Integrative Transcriptomic and Proteomic Analyses of Molecular Mechanism Responding to Salt Stress during Seed Germination in Hulless Barley
Abstract
1. Introduction
2. Results
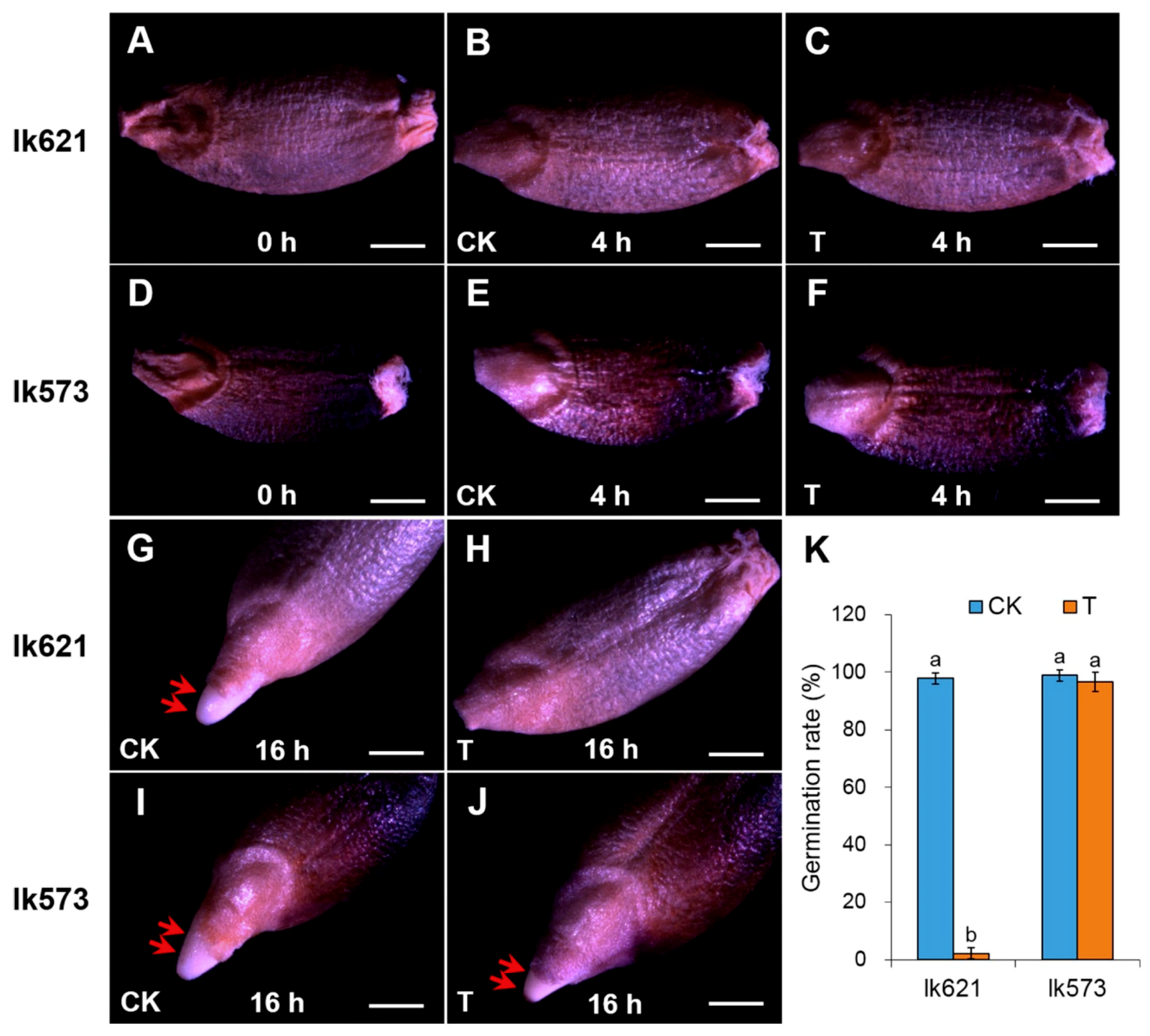
2.1. Differences of Seed Germination between Two Landraces under Salt Stress
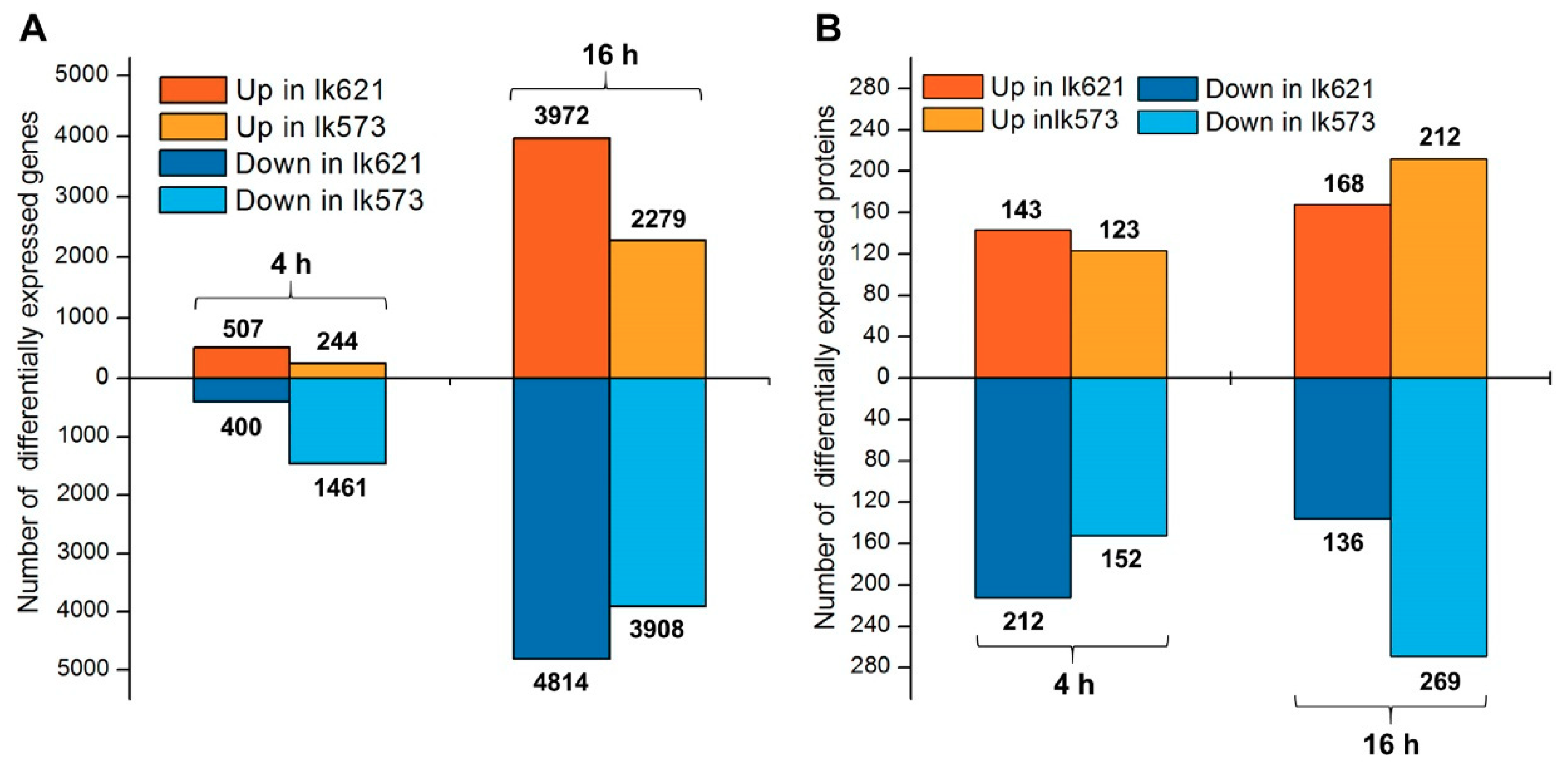
2.2. Overview of Transcriptomic and Quantitative Proteomic Analyses
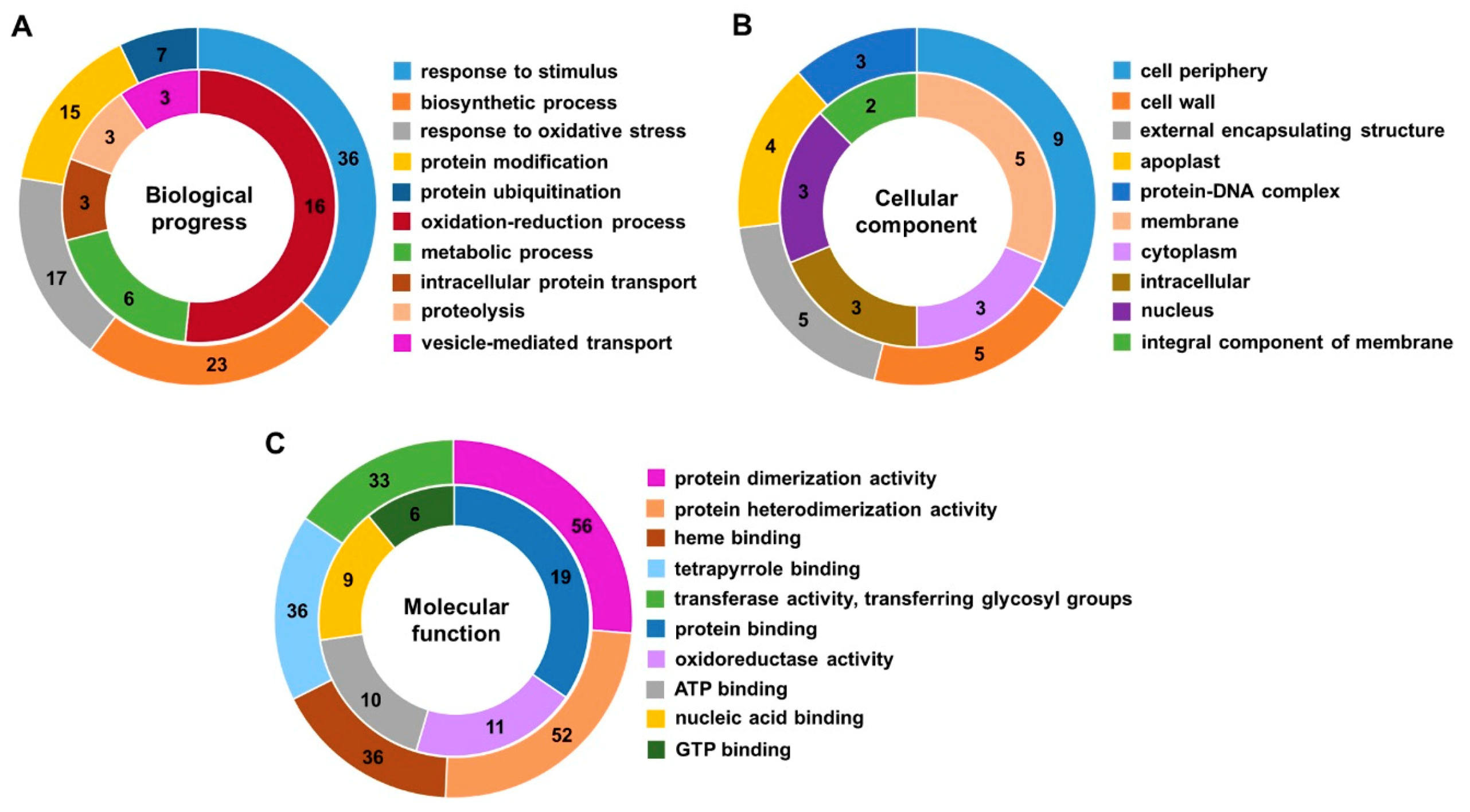
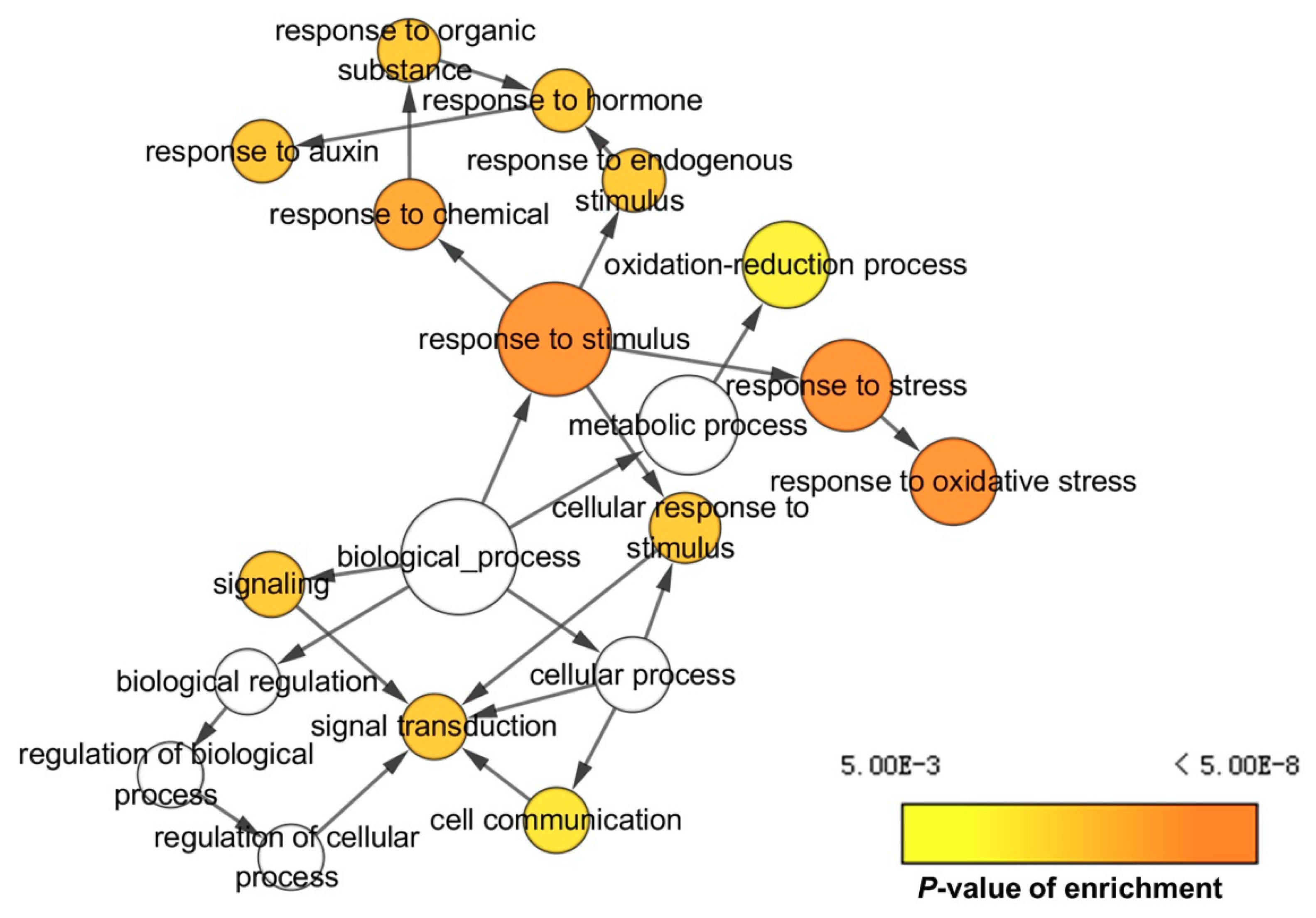
2.3. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Analysis of DEGs and DEPs
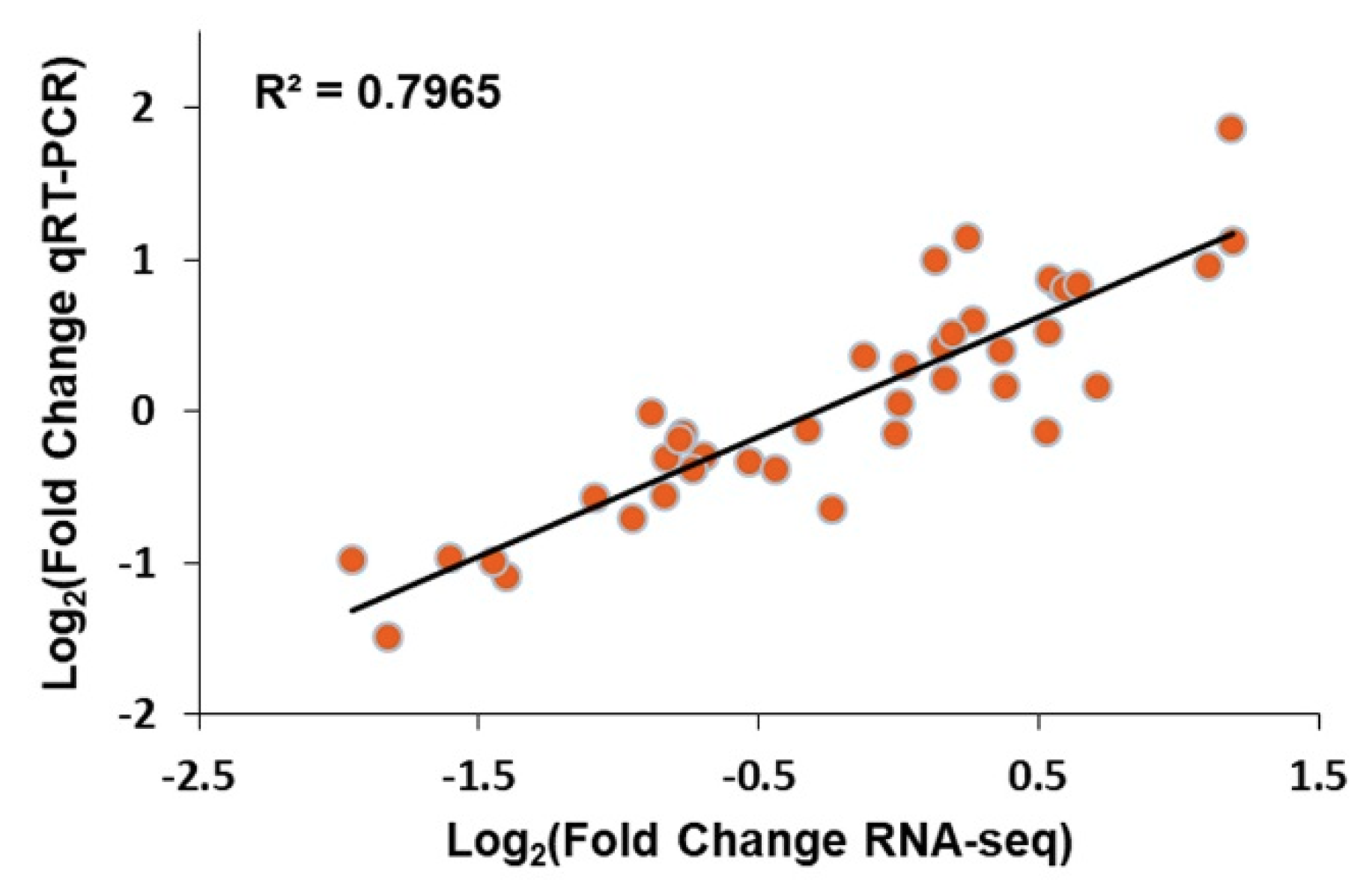
2.4. Correlation between Transcripts and Proteins
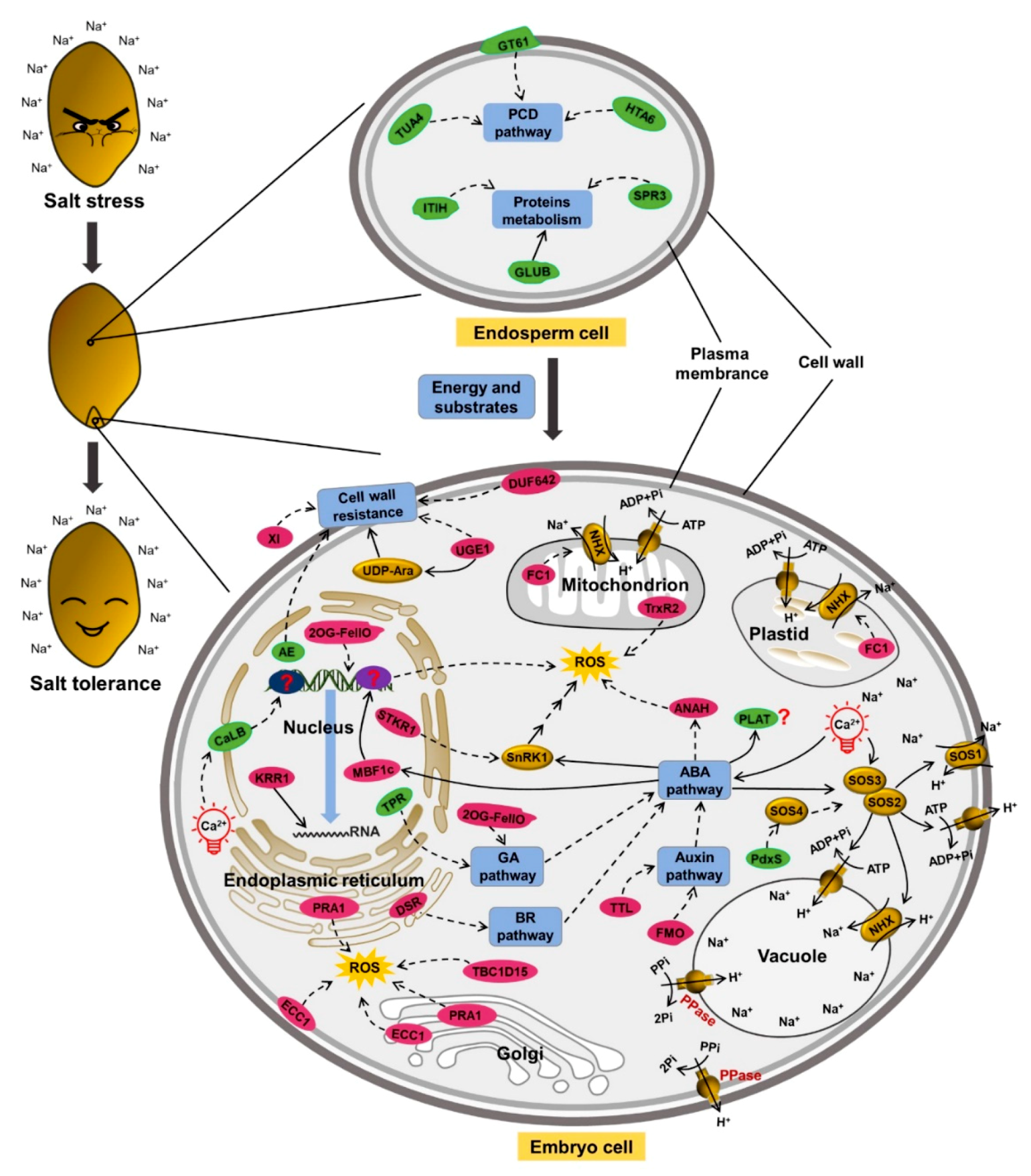
2.5. Genes Related to Salt Tolerance
3. Discussion
3.1. Energy and Substrates Supplied by Endosperm under Salt Stress
3.2. Cell Wall in Salt Tolerance
3.3. Ion Transport for Salt Tolerance
3.4. Ca-Dependent Regulation under Salt Stress
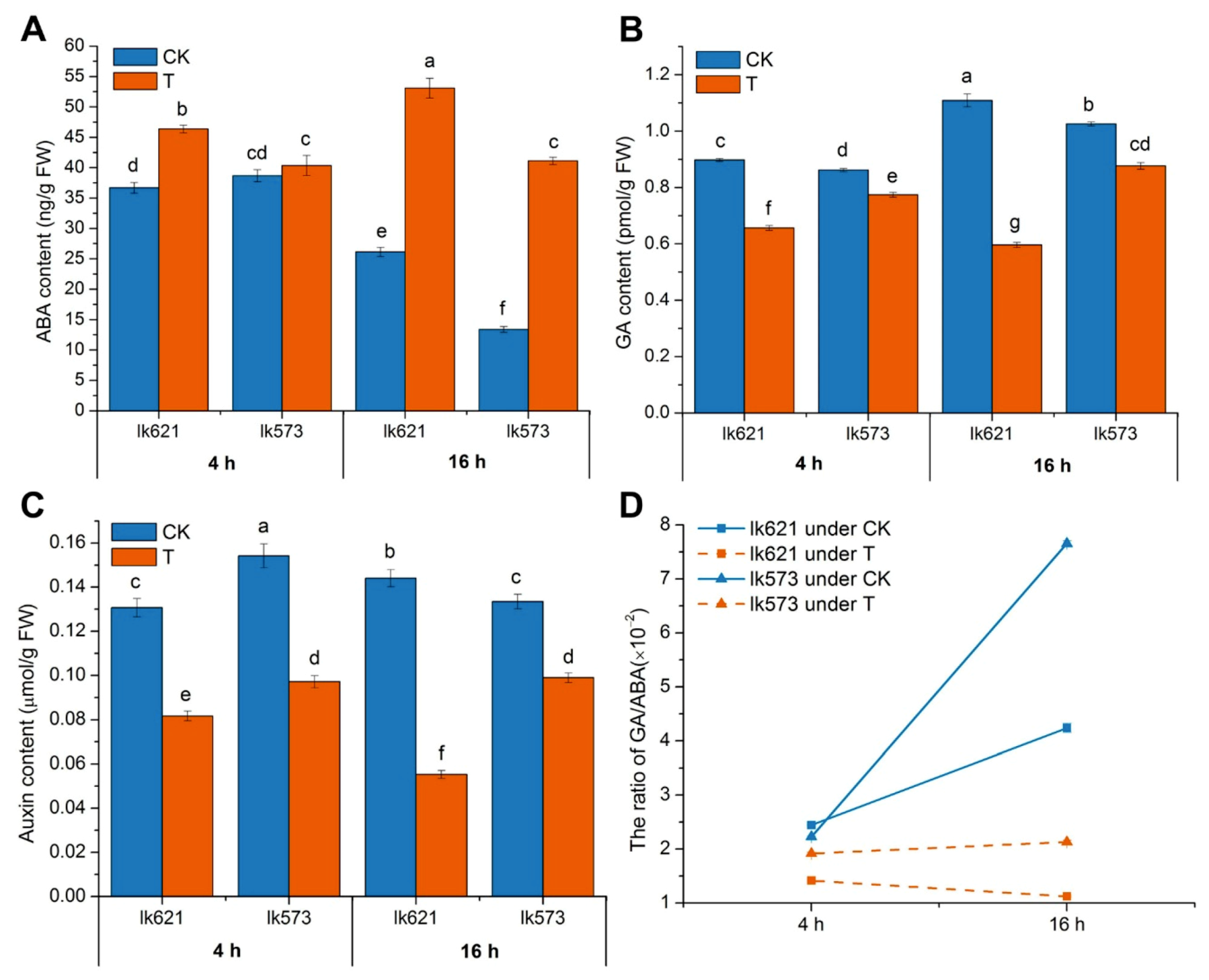
3.5. Phytohormones in Salt Stress
3.6. ROS Reduction for Salt Resistance
3.7. Vesicular Trafficking in Salt Tolerance
4. Materials and Methods
4.1. Seed Germination under Salt Stress
4.2. Plant Hormone Detection
4.3. Transcriptome Sequencing Analysis
4.4. Proteome Analysis
4.5. Bioinformatics Analysis
4.6. Correlation between Transcript and Protein
4.7. Quantitative RT-PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Mascher, M.; Gundlach, H.; Himmelbach, A.; Beier, S.; Twardziok, S.O.; Wicker, T.; Radchuk, V.; Dockter, C.; Hedley, P.E.; Rusell, J.; et al. A chromosome conformation capture ordered sequence of the barley genome. Nature 2017, 544, 427–433. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Long, H.; Wang, Z.; Zhao, S.; Tang, Y.; Huang, Z.; Wang, Y.; Xu, Q.; Mao, L.; Deng, G.; et al. The draft genome of Tibetan hulless barley reveals adaptive patterns to the high stressful Tibetan Plateau. Proc. Natl. Acad. Sci. USA 2015, 112, 1095–1100. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Davies, N.W.; Shabala, L.; Zhou, M.; Brodribb, T.J.; Shabala, S. Residual transpiration as a component of salinity stress tolerance mechanism: A case study for barley. BMC Plant Boil. 2017, 107. [Google Scholar] [CrossRef] [PubMed]
- Hazzouri, K.M.; Khraiwesh, B.; Amiri, K.; Pauli, D.; Blake, T.; Shahid, M.; Mullath, S.K.; Nelson, D.; Mansour, A.L.; Salehi-Ashtiani, K.; et al. Mapping of HKT1; 5 gene in barley using GWAS approach and its implication in salt tolerance mechanism. Front. Plant Sci. 2018, 9, 156. [Google Scholar] [CrossRef] [PubMed]
- Witzel, K.; Matros, A.; Møller, A.L.; Ramireddy, E.; Finnie, C.; Peukert, M.; Rutten, T.; Herzog, A.; Kunze, G.; Melzer, M.; et al. Plasma membrane proteome analysis identifies a role of barley membrane steroid binding protein in root architecture response to salinity. Plant Cell Environ. 2018, 41, 1311–1330. [Google Scholar] [CrossRef]
