DNA Repair and the Stability of the Plant Mitochondrial Genome
Abstract
1. Origin, Size, and Coding Capacity of the Organellar Genomes
2. Structure of Plant Organellar Genomes
3. Repair Mechanisms in the Mitochondria of Plants
3.1. Direct Repair
3.2. Mismatch Repair (MMR)
3.3. Nucleotide Excision Repair (NER)
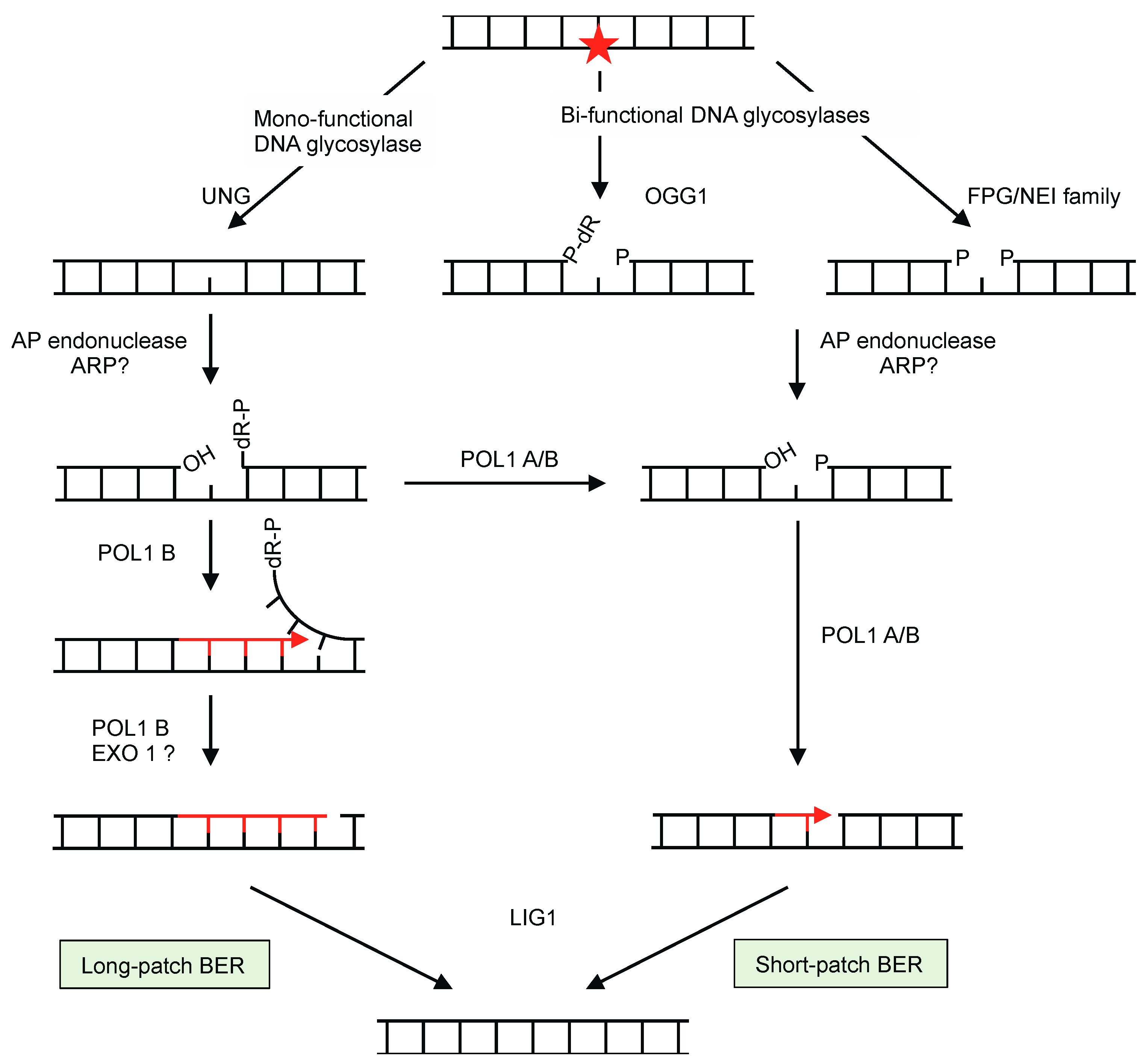
3.4. Base Excision Repair (BER)
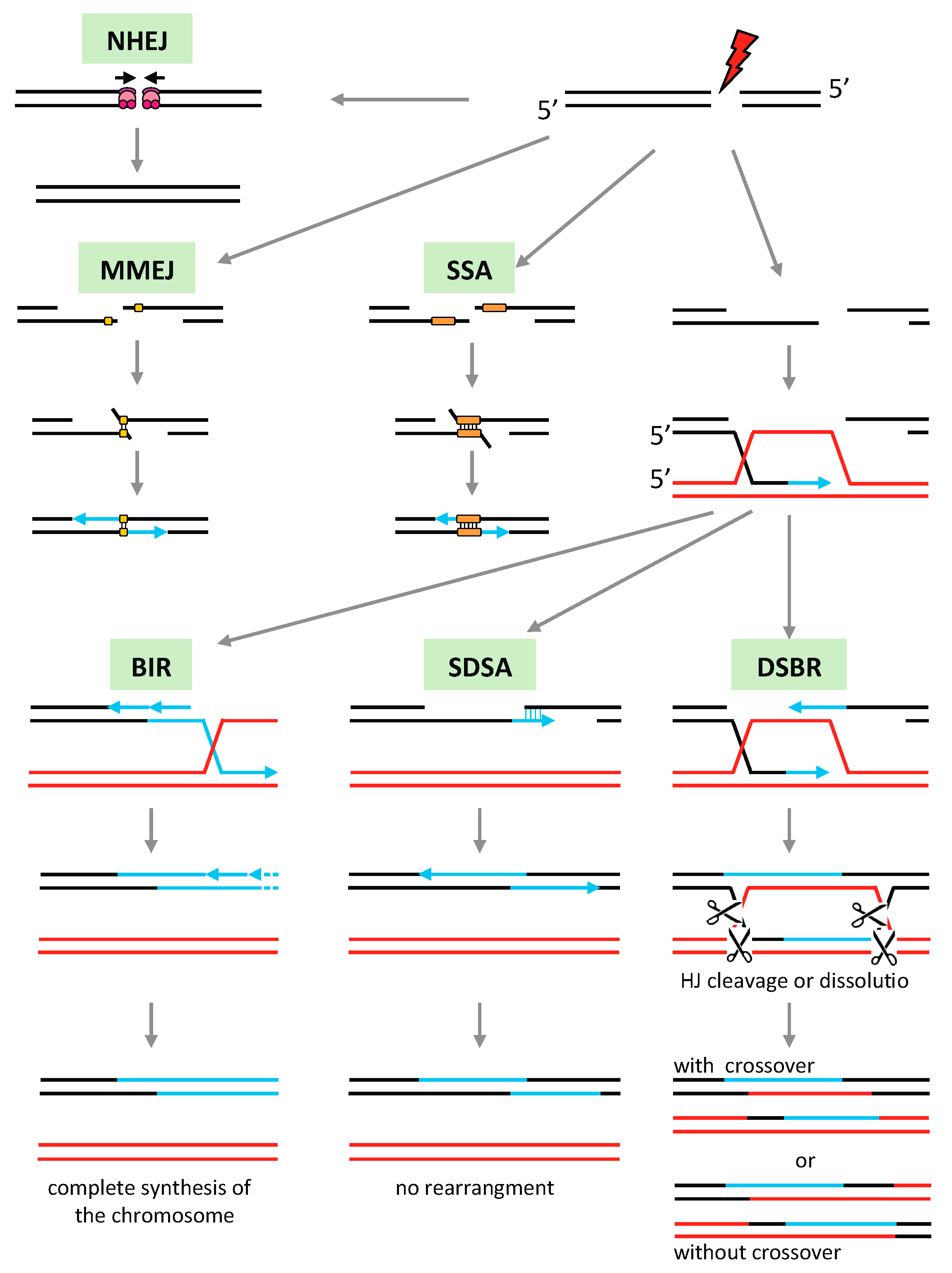
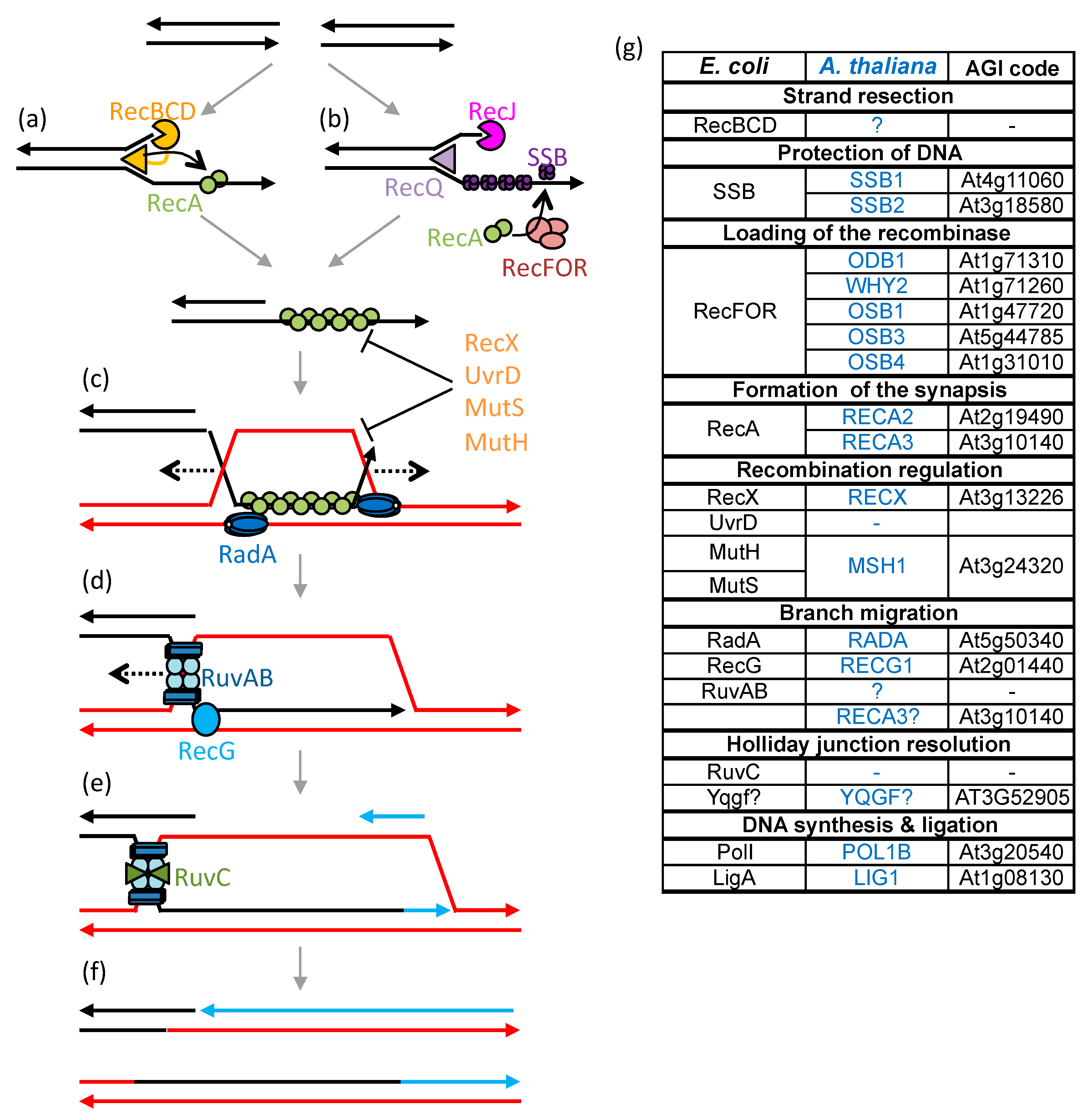
3.5. Recombination Pathways in Plant Mitochondria
4. HR Pathways and the Evolution of the Plant mtDNA
Author Contributions
Funding
Conflicts of Interest
References
- Maréchal, A.; Brisson, N. Recombination and the maintenance of plant organelle genome stability. New Phytol. 2010, 186, 299–317. [Google Scholar] [CrossRef] [PubMed]
- Christensen, A.C. Mitochondrial DNA Repair and Genome Evolution. In Annual Plant Reviews Online; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2018; pp. 11–32. [Google Scholar] [CrossRef]
- Brieba, L.G. Structure-Function Analysis sReveals the Singularity of Plant Mitochondrial DNA Replication Components: A Mosaic and Redundant System. Plants 2019, 8, 533. [Google Scholar] [CrossRef] [PubMed]
- Wynn, E.L.; Christensen, A.C. Repeats of Unusual Size in Plant Mitochondrial Genomes: Identification, Incidence and Evolution. G3 2019, 9, 549–559. [Google Scholar] [CrossRef]
- Gualberto, J.M.; Newton, K.J. Plant Mitochondrial Genomes: Dynamics and Mechanisms of Mutation. Annu. Rev. Plant Biol. 2017, 68, 225–252. [Google Scholar] [CrossRef] [PubMed]
- Sagan, L. On the origin of mitosing cells. J. Theor. Biol. 1967, 14, 255–274. [Google Scholar] [CrossRef]
- Dyall, S.D.; Brown, M.T.; Johnson, P.J. Ancient invasions: From endosymbionts to organelles. Science 2004, 304, 253–257. [Google Scholar] [CrossRef]
- Gray, M.W. Mitochondrial evolution. Cold Spring Harb. Perspect. Biol. 2012, 4, a011403. [Google Scholar] [CrossRef]
- Roger, A.J.; Munoz-Gomez, S.A.; Kamikawa, R. The Origin and Diversification of Mitochondria. Curr. Biol. 2017, 27, R1177–R1192. [Google Scholar] [CrossRef]
- Bouchier, C.; Ma, L.; Créno, S.; Dujon, B.; Fairhead, C. Complete mitochondrial genome sequences of three Nakaseomyces species reveal invasion by palindromic GC clusters and considerable size expansion. FEMS Yeast Res. 2009, 9, 1283–1292. [Google Scholar] [CrossRef]
- Freel, K.C.; Friedrich, A.; Schacherer, J. Mitochondrial genome evolution in yeasts: An all-encompassing view. FEMS Yeast Res. 2015, 15. [Google Scholar] [CrossRef]
- Pramateftaki, P.V.; Kouvelis, V.N.; Lanaridis, P.; Typas, M.A. The mitochondrial genome of the wine yeast Hanseniaspora uvarum: A unique genome organization among yeast/fungal counterparts. FEMS Yeast Res. 2006, 6, 77–90. [Google Scholar] [CrossRef] [PubMed]
- Lang, B.F.; Burger, G.; O’Kelly, C.J.; Cedergren, R.; Golding, G.B.; Lemieux, C.; Sankoff, D.; Turmel, M.; Gray, M.W. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature 1997, 387, 493–497. [Google Scholar] [CrossRef] [PubMed]
- Lukeš, J.; Wheeler, R.; Jirsová, D.; David, V.; Archibald, J.M. Massive mitochondrial DNA content in diplonemid and kinetoplastid protists: Massive mitochondrial DNA in protists. IUBMB Life 2018, 70, 1267–1274. [Google Scholar] [CrossRef] [PubMed]
- Sloan, D.B.; Alverson, A.J.; Chuckalovcak, J.P.; Wu, M.; McCauley, D.E.; Palmer, J.D.; Taylor, D.R. Rapid Evolution of Enormous, Multichromosomal Genomes in Flowering Plant Mitochondria with Exceptionally High Mutation Rates. PLoS Biol. 2012, 10, e1001241. [Google Scholar] [CrossRef]
- Unseld, M.; Marienfeld, J.R.; Brandt, P.; Brennicke, A. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat. Genet. 1997, 15, 57–61. [Google Scholar] [CrossRef]
- Palmer, J.D.; Herbon, L.A. Plant mitochondrial DNA evolves rapidly in structure, but slowly in sequence. J. Mol. Evol. 1988, 28, 87–97. [Google Scholar] [CrossRef]
- Wolfe, K.H.; Li, W.H.; Sharp, P.M. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl. Acad. Sci. USA 1987, 84, 9054–9058. [Google Scholar] [CrossRef]
- Drouin, G.; Daoud, H.; Xia, J. Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. Mol. Phylogenet. Evol. 2008, 49, 827–831. [Google Scholar] [CrossRef]
- Richardson, A.O.; Rice, D.W.; Young, G.J.; Alverson, A.J.; Palmer, J.D. The “fossilized” mitochondrial genome of Liriodendron tulipifera: Ancestral gene content and order, ancestral editing sites, and extraordinarily low mutation rate. BMC Biol. 2013, 11, 29. [Google Scholar] [CrossRef]
- Bakker, F.T.; Breman, F.; Merckx, V. DNA sequence evolution in fast evolving mitochondrial DNA nad1 exons in Geraniaceae and Plantaginaceae. Taxon 2006, 55, 887–896. [Google Scholar] [CrossRef]
- Cho, Y.; Mower, J.P.; Qiu, Y.L.; Palmer, J.D. Mitochondrial substitution rates are extraordinarily elevated and variable in a genus of flowering plants. Proc. Natl. Acad. Sci. USA 2004, 101, 17741–17746. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, C.L.; Mower, J.P.; Qiu, Y.L.; Shirk, A.J.; Song, K.; Young, N.D.; DePamphilis, C.W.; Palmer, J.D. Multiple major increases and decreases in mitochondrial substitution rates in the plant family Geraniaceae. BMC Evol. Biol. 2005, 5, 73. [Google Scholar] [CrossRef] [PubMed]
- Mower, J.P.; Touzet, P.; Gummow, J.S.; Delph, L.F.; Palmer, J.D. Extensive variation in synonymous substitution rates in mitochondrial genes of seed plants. BMC Evol. Biol. 2007, 7, 135. [Google Scholar] [CrossRef] [PubMed]
- Sloan, D.B.; Oxelman, B.; Rautenberg, A.; Taylor, D.R. Phylogenetic analysis of mitochondrial substitution rate variation in the angiosperm tribe Sileneae. BMC Evol. Biol. 2009, 9, 260. [Google Scholar] [CrossRef] [PubMed]
