The Structure of Sucrose-Soaked Levansucrase Crystals from Erwinia tasmaniensis reveals a Binding Pocket for Levanbiose
Abstract
1. Introduction
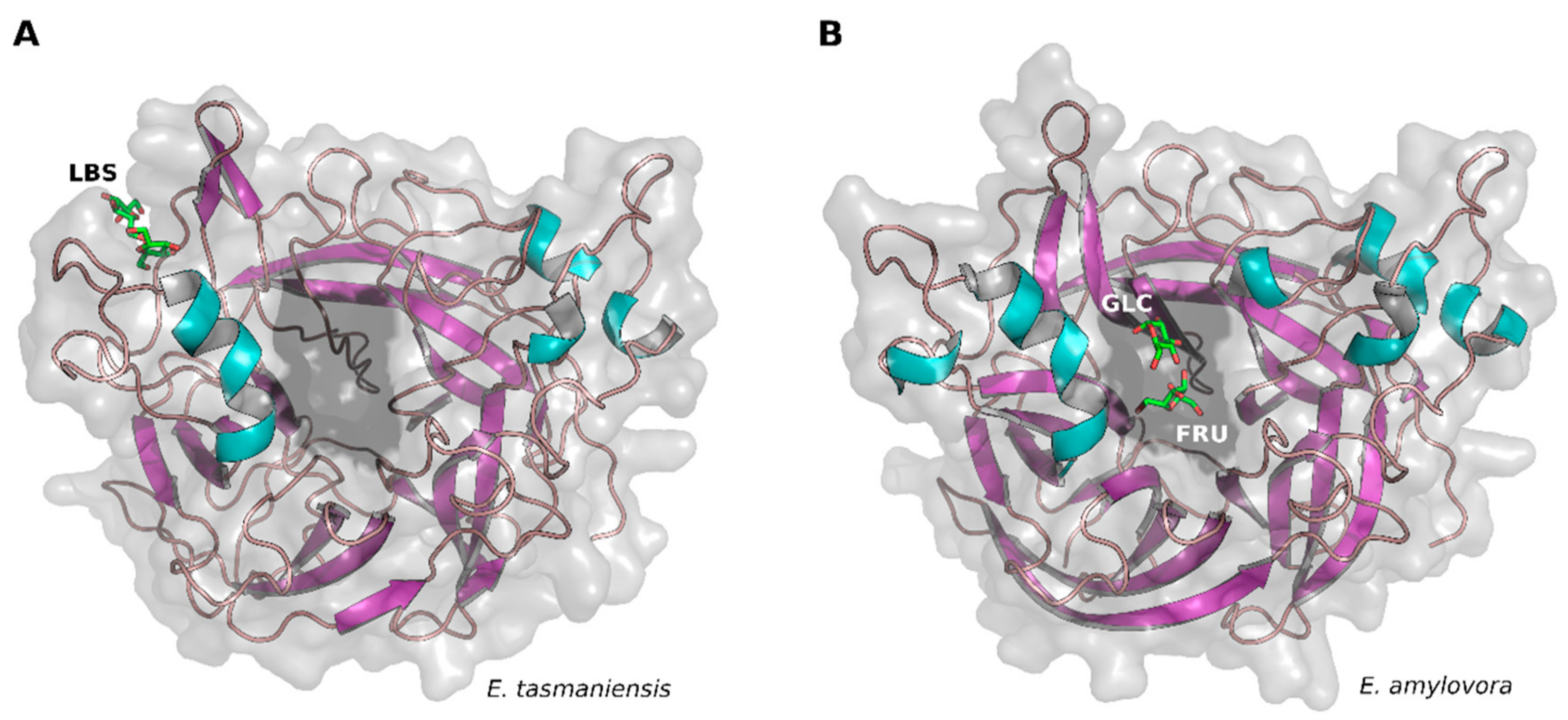
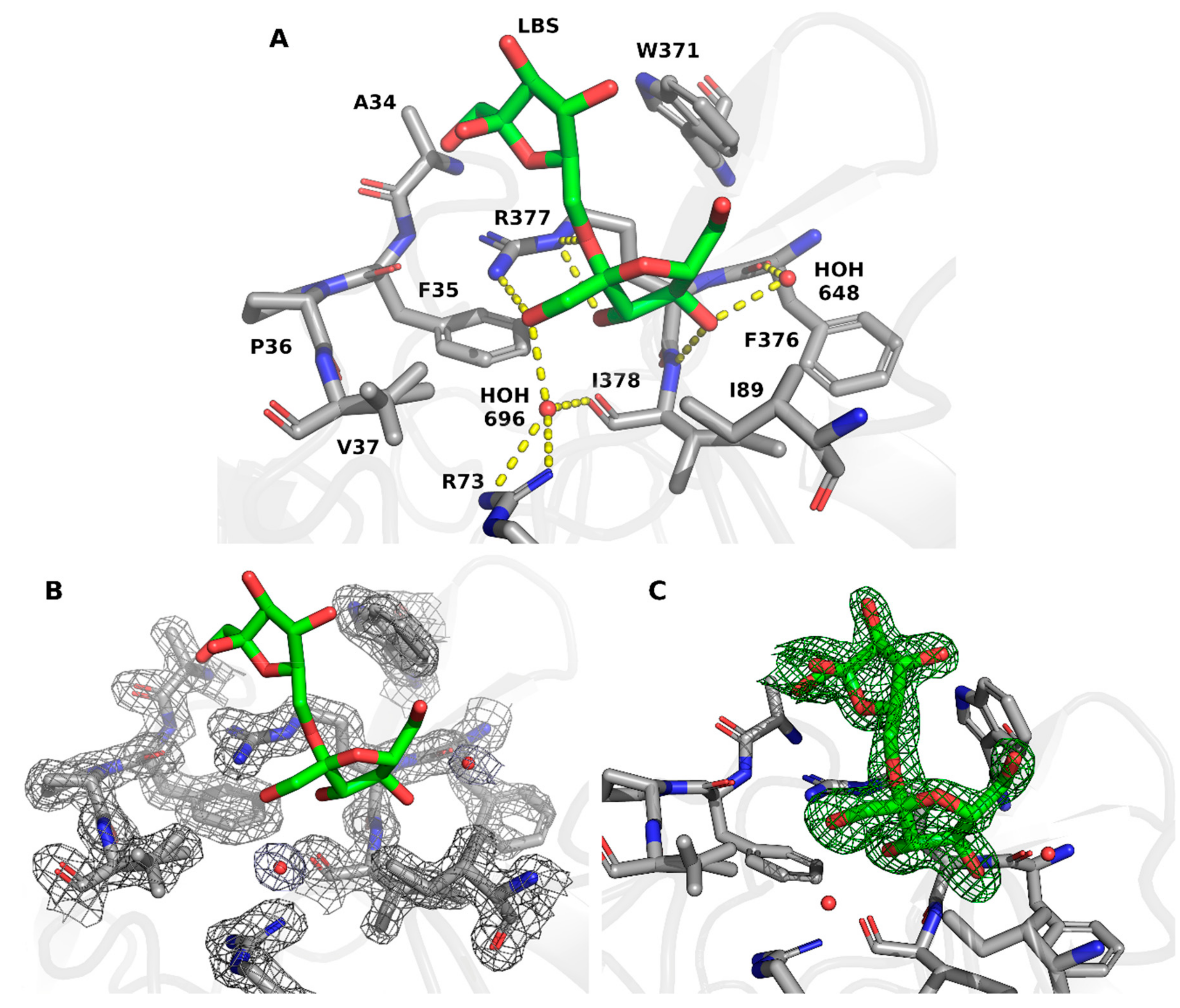
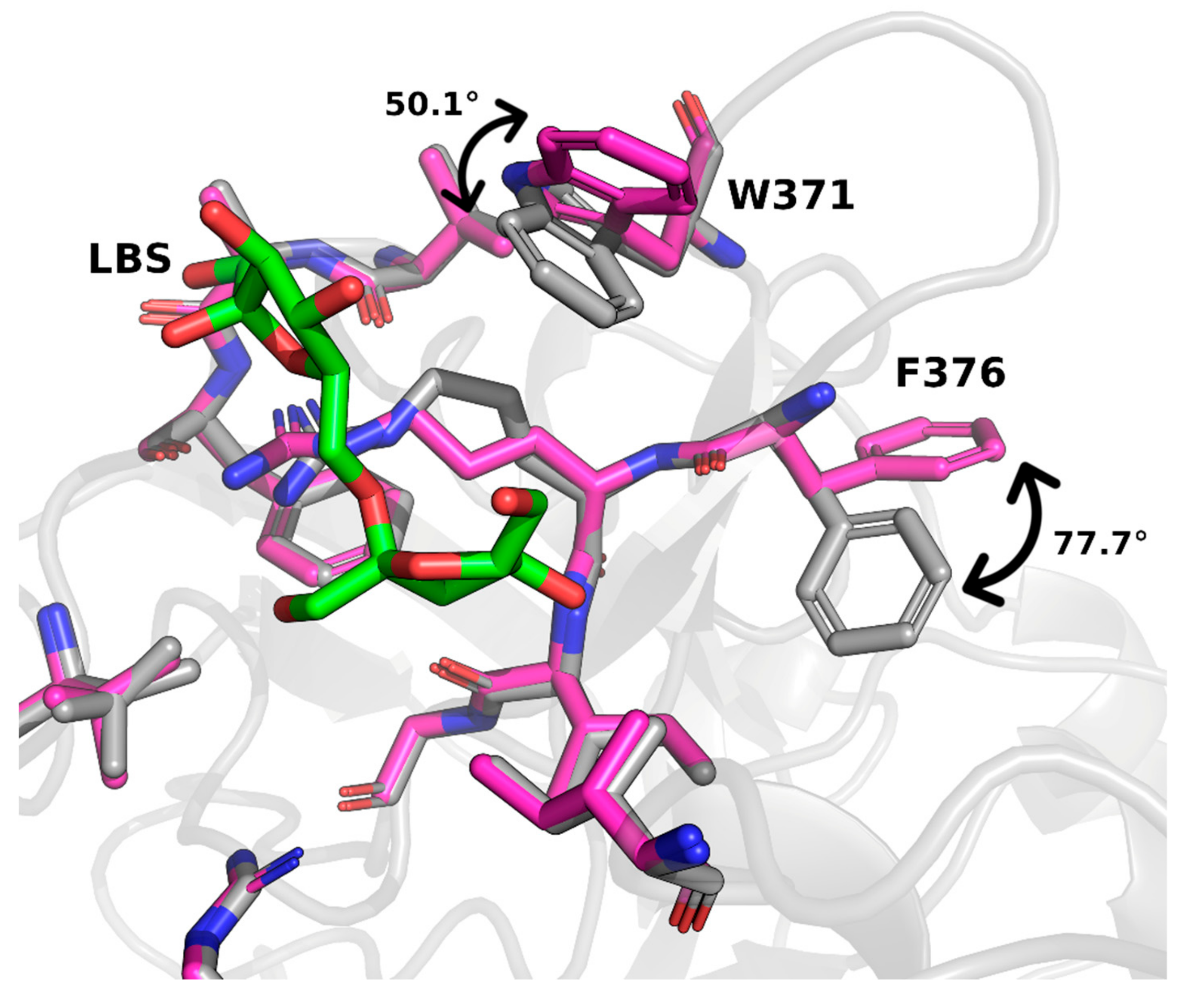
2. Results and Discussion
3. Materials and Methods
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| LSC | Levansucrase |
| FOS | Fructooligosaccharide |
| LBS | Levanbiose |
| INU | Inulosucrase |
| GH68 | Glycosyl hydrolase family 68 |
| HMW | High-molecular weight |
| LMW | Low-molecular weight |
| DP | Degree of polymerization |
References
- Oner, E.T.; Hernandez, L.; Combie, J. Review of Levan Polysaccharide: From a Century of Past Experiences to Future Prospects. Biotechnol. Adv. 2016, 34, 827–844. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Soto, M.E.; Porras-Domínguez, J.R.; Seibel, J.; López-Munguía, A. A Close Look at the Structural Features and Reaction Conditions that Modulate the Synthesis of Low and High Molecular Weight Fructans by Levansucrases. Carbohydr. Polym. 2019, 219, 130–142. [Google Scholar] [CrossRef]
- Combie, J.; Öner, E.T. From Healing Wounds to Resorbable Electronics, Levan can Fill Bioadhesive Roles in Scores of Markets. Bioinspir. Biomim. 2018, 14. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Ni, D.; Zhang, W.; Guang, C.; Zhang, T.; Mu, W. Recent Advances in Levansucrase and Inulosucrase: Evolution, Characteristics, and Application. Crit. Rev. Food Sci. Nutr. 2018, 59, 3630–3647. [Google Scholar] [CrossRef] [PubMed]
- González-Garcinuño, Á.; Tabernero, A.; Domínguez, Á.; Galán, M.A.; Martin del Valle, E.M. Levan and Levansucrases: Polymer, Enzyme, Micro-Organisms and Biomedical Applications. Biocatal. Biotransfor. 2018, 36, 233–244. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes Database (CAZy): An Expert Resource for Glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef]
