GABAA Receptor Ligands Often Interact with Binding Sites in the Transmembrane Domain and in the Extracellular Domain—Can the Promiscuity Code Be Cracked?
Abstract
1. Introduction
2. Results
2.1. Chemistry
2.2. Pharmacological Assessments
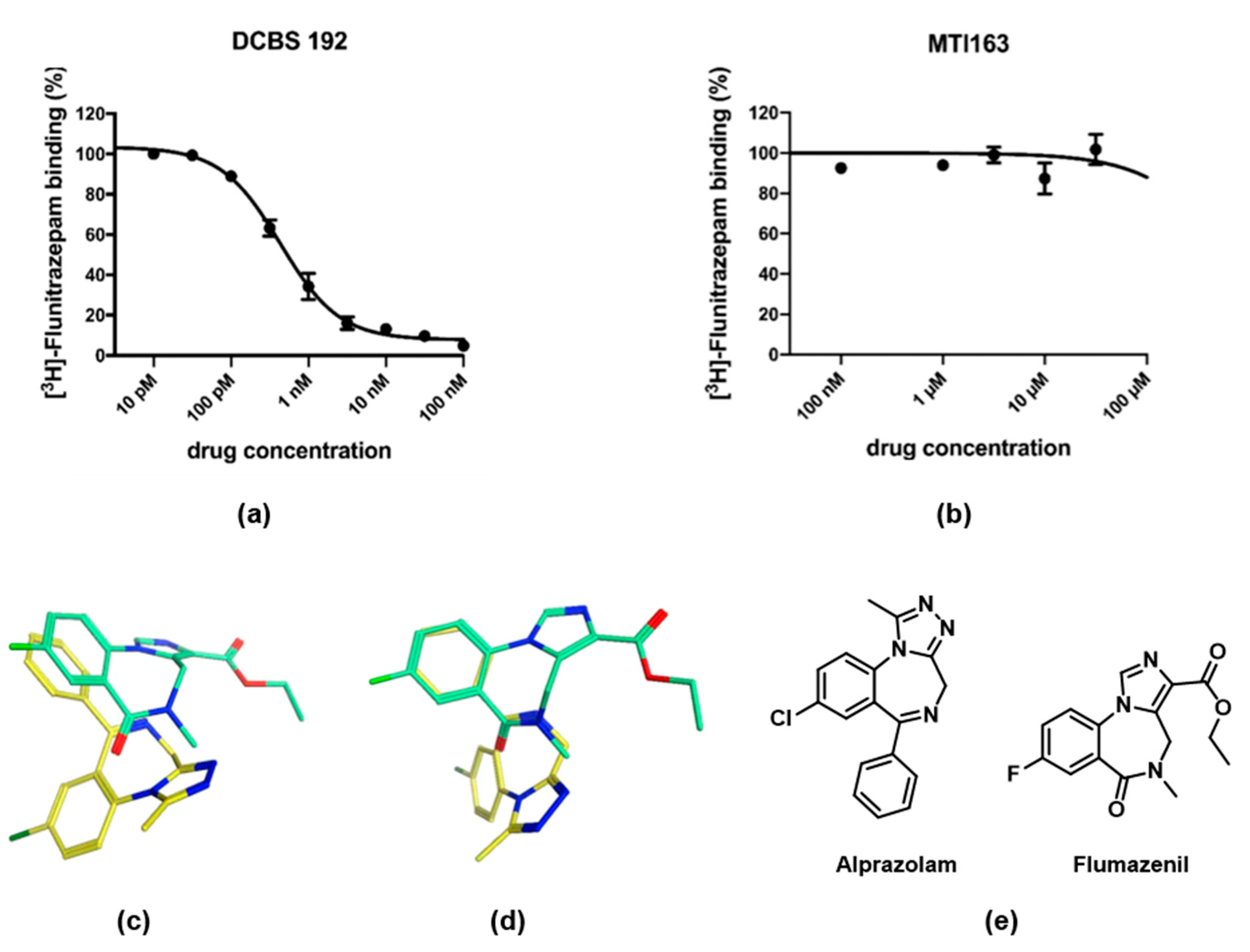
2.2.1. Radioligand Displacement Assays
2.2.2. Functional and Mutational Studies, Beta Isoform Profile
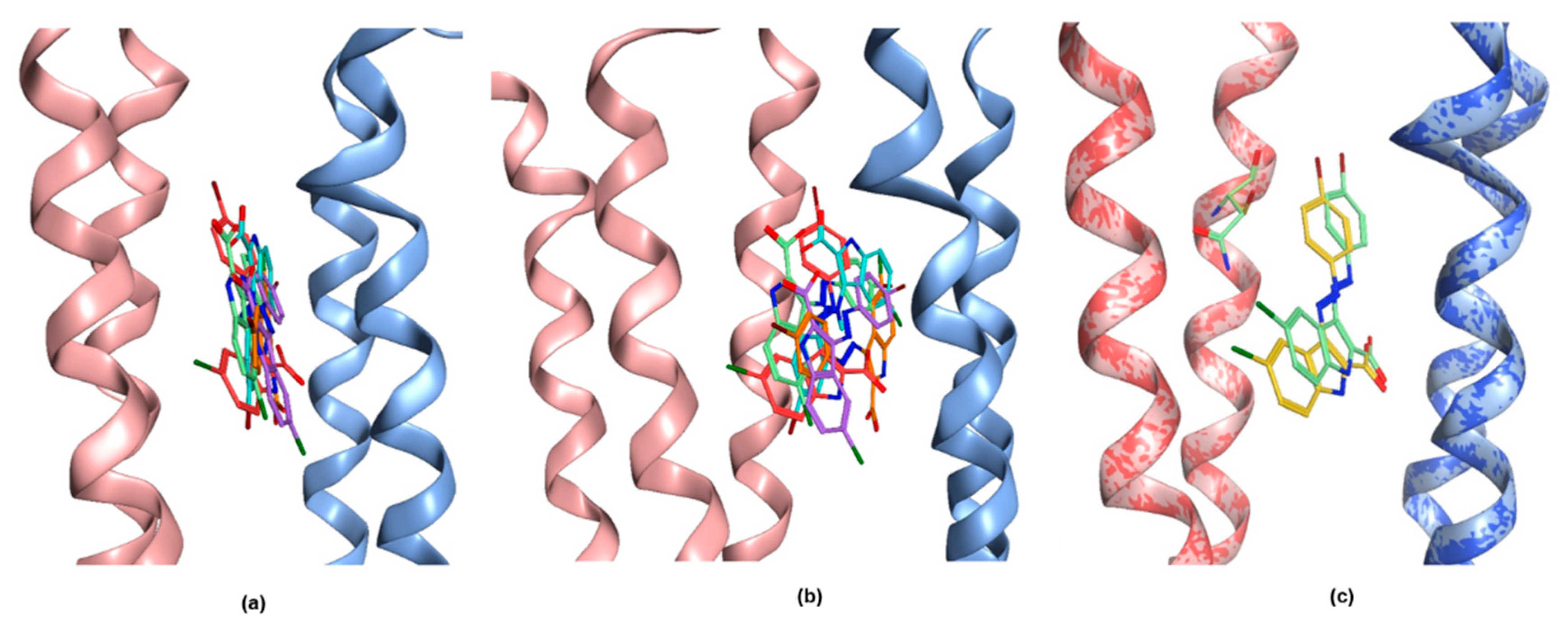
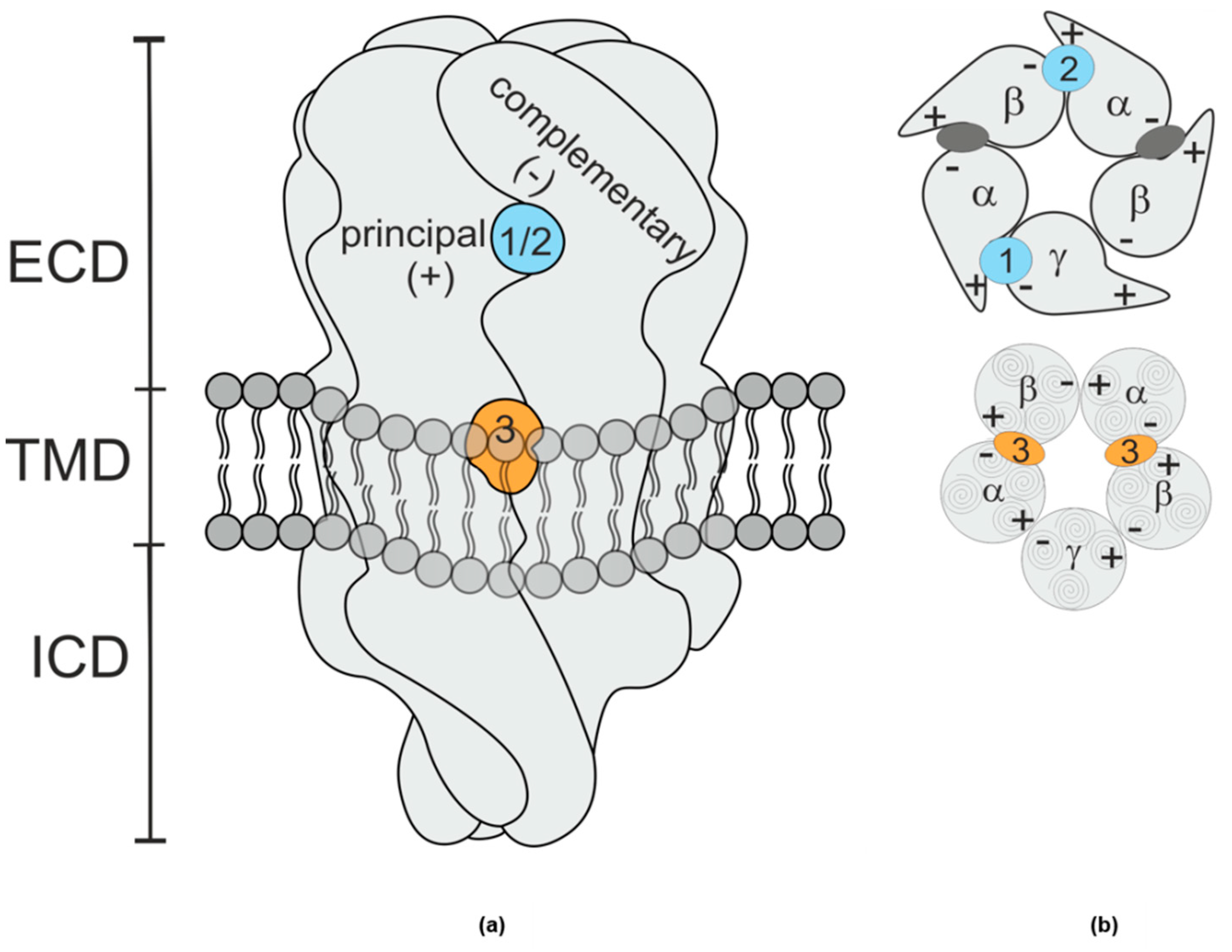
2.3. Structural Hypothesis/Computational Docking
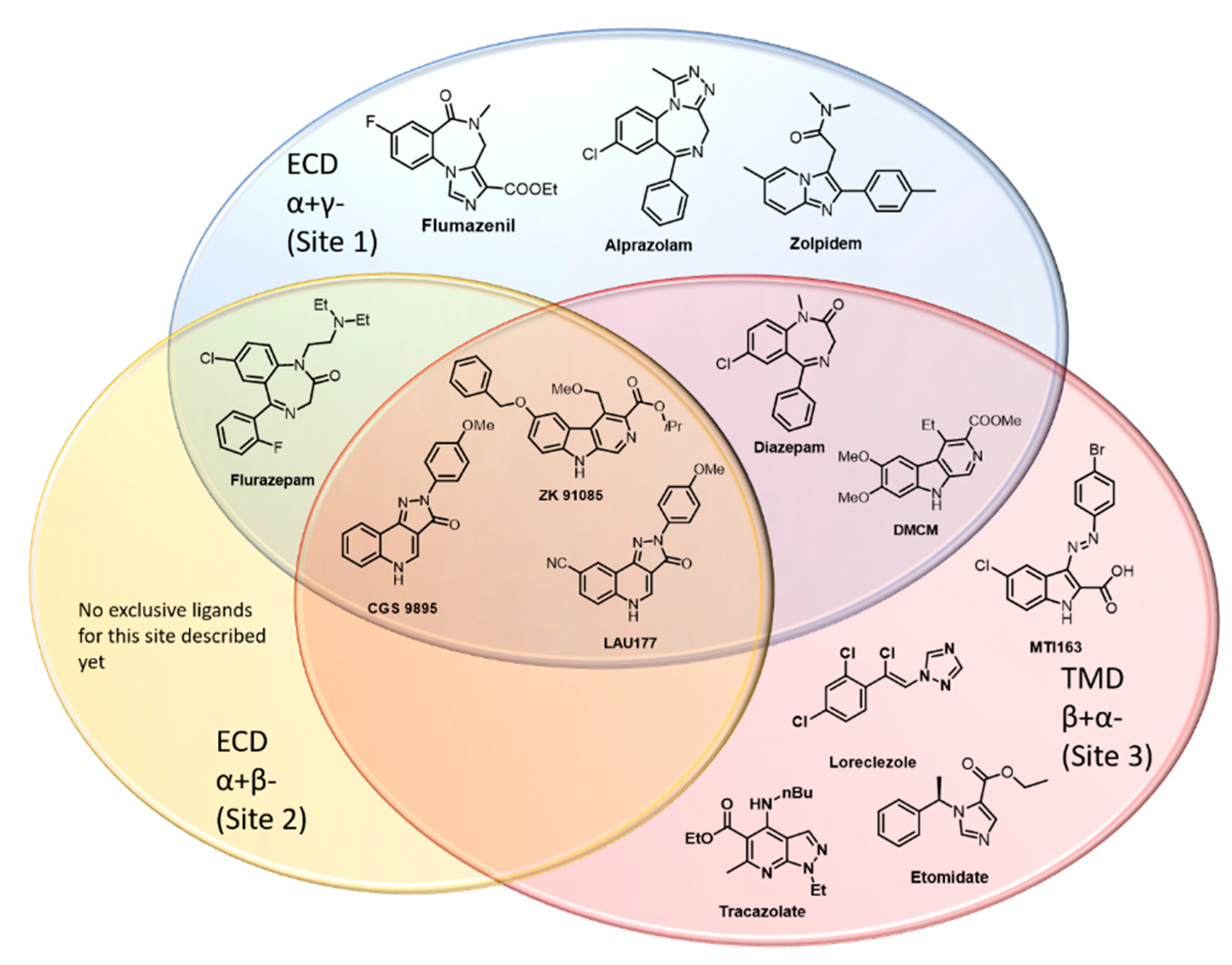
2.4. Ligand Analysis
3. Discussion
4. Materials and Methods
4.1. Compound Synthesis
4.2. Electrophysiology in Xenopus Laevis Oocytes
4.3. Displacement Assays
4.4. Computational Docking
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BZ | Benzodiazepines |
| GABA | Gamma-aminobutyric acid |
| EC | Effective Concentration |
| ECD | Extracellular domain |
| ICD | Intracellular domain |
| TMD | Transmembrane domain |
| PQs | Pyrazoloquinolinone |
| TLC | Thin-layer Chromatography |
Appendix A
Appendix A.1. Compound Synthesis

Appendix A.1.1. Synthesis of ethyl-(Z)-2-(2-phenyl)hydrazineylidene)propanoate 2
Appendix A.1.2. Synthesis of ethyl 1H-indole-2-carboxylate and ethyl 5-chloro-1H-indole-2-carboxylate 3
Appendix A.1.3. Preparation of the Diazonium Salt Solution
Appendix A.1.4. Synthesis of Ethyl (E)-3-((4-bromophenyl)diazenyl)-5-chloro-1H-indole-2-carboxylate 4
Appendix A.1.5. Synthesis of (E)-3-((4-bromophenyl)diazenyl)-5-chloro-1H-indole-2-carboxylic Acid MTI163 (5)
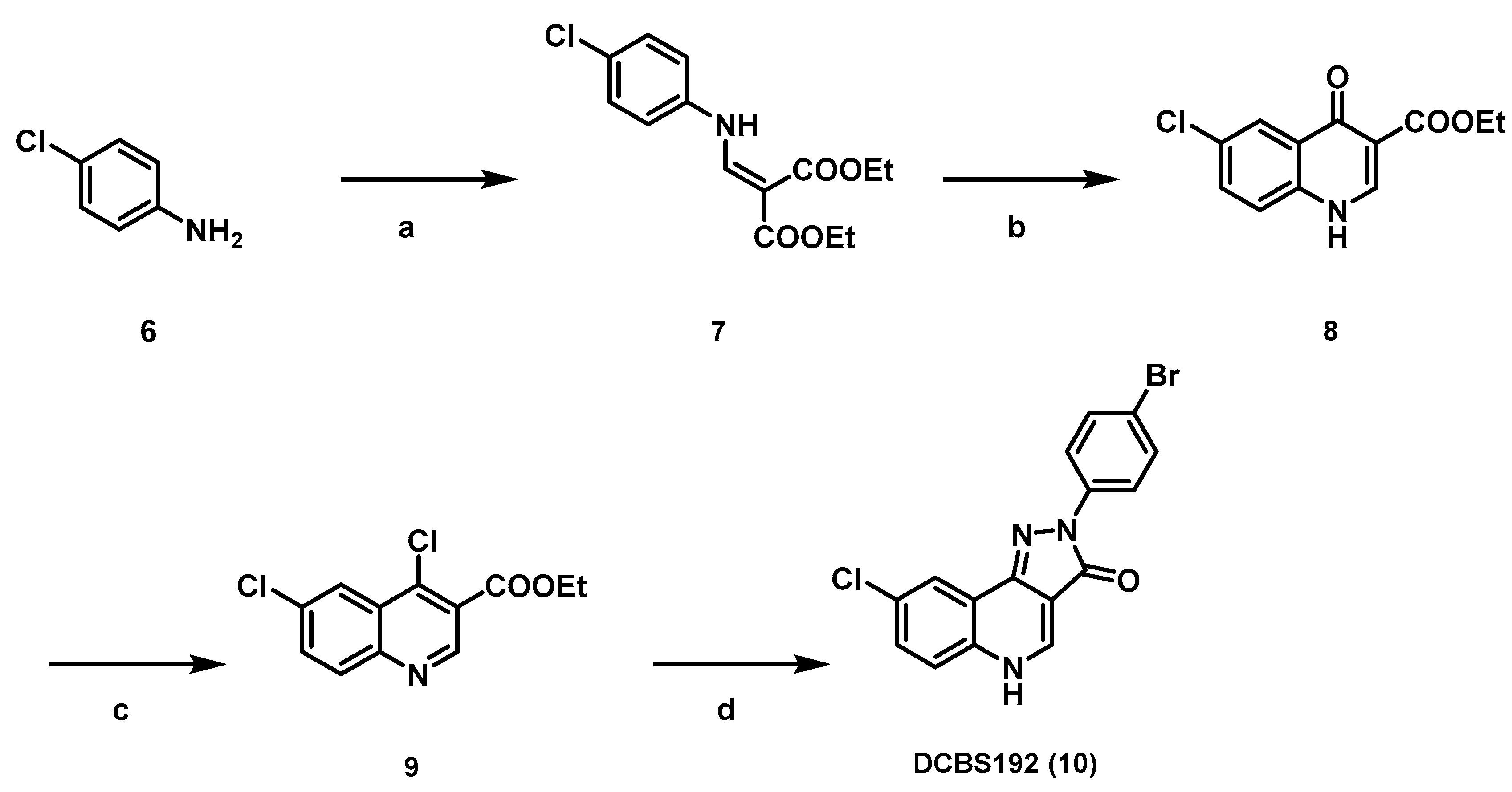
Appendix A.1.6. Diethyl 2-(((4-chlorophenyl)amino)methylene)malonate 7
Appendix A.1.7. Synthesis of Ethyl 6-chloro-4-oxo-1,4-dihydroquinoline-3-carboxylate 8
Appendix A.1.8. Synthesis of Ethyl 4,6-dichloroquinoline-3-carboxylate 9
