Complex Molecular Evolution and Expression of Expansin Gene Families in Three Basic Diploid Species of Brassica
Abstract
1. Introduction
2. Results
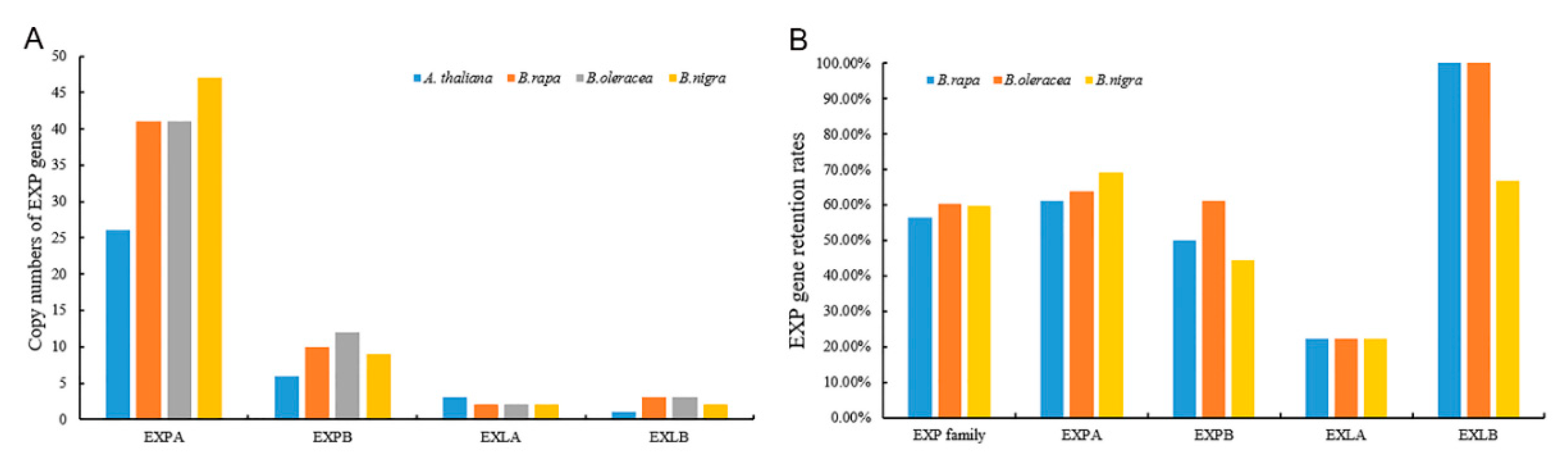
2.1. Genome-Wide Identification of the Expansin Gene Family in Three Basic Diploid Species of Brassica
2.2. Phylogenetic and Structural Analyses of BrEXPs, BolEXPs, and BniEXPs
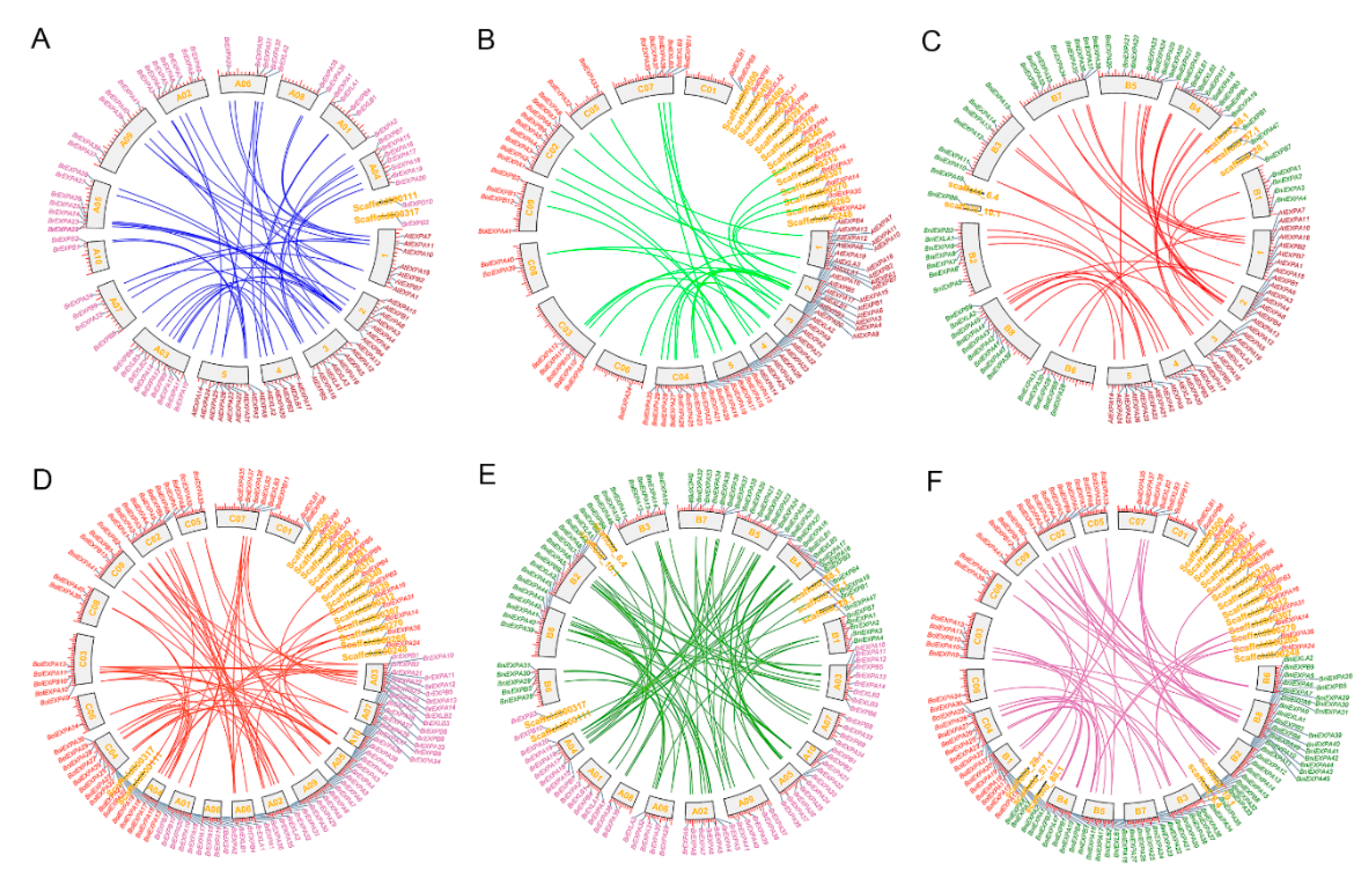
2.3. Chromosomal Distribution, Duplication Mechanism and Retained Proportion in Three Basic Species of Brassica
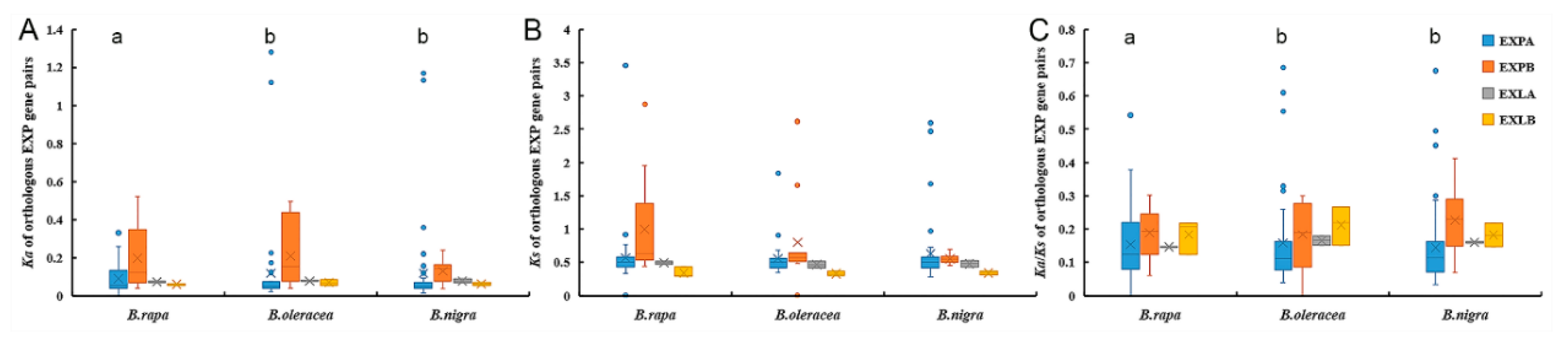
2.4. Coding Sequence and Promoter Evolution Analyses of Expansin Genes in the Three Species of Brassica
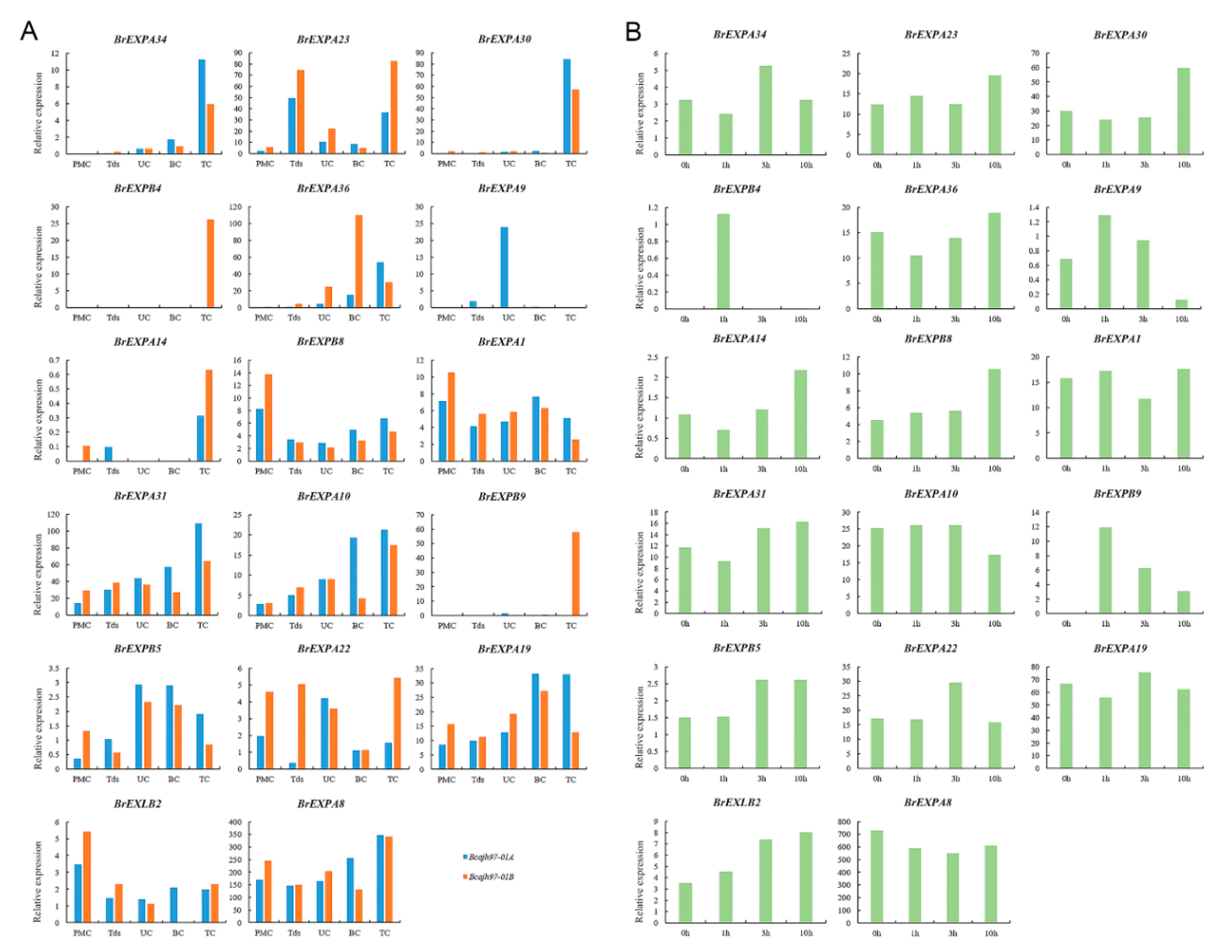
2.5. Expression Patterns of Expansin Family Members in Various Tissues and Organs of B. rapa, B. nigra, and B. oleracea
3. Discussion
3.1. Scale of Expansin Families after WGT in B. rapa, B. oleracea, and B. nigra
3.2. Conservative and Large Family Size of EXLB in Three Basic Species of Brassica
3.3. Promoter Divergence is Closely Related to the Coding Sequence Evolution of Expansin Genes in Three Basic Species of Brassica
3.4. Speculative Roles of Expansin Genes during Reproductive Development
4. Materials and Methods
4.1. Identification and Physicochemical Property Predictions of Expansin Family Members in Three Basic Diploid Species of Brassica
4.2. Phylogenetic Genetic Tree Construction and Structural Analysis
4.3. Chromosome Location, Synteny and Retained Rate Analysis
4.4. Evolution Analysis of Coding Sequences and Promoters
4.5. Expression Analysis of Expansin Family Members
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| BrEXPs | B. rapa expansin genes |
| BolEXPs | B. oleracea expansin genes |
| BniEXPs | B. nigra expansin genes |
| GMS | genic male sterile |
| Ka | nonsynonymous substitution rate |
| Ks | synonymous substitution rate |
| Mw | molecular weights |
| MYA | million years ago |
| PGs | polygalacturonases genes |
| pIs | isoelectric points |
| qRT-PCR | quantitative reverse transcription polymerase chain reaction |
| WGT | whole genome triplication |
References
- Sanchez-Rodriguez, C.; Rubio-Somoza, I.; Sibout, R.; Persson, S. Phytohormones and the cell wall in Arabidopsis during seedling growth. Trends Plant Sci. 2010, 15, 291–301. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Plant expansins: Diversity and interactions with plant cell walls. Curr. Opin. Plant Biol. 2015, 25, 162–172. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 2005, 6, 850–861. [Google Scholar] [CrossRef]
- Cosgrove, D.J.; Li, L.C.; Cho, H.T.; Hoffmann-Benning, S.; Moore, R.C.; Blecker, D. The growing world of expansins. Plant Cell Physiol. 2002, 43, 1436–1444. [Google Scholar] [CrossRef]
- Sampedro, J.; Cosgrove, D.J. The expansin superfamily. Genome Biol. 2005, 6, 242. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kende, H.; Bradford, K.; Brummell, D.; Cho, H.T.; Cosgrove, D.; Fleming, A.; Gehring, C.; Lee, Y.; McQueen-Mason, S.; Rose, J.; et al. Nomenclature for members of the expansin superfamily of genes and proteins. Plant Mol. Biol. 2004, 55, 311–314. [Google Scholar] [CrossRef]
