CaDHN4, a Salt and Cold Stress-Responsive Dehydrin Gene from Pepper Decreases Abscisic Acid Sensitivity in Arabidopsis
Abstract
:1. Introduction
2. Results
2.1. Sub-Cellular Localization of CaDHN4
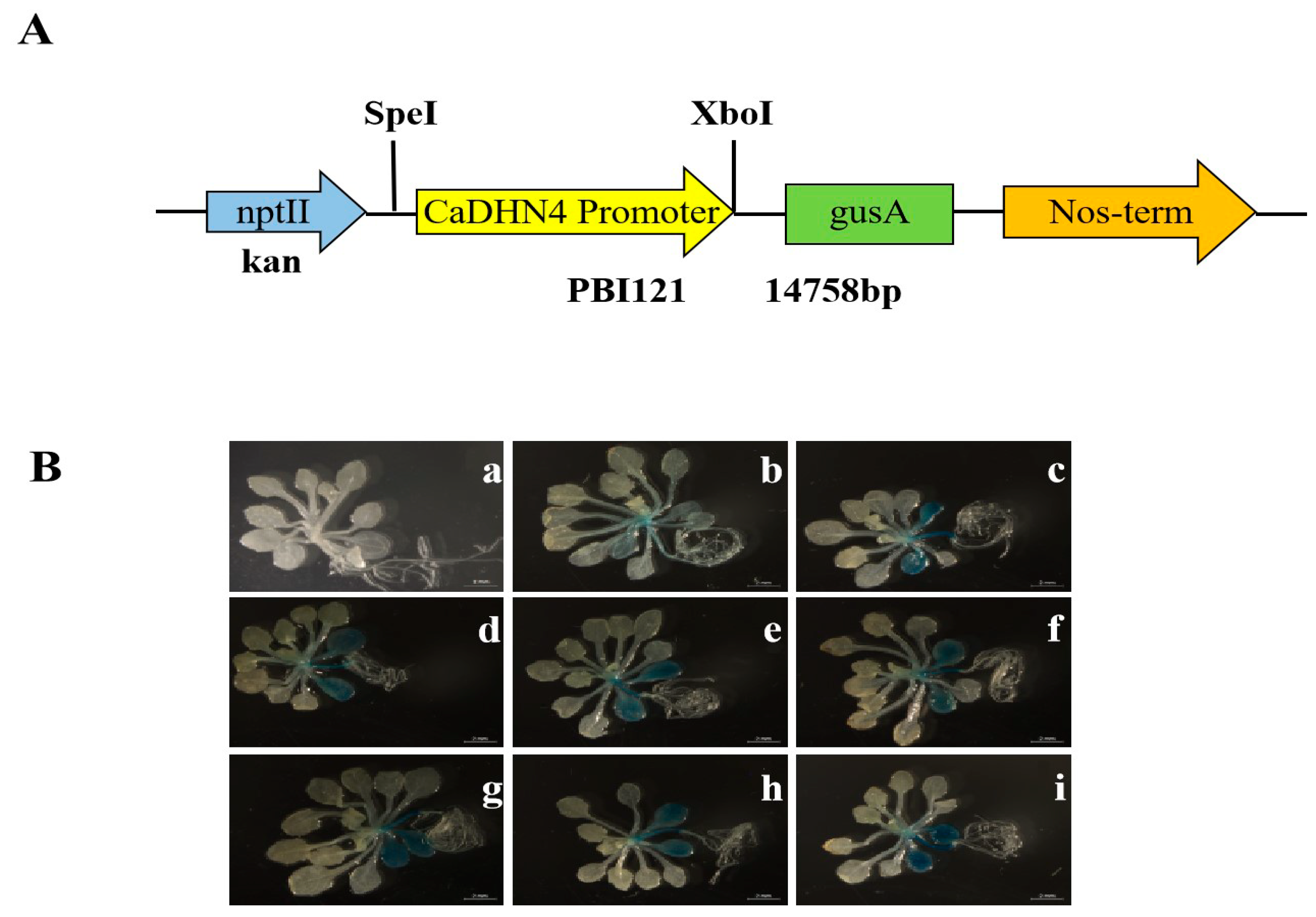
2.2. GUS Histological Assay
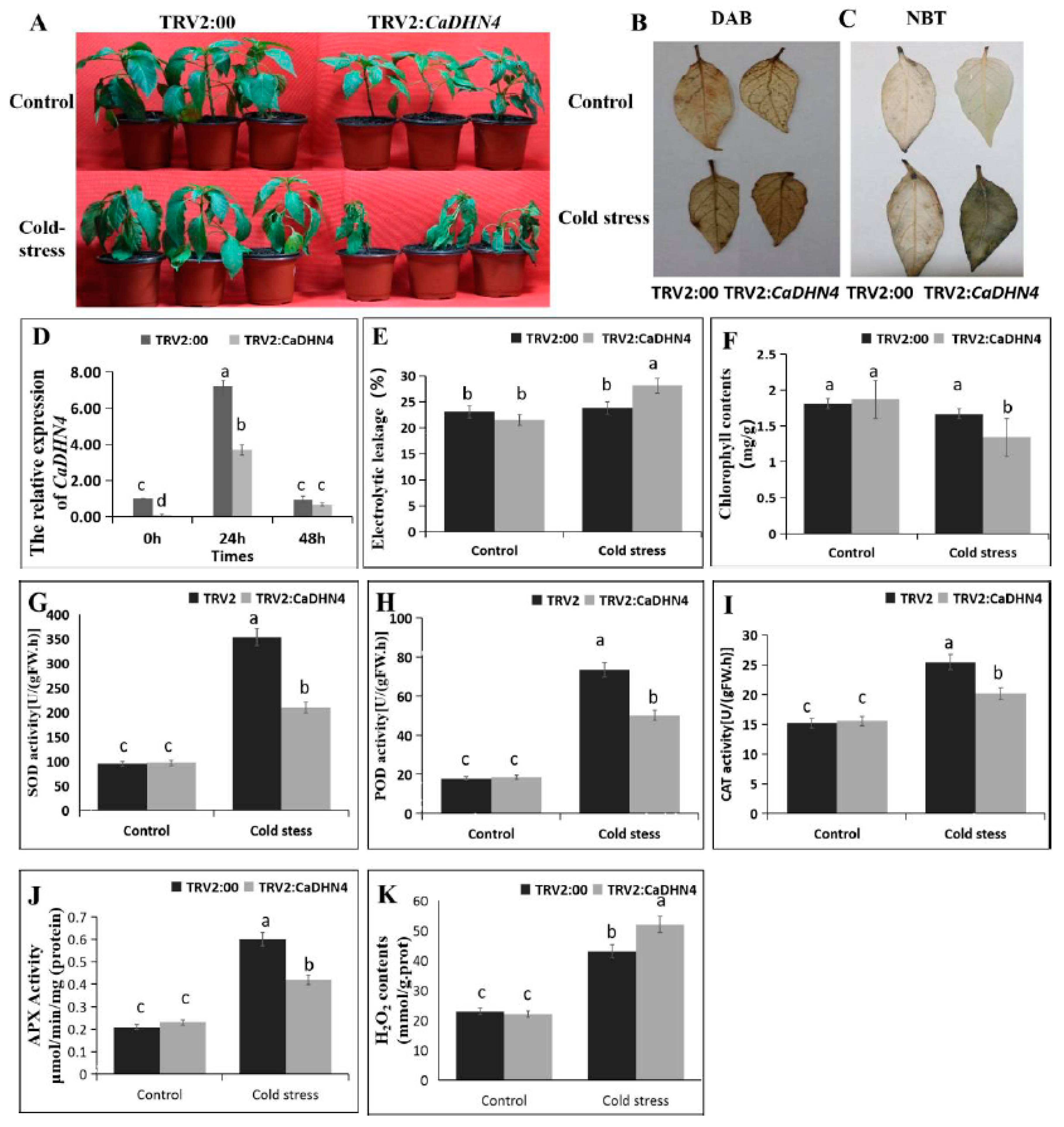
2.3. Virus-Induced Gene Silencing (VIGS) of CaDHN4 Reduces Salt-Stress Tolerance
2.4. Virus-Induced Gene Silencing (VIGS) of CaDHN4 Reduces Tolerance to Salt Stress
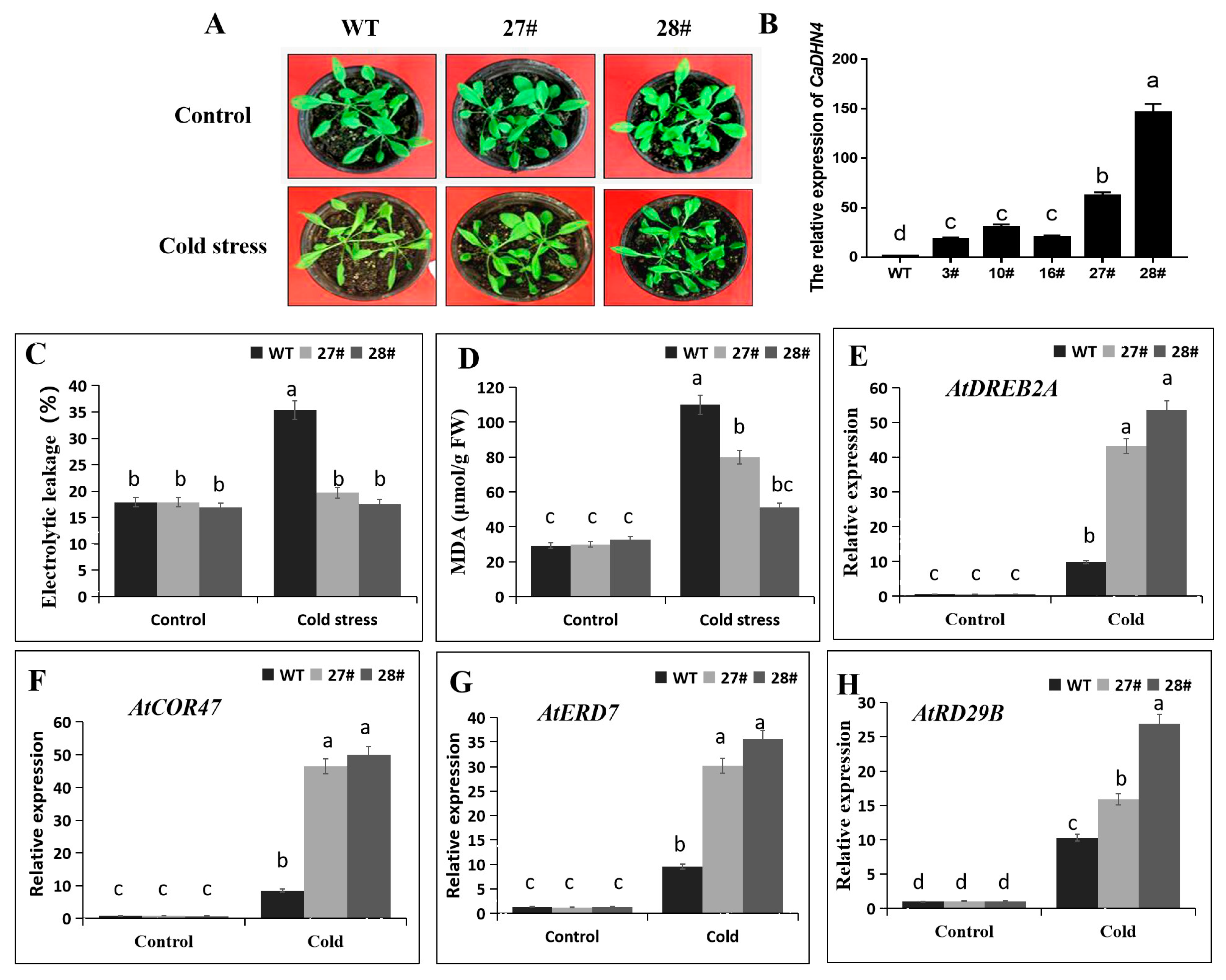
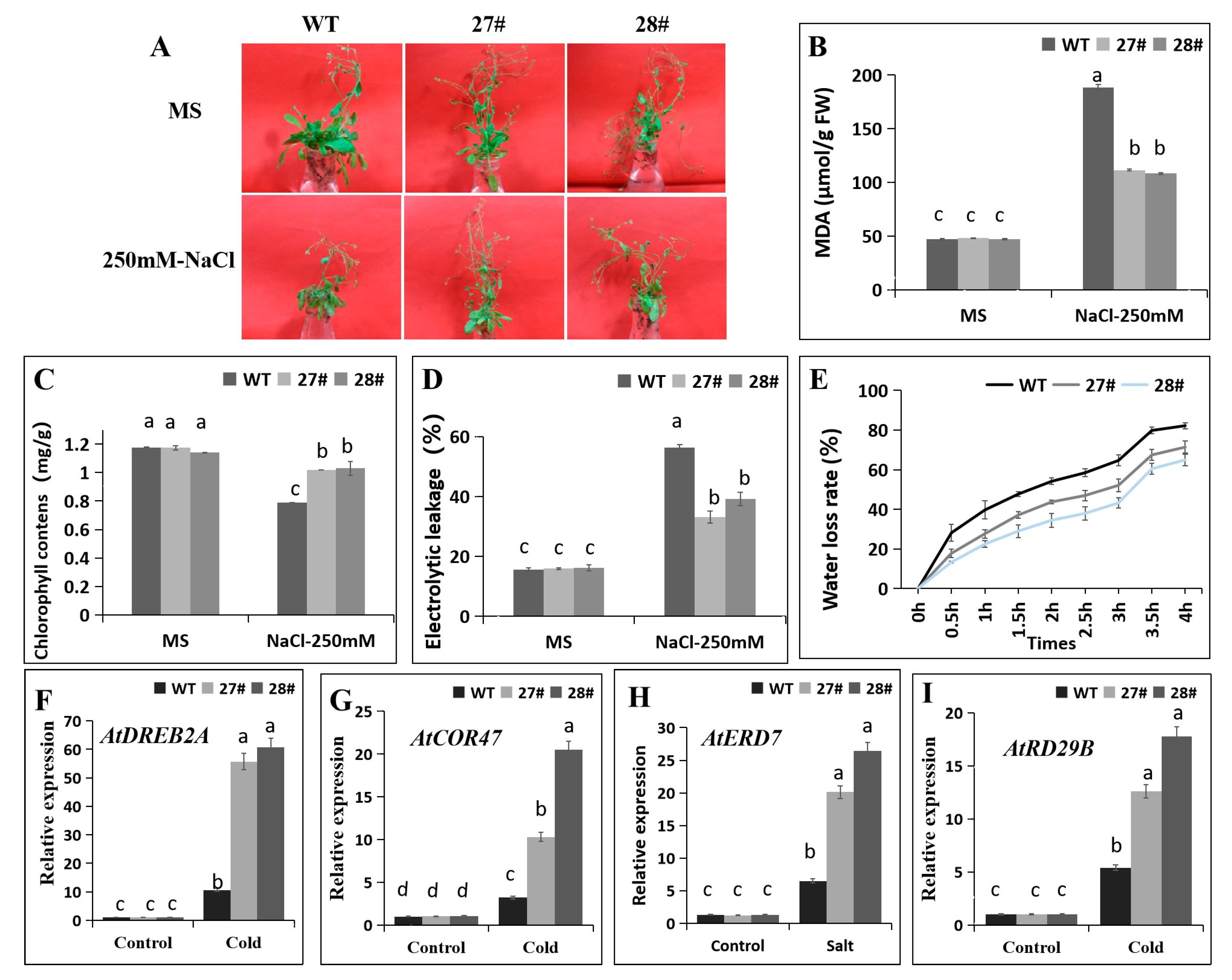
2.5. Overexpression of CaDHN4 in Arabidopsis Increases Cold Stress Tolerance
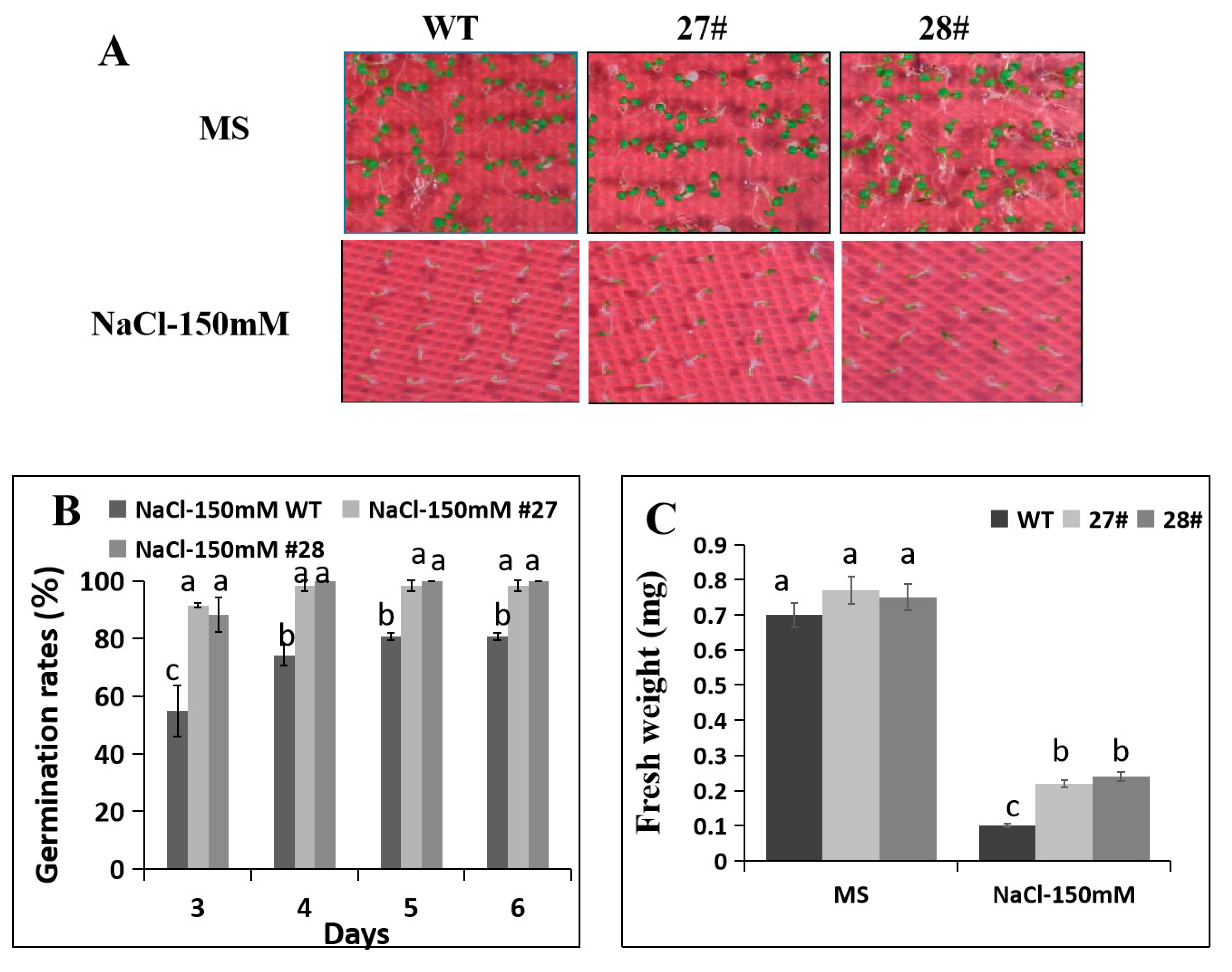
2.6. Overexpression of CaDHN4 in Arabidopsis Increases Salt Stress Tolerance
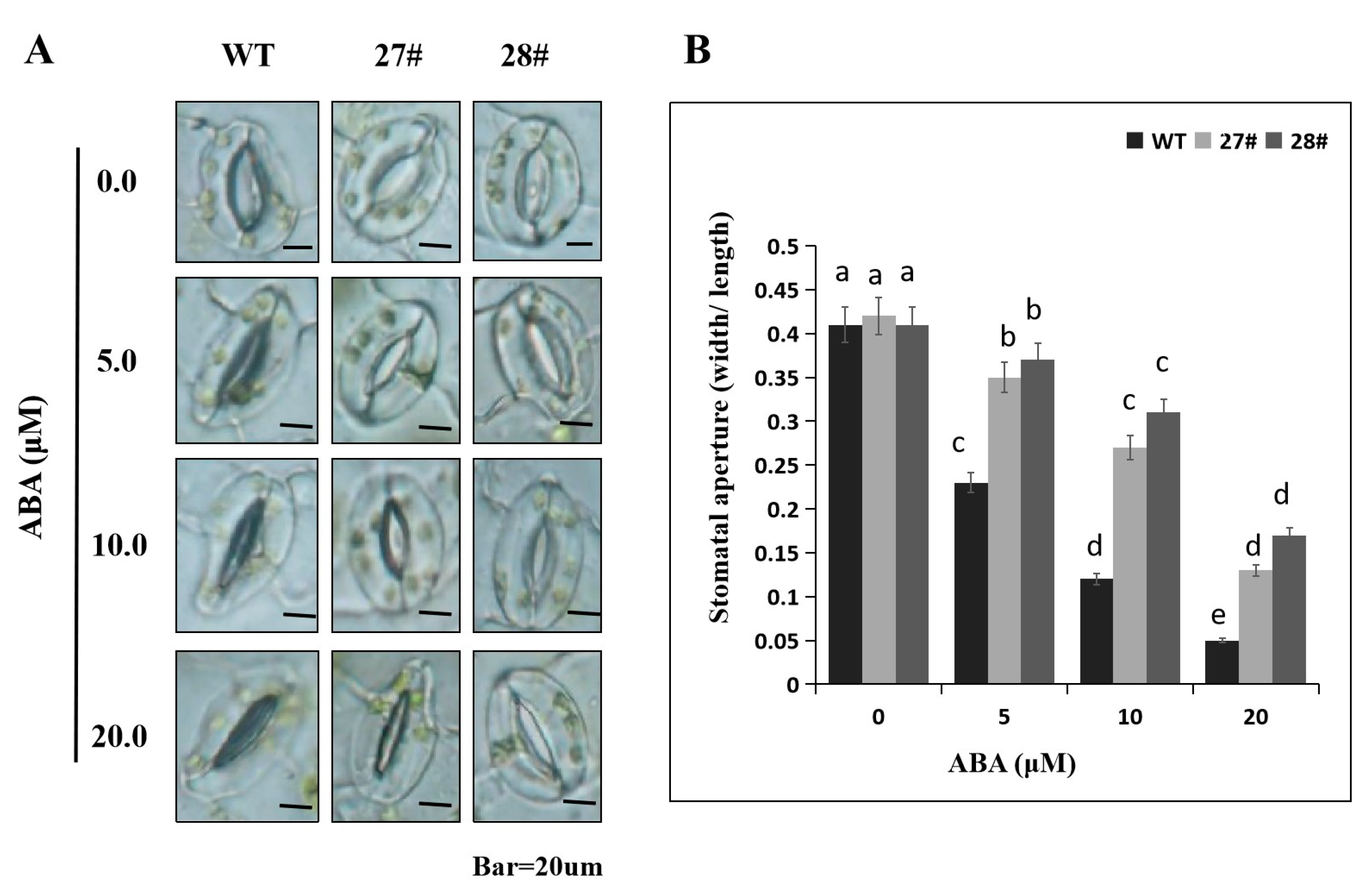
2.7. Overexpression of CaDHN4 in Arabidopsis Decreases ABA Sensitivity
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. RNA Isolation and qRT-qPCR Analysis
4.3. Isolation and Sequence Analysis of CaDHN4
4.4. Promoter Activity Assay Analysis
4.5. Subcellular Localization Assays
4.6. Virus-Induced Gene Silencing (VIGS) of CaDHN4
4.7. Generation of CaDHN4 Transgenic Arabidopsis Plants
4.8. Salt and Cold Stress Tolerance Assays
4.9. Determination of Chlorophyll, MDA, and Relative Electrolyte Contents and Antioxidant Enzyme Activities
4.10. NBT and DAB Staining
4.11. ABA Tolerance Assays
4.12. Measurement of Stomatal Aperture in Response to ABA Treatment
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mittler, R. Abiotic stress, the field environment and stress combination. Trends Plant Sci. 2006, 11, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Jing, H.; Li, C.; Ma, F.; Ma, J.H.; Khan, A.; Wang, X. Genome-Wide Identification, Expression Diversication of Dehydrin Gene Family and Characterization of CaDHN3 in Pepper (Capsicum annuum L.). PLoS ONE 2016, 11, e0161073. [Google Scholar] [CrossRef] [PubMed]