- Özcan, E.; Öner, E.T. Microbial Production of Extracellular Polysaccharides from Biomass Sources. In Polysaccharides; Kishan, G.R., Jean-michel, M., Eds.; Springer International Publishing: Geneva, Switzerland, 2015; pp. 161–184. [Google Scholar]
- Küçükaşik, F.; Kazak, H.; Güney, D.; Finore, I.; Poli, A.; Yenigün, O.; Nicolaus, B.; Öner, E.T. Molasses as Fermentation Substrate for Levan Production by Halomonas Sp. Appl. Microbiol. Biotechnol. 2011, 89, 1729–1740. [Google Scholar] [CrossRef]
- Li, W.; Yu, S.; Zhang, T.; Jiang, B.; Mu, W. Recent Novel Applications of Levansucrases. Appl. Microbiol. Biotechnol. 2015, 99, 6959–6969. [Google Scholar] [CrossRef]
- Visnapuu, T.; Mardo, K.; Mosoarca, C.; Zamfir, A.D.; Vigants, A.; Alamae, T. Levansucrases from Pseudomonas Syringae Pv. Tomato and P. Chlororaphis Subsp. Aurantiaca: Substrate Specificity, Polymerizing Properties and Usage of Different Acceptors for Fructosylation. J. Biotechnol. 2011, 155, 338–349. [Google Scholar] [CrossRef]
- Wu, C.; Zhang, T.; Mu, W.; Miao, M.; Jiang, B. Biosynthesis of Lactosylfructoside by an Intracellular Levansucrase from Bacillus Methylotrophicus SK 21.002. Carbohydr. Res. 2015, 401, 122–126. [Google Scholar] [CrossRef]
- Seibel, J.; Moraru, R.; Gotze, S.; Buchholz, K.; Na’amnieh, S.; Pawlowski, A.; Hecht, H.J. Synthesis of Sucrose Analogues and the Mechanism of Action of Bacillus Subtilis Fructosyltransferase (Levansucrase). Carbohydr. Res. 2006, 341, 2335–2349. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Fu, F.; Zhao, R.; Jin, L.; He, C.; Xu, L.; Xiao, M. A Recombinant Levansucrase from Bacillus Licheniformis 8-37-0-1 Catalyzes Versatile Transfructosylation Reactions. Process Biochem. 2014, 49, 1503–1510. [Google Scholar] [CrossRef]