- Sanchez-Montesino, R.; Bouza-Morcillo, L.; Marquez, J.; Ghita, M.; Duran-Nebreda, S.; Gomez, L.; Holdsworth, M.J.; Bassel, G.; Onate-Sanchez, L. A Regulatory Module Controlling GA-Mediated Endosperm Cell Expansion Is Critical for Seed Germination in Arabidopsis. Mol. Plant 2019, 12, 71–85. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Wen, L.Z.; Guo, Y.H.; Yu, Y.Y.; Sun, C.H.; Fan, H.M.; Ma, F.F.; Zheng, C.S. Expressional and Functional Verification of the Involvement of CmEXPA4 in Chrysanthemum Root Development. J. Plant Growth Regul. 2019, 38, 1375–1386. [Google Scholar] [CrossRef]
- Stamm, P.; Topham, A.T.; Mukhtar, N.K.; Jackson, M.D.; Tome, D.F.; Beynon, J.L.; Bassel, G.W. The Transcription Factor ATHB5 Affects GA-Mediated Plasticity in Hypocotyl Cell Growth during Seed Germination. Plant Physiol. 2017, 173, 907–917. [Google Scholar] [CrossRef] [PubMed]
- Kuluev, B.R.; Knyazev, A.V.; Nikonorov, Y.M.; Chemeris, A.V. Role of the expansin genesNtEXPA1andNtEXPA4in the regulation of cell extension during tobacco leaf growth. Russ. J. Genet. 2013, 50, 489–497. [Google Scholar] [CrossRef]
- Han, Y.C.; Kuang, J.F.; Chen, J.Y.; Liu, X.C.; Xiao, Y.Y.; Fu, C.C.; Wang, J.N.; Wu, K.Q.; Lu, W.J. Banana Transcription Factor MaERF11 Recruits Histone Deacetylase MaHDA1 and Represses the Expression of MaACO1 and Expansins during Fruit Ripening. Plant Physiol. 2016, 171, 1070–1084. [Google Scholar] [CrossRef] [PubMed]
- Lou, Y.; Zhou, H.S.; Han, Y.; Zeng, Q.Y.; Zhu, J.; Yang, Z.N. Positive regulation of AMS by TDF1 and the formation of a TDF1-AMS complex are required for anther development in Arabidopsis thaliana. New Phytol. 2018, 217, 378–391. [Google Scholar] [CrossRef] [PubMed]
- Valdivia, E.R.; Wu, Y.; Li, L.C.; Cosgrove, D.J.; Stephenson, A.G. A group-1 grass pollen allergen influences the outcome of pollen competition in maize. PLoS ONE 2007, 2, e154. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, J.; Chen, G.; Ye, X.; Zhang, L.; Zhu, S.; Yuan, L.; Hou, J.; Wang, C. Functional analysis of a MYB transcription factor BrTDF1 in the tapetum development of Wucai (Brassica rapa ssp.). Sci. Hortic. 2019, 257. [Google Scholar] [CrossRef]
- Krishnamurthy, P.; Muthusamy, M.; Kim, J.A.; Jeong, M.J.; Lee, S.I. Brassica rapa expansin-like B1 gene (BrEXLB1) regulate growth and development in transgenic Arabidopsis and elicits response to abiotic stresses. J. Plant Biochem. Biotechnol. 2019, 28, 437–446. [Google Scholar] [CrossRef]