- Black, M.; Corbineau, F.; Gee, H.; Côme, D. Water content, raffinose, and dehydrins in the induction of desiccation tolerance in immature wheat embryos. Plant Physiol. 1999, 120, 463–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cellier, F.; Conéjéro, G.; Breitler, J.C.; Casse, F. Molecular and physiological responses to water deficit in drought-tolerant and drought-sensitive plants of sunflower accumulation of dehydrin transcripts correlates with tolerance. Plant Physiol. 1998, 116, 319–328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guy, C.; Haskell, D.; Neven, L.; Klein, P.; Smelser, C. Hydration-state-responsive proteins link cold and drought stress in spinach. Planta 1992, 188, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Ingram, J.; Bartels, D. The molecular basis of dehydration tolerance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 377–403. [Google Scholar] [CrossRef] [Green Version]
- Rorat, T.; Szabala, B.M.; Grygorowicz, W.J.; Wojtowicz, B.; Yin, Z.; Rey, P. Expression of sk3-type dehydrin in transporting organs is associated with cold acclimation in solanum species. Planta 2006, 224, 205–221. [Google Scholar] [CrossRef]
- Eriksson, S.K.; Kutzer, M.; Procek, J.; Grobner, G.; Harryson, P. Tunable membrane binding of the intrinsically disordered dehydrin Lti30, a cold-induced plant stress protein. Plant Cell 2011, 23, 2391–2404. [Google Scholar] [CrossRef] [Green Version]
- Hara, M.; Shinoda, Y.; Kubo, M.; Kashima, D.; Takahashi, I.; Kato, T. Biochemical characterization of the Arabidopsis KS-type dehydrin protein whose gene expression is constitutively abundant rather than stress dependent. Acta Physiol. Plant 2011, 33, 2103–2116. [Google Scholar] [CrossRef]
- Close, T.J. Dehydrins: Emergence of a biochemical role of a family of plant dehydration proteins. Physiol. Plant 1996, 97, 795–803. [Google Scholar] [CrossRef]
- Close, T.J. Dehydrins: A commonalty in the response of plants to dehydration and low temperature. Physiol. Plant 1997, 100, 291–296. [Google Scholar] [CrossRef]
- Liii, D. Structural motifs in lea proteins. Curr. Top. Plant Physiol. 1993, 10, 91–103. [Google Scholar]
