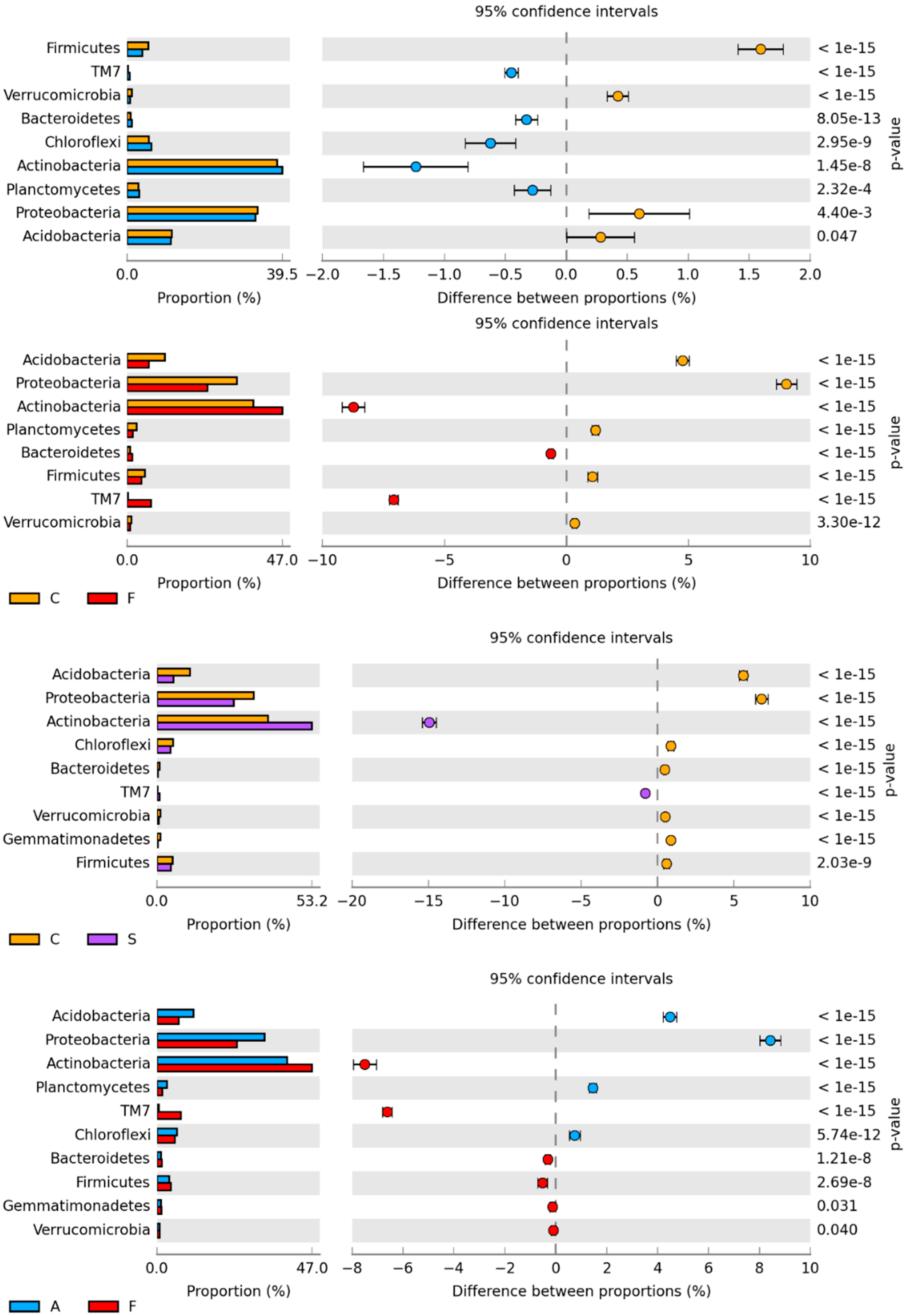
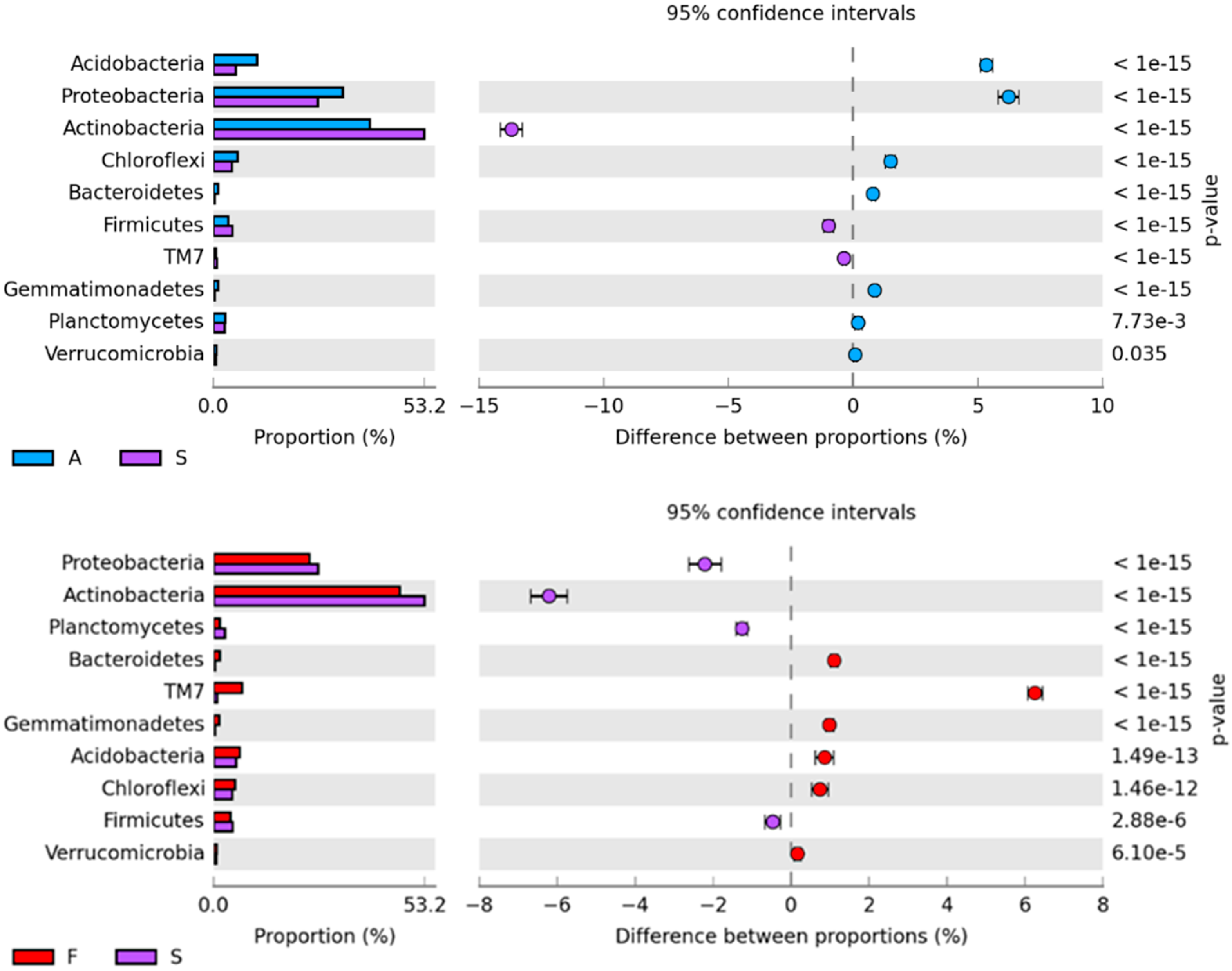
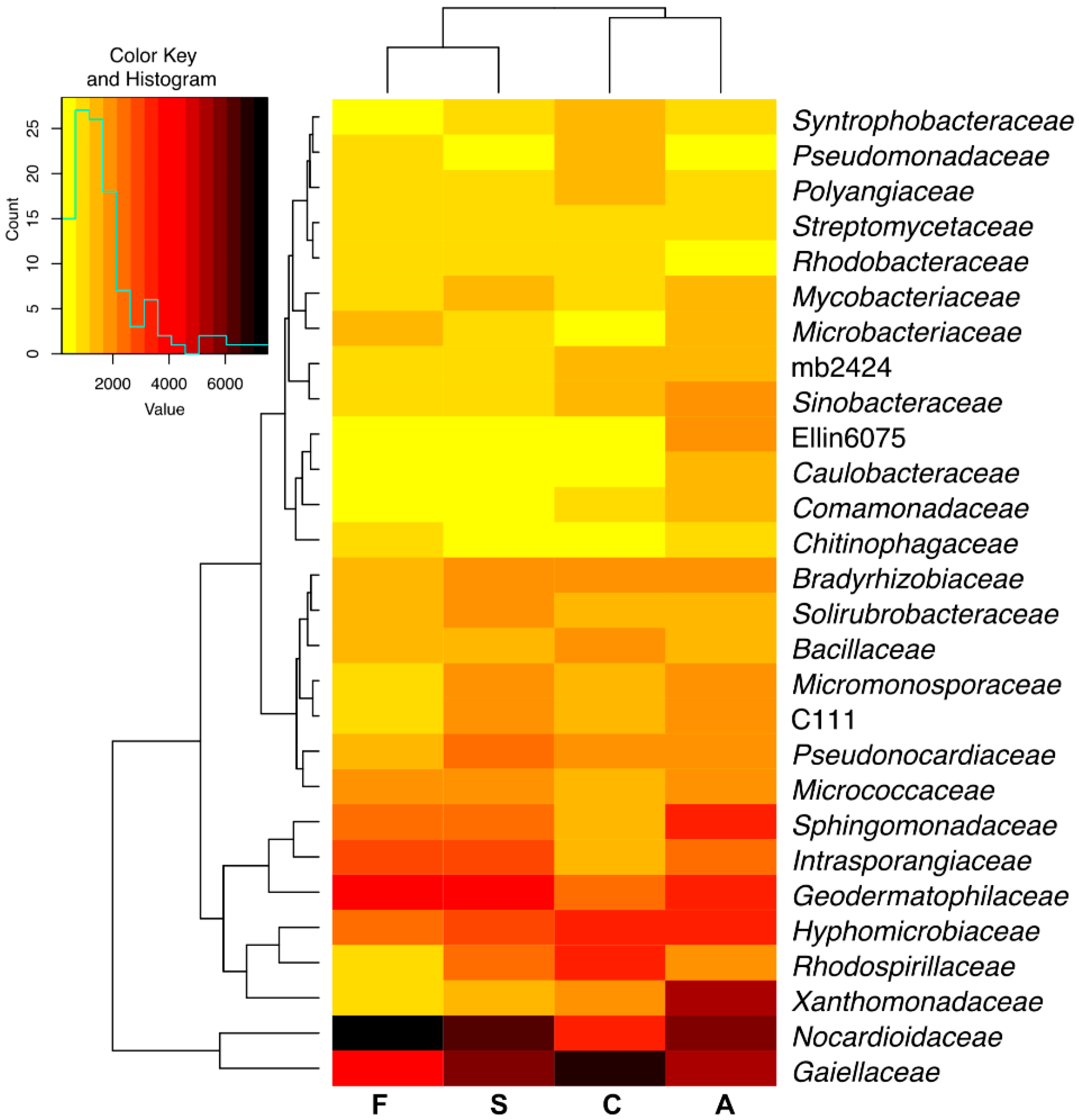
2.1. Counts and Diversity of Microorganisms
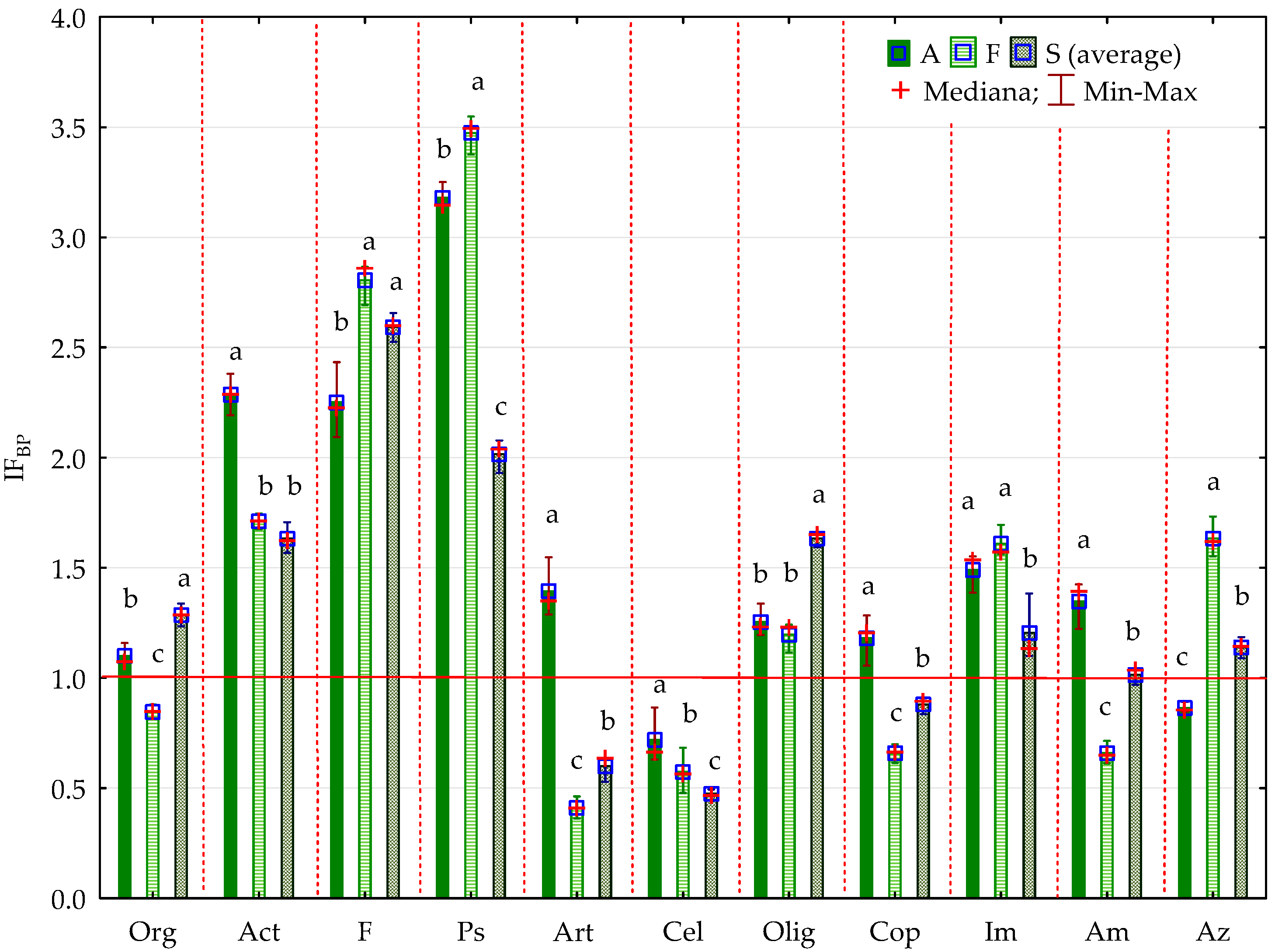
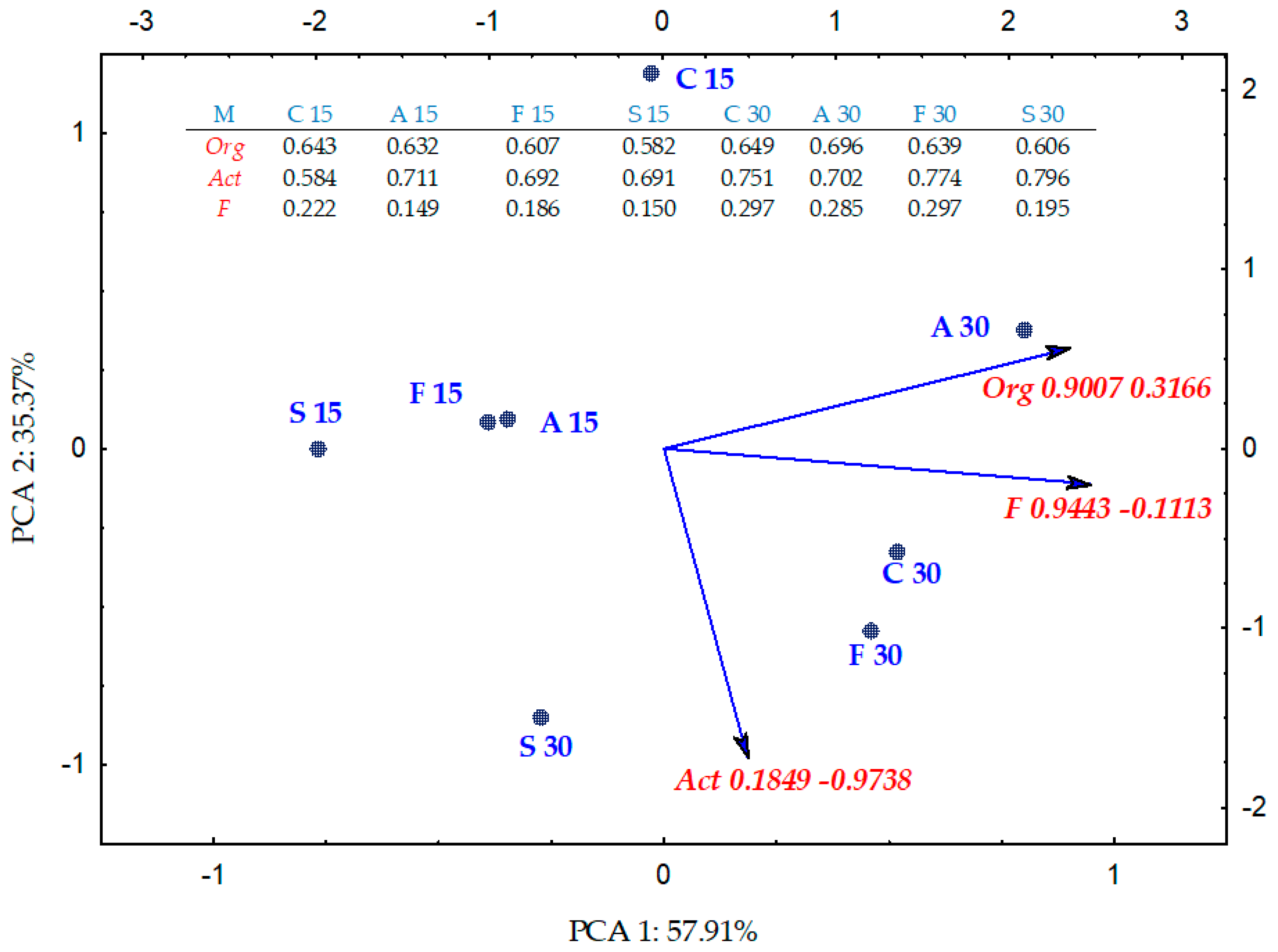
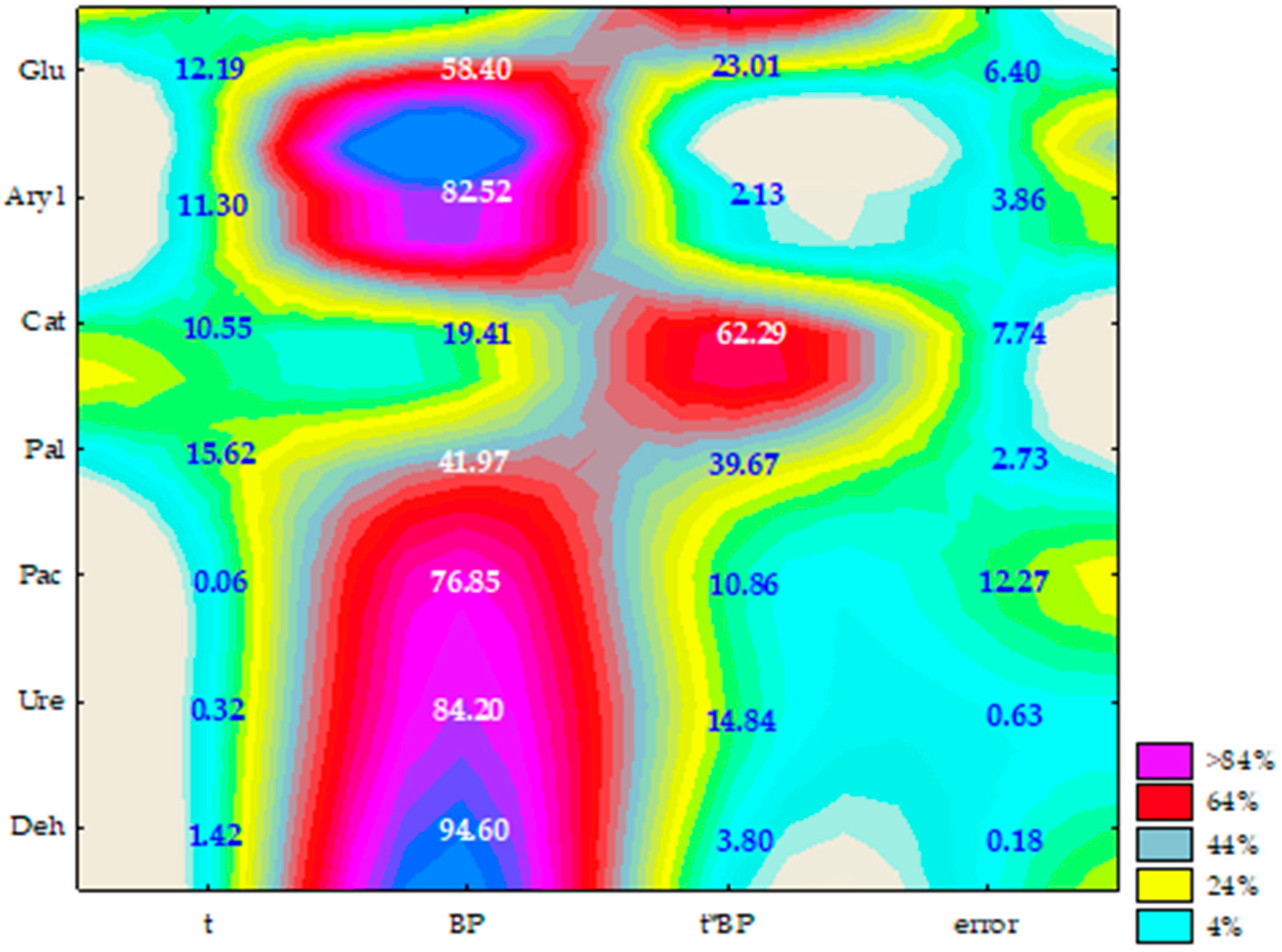
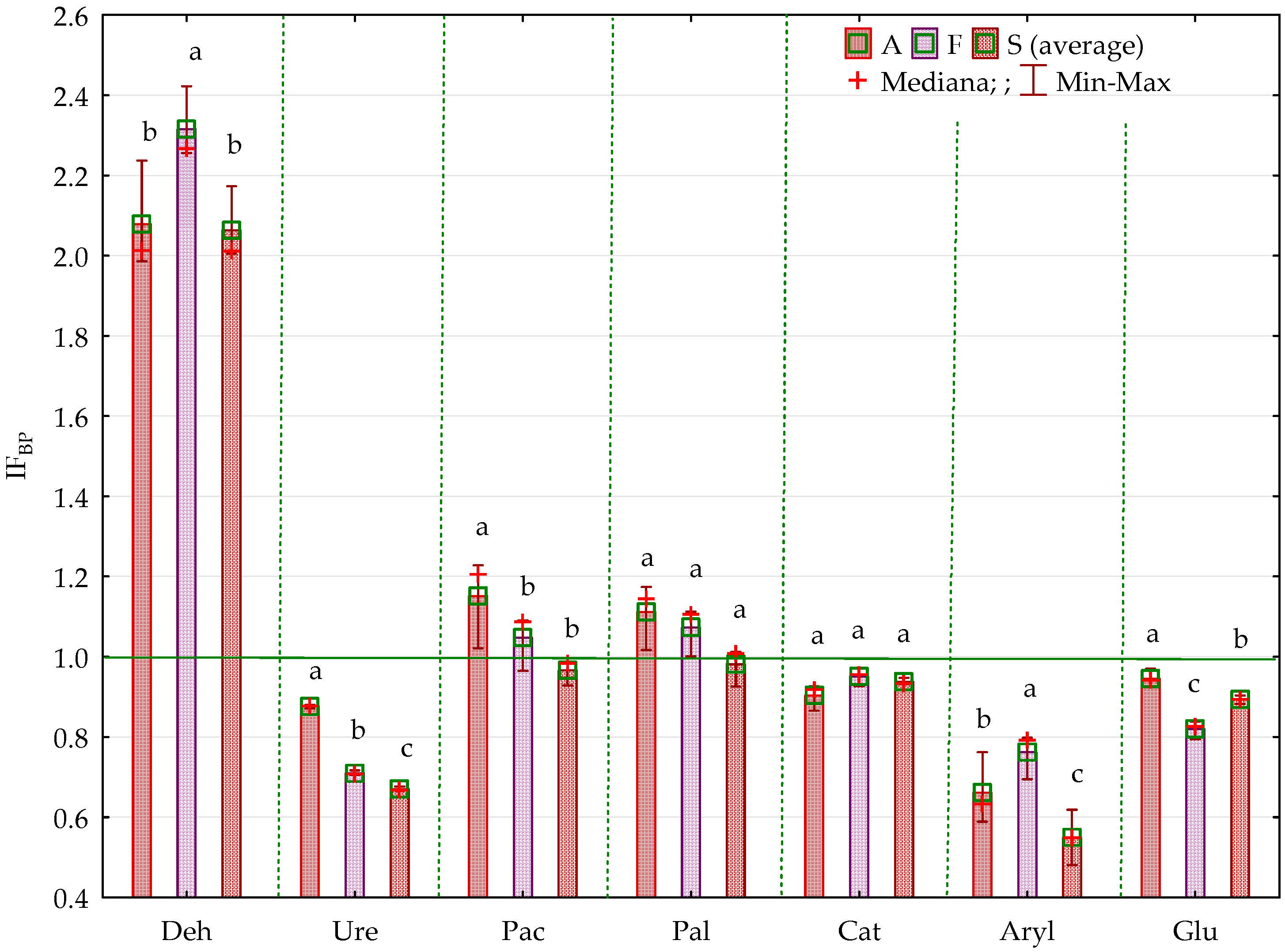
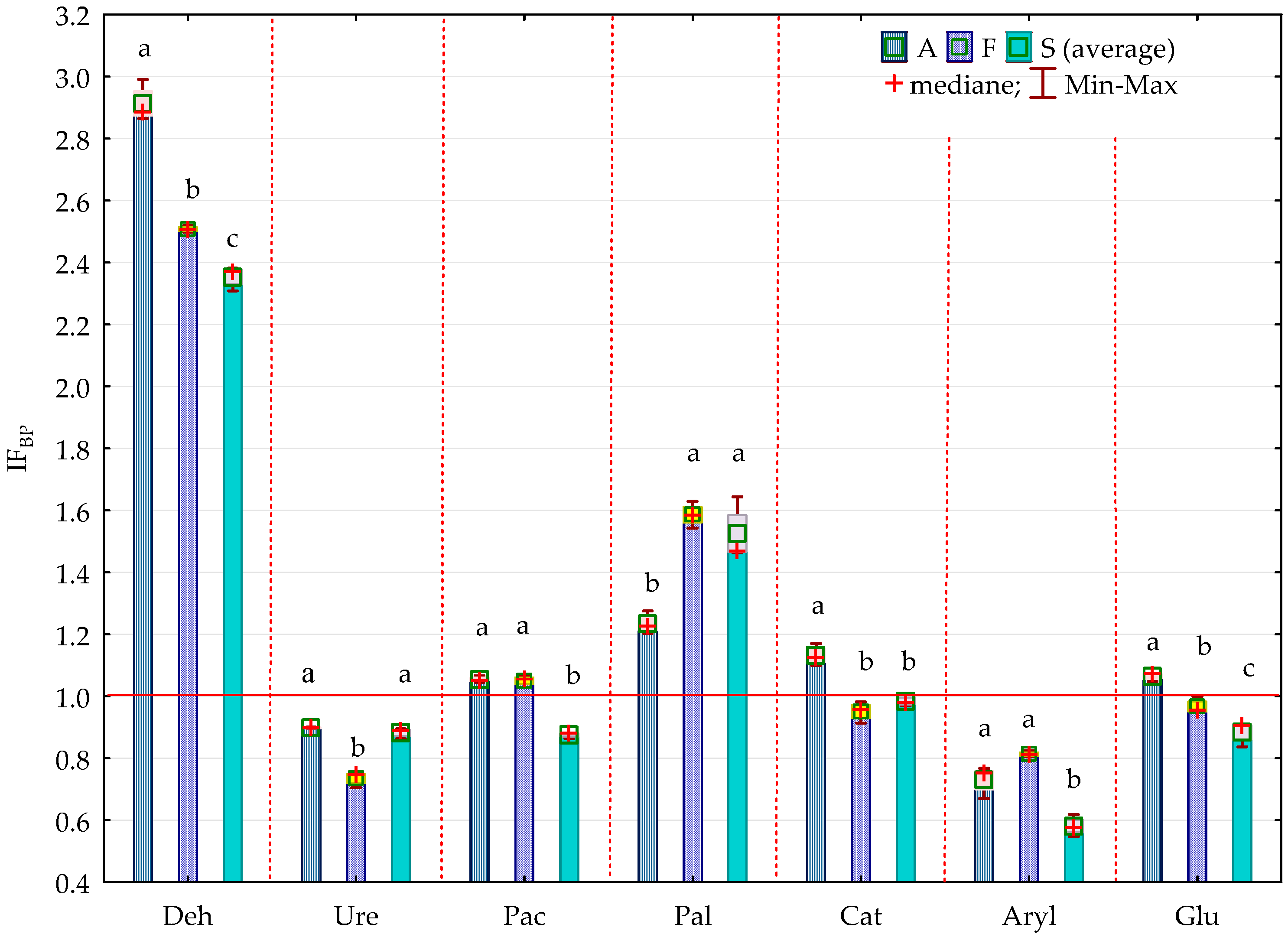
2.2. Enzyme Activity
3. Discussion
3.1. Counts and Diversity of Bacteria
3.2. Soil Enzyme
4. Materials and Methods
4.1. Soil and Facilities
4.2. Experiment Setup
4.3. Characteristics of Bisphenols
4.4. Sample Analysis
4.5. DNA Extraction and Bioinformatic Analysis of Bacteria Taxons
4.6. Determination of Soil Enzyme Activity
4.7. Statistical Data Analysis and Methodology of Calculations
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Salthammer, T. Emerging indoor pollutants. Int. J. Hyg. Environ. Health 2020, 224, 113423. [Google Scholar] [CrossRef]
- Chen, Q.; Zhang, H.; Allgeier, A.; Zhou, Q.; Ouellet, J.D.; Crawford, S.E.; Luo, Y.; Yang, Y.; Shi, H.; Hollert, H. Marine microplastics bound dioxin-like chemicals: Model explanation and risk assessment. J. Hazard Mater. 2019, 364, 82–90. [Google Scholar] [CrossRef]
- Groh, K.J.; Backhaus, T.; Carney-Almroth, B.; Geueke, B.; Inostroza, P.A.; Lennquist, A.; Leslie, H.A.; Maffini, M.; Slunge, D.; Trasande, L.; et al. Overview of known plastic packaging-associated chemicals and their hazards. Sci. Total Environ. 2019, 651, 3253–3268. [Google Scholar] [CrossRef]
- Andaluri, G.; Manickavachagam, M.; Suri, R. Plastic toys as a source of exposure to bisphenol-A and phthalates at childcare facilities. Environ. Monit. Assess. 2018, 190, 65. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Kannan, K.; Tan, H.; Zheng, Z.; Feng, Y.L.; Wu, Y.; Widelka, M. Bisphenol analogues other than BPA: Environmental occurrence, human exposure, and toxicity-A review. Environ. Sci. Technol. 2016, 50, 5438–5453. [Google Scholar] [CrossRef] [PubMed]