- Mu, W.; Chen, Q.; Wang, X.; Zhang, T.; Jiang, B. Current Studies on Physiological Functions and Biological Production of Lactosucrose. Appl. Microbiol. Biotechnol. 2013, 97, 7073–7080. [Google Scholar] [CrossRef] [PubMed]
- Mena-Arizmendi, A.; Alderete, J.; Águila, S.; Marty, A.; Miranda-Molina, A.; López-Munguía, A.; Castillo, E. Enzymatic Fructosylation of Aromatic and Aliphatic Alcohols by Bacillus Subtilis Levansucrase: Reactivity of Acceptors. J. Mol. Catal. B Enzym. 2011, 70, 41–48. [Google Scholar] [CrossRef]
- Núñez-López, G.; Herrera-González, A.; Hernández, L.; Amaya-Delgado, L.; Sandoval, G.; Gschaedler, A.; Arrizon, J.; Remaud-Simeon, M.; Morel, S. Fructosylation of Phenolic Compounds by Levansucrase from Gluconacetobacter Diazotrophicus. Enzym. Microb. Technol. 2019, 122, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Possiel, C.; Ortiz-Soto, M.; Ertl, J.; Münch, A.; Vogel, A.; Schmiedel, R.; Seibel, J. Exploring the Sequence Variability of Polymerization-Involved Residues in the Production of Levan- and Inulin-Type Fructooligosaccharides with a Levansucrase. Sci. Rep. 2019, 9, 7720. [Google Scholar] [CrossRef]
- Ortiz-Soto, M.E.; Possiel, C.; Görl, J.; Vogel, A.; Schmiedel, R.; Seibel, J. Impaired Coordination of Nucleophile and Increased Hydrophobicity in the 1 Subsite Shift Levansucrase Activity Towards Transfructosylation. Glycobiology 2017, 27, 755–765. [Google Scholar] [CrossRef]
- Mardo, K.; Visnapuu, T.; Gromkova, M.; Aasamets, A.; Viigand, K.; Vija, H.; Alamae, T. High-Throughput Assay of Levansucrase Variants in Search of Feasible Catalysts for the Synthesis of Fructooligosaccharides and Levan. Molecules 2014, 19, 8434–8455. [Google Scholar] [CrossRef]