- Battaglia, M. The enigmatic lea proteins and other hydrophilins. Plant Physiol. 2008, 148, 6–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dure, L.; Crouch, M.; Harada, J.; Ho, T.H.D.; Mundy, J.; Quatrano, R. Common amino acid sequence domains among the lea proteins of higher plants. Plant Mol. Biol. 1989, 12, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Hughes, S.L.; Schart, V.; Malcolmson, J.; Hogarth, K.A.; Martynowicz, D.M.; Tralman-Baker, E. The importance of size and disorder in the cryoprotective effects of dehydrins. Plant Physiol. 2013, 163, 1376–1386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nylander, M.; Svensson, J.; Palva, E.T.; Welin, B.V. Stress-induced accumulation and tissue-specific localization of dehydrins in Arabidopsis thaliana. Plant Mol. Biol. 2001, 45, 263–279. [Google Scholar] [CrossRef]
- Hundertmar, K.; Hincha, D.K. LEA (Late Embryogenesis Abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics 2008, 9, 118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hara, M.; Shinoda, Y.; Tanaka, Y.; Kuboi, T. DNA binding of citrus dehydrin promoted by zinc ion. Plant Cell Environ. 2009, 32, 532–541. [Google Scholar] [CrossRef]
- Koag, M.C. The binding of Maize DHN1 to lipid vesicles. Gain of structure and lipid specificity. Plant Physiol. 2003, 131, 309–316. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.H.; Peng, P.H.; Ko, C.Y.; Markhart, A.H.; Lin, T.Y. Characterization of a novel Y2K-type dehydrin VrDhn1 from Vigna radiata. Plant Cell Physiol. 2012, 53, 930–942. [Google Scholar] [CrossRef] [Green Version]
- Rorat, T.; Grygorowicz, W.J.; Irzykowski, W.; Rey, P. Expression of KS-type dehydrins is primarily regulated by factors related to organ type and leaf developmental stage during vegetative growth. Planta 2004, 218, 878–885. [Google Scholar] [CrossRef] [PubMed]
- Takebayashi, N.; Brewer, P.B.; Newbigin, E.; Uyenoyama, M.K. Patterns of variation within self-incompatibility loci. Mol. Biol. Evol. 2003, 20, 1778–1794. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Xiang, X.; Geng, M.; You, Q.; Huang, X. Effect of HbDHN1 and HbDHN2 Genes on Abiotic Stress Responses in Arabidopsis. Front. Plant Sci. 2017, 8, 470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mundy, J.; Chua, N.H. Abscisic acid and water-stress induce the expression of a novel rice gene. EMBO J. 1988, 7, 2279–2286. [Google Scholar] [CrossRef] [PubMed]