- Molina-Molina, J.M.; Amaya, E.; Grimaldi, M.; Saenz, J.M.; Real, M.; Fernandez, M.F. In vitro study on the agonistic and antagonistic activities of bisphenol-S and other bisphenol-A congeners and derivatives via nuclear receptors. Toxicol. Appl. Pharmacol. 2013, 272, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wu, Z.M.; Li, Y.F.; Tan, Y.; Liu, N.; Liu, Y.J. The efficienthydroxyalkylation of phenol with formaldehyde to bisphenol F over athermoregulated phase-separable reaction system containing a water-solubleBrønsted acidic ionic liquid. RSC Adv. 2014, 4, 33466–33473. [Google Scholar] [CrossRef]
- Jana, S.K.; Kugita, T.; Namba, S. Bisphenol F synthesis over mesoporousaluminosilicate MCM-41 molecular sieves. Catal. Lett. 2003, 90, 143–147. [Google Scholar] [CrossRef]
- Herrero, Ó.; Aquilino, M.; Sánchez-Argüello, P.; Planelló, R. The BPA-substitute bisphenol S alters the transcription of genes related to endocrine, stress response and biotransformation pathways in the aquatic midge Chironomus riparius (Diptera, Chironomidae). PLoS ONE 2018, 13, e0193387. [Google Scholar] [CrossRef]
- Qiu, W.; Zhan, H.; Hu, J.; Zhang, T.; Xu, H.; Wong, M.; Xu, B.; Zheng, C. The occurrence, potential toxicity, and toxicity mechanism of bisphenol S, a substitute of bisphenol A: A critical review of recent progress. Ecotox. Environ. Safe. 2019, 173, 192–202. [Google Scholar] [CrossRef]
- Rubin, B.S. Bisphenol A: An endocrine disruptor with widespread exposure and multiple effects. J. Steroid Biochem. Mol. Biol. 2011, 127, 27–34. [Google Scholar] [CrossRef] [PubMed]
- ECHA European Chemicals Agency. 2020. Available online: https://echa.europa.eu/substanceinformation/-/substanceinfo/100.001.133 (accessed on 19 February 2020).
- Cydzik-Kwiatkowska, A.; Bernat, K.; Zielinska, M.; Bułkowska, K.; Wojnowska-Baryla, I. Aerobic granular sludge for bisphenol A (BPA) removal from wastewater. Int. Biodeterior. Biodegrad. 2017, 122, 1–11. [Google Scholar] [CrossRef]
- Danzl, E.; Sei, K.; Soda, S.; Ike, M.; Fujita, M. Biodegradation of bisphenol A, bisphenol F and bisphenol S in seawater. Int. J. Environ. Res. Pub. Health 2009, 6, 1472–1484. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.B.; Liu, J.; Zhang, Q.; Stout, C.D.; Halpert, J.R. Halogen−π interactions in the cytochrome P450 active site: Structural insights into human CYP2B6 substrate selectivity. ACS Chem. Biol. 2017, 12, 1204–1210. [Google Scholar] [CrossRef] [PubMed]