- Zhang, H.; Irving, L.J.; McGill, C.; Matthew, C.; Zhou, D.; Kemp, P. The effects of salinity and osmotic stress on barley germination rate: Sodium as an osmotic regulator. Ann. Bot. 2010, 106, 1027–1035. [Google Scholar] [CrossRef]
- Witzel, K.; Weidner, A.; Surabhi, G.K.; Varshney, R.K.; Kunze, G.; Buck-sorlin, G.H.; Börner, A.; Mock, H.P. Comparative analysis of the grain proteome fraction in barley genotypes with contrasting salinity tolerance during germination. Plant Cell Environ. 2010, 33, 211–222. [Google Scholar] [CrossRef]
- Adem, G.D.; Roy, S.J.; Zhou, M.; Bowman, J.P.; Shabala, S. Evaluating contribution of ionic, osmotic and oxidative stress components towards salinity tolerance in barley. BMC Plant Biol. 2014, 14, 113. [Google Scholar] [CrossRef]
- Ismail, A.M.; Horie, T. Genomics, physiology, and molecular breeding approaches for improving salt tolerance. Annu. Rev. Plant Biol. 2017, 68, 405–434. [Google Scholar] [CrossRef]
- Foolad, M.R.; Lin, G.Y. Absence of a genetic relationship between salt tolerance during seed germination and vegetative growth in tomato. Plant Breed. 1997, 116, 363–367. [Google Scholar] [CrossRef]
- Rajjou, L.; Duval, M.; Gallardo, K.; Catusse, J.; Bally, J.; Job, C.; Job, D. Seed germination and vigor. Annu. Rev. Plant Biol. 2012, 63, 507–533. [Google Scholar] [CrossRef] [PubMed]
- Shu, K.; Liu, X.D.; Xie, Q.; He, Z.H. Two faces of one seed: Hormonal regulation of dormancy and germination. Mol. Plant 2016, 9, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Miransari, M.; Smith, D.L. Plant hormones and seed germination. Environ. Exp. Bot. 2014, 99, 110–121. [Google Scholar] [CrossRef]
- Kong, D.; Ju, C.; Parihar, A.; Kim, S.; Cho, D.; Kwak, J.M. Arabidopsis glutamate receptor homolog3.5 modulates cytosolic Ca2+ level to counteract effect of abscisic acid in seed germination. Plant Physiol. 2015, 167, 1630–1642. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, E.A. Seed priming to alleviate salinity stress in germinating seeds. J. Plant Physiol. 2016, 192, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018, 217, 523–539. [Google Scholar] [CrossRef]
- He, Y.; Yang, B.; He, Y.; Zhan, C.; Cheng, Y.; Zhang, J.; Zhang, H.; Cheng, J.; Wang, Z. A quantitative trait locus, qSE 3, promotes seed germination and seedling establishment under salinity stress in rice. Plant J. 2019, 97, 1089–1104. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, C.; Xue, Y.; Liu, X.; Chen, S.; Song, C.; Yang, Y.; Guo, Y. Calcium-activated 14-3-3 proteins as a molecular switch in salt stress tolerance. Nat. Commun. 2019, 10, 1199. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Zhang, H.J.; Sun, Q.Q.; Cao, Y.Y.; Li, X.; Zhao, B.; Wu, P.; Guo, Y. Proteomic analysis reveals a role of melatonin in promoting cucumber seed germination under high salinity by regulating energy production. Sci. Rep. 2017, 7, 503. [Google Scholar] [CrossRef] [PubMed]