- Allagulova, C.R.; Gimalov, F.R.; Shakirova, F.M.; Vakhitov, V.A. The plant dehydrins: structure and putative functions. Biochemistry (Moscow) 2003, 68, 945–951. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; An, Y.; Wang, Z.; Du, H.; Huang, B. Characterization of gene expression associated with drought avoidance and tolerance traits in a perennial grass species. PLoS ONE 2014, 9, e103611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hara, M.; Terashima, S.; Fukaya, T.; Kuboi, T. Enhancement of cold tolerance and inhibition of lipid peroxidation by citrus dehydrin in transgenic tobacco. Planta 2003, 217, 290–298. [Google Scholar] [CrossRef]
- Shekhawat, U.K.S.; Srinivas, L.; Ganapathi, T.R. MusaDHN-1, a novel multiple stress-inducible SK3-type dehydrin gene, contributes affirmatively to drought- and salt-stress tolerance in banana. Planta 2011, 234, 915–932. [Google Scholar] [CrossRef]
- Qin, Y.X.; Qin, F. Dehydrins from wheat x Thinopyrum ponticum amphiploid increase salinity and drought tolerance under their own inducible promoters without growth retardation. Plant Physiol. Biochem. 2016, 99, 142–149. [Google Scholar] [CrossRef]
- Danyluk, J.; Perron, A.; Houde, M.; Limin, A.; Fowler, B.; Benhamou, N.; Sarhan, F. Accumulation of an acidic dehydrin in the vicinity of the plasma membrane during cold acclimation of wheat. Plant Cell 1998, 10, 623–638. [Google Scholar] [CrossRef] [Green Version]
- Halder, T.; Agarwal, T.; Ray, S. Isolation, cloning, and characterization of a novel Sorghum dehydrin (SbDhn2) protein. Protoplasma 2016, 253, 1475–1488. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, T.; Upadhyaya, G.; Halder, T.; Mukherjee, A.; Majumder, A.L.; Ray, S. Different dehydrins perform separate functions in Physcomitrella patens. Planta 2017, 245, 101–118. [Google Scholar] [CrossRef] [PubMed]
- Moore, K.; Roberts, L.J. Measurement of lipid peroxidation. Free Radic. Res. 1998, 28, 659–671. [Google Scholar] [CrossRef] [PubMed]
- Bajji, M.; Kinet, J.M.; Lutts, S. The use of the electrolyte leakage method for assessing cell membrane stability as a water stress tolerance test in durum wheat. Plant Growth Regul. 2002, 36, 61–70. [Google Scholar] [CrossRef]