- Kudłak, B.; Wieczerzak, M.; Namieśnik, J. Bisphenols (A, S, and F) affect the basic hormonal activity determined for pharmaceuticals-Study of Saccharomyces cerevisiae. Environ. Pollut. 2019, 246, 914–920. [Google Scholar] [CrossRef]
- EFSA. Panel on Food Contact Materials, E. Flavourings Aids, P. Scientific opinion on the risks to public health related to the presence of bisphenol A (BPA) in foodstuffs. Efsa J. 2015, 13, 3978. [Google Scholar] [CrossRef]
- Niu, Y.; Zhang, J.; Duan, H.; Wu, Y.; Shao, B. Bisphenol A and nonylphenol in foodstuffs: Chinese dietary exposure from the 2007 total diet study and infant health risk from formulas. Food Chem. 2015, 167, 320–325. [Google Scholar] [CrossRef]
- Rochester, J.R.; Bolden, A.L. Bisphenol S and F: A systematic review and comparison of the hormonal activity of bisphenol A substitutes. Environ. Health Perspect. 2015, 123, 643–650. [Google Scholar] [CrossRef]
- Zhao, C.; Xie, P.; Wang, H.; Cai, Z. Liquid chromatography-mass spectrometry-based metabolomics and lipidomics reveal toxicological mechanisms of bisphenol F in breast cancer xenografts. J. Hazard. Mater. 2018, 358, 503–507. [Google Scholar] [CrossRef]
- Stossi, F.; Bolt, M.J.; Ashcroft, F.J.; Lamerdin, J.E.; Melnick, J.S.; Powell, R.T.; Dandekar, R.D.; Mancini, M.D.; Walker, C.L.; Westwick, J.K.; et al. Defining estrogenic mechanisms of bisphenol A analogs through high throughput microscopy-based contextual assays. Chem. Biol. 2014, 21, 743–753. [Google Scholar] [CrossRef]
- Cabaton, N.; Dumont, C.; Severinm, I.; Perdu, E.; Zalko, D.; Cherkaoui-Malki, M.; Chagnon, M.C. Genotoxic and endocrine activities of bis(hydroxyphenyl) methane (bisphenol F) and its derivatives in the HepG2 cell line. Toxicology 2009, 255, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Castro, B.; Sánchez, P.; Torres, J.M.; Ortega, E. Bisphenol A, bisphenol F and bisphenol S affect differently 5a-reductase expression and dopamine-serotonin systems in the prefrontal cortex of juvenile female rats. Environ. Res. 2015, 142, 281–287. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Shu, L.; Qiu, Z.; Lee, D.Y.; Settle, S.J.; Que, H.S.; Telesca, D.; Yang, X.; Allard, P. Exposure to the BPA-substitute bisphenol S causes unique alterations of germline function. PLoS Genet. 