- Liu, Y.P.; Zhang, L.; Hao, W.F.; Zhang, L.; Liu, Y.; Chen, L.Q. Expression of Two alpha-Type Expansins from Ammopiptanthus nanus in Arabidopsis thaliana Enhance Tolerance to Cold and Drought Stresses. Int. J. Mol. Sci. 2019, 20, 19. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Liu, Y.; Deng, X.; Liu, D.; Liu, Y.; Hu, Y.; Yan, Y. Genome-wide identification and expression analysis of expansin gene family in common wheat (Triticum aestivum L.). BMC Genom. 2019, 20, 101. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, B.; Li, C.; Lei, C.; Kong, C.; Yang, Y.; Gong, M. A comprehensive expression analysis of the expansin gene family in potato (Solanum tuberosum) discloses stress-responsive expansin-like B genes for drought and heat tolerances. PLoS ONE 2019, 14, e0219837. [Google Scholar] [CrossRef]
- Otulak-Koziel, K.; Koziel, E.; Lockhart, B.E.L.; Bujarski, J.J. The Expression of Potato Expansin A3 (StEXPA3) and Extensin4 (StEXT4) Genes with Distribution of StEXPAs and HRGPs-Extensin Changes as an Effect of Cell Wall Rebuilding in Two Types of PVY(NTN)-Solanum tuberosum Interactions. Viruses 2020, 12, 66. [Google Scholar] [CrossRef]
- Fernando, G.J.; Sebastian, L.; Heike, T.; Britt-Louise, L.; Thomas, K.; David, G.; Edgar, M.; Mark, V.; Pathology, S.E.I.J.M.P. Massive up-regulation of LBD transcription factors and EXPANSINs highlights the regulatory programs of rhizomania disease. Mol. Plant Pathol. 2018, 19, 2333–2348. [Google Scholar] [CrossRef]
- Tan, J.; Wang, M.; Shi, Z.; Miao, X. OsEXPA10 mediates the balance between growth and resistance to biotic stress in rice. Plant Cell Rep. 2018, 37, 993–1002. [Google Scholar] [CrossRef] [PubMed]
- Vannerum, K.; Huysman, M.J.; De Rycke, R.; Vuylsteke, M.; Leliaert, F.; Pollier, J.; Lutz-Meindl, U.; Gillard, J.; De Veylder, L.; Goossens, A.; et al. Transcriptional analysis of cell growth and morphogenesis in the unicellular green alga Micrasterias (Streptophyta), with emphasis on the role of expansin. BMC Plant Biol. 2011, 11, 128. [Google Scholar] [CrossRef] [PubMed]
- Carey, R.E.; Cosgrove, D.J. Portrait of the expansin superfamily in Physcomitrella patens: Comparisons with angiosperm expansins. Ann. Bot. 2007, 99, 1131–1141. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Yan, H.; Chen, W.; Liu, J.; Jiang, C.; Jiang, H.; Zhu, S.; Cheng, B. Genome-wide identification and characterization of maize expansin genes expressed in endosperm. Mol. Genet. Genom. 2014, 289, 1061–1074. [Google Scholar] [CrossRef]
- Zhu, Y.; Wu, N.; Song, W.; Yin, G.; Qin, Y.; Yan, Y.; Hu, Y. Soybean (Glycine max) expansin gene superfamily origins: Segmental and tandem duplication events followed by divergent selection among subfamilies. BMC Plant Biol. 2014, 14, 93. [Google Scholar] [CrossRef]
- Ding, A.; Marowa, P.; Kong, Y. Genome-wide identification of the expansin gene family in tobacco (Nicotiana tabacum). Mol. Genet. Genom. 2016, 291, 1891–1907. [Google Scholar] [CrossRef]
- Zhang, S.; Xu, R.; Gao, Z.; Chen, C.; Jiang, Z.; Shu, H.J.M.G.; Genomics. A genome-wide analysis of the expansin genes in Malus×Domestica. Mol. Genet. Genom. 2014, 289, 225–236. [Google Scholar] [CrossRef]