- Zhu, A.D.; Guo, W.H.; Jain, K.; Mower, J.P. Unprecedented Heterogeneity in the Synonymous Substitution Rate within a Plant Genome. Mol. Biol. Evol. 2014, 31, 1228–1236. [Google Scholar] [CrossRef] [PubMed]
- Timmis, J.N.; Ayliffe, M.A.; Huang, C.Y.; Martin, W. Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 2004, 5, 123–135. [Google Scholar] [CrossRef]
- Kucej, M.; Butow, R.A. Evolutionary tinkering with mitochondrial nucleoids. Trends Cell Biol. 2007, 17, 586–592. [Google Scholar] [CrossRef]
- Sato, N.; Terasawa, K.; Miyajima, K.; Kabeya, Y. Organization, developmental dynamics, and evolution of plastid nucleoids. Int. Rev. Cytol. 2003, 232, 217–262. [Google Scholar]
- Krupinska, K.; Melonek, J.; Krause, K. New insights into plastid nucleoid structure and functionality. Planta 2013, 237, 653–664. [Google Scholar] [CrossRef]
- Bohne, A.V. The nucleoid as a site of rRNA processing and ribosome assembly. Front. Plant Sci. 2014, 5, 257. [Google Scholar] [CrossRef]
- Arimura, S.; Yamamoto, J.; Aida, G.P.; Nakazono, M.; Tsutsumi, N. Frequent fusion and fission of plant mitochondria with unequal nucleoid distribution. Proc. Natl. Acad. Sci. USA 2004, 101, 7805–7808. [Google Scholar] [CrossRef] [PubMed]
- Kukat, C.; Wurm, C.A.; Spahr, H.; Falkenberg, M.; Larsson, N.G.; Jakobs, S. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc. Natl. Acad. Sci. USA 2011, 108, 13534–13539. [Google Scholar] [CrossRef] [PubMed]
- Kukat, C.; Davies, K.M.; Wurm, C.A.; Spahr, H.; Bonekamp, N.A.; Kuhl, I.; Joos, F.; Polosa, P.L.; Park, C.B.; Posse, V.; et al. Cross-strand binding of TFAM to a single mtDNA molecule forms the mitochondrial nucleoid. Proc. Natl. Acad. Sci. USA 2015, 112, 11288–11293. [Google Scholar] [CrossRef] [PubMed]
- Majeran, W.; Friso, G.; Asakura, Y.; Qu, X.; Huang, M.; Ponnala, L.; Watkins, K.P.; Barkan, A.; van Wijk, K.J. Nucleoid-enriched proteomes in developing plastids and chloroplasts from maize leaves: A new conceptual framework for nucleoid functions. Plant Physiol. 2012, 158, 156–189. [Google Scholar] [CrossRef] [PubMed]
- Parent, J.S.; Lepage, E.; Brisson, N. Divergent roles for the two PolI-like organelle DNA polymerases of Arabidopsis. Plant Physiol. 2011, 156, 254–262. [Google Scholar] [CrossRef]
- Pfalz, J.; Liere, K.; Kandlbinder, A.; Dietz, K.J.; Oelmuller, R. pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 2006, 18, 176–197. [Google Scholar] [CrossRef]
- Melonek, J.; Oetke, S.; Krupinska, K. Multifunctionality of plastid nucleoids as revealed by proteome analyses. Biochim. Biophys. Acta Proteins Proteom. 2016, 1864, 1016–1038. [Google Scholar] [CrossRef]
- Melonek, J.; Matros, A.; Trosch, M.; Mock, H.-P.; Krupinska, K. The Core of Chloroplast Nucleoids Contains Architectural SWIB Domain Proteins. Plant Cell 2012, 24, 3060–3073. [Google Scholar] [CrossRef]
- Greiner, S.; Golczyk, H.; Malinova, I.; Pellizzer, T.; Bock, R.; Börner, T.; Herrmann, R.G. Chloroplast nucleoids are highly dynamic in ploidy, number, and structure during angiosperm leaf development. bioRxiv 2019, 632240. [Google Scholar] [CrossRef]
- Gualberto, J.M.; Kuhn, K. DNA-binding proteins in plant mitochondria: Implications for transcription. Mitochondrion 2014, 19, 323–328. [Google Scholar] [CrossRef]
- Xu, Y.-Z.; Arrieta-Montiel, M.P.; Virdi, K.S.; de Paula, W.B.M.; Widhalm, J.R.; Basset, G.J.; Davila, J.I.; Elthon, T.E.; Elowsky, C.G.; Sato, S.J.; et al. MutS HOMOLOG1 is a Nucleoid Protein that Alters Mitochondrial and Plastid Properties and Plant Response to High Light. Plant Cell 2011, 23, 3428–3441. [Google Scholar] [CrossRef] [PubMed]
- Oda, K.; Kohchi, T.; Ohyama, K. Mitochondrial DNA of Marchantia polymorpha as a single circular form with no incorporation of foreign DNA. Biosci. Biotechnol. Biochem. 1992, 56, 132–135. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Negruk, V.I.; Eisner, G.I.; Redichkina, T.D.; Dumanskaya, N.N.; Cherny, D.I.; Alexandrov, A.A.; Shemyakin, M.F.; Butenko, R.G. Diversity of Vicia faba circular mtDNA in whole plants and suspension cultures. Theor. Appl. Genet. 1986, 72, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Oldenburg, D.J.; Bendich, A.J. The structure of mitochondrial DNA from the liverwort. Marchantia Polymorpha J. Mol. Biol. 1998, 276, 745–758. [Google Scholar] [CrossRef] [PubMed]
- Backert, S.; Lurz, R.; Borner, T. Electron microscopic investigation of mitochondrial DNA from Chenopodium album (L.). Curr. Genet. 1996, 29, 427–436. [Google Scholar] [CrossRef]
- Backert, S.; Nielsen, B.L.; Borner, T. The mystery of the rings: Structure and replication of mitochondrial genomes from higher plants. Trends Plant Sci. 1997, 2, 477–483. [Google Scholar] [CrossRef]
- Backert, S.; Lurz, R.; Oyarzabal, O.A.; Börner, T. High content, size and distribution of single-stranded DNA in the mitochondria of Chenopodium album (L.). Plant Mol. Biol. 1997, 33, 1037–1050. [Google Scholar] [CrossRef]
- Bendich, A.J. Structural analysis of mitochondrial DNA molecules from fungi and plants using moving pictures and pulsed-field gel electrophoresis. J. Mol. Biol. 1996, 255, 564–588. [Google Scholar] [CrossRef]
- Kozik, A.; Rowan, B.A.; Lavelle, D.; Berke, L.; Schranz, M.E.; Michelmore, R.W.; Christensen, A.C. The alternative reality of plant mitochondrial DNA: One ring does not rule them all. PLoS Genet. 2019, 15, 1008373. [Google Scholar] [CrossRef]
- Oldenburg, D.J.; Bendich, A.J. DNA maintenance in plastids and mitochondria of plants. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef]