2016, 12, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Mokra, K.; Kuźmińska-Surowaniec, A.; Woźniak, K.; Michałowicz, J. Evaluation of DNA-damaging potential of bisphenol A and its selected analogs in human peripheral blood mononuclear cells (in vitro study). Food Chem. Toxicol. 2017, 100, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Xue, J.; Gao, C.-Z.; Qiu, R.L.; Li, Y.X.; Huang, M.Z.; Kannan, K. Urinary concentrations of bisphenols and their association with biomarkers of oxidative stress in people living near e-waste recycling facilities in China. Environ. Sci. Toxicol. 2016, 50, 4045–4053. [Google Scholar] [CrossRef] [PubMed]
- Ho, S.M.; Rao, R.S.T.; Schoch, W.; Tarapore, P. Bisphenol A and its analogues disrupt centrosome cycle and microtubule dynamics in prostate cancer. Endocr. Relat. Cancer 2017, 24, 83–96. [Google Scholar] [CrossRef]
- ATSDR. Substance Priority List; Agency for Toxic Substances and Disease Registry: Atlanta, GA, USA, 2017. [Google Scholar]
- Wang, W.; Abualnaja, K.O.; Asimakopoulos, A.G.; Covaci, A.; Gevao, B.; Johnson-Restrepo, B.; Kumosani, T.A.; Malarvannan, G.; Minh, T.B.; Moon, H.B.; et al. A comparative assessment of human exposure to tetrabromobisphenol A and eight bisphenols including bisphenol A via indor dust ingestion in twelve countries. Environ. Int. 2015, 83, 183–191. [Google Scholar] [CrossRef]
- Yamazaki, E.; Yamashita, N.; Taniyasu, S.; Lam, J.; Lam, P.K.S.; Moon, H.B.; Jeong, Y.; Kannan, P.; Achyuthan., H.; Munuswamy, N.; et al. Bisphenol A and other bisphenol analogues including BPS and BPF in surface water samples from Japan, China, Korea and India. Ecotox. Environ. Safe 2015, 122, 565–572. [Google Scholar] [CrossRef]
- Huang, C.; Wu, L.H.; Liu, G.Q.; Shi, L.; Guo, Y. Occurrence and Ecological Risk 408 Assessment of Eight Endocrine-Disrupting Chemicals in Urban River Water and Sediments of South China. Arch. Environ. Con. Toxicol. 2018, 75, 224–235. [Google Scholar] [CrossRef]
- Huang, T. Chemical Profile: Asia Phenol. 2016. Available online: https://www.icis.com/resources/news/2016/06/30/10012759/chemical-profile-asia-phenol/ (accessed on 20 February 2020).
