Occurrence of Thermophilic Microorganisms in Different Full Scale Biogas Plants
Abstract
1. Introduction
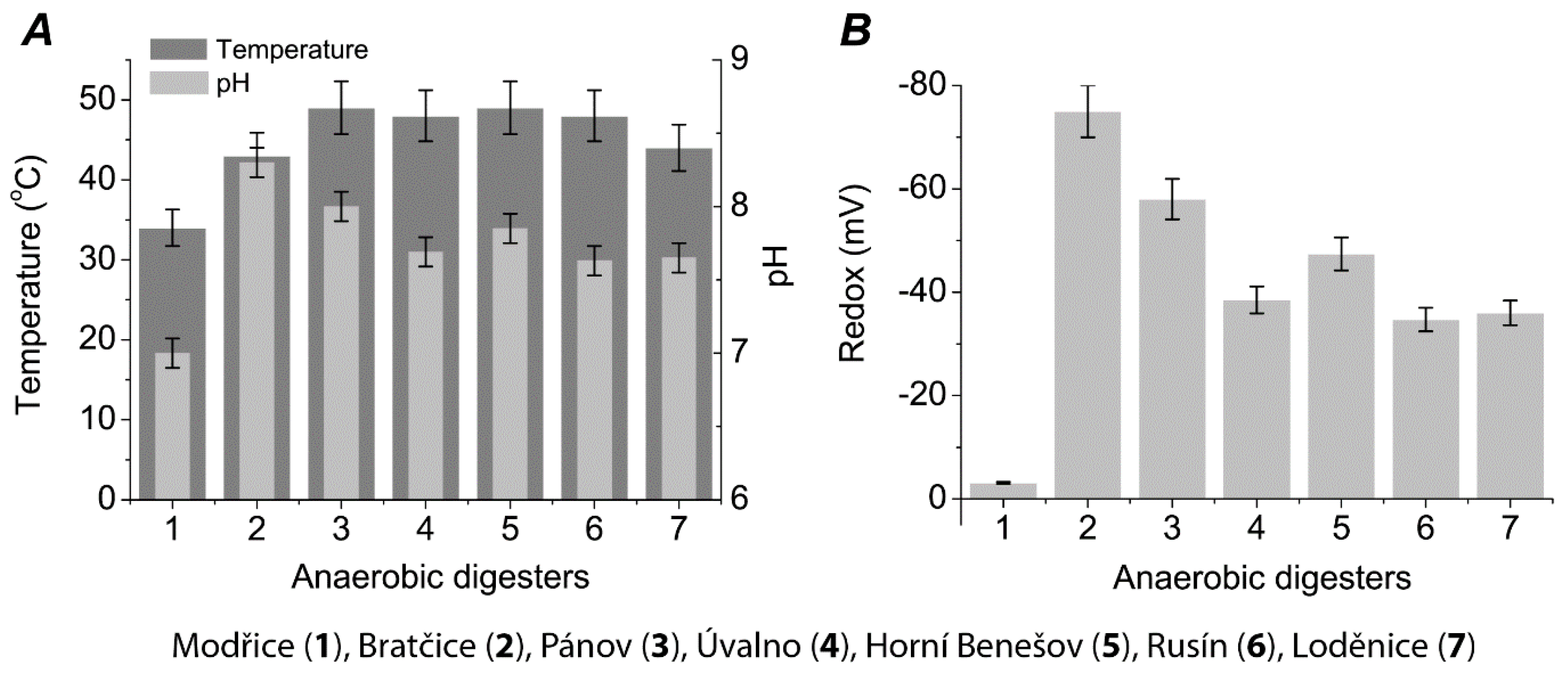
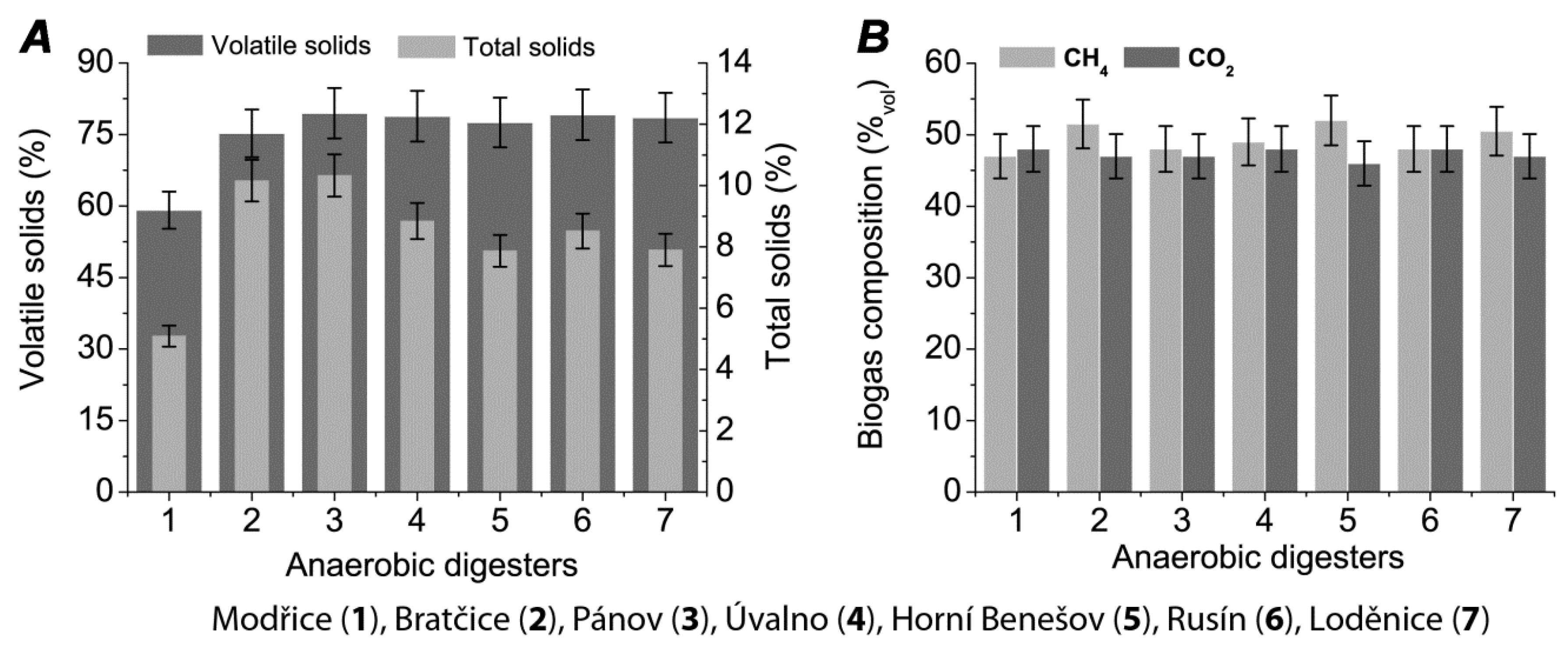
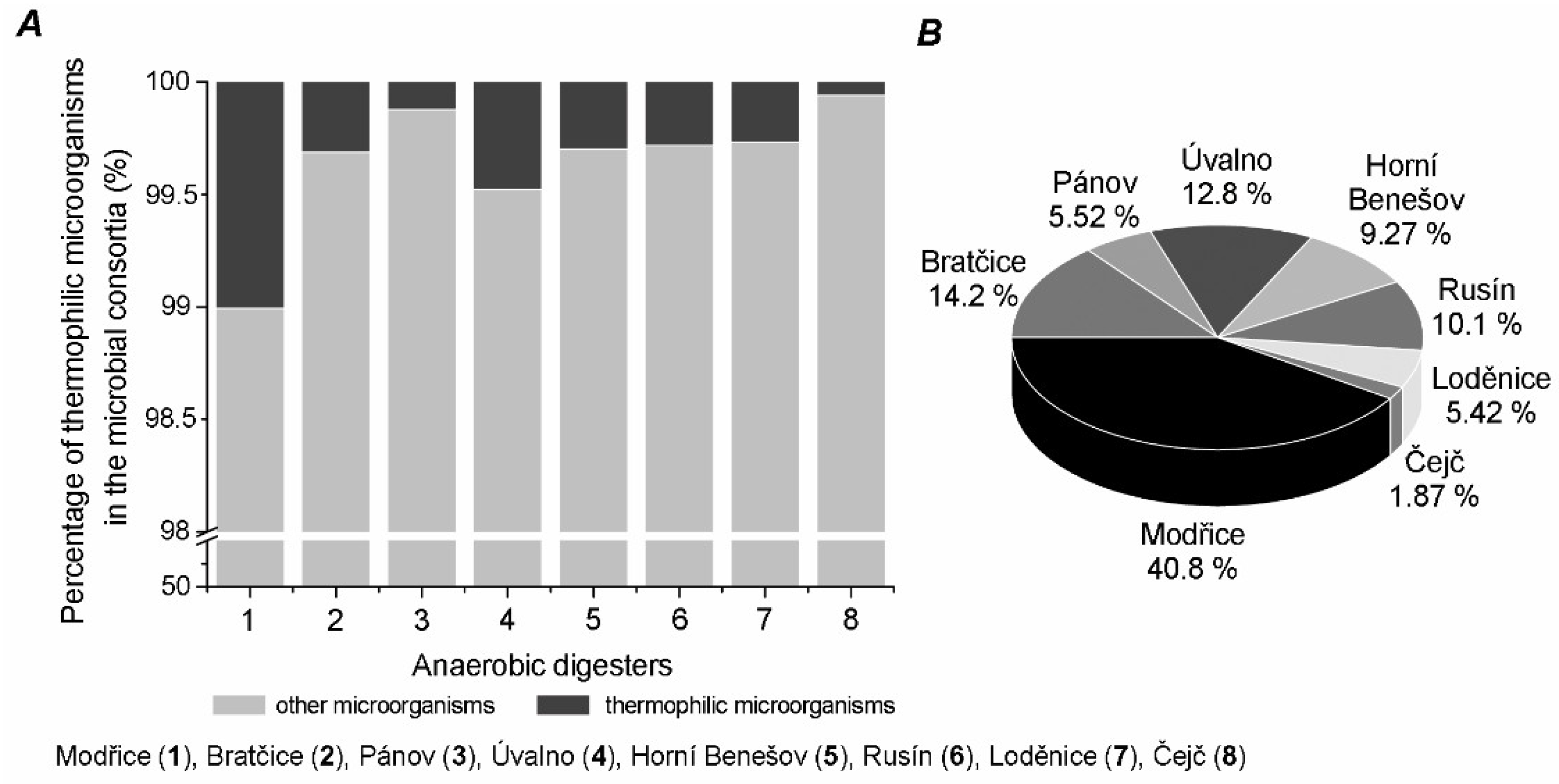
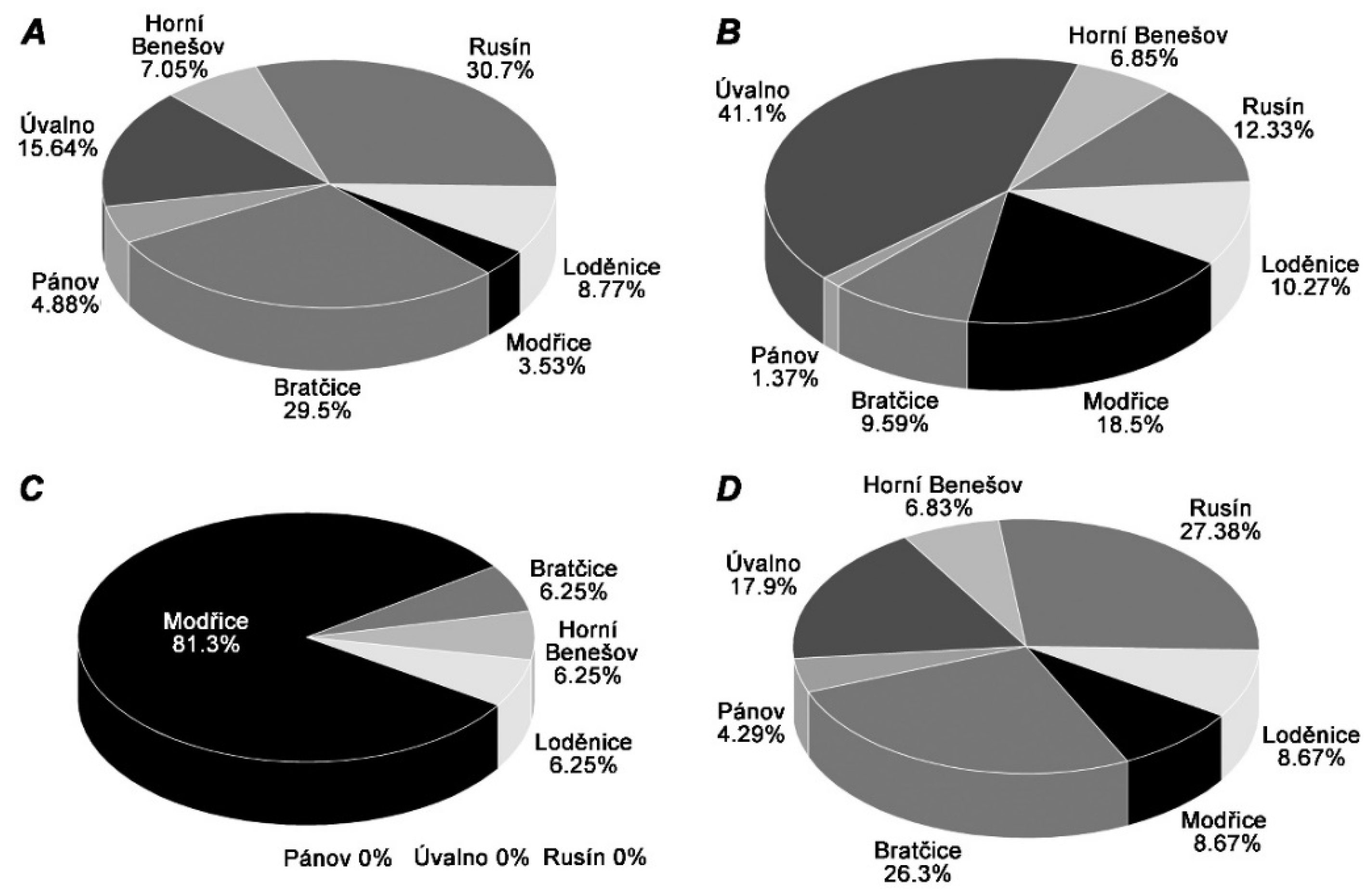
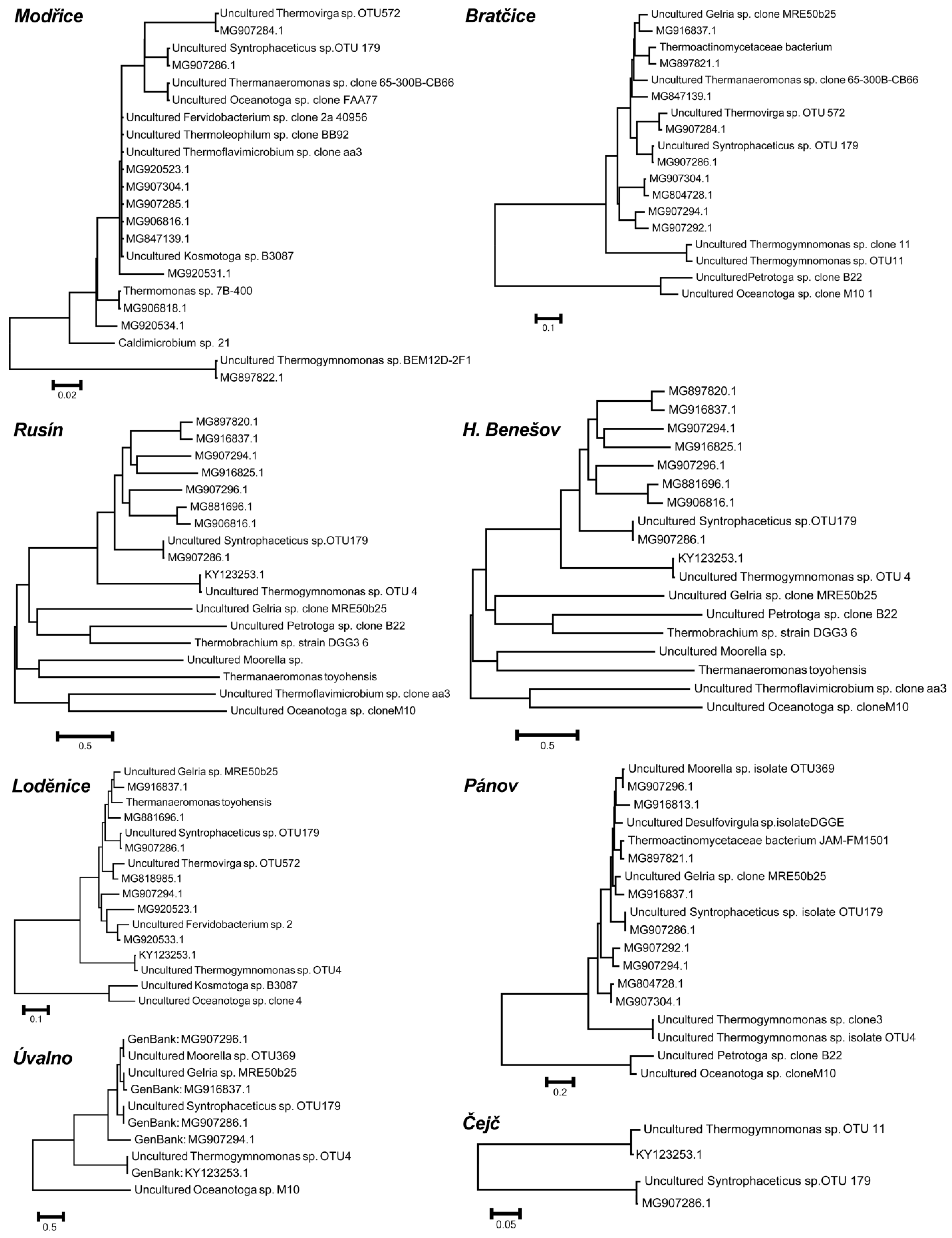
2. Results
3. Discussion
4. Materials and Methods
4.1. Diversity of Biogas Plants
4.2. Sampling and Analytical Methods
4.3. DNA Isolation, Amplification, and Sequencing