- Tenhaken, R. Cell wall remodeling under abiotic stress. Front. Plant Sci. 2015, 5, 771. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Li, Y.; Hua, J. The C2 domain protein BAP1 negatively regulates defense responses in Arabidopsis. Plant J. 2006, 48, 238–248. [Google Scholar] [CrossRef] [PubMed]
- de Silva, K.; Laska, B.; Brown, C.; Sederoff, H.W.; Khodakovskaya, M. Arabidopsis thaliana calcium-dependent lipid-binding protein (AtCLB): A novel repressor of abiotic stress response. J. Exp. Bot. 2011, 62, 2679–2689. [Google Scholar] [CrossRef]
- Colebrook, E.H.; Thomas, S.G.; Phillips, A.L.; Hedden, P. The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Boil. 2014, 217, 67–75. [Google Scholar] [CrossRef]
- Krishna, P. Brassinosteroid-mediated stress responses. J. Plant Growth Regul. 2003, 22, 289–297. [Google Scholar] [CrossRef]
- Zhang, X.M.; Walsh, B.; Mitchell, C.A.; Rowe, T. TBC domain family, member 15 is a novel mammalian Rab GTPase-activating protein with substrate preference for Rab7. Biochem. Biophys. Res. Commun. 2005, 335, 154–161. [Google Scholar] [CrossRef]
- Mazel, A.; Leshem, Y.; Tiwari, B.S.; Levine, A. Induction of salt and osmotic stress tolerance by overexpression of an intracellular vesicle trafficking protein AtRab7 (AtRabG3e). Plant Physiol. 2004, 134, 118–128. [Google Scholar] [CrossRef]
- Zhang, Y.; Immink, R.; Liu, C.M.; Emons, A.M.; Ketelaar, T. The Arabidopsis exocyst subunit SEC3A is essential for embryo development and accumulates in transient puncta at the plasma membrane. New Phytol. 2013, 199, 74–88. [Google Scholar] [CrossRef]
- Geng, G.; Lv, C.; Stevanato, P.; Li, R.; Liu, H.; Yu, L.; Wang, Y. Transcriptome analysis of salt-sensitive and tolerant genotypes reveals salt-tolerance metabolic pathways in Sugar Beet. Int. J. Mol. Sci. 2019, 20, 5910. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Jin, F.; Shan, X.; Li, Y. iTRAQ-based proteomic analysis reveals several strategies to cope with drought stress in maize seedlings. Int. J. Mol. Sci. 2019, 20, 5956. [Google Scholar] [CrossRef] [PubMed]
- Xu, E.; Chen, M.; He, H.; Zhan, C.; Cheng, Y.; Zhang, H.; Wang, Z. Proteomic analysis reveals proteins involved in seed imbibition under salt stress in rice. Front. Plant Sci. 2017, 7, 2006. [Google Scholar] [CrossRef] [PubMed]
- Dong, T.; Zhu, M.; Yu, J.; Han, R.; Tang, C.; Li, Z. RNA-Seq and iTRAQ reveal multiple pathways involved in storage root formation and development in sweet potato (Ipomoea batatas L.). BMC Plant Boil. 2019, 19, 136. [Google Scholar] [CrossRef]
- Lan, P.; Li, W.; Schmidt, W. Complementary proteome and transcriptome profiling in phosphate-deficient Arabidopsis roots reveals multiple levels of gene regulation. Mol. Cell. Proteom. 2012, 1, 1156–1166. [Google Scholar] [CrossRef]