- Liao, C.; Liu, F.; Moon, H.-B.; Yamashita, N.; Kannan, K. Bisphenol analogues in sediments from industrialized areas in the United States, Japan, and Korea: Spatial and temporal distributions. Environ. Sci. Technol. 2012, 46, 11558–11565. [Google Scholar] [CrossRef]
- Rasheed, T.; Bilal, M.; Iqbal, H.M.; Hu, H.; Zhang, X. Reaction mechanism and degradation pathway of rhodamine 6G by photocatalytic treatment. Water Air Soil Pollut. 2017, 228, 291–301. [Google Scholar] [CrossRef]
- Bilal, M.; Rasheed, T.; Iqbal, H.M.; Yan, Y. Peroxidases-assisted removal of environmentally-related hazardous pollutants with reference to the reaction mechanisms of industrial dyes. Sci. Total Environ. 2018, 644, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Pérez, R.A.; Albero, B.; Ferriz, M.; Tadeo, J.L. Rapid multiresidue determination of bisphenol analogues in soil with on-line derivatization. Anal. Bioanal. Chem. 2017, 409, 4571–4580. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Lin, K.; Gan, J. Oxidation of bisphenol F (BPF) by manganese dioxide. Environ. Pollut. 2011, 159, 2546–2551. [Google Scholar] [CrossRef]
- Li, J.; Zhou, Y.; Jiang, J.; Pang, S.; Gao, Y.; Yang, Y.; Liu, G.; Ma, J.; Jiang, C.; Wang, L. Transformation of phenolic compounds by peroxymonosulfate in the presence of iodide and formation of iodinated aromatic products. Chem. Eng. J. 2018, 335, 855–864. [Google Scholar] [CrossRef]
- Im, J.; Prevatte, C.; Campagna, S.; Löffler, F. Identification of 4-hydroxycumyl alcohol as the major MnO2-mediated bisphenol A transformation product and evaluation of its environmental fate. Environ. Sci. Technol. 2015, 49, 6214–6221. [Google Scholar] [CrossRef]
- Li, J.; Jiang, J.; Zhou, Y.; Pang, S.; Gao, Y.; Jiang, C.; Ma, J.; Jin, Y.; Yang, Y.; Liu, G. Kinetics of oxidation of iodide (I-) and hypoiodous acid (HOI) by peroxymonosulfate (PMS) and formation of iodinated products in the PMS/I-/NOM system. Environ. Sci. Technol. Lett. 2017, 4, 76–82. [Google Scholar] [CrossRef]
- Vikesland, P.; Fiss, E.; Wigginton, K.; McNeill, K.; Arnold, W. Halogenation of bisphenol-A, triclosan, and phenols in chlorinated waters containing iodide. Environ. Sci. Technol. 2013, 47, 6764–6772. [Google Scholar] [CrossRef]
- Żwierełło, W.; Maruszewska, A.; Skórka-Majewicz, M.; Goschorska, M.; Baranowska-Bosiacka, I.; Dec, K.; Styburski, D. The influence of polyphenols on metabolic disorders caused by compounds released from plastics–Review. Chemosphere 2020, 240, 124901. [Google Scholar] [CrossRef]
- Rameshrad, M.; Imenshahidi, M.; Razavi, B.M.; Iranshahi, M.; Hosseinzadeh, H. Bisphenol A vascular toxicity: Protective effect of Vitis vinifera (grape) seed extract and resveratrol. Phyther. Res. 2018, 32, 2396–2407. [Google Scholar] [CrossRef]
- Lee, G.A.; Choi, K.C.; Hwang, K.A. Treatment with phytoestrogens reversed triclosan and bisphenol A-induced anti-apoptosis in breast cancer cells. Biomol. Therap. 2018, 26, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Quintella, C.M.; Mata, A.M.T.; Lima, L.C.P. Overview of bioremediation with technology assessment and emphasis on fungal bioremediation of oil contaminated soils. J. Environ. Manag. 2019, 241, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Kucharski, J.; Tomkiel, M.; Baćmaga, M.; Borowik, A.; Wyszkowska, J. Enzyme activity and microorganisms diversity in soil contaminated with the Boreal 58 WG herbicide. J. Environ. Sci. Health Part B 2016, 51, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Wyszkowska, J.; Borowik, A.; Olszewski, J.; Kucharski, J. Soil bacterial community and soil enzyme activity depending on the cultivation of Triticum aestivum, Brassica napus, and Pisum sativum ssp. arvense. Diversity 2019, 11, 246. [Google Scholar] [CrossRef]
- Zdarta, J.; Meyer, A.; Jesionowski, T.; Pinelo, M. A general overview of suport materials for enzyme immobilization: Characteristics, properties, practical utility. Catalysts 2018, 8, 92. [Google Scholar] [CrossRef]
- Vineh, M.B.; Saboury, A.A.; Poostchi, A.A.; Rashidi, A.M.; Parivar, K. Stability and activity improvement of horseradish peroxidase by covalent immobilization on functionalized reduced graphene oxide and biodegradation of high phenol concentration. Int. J. Biol. Macromol. 2018, 106, 1314–1322. [Google Scholar] [CrossRef]
- Gassara, F.; Brar, S.K.; Verma, M.; Tyagi, R.D. Bisphenol A degradation in water by ligninolytic enzymes. Chemosphere 2013, 92, 1356–1360. [Google Scholar] [CrossRef]
- Luan, P.P.; Jiang, Y.J.; Zhang, S.P.; Gao, J.; Su, Z.G.; Ma, G.H.; Zhang, Y. FChitosan-mediated formation of biomimetic silica nanoparticles: An effective method for manganese peroxidase immobilization and stabilization. J. Biosci. Bioeng. 2014, 118, 575–582. [Google Scholar] [CrossRef]
- Cha, J.Y.; Kim, T.W.; Choi, J.H.; Jang, K.S.; Khaleda, L.; Kim, W.Y.; Jeon, J.R. Fungal laccase-catalyzed oxidation of naturally occurring phenols for enhanced germination and salt tolerance of Arabidopsis thaliana: A green route for synthesizing humic-like fertilizers. J. Agric. Food Chem. 2017, 65, 1167–1177. [Google Scholar] [CrossRef]
- Singh, U.; Arorab, N.K.; Sachan, P. Simultaneous biodegradation of phenol and cyanide present in coke-oven effluent using immobilized Pseudomonas putida and Pseudomonas stutzeri. Braz. J. Microbiol. 2018, 49, 38–44. [Google Scholar] [CrossRef]
- Divyateja, D.; Konapalli, P.; Sridevi, V.; Radhika, P. Cell Phenotyping of Pseudomonas sp.Strain DT-4 Capable of Degrading Phenol Using Gen III.; Optimization. Mater. Today Proc. 2018, 5, 17857–17865. [Google Scholar] [CrossRef]
- Yap, L.F.; Lee, Y.K.; Poh, C.L. Mechanism for phenol tolerance in phenol-degrading Comamonas testosteroni strain. Appl. Microbiol. Biotechnol. 1999, 516, 833–840. [Google Scholar] [CrossRef]
- Nešvera, J.; Rucká, L.; Pátek, M. Chapter Four–Catabolism of Phenol and Its Derivatives in Bacteria: Genes, Their Regulation, and Use in the Biodegradation of Toxic Pollutants. Adv. Appl. Microbiol. 2015, 4, 107–160. [Google Scholar] [CrossRef]
- Delgado-Baquerizo, M.; Oliverio, A.M.; Brewer, T.E.; Benavent-González, A.; Eldridge, D.J.; Bardgett, R.D.; Maestre, F.T.; Singh, B.K.; Fierer, N. A global atlas of the dominant bacteria found in soil. Science 2018, 359, 320–325. [Google Scholar] [CrossRef]
- Rasheed, A.; Kola, R.K.; Yalavarthy, P.D. Assessment of antibacterial activity of bisphenol a (4,4′-Isopropylidenebisphenol). Int. J. Innov. Res. Sci. Eng. Technol. 2013, 2, 6003–6008. [Google Scholar]
- Kolvenbach, B.A.; Helbling, D.E.; Kohler, H.E.; Corvini, P.F. Science Direct Emerging chemicals and the evolution of biodegradation capacities and pathways in bacteria. Curr. Opin. Biotechnol. 2014, 27, 8–14. [Google Scholar] [CrossRef]
- Fischer, J.; Kappelmeyer, U.; Kastner, M.; Schauer, F.; Heipieper, H.J. The degradation of bisphenol A by the newly isolated bacterium Cupriavidus basilensis JF1 can be enhanced by biostimulation with phenol. Int. Biodeterior. Biodegrad. 2010, 64, 324–330. [Google Scholar] [CrossRef]
- Ogata, Y.; Goda, S.; Toyama, T.; Sei, K.; Ike, M. The 4-tert-butylphenol-utilizing bacterium Sphingobium fuliginis OMI can degrade bisphenols via phenolic ring hydroxylation and meta-cleavage pathway. Environ. Sci. Technol. 2013, 47, 1017–1023. [Google Scholar] [CrossRef]