- Sloan, D.B. One ring to rule them all? Genome sequencing provides new insights into the ‘master circle’ model of plant mitochondrial DNA structure. New Phytol. 2013, 200, 978–985. [Google Scholar] [CrossRef] [PubMed]
- Stupar, R.M.; Lilly, J.W.; Town, C.D.; Cheng, Z.; Kaul, S.; Buell, C.R.; Jiang, J. Complex mtDNA constitutes an approximate 620-kb insertion on Arabidopsis thaliana chromosome 2: Implication of potential sequencing errors caused by large-unit repeats. Proc. Natl. Acad. Sci. USA 2001, 98, 5099–5103. [Google Scholar] [CrossRef] [PubMed]
- Woloszynska, M.; Trojanowski, D. Counting mtDNA molecules in Phaseolus vulgaris: Sublimons are constantly produced by recombination via short repeats and undergo rigorous selection during substoichiometric shifting. Plant Mol. Biol. 2009, 70, 511–521. [Google Scholar] [CrossRef] [PubMed]
- Janicka, S.; Kuhn, K.; Le Ret, M.; Bonnard, G.; Imbault, P.; Augustyniak, H.; Gualberto, J.M. A RAD52-like single-stranded DNA binding protein affects mitochondrial DNA repair by recombination. Plant J. 2012, 72, 423–435. [Google Scholar] [CrossRef] [PubMed]
- Small, I.; Suffolk, R.; Leaver, C.J. Evolution of plant mitochondrial genomes via substoichiometric intermediates. Cell 1989, 58, 69–76. [Google Scholar] [CrossRef]
- Preuten, T.; Cincu, E.; Fuchs, J.; Zoschke, R.; Liere, K.; Borner, T. Fewer genes than organelles: Extremely low and variable gene copy numbers in mitochondria of somatic plant cells. Plant J. 2010, 64, 948–959. [Google Scholar] [CrossRef]
- Takanashi, H.; Arimura, S.; Sakamoto, W.; Tsutsumi, N. Different amounts of DNA in each mitochondrion in rice root. Genes Genet. Syst. 2006, 81, 215–218. [Google Scholar] [CrossRef]
- Backert, S.; Börner, T. Phage T4-like intermediates of DNA replication and recombination in the mitochondria of the higher plant Chenopodium album (L.). Curr Genet. 2000, 37, 304–314. [Google Scholar] [CrossRef]
- Oldenburg, D.J.; Bendich, A.J. Size and Structure of Replicating Mitochondrial DNA in Cultured Tobacco Cells. Plant Cell 1996, 8, 447–461. [Google Scholar] [CrossRef]
- Backert, S. R-loop-dependent rolling-circle replication and a new model for DNA concatemer resolution by mitochondrial plasmid mp1. EMBO J. 2002, 21, 3128–3136. [Google Scholar] [CrossRef]
- Kazama, T.; Okuno, M.; Watari, Y.; Yanase, S.; Koizuka, C.; Tsuruta, Y.; Sugaya, H.; Toyoda, A.; Itoh, T.; Tsutsumi, N.; et al. Curing cytoplasmic male sterility via TALEN-mediated mitochondrial genome editing. Nat. Plants 2019, 5, 722–730. [Google Scholar] [CrossRef] [PubMed]
- Wallet, C.; Le Ret, M.; Bergdoll, M.; Bichara, M.; Dietrich, A.; Gualberto, J.M. The RECG1 DNA Translocase is a Key Factor in Recombination Surveillance, Repair, and Segregation of the Mitochondrial DNA in Arabidopsis. Plant Cell 2015, 27, 2907–2925. [Google Scholar] [CrossRef] [PubMed]
- Moriyama, T.; Sato, N. Enzymes involved in organellar DNA replication in photosynthetic eukaryotes. Front. Plant Sci. 2014, 5, 480. [Google Scholar] [CrossRef] [PubMed]
- Morley, S.A.; Peralta-Castro, A.; Brieba, L.G.; Miller, J.; Ong, K.L.; Ridge, P.G.; Oliphant, A.; Aldous, S.; Nielsen, B.L. Arabidopsis thaliana organelles mimic the T7 phage DNA replisome with specific interactions between Twinkle protein and DNA polymerases Pol1A and Pol1B. BMC Plant Biol. 2019, 19, 241. [Google Scholar] [CrossRef]
- Diray-Arce, J.; Liu, B.; Cupp, J.D.; Hunt, T.; Nielsen, B.L. The Arabidopsis At1g30680 gene encodes a homologue to the phage T7 gp4 protein that has both DNA primase and DNA helicase activities. BMC Plant Biol. 2013, 13, 36. [Google Scholar] [CrossRef]
- Peralta-Castro, A.; Baruch-Torres, N.; Brieba, L.G. Plant organellar DNA primase-helicase synthesizes RNA primers for organellar DNA polymerases using a unique recognition sequence. Nucleic Acids Res. 2017, 45, 10764–10774. [Google Scholar] [CrossRef][Green Version]
- Takanashi, H.; Ohnishi, T.; Mogi, M.; Okamoto, T.; Arimura, S.; Tsutsumi, N. Studies of mitochondrial morphology and DNA amount in the rice egg cell. Curr. Genet. 2010, 56, 33–41. [Google Scholar] [CrossRef]
- Wang, D.Y.; Zhang, Q.; Liu, Y.; Lin, Z.F.; Zhang, S.X.; Sun, M.X. The levels of male gametic mitochondrial DNA are highly regulated in angiosperms with regard to mitochondrial inheritance. Plant Cell 2010, 22, 2402–2416. [Google Scholar] [CrossRef]
- Sodmergen; Zhang, Q.; Zhang, Y.; Sakamoto, W.; Kuroiwa, T. Reduction in amounts of mitochondrial DNA in the sperm cells as a mechanism for maternal inheritance in Hordeum vulgare. Planta 2002, 216, 235–244. [Google Scholar] [CrossRef]
- Tang, L.Y.; Sakamoto, W. Tissue-specific organelle DNA degradation mediated by DPD1 exonuclease. Plant Signal. Behav. 2011, 6, 1391–1393. [Google Scholar] [CrossRef]
- Takami, T.; Ohnishi, N.; Kurita, Y.; Iwamura, S.; Ohnishi, M.; Kusaba, M.; Mimura, T.; Sakamoto, W. Organelle DNA degradation contributes to the efficient use of phosphate in seed plants. Nat. Plants 2018, 4, 1044–1055. [Google Scholar] [CrossRef] [PubMed]
- Karol, M.H.; Simpson, M.V. DNA biosynthesis by isolated mitochondria: A replicative rather than a repair process. Science 1968, 162, 470–473. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Demple, B. DNA repair in mammalian mitochondria: Much more than we thought? Environ. Mol. Mutagen. 2010, 51, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Boesch, P.; Weber-Lotfi, F.; Ibrahim, N.; Tarasenko, V.; Cosset, A.; Paulus, F.; Lightowlers, R.N.; Dietrich, A. DNA repair in organelles: Pathways, organization, regulation, relevance in disease and aging. Biochim. Biophys. Acta 2011, 1813, 186–200. [Google Scholar] [CrossRef]