- Li, J.M.; San Huang, X.; Li, L.T.; Zheng, D.M.; Xue, C.; Wu, J. Proteome analysis of pear reveals key genes associated with fruit development and quality. Planta 2015, 241, 1363–1379. [Google Scholar] [CrossRef]
- Phan, J.L.; Tucker, M.R.; Khor, S.F.; Shirley, N.; Lahnstein, J.; Beahan, C.; Bacic, A.; Burton, R.A. Differences in glycosyltransferase family 61 accompany variation in seed coat mucilage composition in Plantago spp. J. Exp. Bot. 2016, 67, 6481–6495. [Google Scholar] [CrossRef]
- Kopczak, S.D.; Haas, N.A.; Hussey, P.J.; Silflow, C.D.; Snustad, D.P. The small genome of Arabidopsis contains at least six expressed alpha-tubulin genes. Plant Cell 1992, 4, 539–547. [Google Scholar] [CrossRef]
- Zhuo, L.; Kimata, K. Structure and function of inter-α-trypsin inhibitor heavy chains. Connect. Tissue Res. 2008, 49, 311–320. [Google Scholar] [CrossRef]
- Fluhr, R.; Lampl, N.; Roberts, T.H. Serpin protease inhibitors in plant biology. Physiol. Plantarum 2012, 145, 95–102. [Google Scholar] [CrossRef]
- Kotake, T.; Takata, R.; Verma, R.; Takaba, M.; Yamaguchi, D.; Orita, T.; Kaneko, S.; Matsuoka, K.; Koyama, T.; Reiter, W.D.; et al. Bifunctional cytosolic UDP-glucose 4-epimerases catalyse the interconversion between UDP-D-xylose and UDP-L-arabinose in plants. Biochem. J. 2009, 424, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Zayed, O.; Zeng, F.; Liu, C.; Zhang, L.; Zhu, P.; Hsu, C.C.; Tuncil, Y.E.; Tao, W.A.; Carpita, N.C.; et al. Arabinose biosynthesis is critical for salt stress tolerance in Arabidopsis. New Phytol. 2019, 224, 274–290. [CrossRef] [PubMed]
- Zhang, Q.; Hrmova, M.; Shirley, N.; Lahnstein, J.; Fincher, G. Gene expression patterns and catalytic properties of UDP-D-glucose 4-epimerases from barley (Hordeum vulgare L.). Biochem. J. 2006, 394, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Raedschelders, G.; Debefve, C.; Goesaert, H.; Delcour, J.A.; Volckaert, G.; Van Campenhout, S. Molecular identification and chromosomal localization of genes encoding Triticum aestivum xylanase inhibitor I-like proteins in cereals. Theor. Appl. Genet. 2004, 109, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Igawa, T.; Ochiai-Fukuda, T.; Takahashi-Ando, N.; Ohsato, S.; Shibata, T.; Yamaguchi, I.; Kimura, M. New TAXI-type xylanase inhibitor genes are inducible by pathogens and wounding in hexaploid wheat. Plant Cell Physiol. 2004, 45, 1347–1360. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Lobo, A.; Roujol, D.; Zuñiga-Sánchez, E.; Albenne, C.; Piñero, D.; de Buen, A.G.; Jamet, E. The highly conserved spermatophyte cell wall DUF642 protein family: Phylogeny and first evidence of interaction with cell wall polysaccharides in vitro. Mol. Phylogenet. Evol. 2012, 63, 510–520. [Google Scholar] [CrossRef] [PubMed]