- PN-R-04032. Soil and Mineral Materials-Sampling and Determination of Particle Size Distribution; Polish Committee for Standardization: Warszawa, Poland, 1998. [Google Scholar]
- ISO 10390. Soil Quality-Determination of Ph; ISO: Geneva, Switzerland, 2005. [Google Scholar]
- Klute, A. Methods of Soil Analysis; Monograph 9; American Society of Agronomy: Madison, WI, USA, 1986. [Google Scholar]
- Nelson, D.W.; Sommers, L.E. Total carbon, organic carbon, and organic matter. In Method of Soil Analysis: Chemical Methods; Sparks, D.L., Page, A.L., Helmke, P.A., Loeppert, R.H., Soltanpour, P.N., Tabatabai, M.A., Johnston, C.T., Sumner, M.E., Eds.; American Society of Agronomy: Madison, WI, USA, 1996; pp. 1201–1229. [Google Scholar]
- ISO 11261. Soil quality-determination of total nitrogen-modified Kjeldahl method; ISO: Geneva, Switzerland, 2005. [Google Scholar]
- Egner, H.; Riehm, H.; Domingo, W.R. Untersuchun-gen über die chemische Bodenanalyse als Grundlage für die Beurteilung des Nährstoffzustandes der Böden. II. Chemische Extractionsmethoden zur Phospor- und Kaliumbestimmung. Ann. R. Agric. Coll. Swed. 1960, 26, 199–215. [Google Scholar]
- Schlichting, E.; Blume, H.P.; Stahr, K. Bodenkundliches Praktikum. Pareys Studientexte 81; Blackwell Publishing: Berlin, Germany, 1995. [Google Scholar]
- Öhlinger, R. Dehydrogenase activity with the substrate TTC. In Methods in Soil Biology; Schinner, F., Ohlinger, R., Kandler, E., Margesin, R., Eds.; Springer: Berlin, Germany, 1996; pp. 241–243. [Google Scholar]
- Alef, K.; Nannipieri, P. Methods in Applied Soil Microbiology and Biochemistry; Academic Press: Cambridge, MA, USA, 1998; pp. 316–365. [Google Scholar]
- Bunt, J.S.; Rovira, A.D. Microbiological studies of some subantarctic soil. J. Soil Sci. 1955, 6, 119–128. [Google Scholar] [CrossRef]
- Parkinson, D.; Gray, F.R.G.; Williams, S.T. Methods for Studying the Ecology of Soil Micro-Organism. IBP Handbook 19; Blackwell Science. Publishing: Oxford, UK, 1971. [Google Scholar]
- Martin, J. Use of acid rose bengal and streptpmycin in the plate method for estimating soil fungi. Soil Sci. 1950, 69, 215–233. [Google Scholar] [CrossRef]
- Mulder, E.G.; Antheumisse, J. Morphologie, Physiologie et Ecologie des Arthrobacter; Annales de l’Institut Pasteur: Paris, France, 1963; Volume 105, pp. 46–74. [Google Scholar]
- Ohta, H.; Hattori, T. Oligotrophic bacteria on organic debris and plant roots in paddy field. Soil Biol. Biochem. 1983, 1, 1–8. [Google Scholar]
- Winogradski, S. Soil Microbiology; PWRiL: Warszawa, Poland, 1953. (In Polish) [Google Scholar]
- Fenglerowa, W. Simple method for counting Azotobacter in soil samples. Acta Microbiol. Pol. 1965, 14, 203–206. [Google Scholar]
- Chuanchuen, R.; Karkhoff-Schweizer, R.R.; Schweizer, H.P. High-level triclosan resistance in Pseudomonas aeruginosa is solely a result of efflux. Am. J. Infect. Control. 2003, 31, 124–127. [Google Scholar] [CrossRef]
- Xue, F.; Ya, X.; Tong, Q.; Xiu, Y.; Huang, H. Heterologous overexpression of Pseudomonas umsongens is halohydrin dehalogenase in Escherichia coli and its application in epoxide asymmetric ring opening reactions. Process Biochem. 2018, 75, 139–145. [Google Scholar] [CrossRef]
- Kalyani, D.C.; Telke, A.A.; Surwase, S.N.; Jadhav, S.B.; Lee, J.K.; Jadhav, J.P. Effectual decolorization and detoxification of triphenylmethane dye Malachite Green (MG) by Pseudomonas aeruginosa NCIM 2074 and its enzyme system. Clean Techn. Environ. Policy 2012, 14, 989–1001. [Google Scholar] [CrossRef]
- Shedbalkar, U.; Dhanve, R.; Jadhav, J. Biodegradation of triphenylmethane dye cotton blue by Penicillium ochrochloron MTCC 517. J. Hazard. Mater. 2008, 157, 472–479. [Google Scholar] [CrossRef]
- Hugentobler, K.G.; Müller, M. Towards semisynthetic natural compounds with a biaryl axis: Oxidative phenol coupling in Aspergillus niger. Bioorg. Med. Chem. 2018, 26, 1374–1377. [Google Scholar] [CrossRef]
- Wyszkowska, J.; Boros-Lajszner, E.; Kucharski, J. Microbiological and biochemical properties of soil contaminated with zinc and fertilized with fermented bark, keratin and cellulose. Fresen. Environ. Bull. 2015, 24, 4519–4526. [Google Scholar]
- Utobo, E.B.; Tewari, L. Soil enzymes as bioindicators of soil ecosystem statut. Appl. Ecol. Environ. Res. 2015, 13, 147–169. [Google Scholar]
- Zhu, X.; Wu, X.; Yao, J.; Wang, F.; Liu, W.; Luo, Y.; Jiang, X. Toxic effects of binary toxicants of cresol frother and Cu (II) on soil microorganisms. Int. Biodeter. Biodegrad. 2018, 128, 155–163. [Google Scholar] [CrossRef]
- Patil, J.S.; Anil, A.C. Efficiency of copper and cupronickel substratum to resist development of diatom biofilms. Int. Biodeter. Biodegrad. 2015, 15, 203–214. [Google Scholar] [CrossRef]
- Koeck, D.E.; Pechtl, A.; Zverlov, V.V.; Schwarz, W.H. Genomics of cellulolytic bacteria. Curr. Opin. Biotech. 2014, 29, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Mai, G.; Wang, Z.; Liu, Q.; Zhou, Y.; Ma, Y.; Liu, C. Metagenomic Insights Into a Cellulose-Rich Niche Reveal Microbial Cooperation in Cellulose Degradation. Front. Microbiol. 2019, 10, 1–12. [Google Scholar] [CrossRef]
- Yacoumas, A.; Honvault, N.; Houben, D.; Fontaine, J.; Meglouli, H.; Laruelle, F.; Tisserant, B.; Faucon, M.-P.; Lounès-Hadj Sahraoui, A.; Firmin, S. Contrasting Response of Nutrient Acquisition Traits in Wheat Grown on Bisphenol A-Contaminated Soils. Water. Air Soil Pollut. 2020, 231. [Google Scholar] [CrossRef]
- Jin, J.; Wang, M.; Lua, W.; Zhang, L.; Jiang, Q.; Yeqing, J.; Lu, K.; Sun, S.; Cao, Q.; Wang, Y.; et al. Effect of plants and their root exudate on bacterial activities during rhizobacterium–plant remediation of phenol from water. Environ. Inter. 2019, 127, 114–124. [Google Scholar] [CrossRef]
- Borowik, A.; Wyszkowska, J. Remediation of soil contaminated with diesel oil. J. Elem. 2018, 23, 767–788. [Google Scholar] [CrossRef]
- Bhatti, A.A.; Haq, S.; Bhat, R.A. Actinomycetes benefaction role in soil and plant health. Microb. Pathog. 2017, 111, 458–467. [Google Scholar] [CrossRef]
- Gulve, R.M.; Deshmukh, A.M. Antimicrobial activity of the marine actinomycetes. Inter. Multidiscipl. Res. J. 2012, 2, 16–22. [Google Scholar]
- Sarathchandra, S.U.; Burch, G.; Cox, N.R. Growth patterns of bacterial communities in the rhizoplane and rhizosphere of with clover (Trifolium repens L.) and perennial ryegrass (Lolium perenne L.) in long–term pasture. Appl. Soil Ecol. 1997, 6, 293–299. [Google Scholar] [CrossRef]
- Li, C.; Lu, Q.; Ye, J.; Qin, H.; Long, Y.; Wang, L.; Ou, H. Metabolic and proteomic mechanism of bisphenol A degradation by Bacillus Thuring. Sci. Tot. Environ. 2018, 640–641, 714–725. [Google Scholar] [CrossRef] [PubMed]
- Michałowicz, J.; Duda, W. Phenols–Sources and Toxicity. Pol. J. Environ. Stud. 2007, 16, 347–362. [Google Scholar]
- Dercová, K.; Certík, M.; Malová, A.; Sejáková, Z. Effect of chlorophenols on the membrane lipids of bacterial cells. Int. Biodeter. Biodegr. 2004, 54, 251–254. [Google Scholar] [CrossRef]
- Nannipieri, P.; Ascher-Jenull, J.; Ceccherini, M.T.; Pietramellara, G.; Renella, G.; Schloter, M. Beyond microbial diversity for predicting soil functions: A mini review. Pedosphere 2020, 30, 5–17. [Google Scholar] [CrossRef]