- Brown, W.M.; George, M.; Wilson, A.C. Rapid evolution of animal mitochondrial DNA. Proc. Natl. Acad. Sci. USA 1979, 76, 1967–1971. [Google Scholar] [CrossRef]
- Christensen, A.C. Plant mitochondrial genome evolution can be explained by DNA repair mechanisms. Genome Biol. Evol. 2013, 5, 1079–1086. [Google Scholar] [CrossRef]
- Davila, J.I.; Arrieta-Montiel, M.P.; Wamboldt, Y.; Cao, J.; Hagmann, J.; Shedge, V.; Xu, Y.Z.; Weigel, D.; Mackenzie, S.A. Double-strand break repair processes drive evolution of the mitochondrial genome in Arabidopsis. BMC Biol. 2011, 9, 64. [Google Scholar] [CrossRef]
- Spampinato, C.P. Protecting DNA from errors and damage: An overview of DNA repair mechanisms in plants compared to mammals. Cell Mol. Life Sci. 2017, 74, 1693–1709. [Google Scholar] [CrossRef]
- Sancar, A.; Sancar, G.B. DNA repair enzymes. Annu. Rev. Biochem. 1988, 57, 29–67. [Google Scholar] [CrossRef]
- Brettel, K.; Byrdin, M. Reaction mechanisms of DNA photolyase. Curr. Opin. Struct. Biol. 2010, 20, 693–701. [Google Scholar] [CrossRef]
- Chen, J.J.; Jiang, C.Z.; Britt, A.B. Little or no repair of cyclobutyl pyrimidine dimers is observed in the organellar genomes of the young Arabidopsis seedling. Plant Physiol. 1996, 111, 19–25. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Small, G.D. Repair systems for nuclear and chloroplast DNA in Chlamydomonas reinhardtii. Mutat. Res. 1987, 181, 31–35. [Google Scholar] [CrossRef]
- Yasui, A.; Yajima, H.; Kobayashi, T.; Eker, A.P.; Oikawa, A. Mitochondrial DNA repair by photolyase. Mutat. Res. 1992, 273, 231–236. [Google Scholar] [CrossRef]
- Hada, M.; Hino, K.; Buchholz, G.; Goss, J.; Wellmann, E.; Shin, M. Assay of DNA Photolyase Activity in Spinach Leaves in Relation to Cell Compartmentation-Evidence for Lack of DNA Photolyase in Chloroplasts. Biosci. Biotechnol. Biochem. 2000, 64, 1288–1291. [Google Scholar] [CrossRef]
- Kleine, T.; Lockhart, P.; Batschauer, A. An Arabidopsis protein closely related to Synechocystis cryptochrome is targeted to organelles. Plant J. 2003, 35, 93–103. [Google Scholar] [CrossRef]
- Takahashi, M.; Teranishi, M.; Ishida, H.; Kawasaki, J.; Takeuchi, A.; Yamaya, T.; Watanabe, M.; Makino, A.; Hidema, J. Cyclobutane pyrimidine dimer (CPD) photolyase repairs ultraviolet-B-induced CPDs in rice chloroplast and mitochondrial DNA: CPD photolyase in rice chloroplasts and mitochondria. Plant J. 2011, 66, 433–442. [Google Scholar] [CrossRef]
- Schofield, M.J.; Hsieh, P. DNA mismatch repair: Molecular mechanisms and biological function. Annu. Rev. Microbiol. 2003, 57, 579–608. [Google Scholar] [CrossRef]
- Spampinato, C.P.; Gomez, R.L.; Galles, C.; Lario, L.D. From bacteria to plants: A compendium of mismatch repair assays. Mutat. Res. 2009, 682, 110–128. [Google Scholar] [CrossRef]
- Abdelnoor, R.V.; Christensen, A.C.; Mohammed, S.; Munoz-Castillo, B.; Moriyama, H.; Mackenzie, S.A. Mitochondrial Genome Dynamics in Plants and Animals: Convergent Gene Fusions of a MutS Homologue. J. Mol. Evol. 2006, 63, 165–173. [Google Scholar] [CrossRef]
- Fukui, K.; Harada, A.; Wakamatsu, T.; Minobe, A.; Ohshita, K.; Ashiuchi, M.; Yano, T. The GIY-YIG endonuclease domain of Arabidopsis MutS homolog 1 specifically binds to branched DNA structures. FEBS Lett. 2018, 592, 4066–4077. [Google Scholar] [CrossRef]
- Shedge, V.; Arrieta-Montiel, M.; Christensen, A.C.; Mackenzie, S.A. Plant mitochondrial recombination surveillance requires unusual RecA and MutS homologs. Plant Cell 2007, 19, 1251–1264. [Google Scholar] [CrossRef] [PubMed]
- Sancar, A.; Reardon, J.T. Nucleotide excision repair in E. coli and man. Adv. Protein Chem. 2004, 69, 43–71. [Google Scholar] [CrossRef] [PubMed]
- Melis, J.P.M.; van Steeg, H.; Luijten, M. Oxidative DNA Damage and Nucleotide Excision Repair. Antioxid. Redox Signal. 2013, 18, 2409–2419. [Google Scholar] [CrossRef] [PubMed]
- Selby, C.P. Mfd Protein and Transcription-Repair Coupling in E. coli. Photochem. Photobiol. 2017, 93, 280–295. [Google Scholar] [CrossRef]
- Kamenisch, Y.; Fousteri, M.; Knoch, J.; von Thaler, A.-K.; Fehrenbacher, B.; Kato, H.; Becker, T.; Dollé, M.E.T.; Kuiper, R.; Majora, M.; et al. Proteins of nucleotide and base excision repair pathways interact in mitochondria to protect from loss of subcutaneous fat, a hallmark of aging. J. Exp. Med. 2010, 207, 379–390. [Google Scholar] [CrossRef]
- Liu, J.; Fang, H.; Chi, Z.; Wu, Z.; Wei, D.; Mo, D.; Niu, K.; Balajee, A.S.; Hei, T.K.; Nie, L.; et al. XPD localizes in mitochondria and protects the mitochondrial genome from oxidative DNA damage. Nucleic Acids Res. 2015, 43, 5476–5488. [Google Scholar] [CrossRef]
- Stevnsner, T.; Nyaga, S.; de Souza-Pinto, N.C.; van der Horst, G.T.J.; Gorgels, T.G.M.F.; Hogue, B.A.; Thorslund, T.; Bohr, V.A. Mitochondrial repair of 8-oxoguanine is deficient in Cockayne syndrome group B. Oncogene 2002, 21, 8675–8682. [Google Scholar] [CrossRef]
- Van der Veen, S.; Tang, C.M. The BER necessities: The repair of DNA damage in human-adapted bacterial pathogens. Nat. Rev. Microbiol. 2015, 13, 83–94. [Google Scholar] [CrossRef]
- Krokan, H.E.; Standal, R.; Slupphaug, G. DNA glycosylases in the base excision repair of DNA. Biochem. J. 1997, 325, 1–16. [Google Scholar] [CrossRef]
- Prakash, A.; Doublie, S. Base Excision Repair in the Mitochondria. J. Cell. Biochem. 2015, 116, 1490–1499. [Google Scholar] [CrossRef]