- Krishnamurthy, P.; Hong, J.K.; Kim, J.A.; Jeong, M.J.; Lee, Y.H.; Lee, S.I. Genome-wide analysis of the expansin gene superfamily reveals Brassica rapa-specific evolutionary dynamics upon whole genome triplication. Mol. Genet. Genom. 2015, 290, 521–530. [Google Scholar] [CrossRef]
- Town, C.D.; Cheung, F.; Maiti, R.; Crabtree, J.; Haas, B.J.; Wortman, J.R.; Hine, E.E.; Althoff, R.; Arbogast, T.S.; Tallon, L.J.; et al. Comparative genomics of Brassica oleracea and Arabidopsis thaliana reveal gene loss, fragmentation, and dispersal after polyploidy. Plant Cell 2006, 18, 1348–1359. [Google Scholar] [CrossRef]
- Yu, J.; Tehrim, S.; Zhang, F.; Tong, C.; Huang, J.; Cheng, X.; Dong, C.; Zhou, Y.; Qin, R.; Hua, W.; et al. Genome-wide comparative analysis of NBS-encoding genes between Brassica species and Arabidopsis thaliana. BMC Genom. 2014, 15, 3. [Google Scholar] [CrossRef]
- Yang, T.J.; Kim, J.S.; Kwon, S.J.; Lim, K.B.; Cell, B.-S.P.J.P. Sequence-level analysis of the diploidization process in the triplicated FLOWERING LOCUS C region of Brassica rapa. Plant Cell 2006, 18, 1339–1347. [Google Scholar] [CrossRef] [PubMed]
- Lysak, M.A.; Koch, M.A.; Pecinka, A.; Schubert, I. Chromosome triplication found across the tribe Brassiceae. Genome Res. 2005, 15, 516–525. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Shao, Z.Q.; Wu, X.Z.; Wang, Q.; Wang, B.; Chen, J.Q.; Hang, Y.Y.; Xue, J.Y. Loss/retention and evolution of NBS-encoding genes upon whole genome triplication of Brassica rapa. Gene 2014, 540, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Carey, R.E.; Hepler, N.K.; Cosgrove, D.J. Selaginella moellendorffii has a reduced and highly conserved expansin superfamily with genes more closely related to angiosperms than to bryophytes. BMC Plant Biol. 2013, 13, 4. [Google Scholar] [CrossRef]
- Li, N.; Pu, Y.; Gong, Y.; Yu, Y.; Ding, H. Genomic location and expression analysis of expansin gene family reveals the evolutionary and functional significance in Triticum aestivum. Genes Genom. 2016, 38, 1021–1030. [Google Scholar] [CrossRef]
- Lu, Y.; Liu, L.; Wang, X.; Han, Z.; Ouyang, B.; Zhang, J.; Li, H. Genome-wide identification and expression analysis of the expansin gene family in tomato. Mol. Genet. Genom. 2016, 291, 597–608. [Google Scholar] [CrossRef]
- Sampedro, J.; Carey, R.E.; Cosgrove, D.J. Genome histories clarify evolution of the expansin superfamily: New insights from the poplar genome and pine ESTs. J. Plant Res. 2006, 119, 11–21. [Google Scholar] [CrossRef]
- Seader, V.H.; Thornsberry, J.M.; Carey, R.E. Utility of the Amborella trichopoda expansin superfamily in elucidating the history of angiosperm expansins. J. Plant Res. 2016, 129, 199–207. [Google Scholar] [CrossRef]
- Dal Santo, S.; Vannozzi, A.; Tornielli, G.B.; Fasoli, M.; Venturini, L.; Pezzotti, M.; Zenoni, S. Genome-wide analysis of the expansin gene superfamily reveals grapevine-specific structural and functional characteristics. PLoS ONE 2013, 8, e62206. [Google Scholar] [CrossRef]