- Ortiz-Soto, M.E.; Rivera, M.; Rudiño-Piñera, E.; Olvera, C.; López-Munguía, A. Selected Mutations in Bacillus Subtilis Levansucrase Semi-Conserved Regions Affecting its Biochemical Properties. Protein Eng. Des. Sel. 2008, 21, 589–595. [Google Scholar] [CrossRef]
- Polsinelli, I.; Caliandro, R.; Salomone-Stagni, M.; Demitri, N.; Rejzek, M.; Field, R.A.; Benini, S. Comparison of the Levansucrase from the Epiphyte Erwinia Tasmaniensis Vs its Homologue from the Phytopathogen Erwinia Amylovora. Int. J. Biol. Macromol. 2019, 127, 496–501. [Google Scholar] [CrossRef]
- Caputi, L.; Nepogodiev, S.A.; Malnoy, M.; Rejzek, M.; Field, R.A.; Benini, S. Biomolecular Characterization of the Levansucrase of Erwinia Amylovora, a Promising Biocatalyst for the Synthesis of Fructooligosaccharides. J. Agric. Food Chem. 2013, 61, 12265–12273. [Google Scholar] [CrossRef] [PubMed]
- Caputi, L.; Cianci, M.; Benini, S. Cloning, Expression, Purification, Crystallization and Preliminary X-Ray Analysis of EaLSC, a Levansucrase from Erwinia Amylovora. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2013, 69, 570–573. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Fleites, C.; Ortiz-Lombardia, M.; Pons, T.; Tarbouriech, N.; Taylor, E.J.; Arrieta, J.G.; Hernandez, L.; Davies, G.J. Crystal Structure of Levansucrase from the Gram-Negative Bacterium Gluconacetobacter Diazotrophicus. Biochem. J. 2005, 390, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Wuerges, J.; Caputi, L.; Cianci, M.; Boivin, S.; Meijers, R.; Benini, S. The Crystal Structure of Erwinia Amylovora Levansucrase Provides a Snapshot of the Products of Sucrose Hydrolysis Trapped into the Active Site. J. Struct. Biol. 2015, 191, 290–298. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, L.; Arrieta, J.; Menendez, C.; Vazquez, R.; Coego, A.; Suarez, V.; Selman, G.; Petit-Glatron, M.F.; Chambert, R. Isolation and Enzymic Properties of Levansucrase Secreted by Acetobacter Diazotrophicus SRT4, a Bacterium Associated with Sugar Cane. Biochem. J. 1995, 309 (Pt 1), 113–118. [Google Scholar] [CrossRef]