- Bakker, M.G.; Chaparro, J.M.; Manter, D.K.; Vivanco, J.M. Impacts of bulk soil microbial community structure on rhizosphere microbiomes of Zea mays. Plant Soil. 2015, 392, 115–126. [Google Scholar] [CrossRef]
- Salam, M.; Varma, A. Bacterial community structure in soils contaminated with electronic waste pollutants from Delhi NCR, India. Electron. J. Biotechn. 2019, 41, 72–80. [Google Scholar] [CrossRef]
- Siczek, A.; Frąc, M.; Gryta, A.; Kalembasa, S.; Kalembasa, D. Variation in soil microbial population and enzyme activities under faba bean as affected by pentachlorophenol. Appl. Soil Ecol. 2020, 150, 103466. [Google Scholar] [CrossRef]
- Hassen, W.; Neifar, M.; Cherif, H.; Mahjoubi, M.; Souissi, Y.; Raddadi, N.; Fava, F.; Cherif, A. Assessment of genetic diversity and bioremediation potential of pseudomonads isolated from pesticide-contaminated artichoke farm soils. 3 Biotech. 2018, 8, 263. [Google Scholar] [CrossRef]
- Tian, M.; Dub, D.; Zhou, W.; Zeng, X.; Cheng, G. Phenol degradation and genotypic analysis of dioxygenase genes in bacteria isolated from sediments. Braz. J. Microbiol. 2017, 48, 305–313. [Google Scholar] [CrossRef]
- Spivack, J.; Leib, T.K.; Lobos, J.H. Novel pathway for bacterial metabolism of bisphenol A. J. Biol. Chem. 1994, 269, 7323–7329. [Google Scholar]
- Lin, Z.; Zhen, Z.; Wu, Z.; Yang, J.; Zhong, L.; Hu, H.; Luo, C.; Bai, J.; Li, Y.; Zhang, D. The impact on the soil microbial community and enzyme activity of two earthworm species during the bioremediation of pentachlorophenol-contaminated soils. J. Hazard. Mater. 2016, 301, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Zu, L.; Wong, P.K.; Hui, X.; Lu, Y.; Xiong, J.; An, T. Biodegradation and detoxification of bisphenol A with one newly-isolated strain Bacillus sp. GZB: Kinetics, mechanism and estrogenic transition. Bioresour. Technol. 2012, 114, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Szaleniec, M.; Borowski, T.; Schühle, K.; Witko, M.; Heider, J. Ab inito modeling of ethylbenzene dehydrogenase reaction mechanism. J. Am. Chem. Soc. 2010, 132, 6014–6024. [Google Scholar] [CrossRef] [PubMed]
- Daudzai, Z.; Treesubsuntorn, C.; Thiravetyan, P. Inoculated Clitoria ternatea with Bacillus cereus ERBP for enhancing gaseous ethylbenzene phytoremediation: Plant metabolites and expression of ethylbenzene degradation genes. Ecotox. Environ. Safe. 2018, 164, 50–60. [Google Scholar] [CrossRef]
- Bilal, M.; Adeel, M.; Rasheed, T.; Zhao, Y.; Iqbal, H.M.N. Emerging contaminants of high concern and their enzyme-assisted biodegradation–A review. Environ. Int. 2019, 124, 336–353. [Google Scholar] [CrossRef]
- Zaborowska, M.; Wyszkowska, J.; Kucharski, J. Biochemical activity of soil contaminated with BPS, bioaugmented with a mould fungi consortium and a bacteria consortium. Environ. Sci. Pollut. Res. 2019, 26, 37054–37069. [Google Scholar] [CrossRef]
- Zaborowska, M.; Wyszkowska, J.; Kucharski, J. Soil enzyme response to bisphenol F contamination in the soil bioaugmented using bacterial and mould fungal consortium. Environ. Monit. Assess. 2020, 192, 1–18. [Google Scholar] [CrossRef]
- Carvalho, M.B.; Tavares, S.; Medeiros, J.; Núñez, O.; Gallart-Ayala, H.; Leitão, M.C.; Galceran, M.T.; Hursthouse, A.; Pereira, C.S. Degradation pathway of pentachlorophenol by Mucor plumbeus involves phase II conjugation and oxidation–reduction reactions. J. Hazard. Mater. 2011, 198, 133–142. [Google Scholar] [CrossRef]
- Khadem, A.; Raiesi, F. Response of soil alkaline phosphatase to biochar amendments: Changes in kinetic and thermodynamic Characteristics. Geoderma 2019, 337, 44–54. [Google Scholar] [CrossRef]
- Tang, Z.; Chen, H.; He, H.; Ma, C. Assays for alkaline phosphatase activity: Progress and prospects. TrAC Trend. Anal. Chem. 2019, 113, 32–43. [Google Scholar] [CrossRef]
- Horta, L.P.; Mota, Y.C.C.; Barbosa, G.M.; Braga, T.; Marriel, I.E.; Fátima, A.; Modolo, L.V. Urease inhibitors of agricultural interest inspired by structures of plant phenolic aldehydes. J. Braz. Chem. Soc. 2016, 27, 1512–1519. [Google Scholar] [CrossRef]
- Modolo, L.V.; Silva, C.J.; Brandão, D.S.; Chaves, I.S. Minireview on what we have learned about urease inhibitors of agricultural interest since mid-2000sq. J. Adv. Res. 2018, 13, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Kot, M.; Zaborska, W. Irreversible inhibition of jack bean urease by pyrocatechol. J. Enzym. Inhib. Med. Chem. 2008, 18, 413–417. [Google Scholar] [CrossRef] [PubMed]
- Mazzei, L.; Cianci, M.; Musiani, F.; Lente, G.; Palombo, M.; Ciurli, S. Inactivation of urease by catechol: Kinetics and structure. J. Inorg. Biochem. 2017, 166, 182–189. [Google Scholar] [CrossRef]
- Herter, S.; Schmidt, M.; Thompson, M.L.; Mikolasch, A.; Schauer, F. Study of enzymatic properties of phenol oxidase from nitrogen-fixing Azotobacter chroococcum. AMB Express 2011, 1, 14. [Google Scholar] [CrossRef]
- Mustafa, S.; Perveen, S.; Khan, A. Synthesis, enzyme inhibition and anticancer investigations of unsymmetrical 1,3-disubstituted urease. J. Serb. Chem. Soc. 2014, 79, 1–10. [Google Scholar] [CrossRef]
- Perveen, S.; Khan, K.M.; Lodhi, M.A.; Choudhary, M.I.; Choudhary, M.I.; Rahman, A.; Voelter, W. Urease and a-chymotrypsin inhibitory effects of selected urea derivatives. Lett. Drug. Des. Discov. 2008, 5, 401–405. [Google Scholar] [CrossRef]
- Xiao, Z.P.; Shi, D.H.; Li, H.Q.; Zhang, L.N.; Xu, C.; Zhu, H.L. Polyphenols based on isoflavones as inhibitors of Helicobacter pylori urease. Bioorg. Med. Chem. 2007, 15, 3703–3710. [Google Scholar] [CrossRef]
- Rego, Y.F.; Queiroz, M.P.; Brito, T.O.; Carvalho, P.G.; de Queiroz, V.T.; de Fátima, Â.; Macedo, J.F. A review on the development of urease inhibitors as antimicrobial agents against pathogenic bacteria. J. Adv. Res. 2018, 13, 69–100. [Google Scholar] [CrossRef]
- Taha, M.; Ismail, N.H.; Imran, S.; Wadood, A.; Rahim, F.; Riaz, M. Synthesis of potent urease inhibitors based on disulfide scaffold and their molecular docking studies. Bioorg. Med. Chem. 2015, 23, 7211–7218. [Google Scholar] [CrossRef]
- Burns, R.G.; De Forest, J.L.; Marxsen, J.; Sinsabaugh, R.L.; Stromberger, M.E.; Wallenstein, M.D.; Weintraub, M.N.; Zoppini, A. Soil enzymes in a changing environment: Current knowledge and future directions. Soil Biol. Biochem. 2013, 58, 216–234. [Google Scholar] [CrossRef]
- Harwood, C.S.; Burchhardt, G.; Herrmann, H.; Fuchs, G. Anaerobic metabolism of aromatic compounds via the benzoyl-CoA pathway. FEMS Microbiol. Rev. 1999, 22, 439–458. [Google Scholar] [CrossRef]
- World Reference Base of Soil Resources. International Soil Classification System for Naming Soils and Creating Legends for Soil Maps; World Soils Resources Raports 106; FAO: Roma, Italy, 2014. [Google Scholar]
- Borowik, A.; Wyszkowska, J.; Wyszkowski, M. Resistance of aerobic microorganisms and soil enzyme response to soil contamination with Ekodiesel Ultra fuel. Environ. Sci. Pollut. Res. 2017, 24, 24346–24363. [Google Scholar] [CrossRef] [PubMed]
- Regulation of the Minister of the Environment of September 1, 2016 on the way of assessing the pollution of the earth’s surface; Item 1395. J. Laws 2016. No. 1.