Appendix A.1.9. Synthesis of 2-(4-Bromophenyl)-8-chloro-2,5-dihydro-3H-pyrazolo[4,3-c]quinolin-3-one DCBS192 (10)
Appendix A.2. GAUSSIAN Computational Protocol
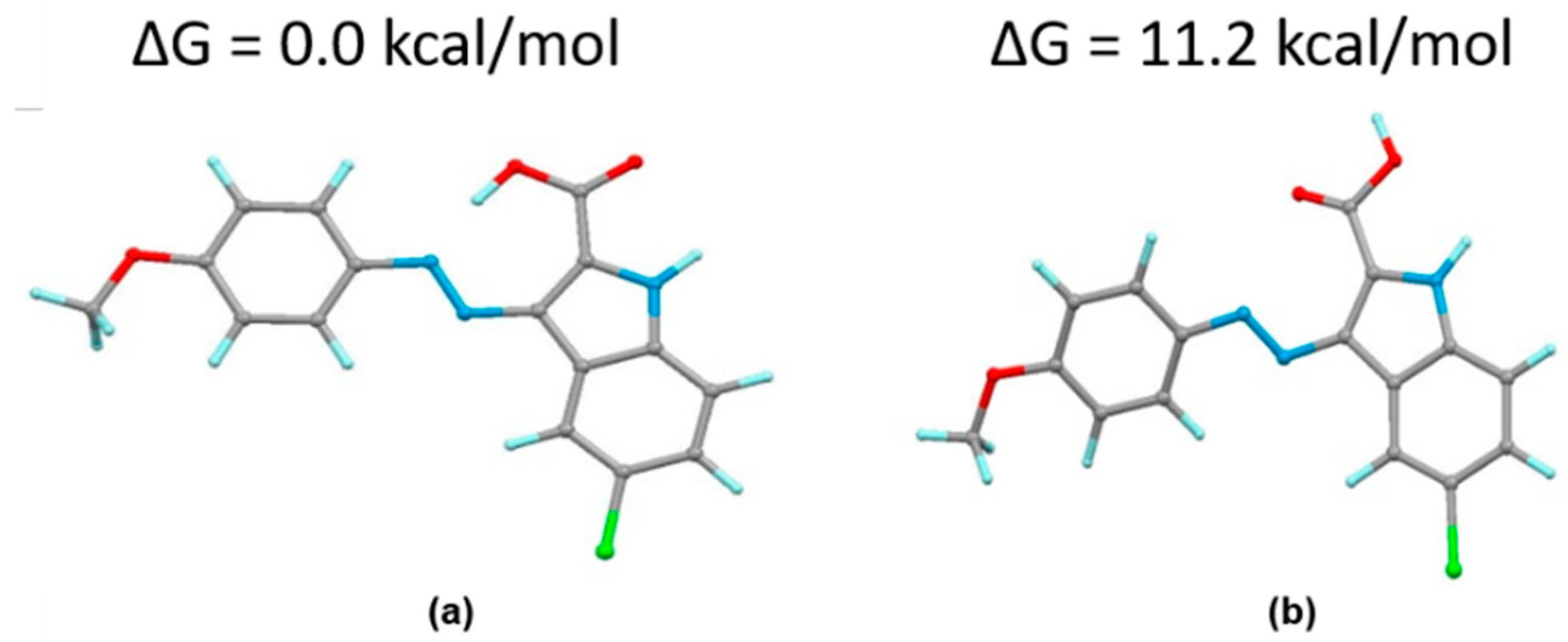
Appendix A.3. Supplementary Figures from Computational Docking


Appendix A.4. MTI163 Dose Response Curve in α1β1Receptors

References
- Sieghart, W.; Savić, M.M. International Union of Basic and Clinical Pharmacology. CVI: GABAA Receptor Subtype- and Function-selective Ligands: Key Issues in Translation to Humans. Pharmacol. Rev. 2018, 70, 836–878. [Google Scholar] [CrossRef] [PubMed]
- Jembrek, M.J.; Vlainic, J. GABA Receptors: Pharmacological Potential and Pitfalls. Curr. Pharm. Des. 2015, 21, 4943–4959. [Google Scholar] [CrossRef] [PubMed]
- Sigel, E.; Buhr, A. The benzodiazepine binding site of GABAA receptors. Trends Pharmacol. Sci. 1997, 18, 425–429. [Google Scholar] [CrossRef]
- Masiulis, S.; Desai, R.; Uchański, T.; Martin, I.S.; Laverty, D.; Karia, D.; Malinauskas, T.; Zivanov, J.; Pardon, E.; Kotecha, A.; et al. GABAA receptor signalling mechanisms revealed by structural pharmacology. Nature 2019, 565, 454–459. [Google Scholar] [CrossRef] [PubMed]
- Walters, R.J.; Hadley, S.H.; Morris, K.D.; Amin, J. Benzodiazepines act on GABAA receptors via two distinct and separable mechanisms. Nat. Neurosci. 2000, 3, 1274–1281. [Google Scholar] [CrossRef]
- Maldifassi, M.C.; Baur, R.; Sigel, E. Functional sites involved in modulation of the GABAA receptor channel by the intravenous anesthetics propofol, etomidate and pentobarbital. Neuropharmacology 2016, 105, 207–214. [Google Scholar] [CrossRef]
- Chiara, D.C.; Dostalova, Z.; Jayakar, S.S.; Zhou, X.; Miller, K.W.; Cohen, J.B. Mapping general anesthetic binding site(s) in human α1β3 γ-aminobutyric acid type A receptors with [³H]TDBzl-etomidate, a photoreactive etomidate analogue. Biochemistry 2012, 51, 836–847. [Google Scholar] [CrossRef]