- Homann, A.; Biedendieck, R.; Goetze, S.; Jahn, D.; Seibel, J. Insights into Polymer Versus Oligosaccharide Synthesis: Mutagenesis and Mechanistic Studies of a Novel Levansucrase from Bacillus Megaterium. Biochem. J. 2007, 407, 189–198. [Google Scholar] [CrossRef]
- Raga-Carbajal, E.; López-Munguía, A.; Alvarez, L.; Olvera, C. Understanding the Transfer Reaction Network Behind the Non-Processive Synthesis of Low Molecular Weight Levan Catalyzed by Bacillus Subtilis Levansucrase. Sci. Rep. 2018, 8, 15035. [Google Scholar] [CrossRef]
- Chambert, R.; Petit-Glatron, M.F. Polymerase and Hydrolase Activities of Bacillus Subtilis Levansucrase can be Separately Modulated by Site-Directed Mutagenesis. Biochem. J. 1991, 279, 35–41. [Google Scholar] [CrossRef]
- Meng, G.; Fütterer, K. Structural Framework of Fructosyl Transfer in Bacillus Subtilis Levansucrase. Nat. Struct. Biol. 2003, 10, 935–941. [Google Scholar] [CrossRef]
- Strube, C.P.; Homann, A.; Gamer, M.; Jahn, D.; Seibel, J.; Heinz, D.W. Polysaccharide Synthesis of the Levansucrase SacB from Bacillus Megaterium is Controlled by Distinct Surface Motifs. J. Biol. Chem. 2011, 286, 17593–17600. [Google Scholar] [CrossRef]
- Eschenfeldt, W.H.; Lucy, S.; Millard, C.S.; Joachimiak, A.; Mark, I.D. A Family of LIC Vectors for High-Throughput Cloning and Purification of Proteins. Methods Mol. Biol. 2009, 498, 105–115. [Google Scholar] [PubMed]
- Lausi, A.; Polentarutti, M.; Onesti, S.; Plaisier, J.R.; Busetto, E.; Bais, G.; Barba, L.; Cassetta, A.; Campi, G.; Lamba, D.; et al. Status of the Crystallography Beamlines at Elettra. Eur. Phys. J. Plus 2015, 130, 43. [Google Scholar] [CrossRef]