- Chen, R.G.; Jing, H.; Guo, W.L.; Wang, S.B.; Ma, F.; Pan, B.G.; Gong, Z.H. Silencing of dehydrin CaDHN1 diminishes tolerance to multiple abiotic stresses in Capsicum annuum L. Plant Cell Rep. 2015, 34, 2189–2200. [Google Scholar] [CrossRef]
- Xing, X.; Liu, Y.; Kong, X.; Liu, Y.; Li, D. Overexpression of a maize dehydrin gene, ZmDHN2b, in tobacco enhances tolerance to low temperature. Plant Growth Regul. 2011, 65, 109–118. [Google Scholar] [CrossRef]
- Houde, M.; Dallaire, S.N.; Dong, D.; Sarhan, F. Overexpression of the acidic dehydrin WCOR410 improves freezing tolerance in transgenic strawberry leaves. Plant Biotechnol. J. 2004, 2, 381–387. [Google Scholar] [CrossRef]
- Hossain, M.A. Hydrogen peroxide priming modulates abiotic oxidative stress tolerance: insights from ROS detoxification and scavenging. Front. Plant Sci. 2015, 6, 420. [Google Scholar] [CrossRef] [Green Version]
- Fujita, M.; Fujita, Y.; Maruyama, K.; Seki, M.; Hiratsu, K.; Ohme-Takagi, M.; Tran, L.P.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 2004, 39, 863–876. [Google Scholar] [CrossRef]
- Biłas, R.; Szafran, K.; Hnatuszko-Konka, K.; Kononowicz, A.K. Cis regulatory elements used to control gene expression in plants. Plant Cell Tissue Organ C. 2016, 127, 269–287. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.C.; Kim, S.H.; Kim, S.R. Drought inducible OsDHN1 promoter is activated by OsDREB1A and OsDREB1D. J. Plant Biol. 2013, 56, 115–121. [Google Scholar] [CrossRef]
- Zhu, W.N.; Zhang, D.P.; Lu, X.X.; Zhang, L.S.; Yu, Z.Y.; Lv, H. Characterisation of an SKn-type dehydrin promoter from wheat and its responsiveness to various abiotic and biotic stresses. Plant Mol. Biol. Rep. 2014, 32, 664–678. [Google Scholar] [CrossRef]
- Lee, S.C.; Lee, M.Y.; Kim, S.J.; Jun, S.H.; An, G.; Kim, S.R. Characterisation of an abiotic stress-inducible dehydrin gene, OsDhn1, in rice (Oryza sativa L.). Mol. Cells 2005, 19, 212–218. [Google Scholar] [PubMed]
- Msanne, J.; Lin, J.S.; Stone, J.M.; Awada, T. Characterization of abiotic stress-responsive Arabidopsis thaliana RD29A and RD29B genes and evaluation of transgenes. Planta 2011, 234, 97–107. [Google Scholar] [CrossRef]