- Boesch, P.; Ibrahim, N.; Paulus, F.; Cosset, A.; Tarasenko, V.; Dietrich, A. Plant mitochondria possess a short-patch base excision DNA repair pathway. Nucleic Acids Res. 2009, 37, 5690–5700. [Google Scholar] [CrossRef] [PubMed]
- Bensen, R.J.; Warner, H.R. The Partial Purification and Characterization of Nuclear and Mitochondrial Uracil-DNA Glycosylase Activities from Zea mays Seedlings. Plant Physiol. 1987, 83, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Ferrando, B.; Furlanetto, A.L.D.M.; Gredilla, R.; Havelund, J.F.; Hebelstrup, K.H.; Moller, I.M.; Stevnsner, T. DNA repair in plant mitochondria—A complete base excision repair pathway in potato tuber mitochondria. Physiol. Plant 2019, 166, 494–512. [Google Scholar] [CrossRef] [PubMed]
- Furlanetto, A.L.D.M.; Cadena, S.M.S.C.; Martinez, G.R.; Ferrando, B.; Stevnsner, T.; Moller, I.M. Short-term high temperature treatment reduces viability and inhibits respiration and DNA repair enzymes in Araucaria angustifolia cells. Physiol. Plant 2019, 166, 513–524. [Google Scholar] [CrossRef] [PubMed]
- Cordoba-Canero, D.; Roldan-Arjona, T.; Ariza, R.R. Arabidopsis ZDP DNA 3′-phosphatase and ARP endonuclease function in 8-oxoG repair initiated by FPG and OGG1 DNA glycosylases. Plant J. 2014, 79, 824–834. [Google Scholar] [CrossRef] [PubMed]
- Macovei, A.; Balestrazzi, A.; Confalonieri, M.; Fae, M.; Carbonera, D. New insights on the barrel medic MtOGG1 and MtFPG functions in relation to oxidative stress response in planta and during seed imbibition. Plant Physiol. Biochem. 2011, 49, 1040–1050. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Chu, P.; Zhou, Y.; Li, Y.; Liu, J.; Ding, Y.; Tsang, E.W.; Jiang, L.; Wu, K.; Huang, S. Overexpression of AtOGG1, a DNA glycosylase/AP lyase, enhances seed longevity and abiotic stress tolerance in Arabidopsis. J. Exp. Bot. 2012, 63, 4107–4121. [Google Scholar] [CrossRef]
- Swartzlander, D.B.; Griffiths, L.M.; Lee, J.; Degtyareva, N.P.; Doetsch, P.W.; Corbett, A.H. Regulation of base excision repair: Ntg1 nuclear and mitochondrial dynamic localization in response to genotoxic stress. Nucleic Acids Res. 2010, 38, 3963–3974. [Google Scholar] [CrossRef]
- Ohtsubo, T.; Matsuda, O.; Iba, K.; Terashima, I.; Sekiguchi, M.; Nakabeppu, Y. Molecular cloning of AtMMH, an Arabidopsis thaliana ortholog of the E. coli mutM gene, and analysis of functional domains of its product. Mol. Gen. Genet. 1998, 259, 577–590. [Google Scholar] [CrossRef]
- Murphy, T.M.; Gao, M.J. Multiple forms of formamidopyrimidine-DNA glycosylase produced by alternative splicing in Arabidopsis thaliana. J. Photochem. Photobiol. B 2001, 61, 87–93. [Google Scholar] [CrossRef]
- Duclos, S.; Aller, P.; Jaruga, P.; Dizdaroglu, M.; Wallace, S.S.; Doublie, S. Structural and biochemical studies of a plant formamidopyrimidine-DNA glycosylase reveal why eukaryotic Fpg glycosylases do not excise 8-oxoguanine. DNA Repair 2012, 11, 714–725. [Google Scholar] [CrossRef] [PubMed]
- Kathe, S.D.; Barrantes-Reynolds, R.; Jaruga, P.; Newton, M.R.; Burrows, C.J.; Bandaru, V.; Dizdaroglu, M.; Bond, J.P.; Wallace, S.S. Plant and fungal Fpg homologs are formamidopyrimidine DNA glycosylases but not 8-oxoguanine DNA glycosylases. DNA Repair 2009, 8, 643–653. [Google Scholar] [CrossRef] [PubMed]
- Kimura, S.; Uchiyama, Y.; Kasai, N.; Namekawa, S.; Saotome, A.; Ueda, T.; Ando, T.; Ishibashi, T.; Oshige, M.; Furukawa, T.; et al. A novel DNA polymerase homologous to E. coli DNA polymerase I from a higher plant, rice (Oryza sativa L.). Nucleic Acids Res. 2002, 30, 1585–1592. [Google Scholar] [CrossRef] [PubMed]
- Shutt, T.E.; Gray, M.W. Bacteriophage origins of mitochondrial replication and transcription proteins. Trends Genet. 2006, 22, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Elo, A.; Lyznik, A.; Gonzalez, D.O.; Kachman, S.D.; Mackenzie, S.A. Nuclear genes that encode mitochondrial proteins for DNA and RNA metabolism are clustered in the Arabidopsis genome. Plant Cell 2003, 15, 1619–1631. [Google Scholar] [CrossRef]
- Ono, Y.; Sakai, A.; Takechi, K.; Takio, S.; Takusagawa, M.; Takano, H. NtPolI-like1 and NtPolI-like2, bacterial DNA polymerase I homologs isolated from BY-2 cultured tobacco cells, encode DNA polymerases engaged in DNA replication in both plastids and mitochondria. Plant Cell Physiol. 2007, 48, 1679–1692. [Google Scholar] [CrossRef]
- Christensen, A.C.; Lyznik, A.; Mohammed, S.; Elowsky, C.G.; Elo, A.; Yule, R.; Mackenzie, S.A. Dual-domain, dual-targeting organellar protein presequences in Arabidopsis can use non-AUG start codons. Plant Cell 2005, 17, 2805–2816. [Google Scholar] [CrossRef]
- Mori, Y.; Kimura, S.; Saotome, A.; Kasai, N.; Sakaguchi, N.; Uchiyama, Y.; Ishibashi, T.; Yamamoto, T.; Chiku, H.; Sakaguchi, K. Plastid DNA polymerases from higher plants, Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2005, 334, 43–50. [Google Scholar] [CrossRef]
- Takeuchi, R.; Kimura, S.; Saotome, A.; Sakaguchi, K. Biochemical properties of a plastidial DNA polymerase of rice. Plant Mol. Biol. 2007, 64, 601–611. [Google Scholar] [CrossRef]
- Baruch-Torres, N.; Brieba, L.G. Plant organellar DNA polymerases are replicative and translesion DNA synthesis polymerases. Nucleic Acids Res. 2017, 45, 10751–10763. [Google Scholar] [CrossRef]
- Cupp, J.D.; Nielsen, B.L. Arabidopsis thaliana organellar DNA polymerase IB mutants exhibit reduced mtDNA levels with a decrease in mitochondrial area density. Physiol. Plant 2012. [Google Scholar] [CrossRef]