- Kabsch, W. XDS Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Vagin, A.; Teplyakov, A. Molecular Replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 22–25. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and Development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Murshudov, G.N.; Skubak, P.; Lebedev, A.A.; Pannu, N.S.; Steiner, R.A.; Nicholls, R.A.; Winn, M.D.; Long, F.; Vagin, A.A. REFMAC5 for the Refinement of Macromolecular Crystal Structures. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 355–367. [Google Scholar] [CrossRef]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A Comprehensive Python-Based System for Macromolecular Structure Solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B. MolProbity: More and Better Reference Data for Improved All-atom Structure Validation. Protein Sci. 2018, 27, 293–315. [Google Scholar] [CrossRef]
- Liebschner, D.; Afonine, P.V.; Moriarty, N.W.; Poon, B.K.; Sobolev, O.V.; Terwilliger, T.C.; Adams, P.D. Polder maps: Improving OMIT maps by excluding bulk solvent. Acta Crystallogr. D Struct. Biol. 2017, 73, 148–157. [Google Scholar] [CrossRef]

| 6RV5 Erwinia tasmaniensis levansucrase | |
|---|---|
| Wavelength (Å) | 1.000 |
| Temperature (K) | 100 |
| Resolution range (Å) | 45.21–1.58 (1.64–1.58) |
| Space group | P 41212 |
| a, b, c (Å) and α, β, γ (°) | 127.886, 127.886, 58.268; 90, 90, 90 |
| Total reflections | 552,442 (86,048) |
| Unique reflections | 162,091 (25,949) |
| Multiplicity | 3.4 (3.3) |
| Completeness (%) | 99.47 (99.95) |
| Mean I/sigma(I) | 20.50 (2.91) |
| Wilson B-factor (Å2) | 18.23 |
| R-merge | 0.02668 (0.2867) |
| R-meas | 0.03773 (0.4055) |
| R-pim | 0.02668 (0.2867) |