- Olsen, R.W. GABAA receptor: Positive and negative allosteric modulators. Neuropharmacology 2018, 136, 10–22. [Google Scholar] [CrossRef]
- Solomon, V.R.; Tallapragada, V.J.; Chebib, M.; Johnston, G.; Hanrahan, J.R. GABA allosteric modulators: An overview of recent developments in non-benzodiazepine modulators. Eur. J. Med. Chem. 2019, 171, 434–461. [Google Scholar] [CrossRef]
- Sieghart, W. Chapter Three—Allosteric Modulation of GABAA Receptors via Multiple Drug-Binding Sites. In Advances in Pharmacology; Rudolph, U., Ed.; Academic Press: Cambridge, MA, USA, 2015; pp. 53–96. [Google Scholar]
- Puthenkalam, R.; Hieckel, M.; Simeone, X.; Suwattanasophon, C.; Feldbauer, R.V.; Ecker, G.F.; Ernst, M. Structural Studies of GABAA Receptor Binding Sites: Which Experimental Structure Tells us What? Front. Mol. Neurosci. 2016, 9, 44. [Google Scholar] [CrossRef]
- Iorio, M.T.; Rehman, S.; Bampali, K.; Stoeger, B.; Schnürch, M.; Ernst, M.; Mihovilovic, M.D. Variations on a scaffold—Novel GABAA receptor modulators. Eur. J. Med. Chem. 2019, 180, 340–349. [Google Scholar] [CrossRef] [PubMed]
- Ramerstorfer, J.; Furtmüller, R.; Sarto-Jackson, I.; Varagic, Z.; Sieghart, W.; Ernst, M. The GABAA Receptor α+β− Interface: A Novel Target for Subtype Selective Drugs. J. Neurosci. 2011, 31, 870–877. [Google Scholar] [CrossRef] [PubMed]
- Maldifassi, M.C.; Baur, R.; Sigel, E. Molecular mode of action of CGS 9895 at α1β2γ2 GABAA receptors. J. Neurochem. 2016, 138, 722–730. [Google Scholar] [CrossRef] [PubMed]