- Shen, X.; Xu, L.; Liu, Y.; Dong, H.; Zhou, D.; Zhang, Y.; Lin, S.; Cao, J.; Huang, L. Comparative transcriptome analysis and ChIP-sequencing reveals stage-specific gene expression and regulation profiles associated with pollen wall formation in Brassica rapa. BMC Genom. 2019, 20, 264. [Google Scholar] [CrossRef]
- Freeling, M. The evolutionary position of subfunctionalization, downgraded. Genome Dyn. 2008, 4, 25–40. [Google Scholar] [CrossRef] [PubMed]
- Freeling, M. Bias in plant gene content following different sorts of duplication: Tandem, whole-genome, segmental, or by transposition. Annu Rev. Plant Biol. 2009, 60, 433–453. [Google Scholar] [CrossRef] [PubMed]
- Papp, B.; Pal, C.; Hurst, L.D. Dosage sensitivity and the evolution of gene families in yeast. Nature 2003, 424, 194–197. [Google Scholar] [CrossRef] [PubMed]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef]
- Moore, R.C.; Purugganan, M.D. The early stages of duplicate gene evolution. Proc. Natl. Acad. Sci. USA 2003, 100, 15682–15687. [Google Scholar] [CrossRef]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef]
- Force, A.; Lynch, M.; Pickett, F.B.; Amores, A.; Postlethwait, J.J.G. Preservation of Duplicate Genes by Complementary, Degenerative Mutations. Genetics 1999, 151, 1531–1545. [Google Scholar]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Guimaraes, L.A.; Mota, A.P.Z.; Araujo, A.C.G.; de Alencar Figueiredo, L.F.D.; Pereira, B.M.; de Passos Saraiva, M.A.; Silva, R.B.; Danchin, E.G.J.; Guimaraes, P.M.; Brasileiro, A.C.M. Genome-wide analysis of expansin superfamily in wild Arachis discloses a stress-responsive expansin-like B gene. Plant Mol.Biol. 2017, 94, 79–96. [Google Scholar] [CrossRef]
- Wang, B.; Du, H.; Li, W.L.; Li, X.H.; Zhang, C.Y. GmEXLB1, a Soybean Expansin-Like B Gene, Alters Root Architecture to Improve Phosphorus Acquisition in Arabidopsis. Front. Plant Sci. 2019, 10, 12. [Google Scholar] [CrossRef]
- Wray, G.A.; Hahn, M.W.; Abouheif, E.; Balhoff, J.P.; Pizer, M.; Rockman, M.V.; Romano, L.A. The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol. 2003, 20, 1377–1419. [Google Scholar] [CrossRef] [PubMed]
- Castillo-Davis, C.I.; Hartl, D.L.; Achaz, G. cis-Regulatory and protein evolution in orthologous and duplicate genes. Genome Res. 2004, 14, 1530–1536. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Haberer, G.; Hindemitt, T.; Meyers, B.C.; Mayer, K.F. Transcriptional similarities, dissimilarities, and conservation of cis-elements in duplicated genes of Arabidopsis. Plant Physiol. 2004, 136, 3009–3022. [Google Scholar] [CrossRef] [PubMed]
- Casneuf, T.; De Bodt, S.; Raes, J.; Maere, S.; Van de Peer, Y. Nonrandom divergence of gene expression following gene and genome duplications in the flowering plant Arabidopsis thaliana. Genome Biol. 2006, 7, R13. [Google Scholar] [CrossRef]
- Ganko, E.W.; Meyers, B.C.; Vision, T.J. Divergence in expression between duplicated genes in Arabidopsis. Mol. Biol. Evol. 2007, 24, 2298–2309. [Google Scholar] [CrossRef]