- Trasvina-Arenas, C.H.; Baruch-Torres, N.; Cordoba-Andrade, F.J.; Ayala-Garcia, V.M.; Garcia-Medel, P.L.; Diaz-Quezada, C.; Peralta-Castro, A.; Ordaz-Ortiz, J.J.; Brieba, L.G. Identification of a unique insertion in plant organellar DNA polymerases responsible for 5′-dRP lyase and strand-displacement activities: Implications for Base Excision Repair. DNA Repair 2018, 65, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Cordoba-Canero, D.; Roldan-Arjona, T.; Ariza, R.R. Arabidopsis ARP endonuclease functions in a branched base excision DNA repair pathway completed by LIG1. Plant J. 2011, 68, 693–702. [Google Scholar] [CrossRef] [PubMed]
- Sunderland, P.A.; West, C.E.; Waterworth, W.M.; Bray, C.M. An evolutionarily conserved translation initiation mechanism regulates nuclear or mitochondrial targeting of DNA ligase 1 in Arabidopsis thaliana. Plant J. 2006, 47, 356–367. [Google Scholar] [CrossRef] [PubMed]
- Heyer, W.D.; Ehmsen, K.T.; Liu, J. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 2010, 44, 113–139. [Google Scholar] [CrossRef] [PubMed]
- Rocha, E.P.; Cornet, E.; Michel, B. Comparative and evolutionary analysis of the bacterial homologous recombination systems. PLoS Genet. 2005, 1, e15. [Google Scholar] [CrossRef]
- Kreuzer, K.N. Recombination-dependent DNA replication in phage T4. Trends Biochem. Sci. 2000, 25, 165–173. [Google Scholar] [CrossRef]
- Yeeles, J.T.; Poli, J.; Marians, K.J.; Pasero, P. Rescuing stalled or damaged replication forks. Cold Spring Harb. Perspect. Biol. 2013, 5, a012815. [Google Scholar] [CrossRef]
- Thomas, C.M.; Nielsen, K.M. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 2005, 3, 711–721. [Google Scholar] [CrossRef]
- Orthwein, A.; Noordermeer, S.M.; Wilson, M.D.; Landry, S.; Enchev, R.I.; Sherker, A.; Munro, M.; Pinder, J.; Salsman, J.; Dellaire, G.; et al. A mechanism for the suppression of homologous recombination in G1 cells. Nature 2015, 528, 422–426. [Google Scholar] [CrossRef]
- Puchta, H. Double-strand break-induced recombination between ectopic homologous sequences in somatic plant cells. Genetics 1999, 152, 1173–1181. [Google Scholar] [PubMed]
- Chang, H.H.Y.; Pannunzio, N.R.; Adachi, N.; Lieber, M.R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017, 18, 495–506. [Google Scholar] [CrossRef] [PubMed]
- Cappadocia, L.; Marechal, A.; Parent, J.S.; Lepage, E.; Sygusch, J.; Brisson, N. Crystal structures of DNA-Whirly complexes and their role in Arabidopsis organelle genome repair. Plant Cell 2010, 22, 1849–1867. [Google Scholar] [CrossRef] [PubMed]
- Zaegel, V.; Guermann, B.; Le Ret, M.; Andres, C.; Meyer, D.; Erhardt, M.; Canaday, J.; Gualberto, J.M.; Imbault, P. The plant-specific ssDNA binding protein OSB1 is involved in the stoichiometric transmission of mitochondrial DNA in Arabidopsis. Plant Cell 2006, 18, 3548–3563. [Google Scholar] [CrossRef]
- Arrieta-Montiel, M.P.; Shedge, V.; Davila, J.; Christensen, A.C.; Mackenzie, S.A. Diversity of the Arabidopsis mitochondrial genome occurs via nuclear-controlled recombination activity. Genetics 2009, 183, 1261–1268. [Google Scholar] [CrossRef]
- Miller-Messmer, M.; Kuhn, K.; Bichara, M.; Le Ret, M.; Imbault, P.; Gualberto, J.M. RecA-dependent DNA repair results in increased heteroplasmy of the Arabidopsis mitochondrial genome. Plant Physiol. 2012, 159, 211–226. [Google Scholar] [CrossRef]
- Galletto, R.; Amitani, I.; Baskin, R.J.; Kowalczykowski, S.C. Direct observation of individual RecA filaments assembling on single DNA molecules. Nature 2006, 443, 875–878. [Google Scholar] [CrossRef]
- Handa, N.; Amitani, I.; Gumlaw, N.; Sandler, S.J.; Kowalczykowski, S.C. Single Molecule Analysis of a Red Fluorescent RecA Protein Reveals a Defect in Nucleoprotein Filament Nucleation That Relates to its Reduced Biological Functions. J. Biol. Chem. 2009, 284, 18664–18673. [Google Scholar] [CrossRef]
- Morimatsu, K.; Kowalczykowski, S.C. RecQ helicase and RecJ nuclease provide complementary functions to resect DNA for homologous recombination. Proc. Natl. Acad. Sci. USA 2014, 111, e5133–e5142. [Google Scholar] [CrossRef]
- Roy, R.; Kozlov, A.G.; Lohman, T.M.; Ha, T. SSB protein diffusion on single-stranded DNA stimulates RecA filament formation. Nature 2009, 461, 1092–1097. [Google Scholar] [CrossRef]
- Umezu, K.; Kolodner, R.D. Protein interactions in genetic recombination in E. coli. Interactions involving RecO and RecR overcome the inhibition of RecA by single-stranded DNA-binding protein. J. Biol. Chem. 1994, 269, 30005–30013. [Google Scholar] [PubMed]
- Sandler, S.J.; Clark, A.J. RecOR suppression of recF mutant phenotypes in E. coli K-12. J. Bacteriol. 1994, 176, 3661–3672. [Google Scholar] [CrossRef] [PubMed]
- McGlynn, P.; Lloyd, R.G. Recombinational repair and restart of damaged replication forks. Nat. Rev. Mol. Cell Biol. 2002, 3, 859–870. [Google Scholar] [CrossRef] [PubMed]
- Forget, A.L.; Kowalczykowski, S.C. Single-molecule imaging of DNA pairing by RecA reveals a three-dimensional homology search. Nature 2012, 482, 423–427. [Google Scholar] [CrossRef] [PubMed]
- Ragunathan, K.; Liu, C.; Ha, T. RecA filament sliding on DNA facilitates homology search. Elife 2012, 1, e00067. [Google Scholar] [CrossRef] [PubMed]
- Cárdenas, P.P.; Carrasco, B.; Defeu Soufo, C.; César, C.E.; Herr, K.; Kaufenstein, M.; Graumann, P.L.; Alonso, J.C. RecX Facilitates Homologous Recombination by Modulating RecA Activities. PLoS Genet. 2012, 8, e1003126. [Google Scholar] [CrossRef] [PubMed]
- Petrova, V.; Chen, S.H.; Molzberger, E.T.; Tomko, E.; Chitteni-Pattu, S.; Jia, H.; Ordabayev, Y.; Lohman, T.M.; Cox, M.M. Active displacement of RecA filaments by UvrD translocase activity. Nucleic Acids Res. 2015, 43, 4133–4149. [Google Scholar] [CrossRef]