- Sakuma, Y.; Maruyama, K.; Osakabe, Y.; Qin, F.; Seki, M.; Shinozaki, K.; YamaguchiShinozaki, K. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 2006, 18, 1292–1309. [Google Scholar] [CrossRef] [Green Version]
- Huang, R.W.; Liu, D.F.; Huang, M.; Ma, J.; Li, Z.M.; Li, M.Y.; Sui, S.Z. CpWRKY71, a WRKY Transcription Factor Gene of Wintersweet (Chimonanthus praecox), Promotes Flowering and Leaf Senescence in Arabidopsis. Int. J. Mol. Sci. 2019, 20, 5325. [Google Scholar] [CrossRef] [Green Version]
- Guan, H.; Liu, X.; Niu, F.; Zhao, Q.; Fan, N.; Cao, D.; Meng, D.; He, W.; Guo, B.; Wei, Y.; et al. OoNAC72, a NAC-Type Oxytropis ochrocephala Transcription Factor, Conferring Enhanced Drought and Salt Stress Tolerance in Arabidopsis. Front. Plant Sci. 2019, 10, 890. [Google Scholar] [CrossRef] [Green Version]
- Shi, X.P.; Ren, J.J.; Yu, Q.; Zhou, S.M.; Ren, Q.P.; Kong, L.J.; Wang, X.L. Overexpression of SDH confers tolerance to salt and osmotic stress, but decreases ABA sensitivity in Arabidopsis. Plant Biol. 2018, 20, 327–337. [Google Scholar] [CrossRef]
- Sah, S.K.; Reddy, K.R.; Li, J.X. Abscisic acid and abiotic stress tolerance in crop plants. Front. Plant Sci. 2016, 7, 571. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.H.; Jia, W.S.; Yang, J.C.; Ismail, A.M. Role of ABA in integrating plant responses to drought and salt stresses. Field Crops Res. 2006, 97, 111–119. [Google Scholar] [CrossRef]
- Hu, H.H.; You, J.; Fang, Y.J.; Zhu, X.Y.; Qi, Z.Y.; Xiong, L. Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol. Biol. 2008, 67, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Aubert, Y.; Vile, D.; Pervent, M.; Aldon, D.; Ranty, B.; Simonneau, T. RD20, a stress-inducible caleosin, participates in stomatal control, transpiration and drought tolerance in Arabidopsis thaliana. Plant Cell Physiol. 2010, 51, 1975–1987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhai, Y.F.; Wang, H.; Liang, M.M.; Lu, M.H. Both silencing- and over-expression of pepper CaATG8c gene compromise plant tolerance to heat and salt stress. Environ. Exp. Bot. 2017, 141, 10–18. [Google Scholar] [CrossRef]