- Zúñiga-Sánchez, E.; Soriano, D.; Martínez-Barajas, E.; Orozco-Segovia, A.; Gamboa-deBuen, A. BIIDXI, the At4g32460 DUF642 gene, is involved in pectin methyl esterase regulation during Arabidopsis thaliana seed germination and plant development. BMC Plant Biol. 2014, 14, 338. [Google Scholar] [CrossRef]
- Sheshukova, E.V.; Komarova, T.V.; Pozdyshev, D.V.; Ershova, N.M.; Shindyapina, A.V.; Tashlitsky, V.N.; Sheval, E.V.; Dorokhov, Y.L. The intergenic interplay between aldose 1-epimerase-like protein and pectin methylesterase in abiotic and biotic stress control. Front. Plant Sci. 2017, 8, 1646. [Google Scholar] [CrossRef]
- Dorokhov, Y.L.; Sheshukova, E.V.; Komarova, T.V. Methanol in plant life. Front. Plant Sci. 2018, 9, 1623. [Google Scholar] [CrossRef]
- Shi, H.; Xiong, L.; Stevenson, B.; Lu, T.; Zhu, J.K. The Arabidopsis salt overly sensitive 4 mutants uncover a critical role for vitamin B6 in plant salt tolerance. Plant Cell 2002, 14, 575–588. [Google Scholar] [CrossRef]
- Zhao, W.T.; Feng, S.J.; Li, H.; Faust, F.; Kleine, T.; Li, L.N.; Yang, Z. Salt stress-induced FERROCHELATASE 1 improves resistance to salt stress by limiting sodium accumulation in Arabidopsis thaliana. Sci. Rep. 2017, 7, 14737. [Google Scholar] [CrossRef] [PubMed]
- Diaz, M.; Sanchez-Barrena, M.J.; Gonzalez-Rubio, J.M.; Rodriguez, L.; Fernandez, D.; Antoni, R.; Yunta, C.; Belda-Palazon, B.; Gonzalez-Guzman, M.; Peirats-Liobet, M.; et al. Calcium-dependent oligomerization of CAR proteins at cell membrane modulates ABA signaling. Proc. Natl. Acad. Sci. USA 2016, 113, E396–E405. [Google Scholar] [CrossRef] [PubMed]
- Nalefski, E.A.; Falke, J.J. The C2 domain calcium-binding motif: Structural and functional diversity. Protein Sci. 1996, 5, 2375–2390. [Google Scholar] [CrossRef] [PubMed]
- Rizo, J.; Südhof, T.C. C2-domains, structure and function of a universal Ca2+-binding domain. J. Biol. Chem. 1998, 273, 15879–15882. [Google Scholar] [CrossRef]
- Rodriguez, L.; Gonzalez-Guzman, M.; Diaz, M.; Rodrigues, A.; Izquierdo-Garcia, A.C.; Peirats-Llobet, M.; Fernandez, M.A.; Antoni, R.; Fernandez, D.; Marquez, J.A.; et al. C2-domain abscisic acid-related proteins mediate the interaction of PYR/PYL/RCAR abscisic acid receptors with the plasma membrane and regulate abscisic acid sensitivity in Arabidopsis. Plant Cell 2014, 26, 4802–4820. [Google Scholar] [CrossRef]
- Nietzsche, M.; Guerra, T.; Alseekh, S.; Wiermer, M.; Sonnewald, S.; Fernie, A.R.; Börnke, F. STOREKEEPER RELATED1/G-element binding protein (STKR1) interacts with protein kinase SnRK1. Plant Physiol. 2018, 176, 1773–1792. [Google Scholar] [CrossRef]
- Hyun, T.K.; van der Graaff, E.; Albacete, A.; Eom, S.H.; Großkinsky, D.K.; Böhm, H.; Janschek, U.; Rim, Y.; Ali, W.W.; Kim, S.Y.; et al. The Arabidopsis PLAT domain protein1 is critically involved in abiotic stress tolerance. PLoS ONE 2014, 9, e112946. [Google Scholar] [CrossRef]