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Griffin, M.E.; McMahon, K.D.; Mackie, R.I.; Raskin, L. Methanogenic population dynamics during start-up of anaerobic digesters treating municipal solid waste and biosolids. Biotechnol. Eng. 2000, 57, 342–355. [Google Scholar] [CrossRef]
- Grothenhuis, J.T.; Smith, M.; Plugge, C.M.; Yuansheng, X.; Lammeren, A.A.; Stams, A.J. Bacteriological composition and structure of granular sludge adapted to different substrates. Appl. Environ. Microbiol. 1991, 57, 1942–1949. [Google Scholar]
- Ilyin, V.K.; Korniushenkova, I.N.; Starkova, L.V.; Lauriniavichius, K.S. Study of methanogenesis during bioutilization of plant residuals. Acta Astronaut. 2005, 56, 465–470. [Google Scholar] [CrossRef] [PubMed]
- Jäckel, U.; Thummes, K.; Kämpfer, P. Thermophilic methane production and oxidation in compost. FEMS Microbiol. Ecol. 2005, 52, 175–184. [Google Scholar] [CrossRef]
- Sreekrishnan, T.R.; Kohli, S.; Rana, V. Enhancement of biogas production from solid substrates using different techniques––A review. Bioresour. Technol. 2004, 95, 1–10. [Google Scholar]
- Krich, K.; Augenstein, D.; Batmale, J.P.; Benemann, J.; Rutledge, B.; Salour, D. Biomethane from Dairy Waste: A Sourcebook for the Production and Use of Renewable Natural Gas in California; USDA Rural Development: Washington, DC, USA, 2005.