- Koom, J.M.; Matzke, M.A.; Meyer, P. Listening to the silent genes: Transgene silencing, gene regulation and pathogen contro1. Trends Plant Sci. 1999, 4, 340–347. [Google Scholar]
- Elmayan, T.; Proux, F.; Vaucheret, H. Arabidopsis RPA2: A genetic link among transcriptional gene silencing, DNA repair, and DNA replication. Curr. Biol. 2005, 15, 1919–1925. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.G.; Guo, W.L.; Yin, Y.X.; Gong, Z.H. A novel F-Box protein CaF-Box is involved in responses to plant hormones and abiotic stress in pepper (Capsicum annuum L.). Int. J. Mol. Sci. 2014, 15, 2413–2430. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-11CT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Guo, W.L.; Chen, R.G.; Gong, Z.H.; Yin, Y.X.; Li, D.W. Suppression subtractive hybridization analysis of genes regulated by application of exogenous abscisic acid in pepper plant (Capsicum annuum L.) leaves under chilling stress. PLoS ONE 2013, 8, e66667. [Google Scholar] [CrossRef]
- Dhindsa, R.S.; Dhindsa, P.P.; Thorpe, T.A. Leaf senescence: Correlated with increased levels of membrane permeability and lipid p. J. Exp. Bot. 1981, 32, 93–101. [Google Scholar] [CrossRef]
- Dionisio-Sese, M.L.; Tobita, S. Antioxidant responses of rice seedlings to salinity stress. Plant Sci. 1998, 135, 1–9. [Google Scholar] [CrossRef]
- Arkus, K.A.J.; Cahoon, E.B.; Jez, J.M. Mechanistic analysis of wheat chlorophyllase. Arch. Biochem. Biophys. 2005, 438, 146–155. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.Q.; Ma, N.N.; Wang, G.D.; Meng, X.; Ai, X.Z.; Meng, Q.W. Suppression of SlNAC1 reduces heat resistance in tomato plants. Biol. Plant. 2015, 59, 92–98. [Google Scholar] [CrossRef]
- Mittova, V.O.; Volokita, M.; Guy, M.; Tal, M. Activities of SOD and the ascorbate-glutathione cycle enzymes in subcellular compartments in leaves and roots of the cultivated tomato and its wild salt-tolerant relative Lycopersicon pennellii. Plant Physiol. 2000, 110, 42–51. [Google Scholar] [CrossRef]