- Qin, F.; Kodaira, K.S.; Maruyama, K.; Mizoi, J.; Tran, L.S.P.; Fujita, Y.; Morimoto, K.; Shinozaki, K.; Yamaguchi-Shinozaki, K. SPINDLY, a negative regulator of gibberellic acid signaling, is involved in the plant abiotic stress response. Plant Physiol. 2011, 157, 1900–1913. [Google Scholar] [CrossRef]
- Kawai, Y.; Ono, E.; Mizutani, M. Evolution and diversity of the 2-oxoglutarate-dependent dioxygenase superfamily in plants. Plant J. 2014, 78, 328–343. [Google Scholar] [CrossRef]
- Park, J.; Kim, Y.S.; Kim, S.G.; Jung, J.H.; Woo, J.C.; Park, C.M. Integration of auxin and salt signals by the NAC transcription factor NTM2 during seed germination in Arabidopsis. Plant Physiol. 2011, 156, 537–549. [Google Scholar] [CrossRef]
- Schlaich, N.L. Flavin-containing monooxygenases in plants: Looking beyond detox. Trends Plant Sci. 2007, 12, 412–418. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.D.; Slovin, J.P.; Hendrickson, A.M. Two genetically discrete pathways convert tryptophan to auxin: More redundancy in auxin biosynthesis. Trends Plant Sci. 2003, 8, 197–199. [Google Scholar] [CrossRef]
- Steber, C.M.; McCourt, P. A role for brassinosteroids in germination in Arabidopsis. Plant Physiol. 2001, 125, 763–769. [Google Scholar] [CrossRef] [PubMed]
- Kagale, S.; Divi, U.K.; Krochko, J.E.; Keller, W.A.; Krishna, P. Brassinosteroid confers tolerance in Arabidopsis thaliana and Brassica napus to a range of abiotic stresses. Planta 2007, 225, 353–364. [Google Scholar] [CrossRef]
- Klahre, U.; Noguchi, T.; Fujioka, S.; Takatsuto, S.; Yokota, T.; Nomura, T.; Yoshida, S.; Chua, N.H. The Arabidopsis DIMINUTO/DWARF1 gene encodes a protein involved in steroid synthesis. Plant Cell 1998, 10, 1677–1690. [Google Scholar] [CrossRef]
- Nikolić, R.; Mitić, N.; Miletić, R.; Nešković, M. Effects of cytokinins on in vitro seed germination and early seedling morphogenesis in Lotus corniculatus L. J. Plant Growth Regul. 2006, 25, 187. [Google Scholar] [CrossRef]
- Peleg, Z.; Blumwald, E. Hormone balance and abiotic stress tolerance in crop plants. Curr. Opin. Plant Boil. 2011, 14, 290–295. [Google Scholar] [CrossRef]
- Walia, H.; Wilson, C.; Condamine, P.; Liu, X.; Ismail, A.M.; Close, T.J. Large-scale expression profiling and physiological characterization of jasmonic acid-mediated adaptation of barley to salinity stress. Plant Cell Environ. 2007, 30, 410–421. [Google Scholar] [CrossRef]
- Lennon, B.W.; Williams, C.H.; Ludwig, M.L. Twists in catalysis: Alternating conformations of Escherichia coli thioredoxin reductase. Science 2000, 289, 1190–1194. [Google Scholar] [CrossRef] [PubMed]
- Piaz, F.D.; Braca, A.; Belisario, M.A.; De Tommasi, N. Thioredoxin system modulation by plant and fungal secondary metabolites. Curr. Med. Chem. 2010, 17, 479–494. [Google Scholar] [CrossRef]