- Mawer, J.S.; Leach, D.R. Branch migration prevents DNA loss during double-strand break repair. PLoS Genet. 2014, 10, e1004485. [Google Scholar] [CrossRef]
- Cooper, D.L.; Lovett, S.T. Recombinational branch migration by the RadA/Sms paralog of RecA in E. coli. Elife 2016, 5. [Google Scholar] [CrossRef]
- Marie, L.; Rapisarda, C.; Morales, V.; Bergé, M.; Perry, T.; Soulet, A.-L.; Gruget, C.; Remaut, H.; Fronzes, R.; Polard, P. Bacterial RadA is a DnaB-type helicase interacting with RecA to promote bidirectional D-loop extension. Nat. Commun. 2017, 8, 15638. [Google Scholar] [CrossRef]
- Whitby, M.C.; Vincent, S.D.; Lloyd, R.G. Branch migration of Holliday junctions: Identification of RecG protein as a junction specific DNA helicase. EMBO J. 1994, 13, 5220–5228. [Google Scholar] [CrossRef] [PubMed]
- West, S.C. Processing of recombination intermediates by the RuvABC proteins. Annu. Rev. Genet. 1997, 31, 213–244. [Google Scholar] [CrossRef] [PubMed]
- Edmondson, A.C.; Song, D.; Alvarez, L.A.; Wall, M.K.; Almond, D.; McClellan, D.A.; Maxwell, A.; Nielsen, B.L. Characterization of a mitochondrially targeted single-stranded DNA-binding protein in Arabidopsis thaliana. Mol. Genet. Genom. 2005, 273, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Medel, P.L.; Baruch-Torres, N.; Peralta-Castro, A.; Trasvina-Arenas, C.H.; Torres-Larios, A.; Brieba, L.G. Plant organellar DNA polymerases repair double-stranded breaks by microhomology-mediated end-joining. Nucleic Acids Res. 2019. [Google Scholar] [CrossRef] [PubMed]
- Marechal, A.; Parent, J.S.; Veronneau-Lafortune, F.; Joyeux, A.; Lang, B.F.; Brisson, N. Whirly proteins maintain plastid genome stability in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 14693–14698. [Google Scholar] [CrossRef] [PubMed]
- Samach, A.; Melamed-Bessudo, C.; Avivi-Ragolski, N.; Pietrokovski, S.; Levy, A.A. Identification of plant RAD52 homologs and characterization of the Arabidopsis thaliana RAD52-like genes. Plant Cell 2011, 23, 4266–4279. [Google Scholar] [CrossRef]
- Pfalz, J.; Pfannschmidt, T. Essential nucleoid proteins in early chloroplast development. Trends Plant Sci. 2013, 18, 186–194. [Google Scholar] [CrossRef]
- Wallet, C. L’hélicase RECG1, un Facteur-clé Dans le Maintien et la Ségrégation de l’ADN Mitochondrial d’Arabidopsis thaliana; Université de Strasbourg: Strasbourg, France, 2016. [Google Scholar]
- Drees, J.C.; Lusetti, S.L.; Cox, M.M. Inhibition of RecA protein by the E. coli RecX protein: Modulation by the RecA C terminus and filament functional state. J. Biol. Chem. 2004, 279, 52991–52997. [Google Scholar] [CrossRef]
- Eggler, A.L.; Lusetti, S.L.; Cox, M.M. The C terminus of the E. coli RecA protein modulates the DNA binding competition with single-stranded DNA-binding protein. J. Biol. Chem. 2003, 278, 16389–16396. [Google Scholar] [CrossRef]
- Odahara, M.; Sekine, Y. RECX interacts with mitochondrial RECA to maintain mitochondrial genome stability. Plant Physiol. 2018. [Google Scholar] [CrossRef]
- Odahara, M.; Masuda, Y.; Sato, M.; Wakazaki, M.; Harada, C.; Toyooka, K.; Sekine, Y. RECG maintains plastid and mitochondrial genome stability by suppressing extensive recombination between short dispersed repeats. PLoS Genet. 2015, 11, e1005080. [Google Scholar] [CrossRef] [PubMed]
- Beam, C.E.; Saveson, C.J.; Lovett, S.T. Role for radA/sms in recombination intermediate processing in E. coli. J. Bacteriol. 2002, 184, 6836–6844. [Google Scholar] [CrossRef] [PubMed]
- Chevigny, N.; Nadiras, C.; Raynaud, C.; Le Ret, M.; Bichara, M.; Erhardt, M.; Dietrich, A.; Gualberto, J.M. RADA is the main branch migration factor in plant mitochondrial recombination and its defect leads to mtDNA instability and cell cycle arrest. bioRxiv 2019. [Google Scholar] [CrossRef]
- Cox, M.M. Regulation of bacterial RecA protein function. Crit. Rev. Biochem. Mol. Biol. 2007, 42, 41–63. [Google Scholar] [CrossRef]
- Lusetti, S.L.; Shaw, J.J.; Cox, M.M. Magnesium ion-dependent activation of the RecA protein involves the C terminus. J. Biol. Chem. 2003, 278, 16381–16388. [Google Scholar] [CrossRef]
- Lusetti, S.L.; Wood, E.A.; Fleming, C.D.; Modica, M.J.; Korth, J.; Abbott, L.; Dwyer, D.W.; Roca, A.I.; Inman, R.B.; Cox, M.M. C-terminal deletions of the E. coli RecA protein. Characterization of in vivo and in vitro effects. J. Biol. Chem. 2003, 278, 16372–16380. [Google Scholar] [CrossRef]
- Aravind, L.; Makarova, K.S.; Koonin, E.V. SURVEY AND SUMMARY: Holliday junction resolvases and related nucleases: Identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 2000, 28, 3417–3432. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Misumi, O.; Odahara, M.; Ishibashi, K.; Hirono, M.; Hidaka, K.; Endo, M.; Sugiyama, H.; Iwasaki, H.; Kuroiwa, T.; et al. Holliday junction resolvases mediate chloroplast nucleoid segregation. Science 2017, 356, 631–634. [Google Scholar] [CrossRef]
- Wardrope, L.; Okely, E.; Leach, D. Resolution of joint molecules by RuvABC and RecG following cleavage of the E. coli chromosome by EcoKI. PLoS ONE 2009, 4, e6542. [Google Scholar] [CrossRef]
- Touzet, P.; Meyer, E.H. Cytoplasmic male sterility and mitochondrial metabolism in plants. Mitochondrion 2014, 19, 166–171. [Google Scholar] [CrossRef]
- Woloszynska, M. Heteroplasmy and stoichiometric complexity of plant mitochondrial genomes—Though this be madness, yet there’s method in’t. J. Exp. Bot. 2010, 61, 657–671. [Google Scholar] [CrossRef] [PubMed]
- Christensen, A.C. Genes and junk in plant mitochondria-repair mechanisms and selection. Genome Biol. Evol. 2014, 6, 1448–1453. [Google Scholar] [CrossRef] [PubMed]