| CC1/2 | 0.999 (0.888) |
| Reflections used in refinement | 66,162 (6515) |
| Reflections used for R-free | 3263 (343) |
| R-work | 0.1456 (0.2640) |
| R-free | 0.1929 (0.2902) |
| CC (work) | 0.966 (0.827) |
| CC (free) | 0.951 (0.716) |
| Ligands atoms | 97 |
| Solvent molecules | 431 |
| Protein residues | 412 |
| RMS (bonds) (Å) | 0.017 |
| RMS (angles) (°) | 2.02 |
| Ramachandran favored (%) | 96.34 |
| Ramachandran allowed (%) | 3.41 |
| Ramachandran outliers (%) | 0.24 |
| Rotamer outliers (%) | 1.87 |
| Clashscore | 5.97 |
| Average B-factor (Å2) | 26.33 |
| Macromolecules | 23.87 |
| Ligands | 42.23 |
| Solvent | 41.99 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Polsinelli, I.; Caliandro, R.; Demitri, N.; Benini, S. The Structure of Sucrose-Soaked Levansucrase Crystals from Erwinia tasmaniensis reveals a Binding Pocket for Levanbiose. Int. J. Mol. Sci. 2020, 21, 83. https://doi.org/10.3390/ijms21010083
Polsinelli I, Caliandro R, Demitri N, Benini S. The Structure of Sucrose-Soaked Levansucrase Crystals from Erwinia tasmaniensis reveals a Binding Pocket for Levanbiose. International Journal of Molecular Sciences. 2020; 21(1):83. https://doi.org/10.3390/ijms21010083
Chicago/Turabian StylePolsinelli, Ivan, Rosanna Caliandro, Nicola Demitri, and Stefano Benini. 2020. "The Structure of Sucrose-Soaked Levansucrase Crystals from Erwinia tasmaniensis reveals a Binding Pocket for Levanbiose" International Journal of Molecular Sciences 21, no. 1: 83. https://doi.org/10.3390/ijms21010083
APA StylePolsinelli, I., Caliandro, R., Demitri, N., & Benini, S. (2020). The Structure of Sucrose-Soaked Levansucrase Crystals from Erwinia tasmaniensis reveals a Binding Pocket for Levanbiose. International Journal of Molecular Sciences, 21(1), 83. https://doi.org/10.3390/ijms21010083