- Jung, Y.J.; Melencion, S.M.B.; Lee, E.S.; Park, J.H.; Alinapon, C.V.; Oh, H.T.; Yun, D.J.; Chi, Y.; Lee, S.Y. Universal stress protein exhibits a redox-dependent chaperone function in Arabidopsis and enhances plant tolerance to heat shock and oxidative stress. Front. Plant Sci. 2015, 6, 1141. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Rizhsky, L.; Liang, H.; Shuman, J.; Shulaev, V.; Mittler, R. Enhanced tolerance to environmental stress in transgenic plants expressing the transcriptional coactivator multiprotein bridging factor 1c. Plant Physiol. 2005, 139, 1313–1322. [Google Scholar] [CrossRef] [PubMed]
- Arce, D.P.; Godoy, A.V.; Tsuda, K.; Yamazaki, K.I.; Valle, E.M.; Iglesias, M.J.; Di Mauroa, M.; Casalongué, C.A. The analysis of an Arabidopsis triple knock-down mutant reveals functions for MBF1 genes under oxidative stress conditions. J. Plant Physiol. 2010, 167, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Kamei, C.L.A.; Boruc, J.; Vandepoele, K.; Van den Daele, H.; Maes, S.; Russinova, E.; Inzé, D.; De Veylder, L. The PRA1 gene family in Arabidopsis. Plant Physiol. 2008, 147, 1735–1749. [Google Scholar] [CrossRef]
- Lee, M.H.; Yoo, Y.J.; Kim, D.H.; Hanh, N.H.; Kwon, Y.; Hwang, I. The prenylated rab GTPase receptor PRA1. F4 contributes to protein exit from the Golgi apparatus. Plant Physiol. 2017, 174, 1576–1594. [Google Scholar] [CrossRef] [PubMed]
- Sturm, M.; Cheng, J.; Baßler, J.; Beckmann, R.; Hurt, E. Interdependent action of KH domain proteins Krr1 and Dim2 drive the 40S platform assembly. Nat. Commun. 2017, 8, 2213. [Google Scholar] [CrossRef]
- Lafuente, M.T.; Martínez-Téllez, M.A.; Zacarias, L. Abscisic acid in the response of ‘Fortune’mandarins to chilling. Effect of maturity and high-temperature conditioning. J. Sci. Food Agric. 1997, 73, 494–502. [Google Scholar] [CrossRef]
- Maere, S.; Heymans, K.; Kuiper, M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005, 21, 3448–3449. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
| Gene ID | Tran(log2FC) | Pro(log2FC) | Description |
|---|---|---|---|
| HORVU4Hr1G058810 | −1.59814 | −0.37389 | Histone H2A 6 |
| HORVU6Hr1G066250 | −0.86759 | −0.43371 | Tetratricopeptide repeat (TPR)-like superfamily protein |
| HORVU2Hr1G065120 | −1.22601 | −0.65284 | Pyridoxal 5-phosphate synthase subunit PdxS |
| HORVU3Hr1G085130 | −0.71579 | −0.46692 | Calcium-dependent lipid-binding (CaLB domain) family protein |
| HORVU5Hr1G109880 | −0.6552 | 0.28447 | Protein of unknown function, DUF642 |
| HORVU7Hr1G100810 | −0.58782 | 0.408756 | 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
| HORVU3Hr1G088130 | −1.28183 | 0.329661 | PRA1 (Prenylated rab acceptor) family protein |
| HORVU3Hr1G109590 | −0.79428 | −0.26857 | Glycosyltransferase family 61 protein |