- Varagic, Z.; Ramerstorfer, J.; Huang, S.; Rallapalli, S.; Sarto-Jackson, I.; Cook, J.; Sieghart, W.; Ernst, M. Subtype selectivity of α+β− site ligands of GABAA receptors: Identification of the first highly specific positive modulators at α6β2/3γ2 receptors. Br. J Pharm. 2013, 169, 384–399. [Google Scholar] [CrossRef] [PubMed]
- Siebert, D.C.B.; Bampali, K.; Puthenkalam, R.; Varagic, Z.; Sarto-Jackson, I.; Scholze, P.; Sieghart, W.; Mihovilovic, M.D.; Schnürch, M.; Ernst, M. Engineered Flumazenil Recognition Site Provides Mechanistic Insight Governing Benzodiazepine Modulation in GABAA Receptors. ACS Chem. Boil. 2018, 13, 2040–2047. [Google Scholar] [CrossRef]
- Zhu, S.; Noviello, C.M.; Teng, J.; Walsh, R.M.; Kim, J.J.; Hibbs, R.E. Structure of a human synaptic GABAA receptor. Nature 2018, 559, 67–72. [Google Scholar] [CrossRef]
- Simeone, X.; Siebert, D.C.; Bampali, K.; Varagic, Z.; Treven, M.; Rehman, S.; Pyszkowski, J.; Holzinger, R.; Steudle, F.; Scholze, P.; et al. Molecular tools for GABAA receptors: High affinity ligands for β1-containing subtypes. Sci. Rep. 2017, 7, 5674–5685. [Google Scholar] [CrossRef]
- Thomet, U.; Baur, R.; Scholze, P.; Sieghart, W.; Sigel, E. Dual mode of stimulation by the beta-carboline ZK 91085 of recombinant GABAA receptor currents: Molecular determinants affecting its action. Br. J. Pharmacol. 1999, 127, 1231–1239. [Google Scholar] [CrossRef]
- Wingrove, P.B.; Wafford, K.A.; Bain, C.; Whiting, P.J. The modulatory action of loreclezole at the gamma-aminobutyric acid type A receptor is determined by a single amino acid in the beta 2 and beta 3 subunit. Proc. Natl. Acad. Sci. USA 1994, 91, 4569–4573. [Google Scholar] [CrossRef]
- Desai, R.; Ruesch, D.; Forman, S.A. Gamma-amino butyric acid type A receptor mutations at beta2N265 alter etomidate efficacy while preserving basal and agonist-dependent activity. Anesthesiology 2009, 111, 774–784. [Google Scholar] [CrossRef]