- Conrad, R. Contribution of hydrogen to methane production and control of hydrogen concentration in methanogenic soils and sediments. FEMS Microbiol. Ecol. 1999, 28, 193–202. [Google Scholar] [CrossRef]
- Demirel, B.; Scherer, P. The roles of acetotrophic and hydrogenotrophic methanogens during anaerobic conversion of biomass to methane: A review. Rev. Environ. Sci. Biotechnol. 2008, 7, 173–190. [Google Scholar] [CrossRef]
- Wilkie, A. Biomethane from Biomass. In Biowaste and Biofuels; Harwood, C., Demain, A., Eds.; ASM Press: Washington, DC, USA, 2008; pp. 195–205. [Google Scholar]
- Ahring, B.; Ibrahim, A.A.; Mladenovska, Z. Effect of temperature increase from 55 to 65 °C on performance and microbial population dynamics of an anaerobic reactor treating cattle manure. Water Resour. 2001, 35, 2446–2452. [Google Scholar] [CrossRef]
- Kushkevych, I.; Vítězová, M.; Vítěz, T.; Bartoš, M. Production of biogas: Relationship between methanogenic and sulfate-reducing microorganisms. Open Life Sci. 2017, 12, 82–91. [Google Scholar] [CrossRef]
- Kushkevych, I.; Kováč, J.; Vítězová, M.; Vítěz, T.; Bartoš, M. The diversity of sulfate-reducing bacteria in the seven bioreactors. Arch. Microbial. 2018, 200, 945–950. [Google Scholar] [CrossRef]
- Ziemiński, K.; Frąc, M. Methane fermentation process as anaerobic digestion of biomass: Transformations, stages and microorganisms. Afr. J. Biotechnol. 2012, 11, 4127–4139. [Google Scholar]
- Scherer, P.A.; Vollmer, G.R.; Fakhouri, T.; Martensen, S. Development of methanogenic process to degrade exhaustively the organic fraction of municipal grey waste under thermophilic and hyperthermophilic conditions. Water Sci. Technol. 2000, 41, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Schink, B. Energetics of syntrophic cooperation in methanogenic degradation. Microbiol. Mol. Biol. Rev. 1997, 61, 262–280. [Google Scholar] [PubMed]
- Weiland, P. Biogas production: Current state and perspectives. Appl. Microbiol. Biotechnol. 2010, 85, 849–860. [Google Scholar] [CrossRef] [PubMed]
- Madigan, M.T.; Martino, J.M.; Thomas, D.B. Brock Biology of Microorganisms; Pearson Prentice Hall: Upper Saddle River, NJ, USA, 2006. [Google Scholar]
- Stetter, K. History of discovery of the first hyperthermophiles. Extremophiles 2006, 10, 357–362. [Google Scholar] [CrossRef] [PubMed]
- Satyanarayana, T.; Littlechild, J.; Kawarabayasi, Y. Thermophilic Microbes in Environmental and Industrial Biotechnology. Biotechnol. Thermophiles 2013, 3. [Google Scholar] [CrossRef]
- Barker, H.A. On the biochemistry of methane fermentation. Arch. Microbiol. 1936, 7, 404–419. [Google Scholar] [CrossRef]
- Zinder, S.H.; Koch, M. Non-aceticlastic methanogenesis from acetate: Acetate oxidation by a thermophilic syntrophic coculture. Arch. Microbiol. 1984, 138, 263–272. [Google Scholar] [CrossRef]