- Yang, L.; Takuno, S.; Waters, E.R.; Gaut, B.S. Lowly expressed genes in Arabidopsis thaliana bear the signature of possible pseudogenization by promoter degradation. Mol. Biol. Evol. 2011, 28, 1193–1203. [Google Scholar] [CrossRef][Green Version]
- Berke, L.; Sanchez-Perez, G.F.; Snel, B. Contribution of the epigenetic mark H3K27me3 to functional divergence after whole genome duplication in Arabidopsis. Genome Biol. 2012, 13, R94. [Google Scholar] [CrossRef]
- Liang, Y.; Yu, Y.; Shen, X.; Dong, H.; Lyu, M.; Xu, L.; Ma, Z.; Liu, T.; Cao, J. Dissecting the complex molecular evolution and expression of polygalacturonase gene family in Brassica rapa ssp. chinensis. Plant Mol. Biol. 2015, 89, 629–646. [Google Scholar] [CrossRef]
- Cosgrove, D.J. New genes and new biological roles for expansins. Curr. Opin. Plant Biol. 2000, 3, 73–78. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Loosening of plant cell walls by expansins. Nature 2000, 407, 321–326. [Google Scholar] [CrossRef]
- Cosgrove, D.J.; Bedinger, P.; Durachko, D.M. Group I allergens of grass pollen as cell wall-loosening agents. Proc. Natl. Acad. Sci. USA 1997, 94, 6559–6564. [Google Scholar] [CrossRef] [PubMed]
- Valdivia, E.R.; Stephenson, A.G.; Durachko, D.M.; Cosgrove, D. Class B beta-expansins are needed for pollen separation and stigma penetration. Sex. Plant Reprod. 2009, 22, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Pezzotti, M.; Feron, R.; Mariani, C. Pollination modulates expression of the PPAL gene, a pistil-specific beta-expansin. Plant Mol. Biol. 2002, 49, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Mollet, J.C.; Leroux, C.; Dardelle, F.; Lehner, A. Cell Wall Composition, Biosynthesis and Remodeling during Pollen Tube Growth. Plants 2013, 2, 107–147. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Peng, Z.; Liu, S.; He, Y.; Cheng, L.; Kong, F.; Wang, J.; Lu, G. Genome-wide analysis of Aux/IAA gene family in Solanaceae species using tomato as a model. Mol. Genet. Genom. 2012, 287, 295–311. [Google Scholar] [CrossRef]
- Lee, Y.; Choi, D.; Kende, H. Expansins: Ever-expanding numbers and functions. Curr. Opin. Plant Biol. 2001, 4, 527–532. [Google Scholar] [CrossRef]
- Chen, C.; Xia, R.; Chen, H.; He, Y. TBtools, a Toolkit for Biologists integrating various HTS-data handling tools with a user-friendly interface. bioRxiv 2018. [Google Scholar] [CrossRef]
- Lin, Y.X.; Jiang, H.Y.; Chu, Z.X.; Tang, X.L.; Zhu, S.W.; Cheng, B.J. Genome-wide identification, classification and analysis of heat shock transcription factor family in maize. BMC Genom. 2011, 12, 76. [Google Scholar] [CrossRef]