- Wafford, K.; Bain, C.; Quirk, K.; McKernan, R.; Wingrove, P.; Whiting, P.; Kemp, J. A novel allosteric modulatory site on the GABAA receptor β subunit. Neuron 1994, 12, 775–782. [Google Scholar] [CrossRef]
- Thompson, S.A.; Wingrove, P.B.; Connelly, L.; Whiting, P.J.; Wafford, K.A. Tracazolate Reveals a Novel Type of Allosteric Interaction with Recombinant γ-Aminobutyric AcidA Receptors. Mol. Pharmacol. 2002, 61, 861–869. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, A.; Wingrove, P.B.; Whiting, P.J.; Wafford, K.A. beta-Carboline gamma-aminobutyric acidA receptor inverse agonists modulate gamma-aminobutyric acid via the loreclezole binding site as well as the benzodiazepine site. Mol. Pharmacol. 1995, 48, 965–969. [Google Scholar] [PubMed]
- Fernandez, S.P.; Karim, N.; Mewett, K.N.; Chebib, M.; Johnston, G.A.; Hanrahan, J.R. Flavan-3-ol esters: New agents for exploring modulatory sites on GABAA receptors. Br. J. Pharmacol. 2012, 165, 965–977. [Google Scholar] [CrossRef] [PubMed]
- Belelli, D.; Lambert, J.J.; Peters, J.A.; Wafford, K.; Whiting, P.J. The interaction of the general anesthetic etomidate with the gamma-aminobutyric acid type A receptor is influenced by a single amino acid. Proc. Natl. Acad. Sci. USA 1997, 94, 11031–11036. [Google Scholar] [CrossRef]
- Hanrahan, J.R.; Chebib, M.; Johnston, G.A.R. Chapter Seven—Interactions of Flavonoids with Ionotropic GABA Receptors. In Advances in Pharmacology; Rudolph, U., Ed.; Academic Press: Cambridge, MA, USA, 2015; pp. 189–200. [Google Scholar]
- Kavvadias, D.; Monschein, V.; Sand, P.; Riederer, P.; Schreier, P. Constituents of Sage (Salvia officinalis) with in vitro Affinity to Human Brain Benzodiazepine Receptor. Planta Med. 2003, 69, 113–117. [Google Scholar] [CrossRef]