- Schnurer, A.; Houwen, F.P.; Svensson, B.H. Mesophilic syntrophic acetate oxidation during methane formation by a triculture at high ammonium concentration. Arch. Microbiol. 1994, 162, 70–74. [Google Scholar] [CrossRef]
- Nazina, T.N.; Shestakova, N.M.; Grigor’yan, A.A.; Mikhailova, E.M.; Tourova, T.P.; Poltaraus, A.B.; Feng, C.; Ni, F.; Belyaev, S.S. Phylogenetic diversity and activity of anaerobic microorganism of high-temperature horizons of the Dagang oilfield (P. R. China). Microbiology 2006, 75, 70–81. [Google Scholar] [CrossRef]
- McInerney, M.J.; Struchtemeyer, C.G.; Sieber, J.; Mouttaki, H.; Stams, A.J.M.; Schink, B.; Rohlin, L.; Gunsalus, R.P. Physiology, ecology, phylogeny and genomics of microorganisms capable of syntrophic metabolism. Ann. N. Y. Acad. Sci. USA 2008, 1125, 58–72. [Google Scholar] [CrossRef] [PubMed]
- Westerholm, W.; Roos, S.; Schnurer, A. Syntrophaceticus schinkii gen. nov. sp. nov., an anaerobic, syntrophic acetate-oxidizing bacterium isolated fromamesophilic anaerobic filter. FEMS Microbiol. Lett. 2010, 309, 100–104. [Google Scholar] [PubMed]
- Itoh, T.; Yoshikawa, N.; Takashina, T. Thermogymnomonas acidicola gen. nov. sp. nov. a novel thermoacidophilic, cell wall-less archaeon in the order Thermoplasmatales, isolated from a solfataric soil in Hakone, Japan. Int. J. Syst. Evol. Microbiol. 2007, 57, 2557–2561. [Google Scholar] [CrossRef] [PubMed]
- Dong, P.; Li, L.; Zhen, F.; Kong, X.; Sun, Y.; Zhang, Y. Comparison of dry and wet milling pretreatment methods for improving the anaerobic digestion performance of the Pennisetum hybrid. RSC Adv. 2017, 7/21, 12610–12619. [Google Scholar]
- Plugge, C.M.; Balk, M.; Zoetendal, E.G.; Stams, A.J. Gelria glutamica gen. nov. sp. nov. a thermophilic, obligately syntrophic, glutamate-degrading anaerobe. Int. J. Syst. Evol. Microbiol. 2002, 52, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Jayasinghearachchi, H.S.; Lal, B. Oceanotoga teriensis gen. nov., sp. nov., a thermophilic bacterium isolated from offshore oil-producing wells. Int. J. Syst. Evol. Microbiol. 2011, 61, 554–560. [Google Scholar] [CrossRef] [PubMed]
- Kushkevych, I.; Vítězová, M.; Fedrová, M.; Vochyanová, Z.; Paráková, L.; Hošek, J. Kinetic properties of growth of intestinal sulphate-reducing bacteria isolated from healthy mice and mice with ulcerative colitis. Acta Vet. Brno 2017, 86, 405–411. [Google Scholar] [CrossRef]
- Kushkevych, I.; Vítězová, M.; Vítěz, T.; Kováč, J.; Kaucká, P.; Jesionek, W.; Bartoš, M.; Barton, L. A new combination of substrates: Biogas production and diversity of the methanogenic microorganisms. Open Life Sci. 2018, 13, 119–128. [Google Scholar] [CrossRef]