- Hu, Y.; Zhu, Q.; Yan, X.; Liao, C.; Jiang, G. Occurrence, fate and risk assessment of BPA and its substituents in wastewater treatment plant: A review. Environ. Res. 2019, 178, 108732. [Google Scholar] [CrossRef] [PubMed]
- De Leij, F.A.; Whipps, J.M.; Lynch, J.M. The use for colony development for the characterization of bacterial communities in soil and on roots. Microb. Ecol. 1993, 27, 81–97. [Google Scholar] [CrossRef]
- Ravel, J.; Gajer, P.; Abdo, Z.; Schneider, G.M.; Koenig, S.S.; McCulle, S.L.; Karlebach, S.; Gorle, R.; Russell, J.; Tacket, C.O.; et al. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. USA 2011, 108, 4680–4687. [Google Scholar] [CrossRef]
- Dell Inc. Dell Statistica Data Analysis Software System; Dell Inc.: Tulsa, OK, USA, 2016; version 13.1. [Google Scholar]
- Gregory, R.; Warnes, B.; Bonebakker, L.; Gentleman, R.; Liaw, W.H.A.; Lumley, T.; Maechler, M.; Magnusson, A.; Moeller, S.; Schwartz, M.; et al. Gplots: Various R Programming Tools for Plotting Data. R Package Version 2.17.0. 2020. Available online: https://CRAN.R-project.org/package=gplots (accessed on 23 February 2020).
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef]
- RStudio Team. RStudio: Integrated Development for R; RStudio, Inc.: Boston, MA, USA, 2019; Available online: http://www.rstudio.com/ (accessed on 23 February 2020).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019; Available online: https://www.R-project.org/ (accessed on 23 February 2020).




| Parameters | Unit | Value | References |
|---|---|---|---|
| Granulometric Composition of fractions (%) (d,mm) | sand 2.00 ≥ d > 0.05 | 74.93 | [62] |
| silt 0.05 ≥ d > 0.002 | 22.85 | ||
| clay d ≤ 0.002 | 2.22 | ||
| pHKCl | (mol KCl dm−3) | 6.70 | [63] |
| HAC | (mM (+) kg−1 dry matter of soil) | 11.40 | |
| EBC | 49.00 | [64] | |
| CEC | 0.40 | ||
| BS | (%) | 81.10 | [65] |
| Corg | (g kg−1 dry matter of soil) | 9.30 | |
| Ntot | 0.62 | [66] | |
| Paveilabe | (mg kg−1 dry matter of soil) | 93.68 | [67] |
| Kaveilabe | 141.10 | ||
| Mgaveilabe | 42.00 | [68] | |
| Enzymes activity | |||
| Deh | (μmol triphenyl formazan (TFF) kg−1 dry matter of soil h−1) | 0.546 | [69] |
| Ure | (mmol N-NH4 kg−1 dry matter of soil h−1) | 0.632 | [70] |
| Cat | (mol O2 kg−1 dry matter of soil h−1) | 0.086 | |
| Pal | (mmol 4-nitrofenol (PN) kg−1 dry matter of soil h−1) | 2.309 | |
| Pac | 0.510 | ||
| Aryl | 0.013 | ||
| Glu | 0.422 | ||
| Number of microorganisms | |||
| Org | (cfu 10n kg−1 dry matter of soil) | 27.971 | [71] |
| Act | 116.194 | [72] | |
| F | 802.414 | [73] | |
| Ps | 33.473 | [74] | |
| Art | 37.657 | ||
| Cel | 27.197 | [75] | |
| Olig | 191.771 | ||
| Cop | 166.667 | ||
| Im | 103.208 | [76] | |
| Am | 341.702 | ||
| Az | 20.921 | [77] |
| Kind of BP | Org | Act | F | Ps | Art | Cel | Olig | Cop | Im | Am | Az |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 15th day | |||||||||||
| C | 470.874 a | 482.581 b | 87.664 c | 109.973 c | 145.620 a,b | 169.132 a | 910.884 b | 242.700 b | 1305.271 a | 847.175 a | 139.553 a |
| A | 330.097 b | 464.306 b | 147.630 a | 81.356 d | 85.122 c | 165.725 a | 767.608 d | 223.729 b | 381.168 d | 566.478 b | 164.218 a |
| F | 348.372 b | 441.462 b | 115.934 b,c | 187.547 a | 188.307 a | 67.578 b | 991.648 a | 302.961 a | 647.684 b | 934.700 a | 29.613 b |
| S | 245.288 c | 539.406 a | 147.915 a | 136.434 a,b | 136.434 b | 59.690c | 834.884 c | 333.333 a | 491.473 c | 865.116 a | 31.008 b |
| 30th day | |||||||||||
| C | 297.783 b | 254.561 d | 103.564 d | 68.057 d | 133.919 a | 164.654 a | 488.108 c | 283.937 b | 381.998 c | 607.391 b | 150.018 b,c |
| A | 318.271 b | 581.813 a | 216.952 c | 176.448 a | 169.544 a | 104.334 b | 637.514 b | 333.717 a | 601.458 a | 817.031 a | 119.678 c |
| F | 252.315 b | 435.307 c | 271.401 b | 169.197 b | 42.661 c | 97.614 b | 624.729 b | 185.105 d | 436.732 b | 399.855 c | 245.842 a |
| S | 382.543 a | 457.480 b | 305.641 a | 126.984 c | 80.527 b | 80.527 c | 744.870 a | 239.257 c | 552.846 a | 651.955 b | 160.279 b |
| Kind of BP | Deh | Cat | Pac | Pal | Aryl | Glu | Ure |
|---|---|---|---|---|---|---|---|
| (μMol TFF) | (mol O2) | (mmol 4-Nitrophenol PN) | (mmol N-NH4) | ||||
| 15th day | |||||||
| C | 6.625 c | 0.422 a | 1.606 b | 2.159 b,c | 0.060 a | 1.115 a | 2.435 a |
| A | 13.743 b | 0.381 c | 1.845 a | 2.392 a | 0.040 b,c | 1.051 b | 2.142 b |
| F | 15.320 a | 0.401 b | 1.679 a,b | 2.312 a,b | 0.046 b | 0.914 d | 1.731 c |
| S | 13.648 b | 0.396 b,c | 1.560 b | 2.115 c | 0.033 c | 0.999 c | 1.634 d |
| 30th day | |||||||
| C | 5.233 d | 0.379 b | 1.683 b | 1.892 c | 0.049 a | 0.994 a,b | 2.224 a |
| A | 15.241 a | 0.418 a | 1.774 a | 2.336 b | 0.036 c | 1.050 a | 1.933 b |
| F | 13.121 b | 0.360 b | 1.768 a | 3.000 a | 0.040 b | 0.951 b | 1.630 c |
| S | 12.311 c | 0.373 b | 1.473 c | 2.884 a | 0.028 d | 0.873 c | 1.852 b |
| Compound | Molecular Formula | Molecular Weight | BCF | VP | Log Kow | WS | BP (°C) |
|---|---|---|---|---|---|---|---|
| Bisphenol A | C15H16O2 | 228.29 | 71.85 | 2.27 × 10−7 | 3.64 | 172.7 | 363.54 |
| Bisphenol S | C12H10O4S | 250.27 | 36.97 | 4.72 × 10−10 | 1.65 | 351.8 | 422.52 |
| Bipshenol F | C13H12O2 | 200.23 | 34.73 | 3.72 × 10−7 | 3.06 | 542.8 | 351.92 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zaborowska, M.; Wyszkowska, J.; Borowik, A. Soil Microbiome Response to Contamination with Bisphenol A, Bisphenol F and Bisphenol S. Int. J. Mol. Sci. 2020, 21, 3529. https://doi.org/10.3390/ijms21103529
Zaborowska M, Wyszkowska J, Borowik A. Soil Microbiome Response to Contamination with Bisphenol A, Bisphenol F and Bisphenol S. International Journal of Molecular Sciences. 2020; 21(10):3529. https://doi.org/10.3390/ijms21103529
Chicago/Turabian StyleZaborowska, Magdalena, Jadwiga Wyszkowska, and Agata Borowik. 2020. "Soil Microbiome Response to Contamination with Bisphenol A, Bisphenol F and Bisphenol S" International Journal of Molecular Sciences 21, no. 10: 3529. https://doi.org/10.3390/ijms21103529
APA StyleZaborowska, M., Wyszkowska, J., & Borowik, A. (2020). Soil Microbiome Response to Contamination with Bisphenol A, Bisphenol F and Bisphenol S. International Journal of Molecular Sciences, 21(10), 3529. https://doi.org/10.3390/ijms21103529