- Yang, Z. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997, 13, 555–556. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Gao, F.; Chen, C.; Arab, D.A.; Du, Z.; He, Y.; Ho, S.Y.W. EasyCodeML: A visual tool for analysis of selection using CodeML. Ecol. Evol. 2019, 9, 3891–3898. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Gaut, B.S. Factors that contribute to variation in evolutionary rate among Arabidopsis genes. Mol. Biol. Evol. 2011, 28, 2359–2369. [Google Scholar] [CrossRef] [PubMed]
- Tong, C.; Wang, X.; Yu, J.; Wu, J.; Li, W.; Huang, J.; Dong, C.; Hua, W.; Liu, S. Comprehensive analysis of RNA-seq data reveals the complexity of the transcriptome in Brassica rapa. BMC Genom. 2013, 14, 689. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Ye, W.-Z.; Liu, T.-T.; Cao, J.-S. Characterization of the Male-sterile Line Bcajh97-01A/B and Identification of Candidate Genes for Genic Male Sterility in Chinese Cabbage-pak-choi. J. Am. Soc. Hortic. Sci. 2009, 134, 632–640. [Google Scholar] [CrossRef]
- Lin, S.; Yue, X.; Miao, Y.; Yu, Y.; Dong, H.; Huang, L.; Cao, J. The distinct functions of two classical arabinogalactan proteins BcMF8 and BcMF18 during pollen wall development in Brassica campestris. Plant J. 2018, 94, 60–76. [Google Scholar] [CrossRef]
- Hu, Z.; Shen, X.; Xiang, X.; Cao, J. Evolution of MIR159/319 genes in Brassica campestris and their function in pollen development. Plant Mol. Biol. 2019, 101, 537–550. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
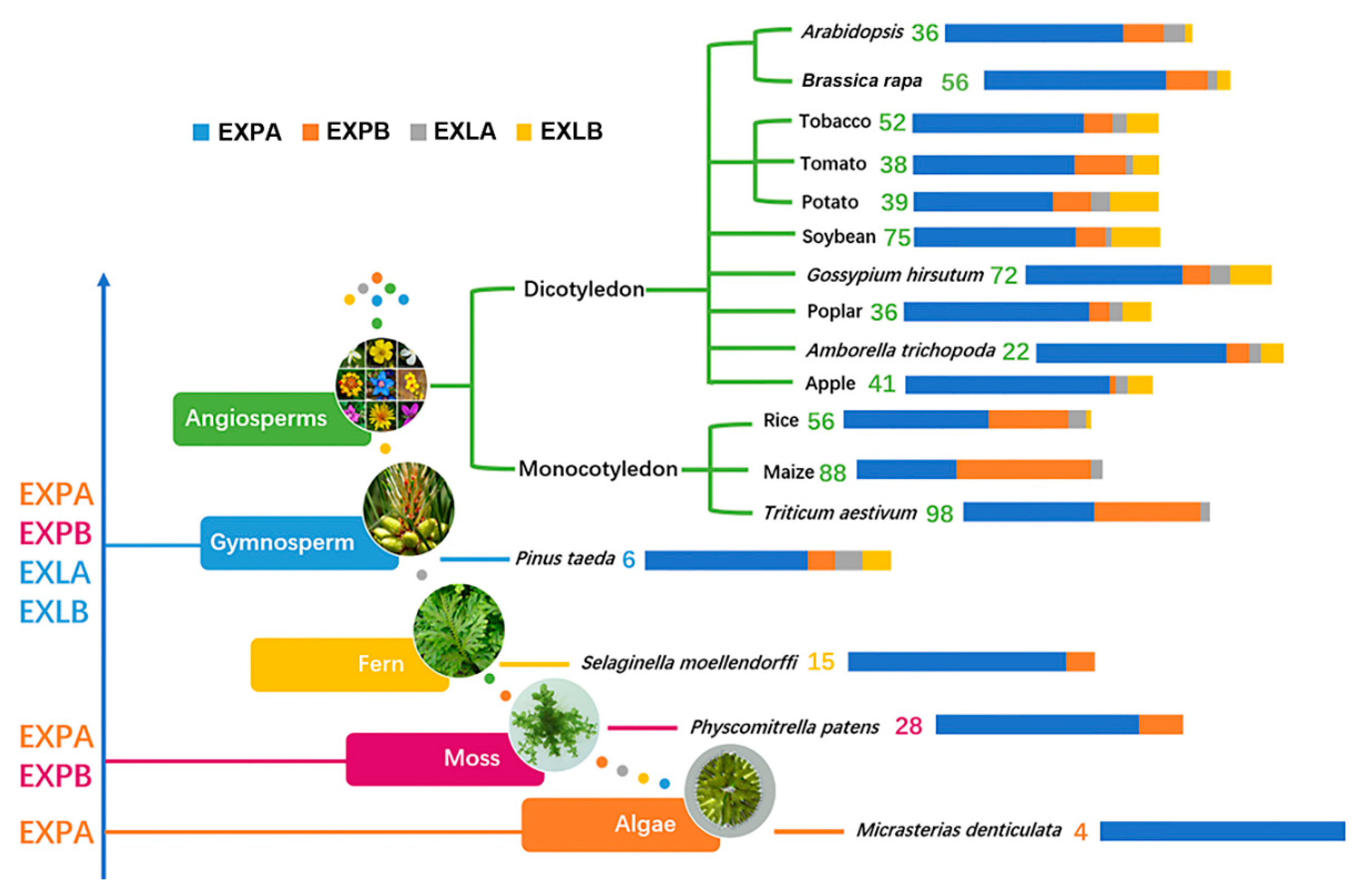
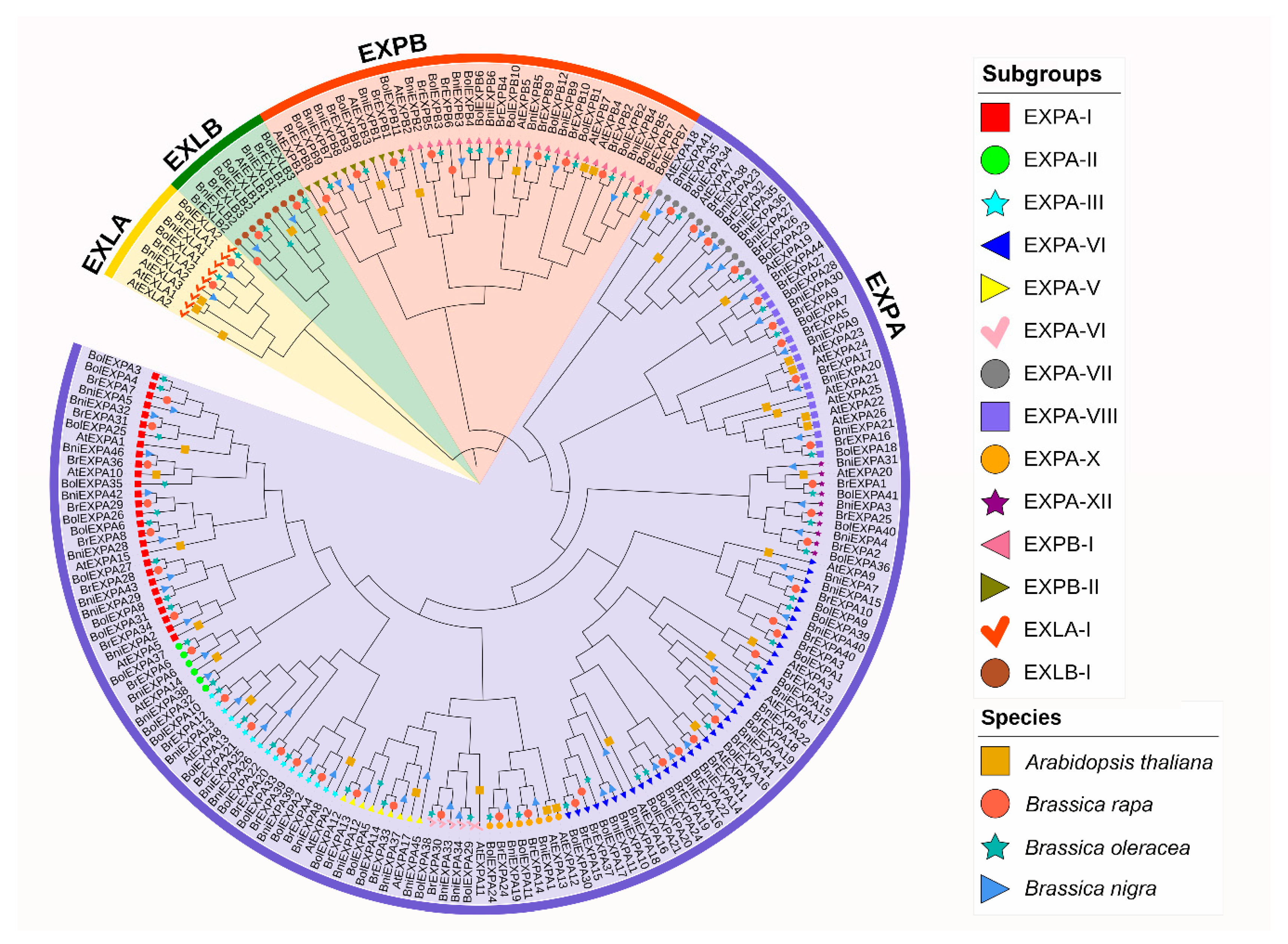
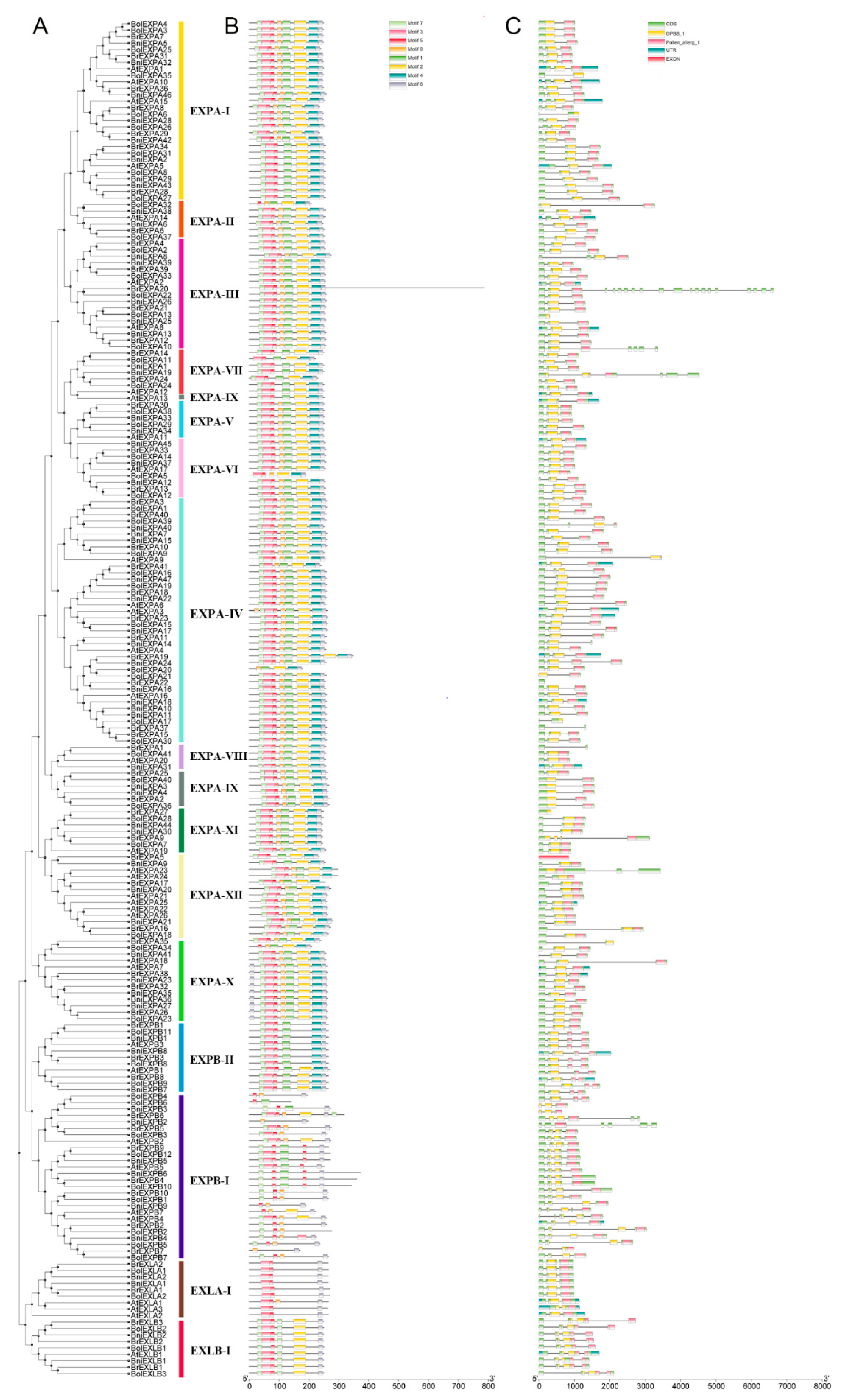
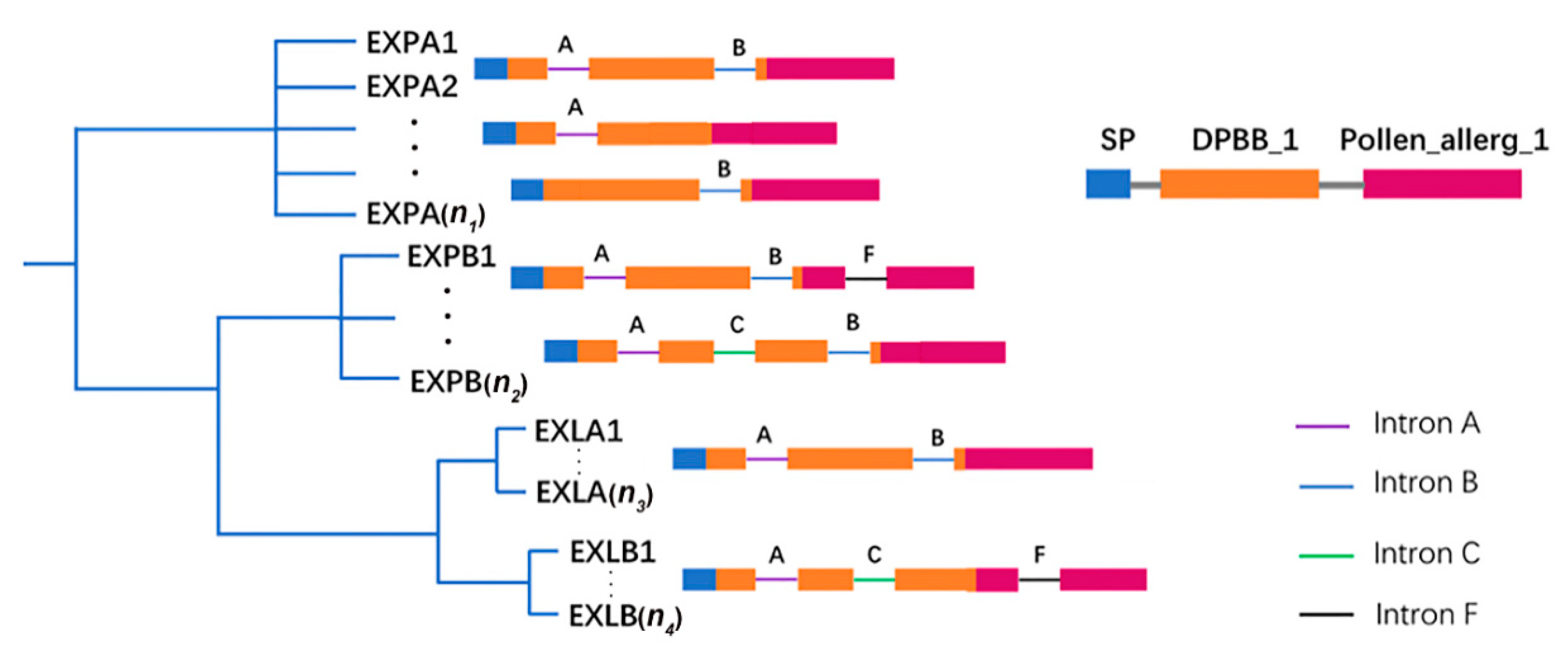



| Species | EXPA | EXPB | EXLA | EXLB | Total | References |
|---|---|---|---|---|---|---|
| Algae | ||||||
| Micrasterias denticulata | 4′EXP’ | [22] | ||||
| Moss | ||||||
| Physcomitrella patens | 28 | 7 | 0 | 0 | 34 | [23] |
| Fern | ||||||
| Selaginella moellendorffi | 15 | 2 | 0 | 0 | 17 | [34] |
| Gymnosperm | ||||||
| Pinus taeda | 6 | 1 | 1 | 1 | 9 | [5] |
| Angiosperms | ||||||
| Monocotyledon | ||||||
| Rice | 33 | 18 | 4 | 1 | 56 | [5] |
| Maize | 36 | 48 | 4 | 0 | 88 | [24] |
| Triticum aestivum | 52 | 42 | 4 | 0 | 98 | [35] |
| Dicotyledon | ||||||
| Cruciferae | ||||||
| Arabidopsis thaliana | 26 | 6 | 3 | 1 | 36 | [5] |
| Brassica rapa | 41 | 10 | 2 | 3 | 56 | |
| Brassica oleracea | 41 | 12 | 2 | 3 | 58 | |
| Brassica nigra | 47 | 9 | 2 | 2 | 60 | |
| Solanaceae | ||||||
| Tobacco | 36 | 6 | 3 | 7 | 52 | [26] |
| Tomato | 25 | 8 | 1 | 4 | 38 | [36] |
| Potato | 24 | 5 | 1 | 6 | 36 | [18] |
| Leguminosae | ||||||
| Soybean | 49 | 9 | 2 | 15 | 75 | [25] |
| Salicaceae | ||||||
| Poplar | 27 | 3 | 2 | 4 | 36 | [37] |
| Amborellaceae | ||||||
| Amborella trichopoda | 17 | 2 | 1 | 2 | 22 | [38] |
| Vitaceae | ||||||
| Grape | 20 | 4 | 1 | 4 | 29 | [39] |
| Rosaceae | ||||||
| Apple | 34 | 1 | 2 | 4 | 41 | [27] |
| Species | Model | Estimates of Parameters | ln L | LRT p-Value |
|---|---|---|---|---|
| Arabidopsis thaliana | One-ratio | ω0 = 0.137 for all subfamilies | −13843.914525 | 0.4417919 |
| Free-ratio | ω1 = 999.000 for EXPA | −13840.170755 | ||
| ω2 = 0.439 for EXPB | ||||