- Kushkevych, I.V. Kinetic Properties of Pyruvate Ferredoxin Oxidoreductase of Intestinal Sulfate-Reducing Bacteria Desulfovibrio piger Vib-7 and Desulfomicrobium sp. Rod-9. Pol. J. Microbiol. 2015, 64, 107–114. [Google Scholar] [CrossRef]
- Kushkevych, I.; Fafula, R.; Parak, T.; Bartos, M. Activity of Na+/K+-activated Mg2+-dependent ATP hydrolase in the cell-free extracts of the sulfate-reducing bacteria Desulfovibrio piger Vib-7 and Desulfomicrobium sp. Rod-9. Acta Vet. Brno 2015, 84, 3–12. [Google Scholar] [CrossRef]
- Kushkevych, I.V. Activity and kinetic properties of phosphotransacetylase from intestinal sulfate-reducing bacteria. Acta Biochim. Pol. 2015, 62, 1037–1108. [Google Scholar] [CrossRef] [PubMed]
- Kushkevych, I.; Dordević, D.; Vítězová, M. Analysis of pH dose-dependent growth of sulfate-reducing bacteria. Open Med. 2019, 14, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Kushkevych, I.; Dordević, D.; Vítězová, M. Toxicity of hydrogen sulfide toward sulfate-reducing bacteria Desulfovibrio piger Vib-7. Arch. Microbiol. 2019, 201, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Kushkevych, I.; Kobzová, E.; Vítězová, M.; Vítěz, T.; Dordević, D.; Bartoš, M. Acetogenic microorganisms in operating biogas plants depending on substrate combinations. Biologia 2019, 74, 1229–1236. [Google Scholar] [CrossRef]
- Kushkevych, I.; Kollar, P.; Suchy, P.; Parak, K.; Pauk, K.; Imramovsky, A. Activity of selected salicylamides against intestinal sulfate-reducing bacteria. Neuroendocrinol Lett. 2015, 36, 106–113. [Google Scholar]
- Kushkevych, I.; Kollar, P.; Ferreira, A.L.; Palma, D. Antimicrobial effect of salicylamide derivatives against intestinal sulfate-reducing bacteria. J. Appl. Biomed. 2016, 14, 125–130. [Google Scholar] [CrossRef]
- Kushkevych, I.; Vítězová, M.; Kos, J.; Kollár, P.; Jampílek, J. Effect of selected 8-hydroxyquinoline-2-carboxanilides on viability and sulfate metabolism of Desulfovibrio piger. J. App. Biomed. 2018, 16, 241–246. [Google Scholar] [CrossRef]
- Kushkevych, I.; Dordević, D.; Kollar, P. Analysis of physiological parameters of Desulfovibrio strains from individuals with colitis. Open Life Sci. 2018, 13, 481–488. [Google Scholar] [CrossRef]
- Kushkevych, I.; Dordević, D.; Vítězová, M.; Kollar, P. Cross-correlation analysis of the Desulfovibrio growth parameters of intestinal species isolated from people with colitis. Biologia 2018, 73, 1137–1143. [Google Scholar] [CrossRef]
- CSN EN 14346 Characterization of Waste–Calculation of Dry Matter by Determination of Dry Residue or Water Content; Czech Standards Institute: Prague, Czech Republic, 2007.
- CSN EN 15169 Characterization of Waste–Determination of Loss on Ignition in Waste, Sludge and Sediments; Czech Standards Institute: Prague, Czech Republic, 2007.