- Brennan, T.; Frenkel, C. Involvement of hydrogen peroxide in the regulation of senescence in pear. Plant Physiol. 1977, 59, 411–416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thordal-Christensen, H.; Zhang, Z.; Wei, Y.; Collinge, D.B. Subcellular localization of H2O2 in plants, H2O2 accumulation in papillae and hypersensitive response during barley-powdery mildew interaction. Plant J. 1997, 11, 1187–1194. [Google Scholar] [CrossRef]
- Jabs, T.; Dietrich, R.A.; Dangl, J.L. Initiation of runaway cell death in an Arabidopsis mutant by extracellular superoxide. Science 1996, 273, 1853–1856. [Google Scholar] [CrossRef]
- Wei, W.; Cui, M.Y.; Hu, Y.; Gao, K.; Xie, Y.G.; Jiang, Y.; Feng, J.Y. Ectopic expression of FvWRKY42, a WRKY transcription factor from the diploid woodland strawberry (Fragaria vesca), enhances resistance to powdery mildew, improves osmotic stress resistance, and increases abscisic acid sensitivity in Arabidopsis. Plant Sci. 2018, 275, 60–74. [Google Scholar] [CrossRef]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.-f.; Liu, S.-y.; Ma, J.-h.; Wang, X.-k.; Haq, S.u.; Meng, Y.-c.; Zhang, Y.-m.; Chen, R.-g. CaDHN4, a Salt and Cold Stress-Responsive Dehydrin Gene from Pepper Decreases Abscisic Acid Sensitivity in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 26. https://doi.org/10.3390/ijms21010026
Zhang H-f, Liu S-y, Ma J-h, Wang X-k, Haq Su, Meng Y-c, Zhang Y-m, Chen R-g. CaDHN4, a Salt and Cold Stress-Responsive Dehydrin Gene from Pepper Decreases Abscisic Acid Sensitivity in Arabidopsis. International Journal of Molecular Sciences. 2020; 21(1):26. https://doi.org/10.3390/ijms21010026
Chicago/Turabian StyleZhang, Hua-feng, Su-ya Liu, Ji-hui Ma, Xin-ke Wang, Saeed ul Haq, Yuan-cheng Meng, Yu-meng Zhang, and Ru-gang Chen. 2020. "CaDHN4, a Salt and Cold Stress-Responsive Dehydrin Gene from Pepper Decreases Abscisic Acid Sensitivity in Arabidopsis" International Journal of Molecular Sciences 21, no. 1: 26. https://doi.org/10.3390/ijms21010026
APA StyleZhang, H.-f., Liu, S.-y., Ma, J.-h., Wang, X.-k., Haq, S. u., Meng, Y.-c., Zhang, Y.-m., & Chen, R.-g. (2020). CaDHN4, a Salt and Cold Stress-Responsive Dehydrin Gene from Pepper Decreases Abscisic Acid Sensitivity in Arabidopsis. International Journal of Molecular Sciences, 21(1), 26. https://doi.org/10.3390/ijms21010026