| ω3 = 0.124 for EXLA | ||||
| ω4 = 0.045 for EXLB | ||||
| Brassica rapa | One-ratio | ω0 = 0.118 for all subfamilies | −13413.618539 | 0.2491653 |
| Free-ratio | ω1 = 999.000 for EXPA | −13408.224099 | ||
| ω2 = 999.000 for EXPB | ||||
| ω3 = 0.225 for EXLA | ||||
| ω4 = 0.005 for EXLB | ||||
| Brassica oleracea | One-ratio | ω0 = 0.107 for all subfamilies | −7890.958102 | 0.7815311 |
| Free-ratio | ω1 = 999.000 for EXPA | −7889.207635 | ||
| ω2 = 999.000 for EXPB | ||||
| ω3 = 0.122 for EXLA | ||||
| ω4 = 0.070 for EXLB | ||||
| Brassica nigra | One-ratio | ω0 = 0.125 for all clades | −11781.483673 | 0.9198657 |
| Free-ratio | ω1 = 999.000 for EXPA | −11780.551263 | ||
| ω2 = 999.000 for EXPB | ||||
| ω3 = 0.154 for EXLA |
| EXPA | EXPB | EXLA | EXLB | ||
|---|---|---|---|---|---|
| A. thaliana vs. B. rapa | Ka | 0.085(0.067) a | 0.106 (0.055) | 0.072 (0.006) | 0.059 (0.005) |
| Ks | 0.508(0.114) | 0.589 (0.114) | 0.493 (0.034) | 0.338 (0.077) | |
| Ka/Ks | 0.149(0.090) a | 0.174 (0.070) | 0.146 (0.002) | 0.183 (0.051) | |
| dSM | 0.600(0.286) | 0.747 (0.315) | 0.803 (0.062) | 0.720 (0.076) | |
| A. thaliana vs. B. oleracea | Ka | 0.053(0.026) b | 0.112 (0.055) | 0.076 (0.002) | 0.069 (0.015) |
| Ks | 0.487(0.093) | 0.568 (0.059) | 0.465 (0.073) | 0.329 (0.032) | |
| Ka/Ks | 0.116(0.057) b | 0.202 (0.083) | 0.165 (0.021) | 0.212 (0.058) | |
| dSM | 0.592(0.256) | 0.762 (0.304) | 0.775 (0.101) | 0.694 (0.059) | |
| A. thaliana vs. B. nigra | Ka | 0.048(0.019) b | 0.127 (0.063) | 0.076 (0.013) | 0.061 (0.010) |
| Ks | 0.486(0.100) | 0.548 (0.068) | 0.478 (0.069) | 0.340 (0.039) | |
| Ka/Ks | 0.114(0.059) b | 0.207 (0.083) | 0.160 (0.003) | 0.182 (0.049) | |
| dSM | 0.573(0.285) | 0.692 (0.285) | 0.738 (0.151) | 0.672 (0.100) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, W.; Lyu, T.; Xu, L.; Hu, Z.; Xiong, X.; Liu, T.; Cao, J. Complex Molecular Evolution and Expression of Expansin Gene Families in Three Basic Diploid Species of Brassica. Int. J. Mol. Sci. 2020, 21, 3424. https://doi.org/10.3390/ijms21103424
Liu W, Lyu T, Xu L, Hu Z, Xiong X, Liu T, Cao J. Complex Molecular Evolution and Expression of Expansin Gene Families in Three Basic Diploid Species of Brassica. International Journal of Molecular Sciences. 2020; 21(10):3424. https://doi.org/10.3390/ijms21103424
Chicago/Turabian StyleLiu, Weimiao, Tianqi Lyu, Liai Xu, Ziwei Hu, Xingpeng Xiong, Tingting Liu, and Jiashu Cao. 2020. "Complex Molecular Evolution and Expression of Expansin Gene Families in Three Basic Diploid Species of Brassica" International Journal of Molecular Sciences 21, no. 10: 3424. https://doi.org/10.3390/ijms21103424
APA StyleLiu, W., Lyu, T., Xu, L., Hu, Z., Xiong, X., Liu, T., & Cao, J. (2020). Complex Molecular Evolution and Expression of Expansin Gene Families in Three Basic Diploid Species of Brassica. International Journal of Molecular Sciences, 21(10), 3424. https://doi.org/10.3390/ijms21103424