| HORVU5Hr1G098960 | −0.83372 | −0.38633 | Tubulin alpha-4 chain |
| HORVU7Hr1G001030 | −1.22656 | −0.37861 | Inter-alpha-trypsin inhibitor heavy chain-related |
| HORVU5Hr1G087760 | −1.60172 | −0.48224 | Glutelin type-B-like protein |
| HORVU1Hr1G071460 | −1.34747 | −0.29342 | Serpin 3 |
| HORVU2Hr1G080100 | −0.92463 | −0.37249 | Aldose 1-epimerase |
| HORVU3Hr1G077000 | −0.58028 | −0.28372 | Adenine nucleotide alpha hydrolases-like superfamily protein |
| HORVU6Hr1G074940 | −0.5826 | −0.38866 | Lipase/lipooxygenase, PLAT/LH2 family protein |
| HORVU5Hr1G067630 | −0.82874 | −0.90215 | DNA-binding storekeeper protein-related transcriptional regulator |
| HORVU3Hr1G100410 | −1.02777 | 0.449302 | Xylanase inhibitor |
| HORVU1Hr1G095430 | 0.561136 | 0.345589 | UDP-glucose 4-epimerase 1 |
| HORVU5Hr1G054060 | 0.639856 | 0.418968 | Ferrochelatase 1 |
| HORVU6Hr1G070120 | −0.58765 | 0.266884 | Thioredoxin reductase 2 |
| HORVU0Hr1G012990 | −1.37722 | 0.861165 | Flavin-containing monooxygenase family protein |
| HORVU7Hr1G085130 | 1.103488 | 0.418055 | Multiprotein bridging factor 1C |
| HORVU3Hr1G067370 | 0.632487 | 0.319481 | KRR1 small subunit processome component homolog |
| HORVU7Hr1G120030 | −0.87435 | 0.474998 | Delta(24)-sterol reductase |
| HORVU5Hr1G025320 | 0.5301 | 0.40109 | Tryptophan--tRNA ligase |
| HORVU4Hr1G011940 | 0.657604 | 0.878678 | Exocyst complex component 1 |
| HORVU5Hr1G079100 | 0.495959 | 0.321981 | TBC1 domain family member 15 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lai, Y.; Zhang, D.; Wang, J.; Wang, J.; Ren, P.; Yao, L.; Si, E.; Kong, Y.; Wang, H. Integrative Transcriptomic and Proteomic Analyses of Molecular Mechanism Responding to Salt Stress during Seed Germination in Hulless Barley. Int. J. Mol. Sci. 2020, 21, 359. https://doi.org/10.3390/ijms21010359
Lai Y, Zhang D, Wang J, Wang J, Ren P, Yao L, Si E, Kong Y, Wang H. Integrative Transcriptomic and Proteomic Analyses of Molecular Mechanism Responding to Salt Stress during Seed Germination in Hulless Barley. International Journal of Molecular Sciences. 2020; 21(1):359. https://doi.org/10.3390/ijms21010359
Chicago/Turabian StyleLai, Yong, Dangquan Zhang, Jinmin Wang, Juncheng Wang, Panrong Ren, Lirong Yao, Erjing Si, Yuhua Kong, and Huajun Wang. 2020. "Integrative Transcriptomic and Proteomic Analyses of Molecular Mechanism Responding to Salt Stress during Seed Germination in Hulless Barley" International Journal of Molecular Sciences 21, no. 1: 359. https://doi.org/10.3390/ijms21010359
APA StyleLai, Y., Zhang, D., Wang, J., Wang, J., Ren, P., Yao, L., Si, E., Kong, Y., & Wang, H. (2020). Integrative Transcriptomic and Proteomic Analyses of Molecular Mechanism Responding to Salt Stress during Seed Germination in Hulless Barley. International Journal of Molecular Sciences, 21(1), 359. https://doi.org/10.3390/ijms21010359