- CSN EN 12176 Characterization of Sludge–Determination of pH-value; Czech Standards Institute: Prague, Czech Republic, 1999.
- Bailey, N.T.J. Statistical Methods in Biology; Cambridge University Press: Cambridge, UK, 1995. [Google Scholar]
- Nossa, C.W.; Oberdorf, W.E.; Yang, L.; Aas, J.A.; Paster, B.J.; Desantis, T.Z. Design of 16S rRNA gene primers for 454 pyrosequencing of the human foregut microbiome. World J. Gastroenterol. 2010, 16, 4135–4144. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Mille, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
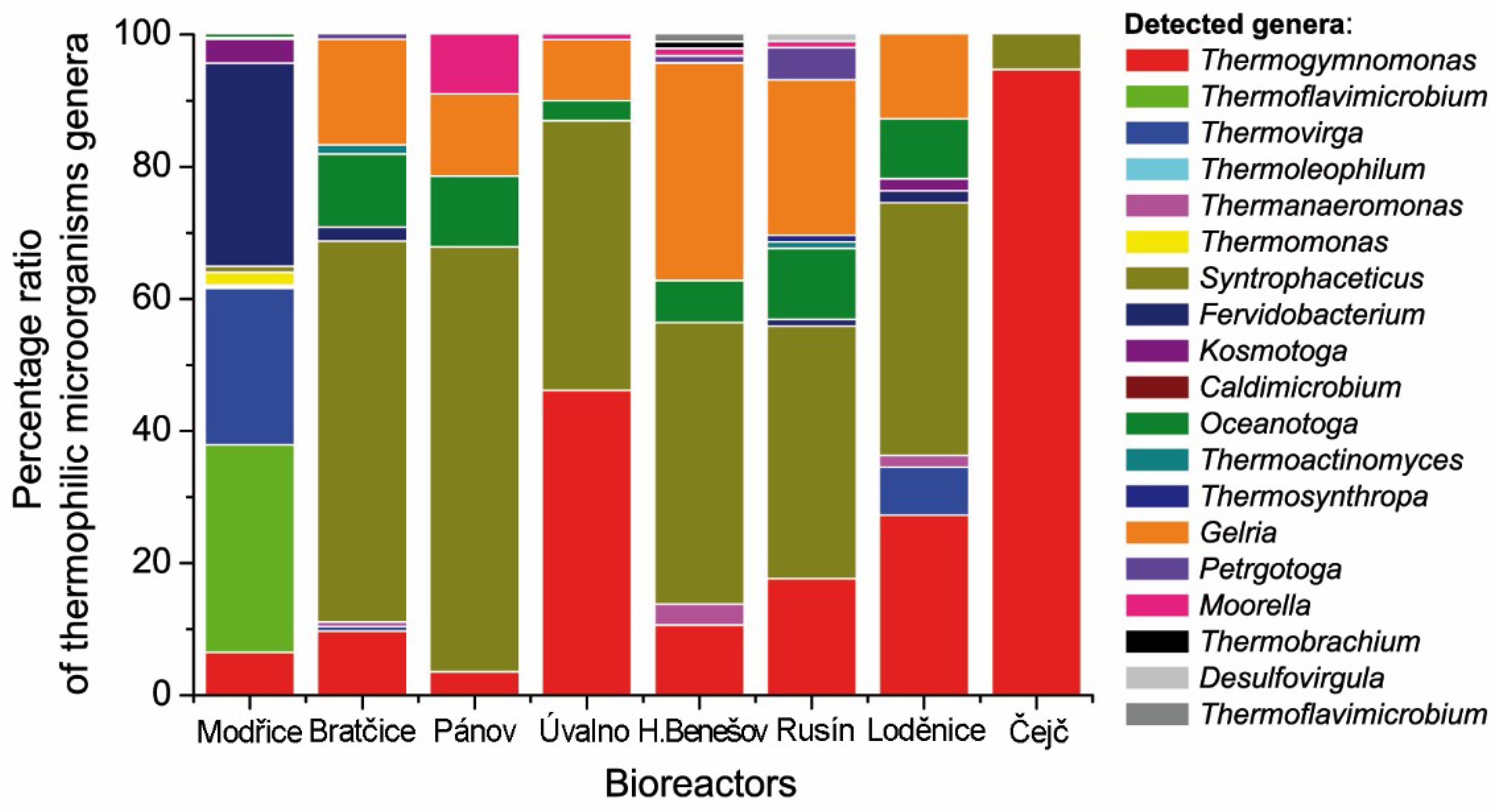
| Fermenter | Installed Power (kWel) | Fermenter Volume (m3) | Process Tempe-rature (°C) | Hydraulic Retention time | Daily Biogas production Rate (Lbiogas·Lferm.vol.−1) | CH4 Content in Biogas (%vol) * | pH in Fermenter (−) * | Solids Content in Fermenter (%) * | Volatile Solids Content in Fermenter (%) * |
|---|---|---|---|---|---|---|---|---|---|
| Modřice | 1000 | 6 × 3000 | 34 | 22 | 0.64 | 47 | 7.02 | 5.09 | 59.13 |
| Bratčice | 750 | 2 × 1040 1 × 1040 | 40 | 86 | 2.77 | 51.5 | 8.3 | 10.16 | 75.23 |
| Pánov | 500 | 2 × 1320 1 × 1630 | 41 | 85 | 1.76 | 48 | 8.03 | 10.33 | 79.46 |
| Úvalno | 750 | 2 × 1040 1 × 1040 | 40 | 78 | 2.77 | 49 | 7.69 | 8.84 | 78.85 |
| Horní Benešov | 750 | 2 × 1040 1 × 1040 | 41 | 85 | 2.77 | 52 | 7.85 | 7.87 | 77.52 |
| Rusín | 750 | 2 × 1970 1 × 1630 | 41 | 85 | 1.56 | 48 | 7.63 | 8.52 | 79.15 |
| Loděnice | 840 | 3 × 1970 | 41 | 90 | 1.64 | 50.5 | 7.65 | 7.9 | 78.51 |
| Čejč | 750 | 2 × 3500 1 × 3800 | 40 | 65 | 0.81 | 50.3 | 7.54 | 4.3 | 78.98 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kushkevych, I.; Cejnar, J.; Vítězová, M.; Vítěz, T.; Dordević, D.; Bomble, Y.J. Occurrence of Thermophilic Microorganisms in Different Full Scale Biogas Plants. Int. J. Mol. Sci. 2020, 21, 283. https://doi.org/10.3390/ijms21010283
Kushkevych I, Cejnar J, Vítězová M, Vítěz T, Dordević D, Bomble YJ. Occurrence of Thermophilic Microorganisms in Different Full Scale Biogas Plants. International Journal of Molecular Sciences. 2020; 21(1):283. https://doi.org/10.3390/ijms21010283
Chicago/Turabian StyleKushkevych, Ivan, Jiří Cejnar, Monika Vítězová, Tomáš Vítěz, Dani Dordević, and Yannick J. Bomble. 2020. "Occurrence of Thermophilic Microorganisms in Different Full Scale Biogas Plants" International Journal of Molecular Sciences 21, no. 1: 283. https://doi.org/10.3390/ijms21010283
APA StyleKushkevych, I., Cejnar, J., Vítězová, M., Vítěz, T., Dordević, D., & Bomble, Y. J. (2020). Occurrence of Thermophilic Microorganisms in Different Full Scale Biogas Plants. International Journal of Molecular Sciences, 21(1), 283. https://doi.org/10.3390/ijms21010283