- Kavvadias, D.; Sand, P.; A Youdim, K.; Qaiser, M.Z.; Rice-Evans, C.; Baur, R.; Sigel, E.; Rausch, W.-D.; Riederer, P.; Schreier, P. The flavone hispidulin, a benzodiazepine receptor ligand with positive allosteric properties, traverses the blood–brain barrier and exhibits anticonvulsive effects. Br. J. Pharmacol. 2004, 142, 811–820. [Google Scholar] [CrossRef]
- Sigel, E.; Baur, R.; Furtmueller, R.; Razet, R.; Dodd, R.H.; Sieghart, W. Differential Cross Talk of ROD Compounds with the Benzodiazepine Binding Site. Mol. Pharmacol. 2001, 59, 1470–1477. [Google Scholar] [CrossRef]
- El Hadri, A.; Abouabdellah, A.; Thomet, U.; Baur, R.; Furtmüller, R.; Sigel, E.; Sieghart, W.; Dodd, R.H. N-Substituted 4-amino-3,3-dipropyl-2(3H)-furanones: New positive allosteric modulators of the GABAA receptor sharing electrophysiological properties with the anticonvulsant loreclezole. J. Med. Chem. 2002, 45, 2824–2831. [Google Scholar] [CrossRef]
- Stornaiuolo, M.; De Kloe, G.E.; Rucktooa, P.; Fish, A.; Van Elk, R.; Edink, E.S.; Bertrand, D.; Smit, A.B.; De Esch, I.J.P.; Sixma, T.K. Assembly of a π–π stack of ligands in the binding site of an acetylcholine-binding protein. Nat. Commun. 2013, 4, 1875. [Google Scholar] [CrossRef]
- Richter, L.; De Graaf, C.; Sieghart, W.; Varagic, Z.; Mörzinger, M.; De Esch, I.J.P.; Ecker, G.F.; Ernst, M. Diazepam-bound GABAA receptor models identify new benzodiazepine binding-site ligands. Nat. Methods 2012, 8, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Thomet, U.; Baur, R.; Razet, R.; Dodd, R.H.; Furtmüller, R.; Sieghart, W.; Sigel, E. A novel positive allosteric modulator of the GABAA receptor: The action of (+)-ROD188. Br. J. Pharmacol. 2000, 131, 843–850. [Google Scholar] [CrossRef] [PubMed]
- Armarego, W.L.F.; Lin Chai, C.L. Purification of Laboratory Chemicals, 7th ed.; Butterworth-Heinemann: Oxford, UK, 2012. [Google Scholar]