- De Paula, W.B.M.; Agip, A.-N.A.; Missirlis, F.; Ashworth, R.; Vizcay-Barrena, G.; Lucas, C.H.; Allen, J.F. Female and Male Gamete Mitochondria Are Distinct and Complementary in Transcription, Structure, and Genome Function. Genome Biol. Evol. 2013, 5, 1969–1977. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Cuthbert, J.M.; Taylor, D.R.; Sloan, D.B. The massive mitochondrial genome of the angiosperm Silene noctiflora is evolving by gain or loss of entire chromosomes. Proc. Natl. Acad. Sci. USA 2015, 112, 10185–10191. [Google Scholar] [CrossRef]
- Lovett, S.T.; Hurley, R.L.; Sutera, V.A., Jr.; Aubuchon, R.H.; Lebedeva, M.A. Crossing over between regions of limited homology in E. coli. RecA-dependent and RecA-independent pathways. Genetics 2002, 160, 851–859. [Google Scholar]
- Vitart, V.; Depaepe, R.; Mathieu, C.; Chetrit, P.; Vedel, F. Amplification of substoichiometric recombinant mitochondrial DNA sequences in a nuclear, male sterile mutant regenerated from protoplast culture in Nicotiana sylvestris. Mol. Gen. Genet. 1992, 233, 193–200. [Google Scholar] [CrossRef]
- Janska, H.; Sarria, R.; Woloszynska, M.; Arrieta-Montiel, M.; Mackenzie, S.A. Stoichiometric shifts in the common bean mitochondrial genome leading to male sterility and spontaneous reversion to fertility. Plant Cell 1998, 10, 1163–1180. [Google Scholar] [CrossRef]
- Bellaoui, M.; Martin-Canadell, A.; Pelletier, G.; Budar, F. Low-copy-number molecules are produced by recombination, actively maintained and can be amplified in the mitochondrial genome of Brassicaceae: Relationship to reversion of the male sterile phenotype in some cybrids. Mol. Gen. Genet. 1998, 257, 177–185. [Google Scholar] [CrossRef]
- Garcia, L.E.; Zubko, M.K.; Zubko, E.I.; Sanchez-Puerta, M.V. Elucidating genomic patterns and recombination events in plant cybrid mitochondria. Plant Mol. Biol. 2019, 100, 433–450. [Google Scholar] [CrossRef]
- Sanchez-Puerta, M.V.; Zubko, M.K.; Palmer, J.D. Homologous recombination and retention of a single form of most genes shape the highly chimeric mitochondrial genome of a cybrid plant. New Phytol. 2015, 206, 381–396. [Google Scholar] [CrossRef]
- Hanson, M.R.; Bentolila, S. Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell 2004, 16, S154–S169. [Google Scholar] [CrossRef] [PubMed]
- Kubo, T.; Newton, K.J. Angiosperm mitochondrial genomes and mutations. Mitochondrion 2008, 8, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Budar, F.; Pelletier, G. Male sterility in plants: Occurrence, determinism, significance and use. C. R. Acad. Sci. III 2001, 324, 543–550. [Google Scholar] [CrossRef]
- Budar, F.; Touzet, P.; De Paepe, R. The nucleo-mitochondrial conflict in cytoplasmic male sterilities revisited. Genetica 2003, 117, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Khakhlova, O.; Bock, R. Elimination of deleterious mutations in plastid genomes by gene conversion. Plant J. 2006, 46, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Kwon, T.; Huq, E.; Herrin, D.L. Microhomology-mediated and nonhomologous repair of a double-strand break in the chloroplast genome of Arabidopsis. Proc. Natl. Acad. Sci. USA 2010, 107, 13954–13959. [Google Scholar] [CrossRef] [PubMed]
- Le Ret, M.; Belcher, S.; Graindorge, S.; Wallet, C.; Koechler, S.; Erhardt, M.; Williams-Carrier, R.; Barkan, A.; Gualberto, J.M. Efficient Replication of the Plastid Genome Requires an Organellar Thymidine Kinase. Plant Physiol. 2018, 178, 1643–1656. [Google Scholar] [CrossRef]
- Zampini, E.; Lepage, E.; Tremblay-Belzile, S.; Truche, S.; Brisson, N. Organelle DNA rearrangement mapping reveals U-turn-like inversions as a major source of genomic instability in Arabidopsis and humans. Genome Res. 2015, 25, 645–654. [Google Scholar] [CrossRef]
- Massouh, A.; Schubert, J.; Yaneva-Roder, L.; Ulbricht-Jones, E.S.; Zupok, A.; Johnson, M.T.; Wright, S.I.; Pellizzer, T.; Sobanski, J.; Bock, R.; et al. Spontaneous Chloroplast Mutants Mostly Occur by Replication Slippage and Show a Biased Pattern in the Plastome of Oenothera. Plant Cell 2016, 28, 911–929. [Google Scholar] [CrossRef]
- Higgs, D.C.; Kuras, R.; Kindle, K.L.; Wollman, F.A.; Stern, D.B. Inversions in the Chlamydomonas chloroplast genome suppress a petD 5′ untranslated region deletion by creating functional chimeric mRNAs. Plant J. 1998, 14, 663–671. [Google Scholar] [CrossRef]
- Odahara, M.; Inouye, T.; Nishimura, Y.; Sekine, Y. RECA plays a dual role in the maintenance of chloroplast genome stability in Physcomitrella patens. Plant J. 2015, 84, 516–526. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chevigny, N.; Schatz-Daas, D.; Lotfi, F.; Gualberto, J.M. DNA Repair and the Stability of the Plant Mitochondrial Genome. Int. J. Mol. Sci. 2020, 21, 328. https://doi.org/10.3390/ijms21010328
Chevigny N, Schatz-Daas D, Lotfi F, Gualberto JM. DNA Repair and the Stability of the Plant Mitochondrial Genome. International Journal of Molecular Sciences. 2020; 21(1):328. https://doi.org/10.3390/ijms21010328
Chicago/Turabian StyleChevigny, Nicolas, Déborah Schatz-Daas, Frédérique Lotfi, and José Manuel Gualberto. 2020. "DNA Repair and the Stability of the Plant Mitochondrial Genome" International Journal of Molecular Sciences 21, no. 1: 328. https://doi.org/10.3390/ijms21010328
APA StyleChevigny, N., Schatz-Daas, D., Lotfi, F., & Gualberto, J. M. (2020). DNA Repair and the Stability of the Plant Mitochondrial Genome. International Journal of Molecular Sciences, 21(1), 328. https://doi.org/10.3390/ijms21010328