- Varagic, Z.; Wimmer, L.; Schnürch, M.; Mihovilovic, M.D.; Huang, S.; Rallapalli, S.; Cook, J.M.; Mirheydari, P.; Ecker, G.F.; Sieghart, W.; et al. Identification of novel positive allosteric modulators and null modulators at the GABAA receptor α+β− interface. Br. J. Pharmacol. 2013, 169, 371–383. [Google Scholar] [CrossRef]
- Minier, F.; Sigel, E. Positioning of the α-subunit isoforms confers a functional signature to γ-aminobutyric acid type A receptors. Proc. Natl. Acad. Sci. USA 2004, 101, 7769–7774. [Google Scholar] [CrossRef] [PubMed]
- Baumann, S.W.; Baur, R.; Sigel, E. Subunit Arrangement of γ-Aminobutyric Acid Type A Receptors. J. Biol. Chem. 2001, 276, 36275–36280. [Google Scholar] [CrossRef] [PubMed]
- Baumann, S.W.; Baur, R.; Sigel, E. Forced Subunit Assembly in α1β2γ2 GABAAReceptors: INSIGHT INTO THE ABSOLUTE ARRANGEMENT. J. Biol. Chem. 2002, 277, 46020–46025. [Google Scholar] [CrossRef] [PubMed]
- Sieghart, W.; Schuster, A. Affinity of various ligands for benzodiazepine receptors in rat cerebellum and hippocampus. Biochem. Pharmacol. 1984, 33, 4033–4038. [Google Scholar] [CrossRef]
- Yung-Chi, C.; Prusoff, W.H. Relationship between the inhibition constant (KI) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 1973, 22, 3099–3108. [Google Scholar] [CrossRef]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and validation of a genetic algorithm for flexible docking11Edited by F. E. Cohen. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef]
- Gaussian, R.A.; Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; et al. Gaussian, Inc., Wallingford, CT, USA. 2009. Available online: http://wild.life.nctu.edu.tw/~jsyu/compchem/g09/g09ur/m_citation.htm (accessed on 3 January 2020).
- Hehre, W.J. Ab initio molecular orbital theory. Accounts Chem. Res. 1976, 9, 399–406. [Google Scholar] [CrossRef]
- Parr, R.G.Y. Density Functional Theory of Atoms and Molecules; Oxford University Press: New York, NY, USA, 1989. [Google Scholar]
- Perdew, J.P. Density-functional approximation for the correlation energy of the inhomogeneous electron gas. Phys. Rev. B 1986, 33, 8822–8824. [Google Scholar] [CrossRef] [PubMed]
- Perdew, J.P.; Burke, K.; Ernzerhof, M. Generalized Gradient Approximation Made Simple. Phys. Rev. Lett. 1996, 78, 1396. [Google Scholar] [CrossRef]
- Binning, R.C., Jr.; Curtiss, L.A. Compact contracted basis sets for third-row atoms: Ga–Kr. J. Chem. Physics. 1990, 11, 1206–1216. [Google Scholar] [CrossRef]
- Hay, P.J. Gaussian basis sets for molecular calculations. The representation of 3d orbitals in transition-metal atoms. J. Chem. Phys. 1977, 66, 4377–4384. [Google Scholar] [CrossRef]
- Krishnan, R.; Binkley, J.S.; Seeger, R.; Pople, J.A. Self-consistent molecular orbital methods. XX. A basis set for correlated wave functions. J. Chem. Phys. 1980, 72, 650–654. [Google Scholar] [CrossRef]
- McGrath, M.P.; Radom, L. Extension of Gaussian-1 (G1) theory to bromine-containing molecules. J. Chem. Phys. 1991, 94, 511–516. [Google Scholar] [CrossRef]
- McLean, A.D.; Chandler, G.S. Contracted Gaussian basis sets for molecular calculations. I. Second row atoms, Z = 11–18. J. Chem. Phys. 1980, 72, 5639–5648. [Google Scholar] [CrossRef]
- Raghavachari, K.; Trucks, G.W. Highly correlated systems. Excitation energies of first row transition metals Sc-Cu. J. Chem. Phys. 1989, 91, 1062–1065. [Google Scholar] [CrossRef]
- Wachters, A.J.H. Gaussian Basis Set for Molecular Wavefunctions Containing Third-Row Atoms. J. Chem. Phys. 1970, 52, 1033–1036. [Google Scholar] [CrossRef]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iorio, M.T.; Vogel, F.D.; Koniuszewski, F.; Scholze, P.; Rehman, S.; Simeone, X.; Schnürch, M.; Mihovilovic, M.D.; Ernst, M. GABAA Receptor Ligands Often Interact with Binding Sites in the Transmembrane Domain and in the Extracellular Domain—Can the Promiscuity Code Be Cracked? Int. J. Mol. Sci. 2020, 21, 334. https://doi.org/10.3390/ijms21010334
Iorio MT, Vogel FD, Koniuszewski F, Scholze P, Rehman S, Simeone X, Schnürch M, Mihovilovic MD, Ernst M. GABAA Receptor Ligands Often Interact with Binding Sites in the Transmembrane Domain and in the Extracellular Domain—Can the Promiscuity Code Be Cracked? International Journal of Molecular Sciences. 2020; 21(1):334. https://doi.org/10.3390/ijms21010334
Chicago/Turabian StyleIorio, Maria Teresa, Florian Daniel Vogel, Filip Koniuszewski, Petra Scholze, Sabah Rehman, Xenia Simeone, Michael Schnürch, Marko D. Mihovilovic, and Margot Ernst. 2020. "GABAA Receptor Ligands Often Interact with Binding Sites in the Transmembrane Domain and in the Extracellular Domain—Can the Promiscuity Code Be Cracked?" International Journal of Molecular Sciences 21, no. 1: 334. https://doi.org/10.3390/ijms21010334
APA StyleIorio, M. T., Vogel, F. D., Koniuszewski, F., Scholze, P., Rehman, S., Simeone, X., Schnürch, M., Mihovilovic, M. D., & Ernst, M. (2020). GABAA Receptor Ligands Often Interact with Binding Sites in the Transmembrane Domain and in the Extracellular Domain—Can the Promiscuity Code Be Cracked? International Journal of Molecular Sciences, 21(1), 334. https://doi.org/10.3390/ijms21010334





