Abstract
The G protein-coupled cannabinoid receptors type 1 (CB1R) and type 2 (CB2R), and their endocannabinoid (eCBs) ligands, have been implicated in several aspects of brain wiring during development. Here we aim to assess whether interfering with CB1R affects development, neuritogenesis and pathfinding of GnRH and AgRP neurons, forebrain neurons that control respectively reproduction and appetite. We pharmacologically and genetically interfered with CB1R in zebrafish strains with fluorescently labeled GnRH3 and the AgRP1 neurons. By applying CB1R antagonists we observed a reduced number of GnRH3 neurons, fiber misrouting and altered fasciculation. Similar phenotypes were observed by CB1R knockdown. Interfering with CB1R also resulted in a reduced number, misrouting and poor fasciculation of the AgRP1 neuron’s axonal projections. Using a bioinformatic approach followed by qPCR validation, we have attempted to link CB1R functions with known guidance and fasciculation proteins. The search identified stathmin-2, a protein controlling microtubule dynamics, previously demonstrated to be coexpressed with CB1R and now shown to be downregulated upon interference with CB1R in zebrafish. Together, these results raise the likely possibility that embryonic exposure to low doses of CB1R-interfering compounds could impact on the development of the neuroendocrine systems controlling sexual maturation, reproduction and food intake.
1. Introduction
Marijuana and other derivatives of the plant Cannabis spp. have been used for thousands of years for their therapeutic and mood-altering/recreational properties. Cannabis-derived psychoactive compounds, mainly ∆9-tetrahydrocannabinol (∆9-THC) and cannabidiol (CBD), can pass the placental barrier and be transferred to the embryo [1]. It is now recognized that Cannabis consumption during pregnancy can exert adverse consequences on the progeny, including anxiety, cognitive and attention deficits, as well as depression [2].
In the central nervous system (CNS), ∆9-THC, CBD and various other synthetic cannabinoids bind the G protein-coupled receptors of the cannabinoid family, CB1R and CB2R [3]. The cognate endogenous ligands, known as endocannabinoids (eCBs), are small signaling lipids synthesized “on demand” from membrane lipids. The most abundant eCB in the CNS is 2-arachidonoylglycerol (2-AG), derived from the cleavage of diacylglycerol by diacyl-glycerol lipase (DAGL) and degraded by the action of monoacylglycerol lipase (MAGL) [4,5,6]. eCBs are mainly recognized by the CB1 and CB2 receptors, although they can also bind to other receptor types (TRP, PPARs, etc.). During CNS development, CB1R expression levels gradually increase and are localized on developing axonal projections, while CB2Rs are rather expressed by uncommitted precursor cells [7]. The localization of the eCB-synthesizing enzyme DAGL usually overlaps with that of CB1Rs in preterminal axons and growth cones, while MAGL is actively excluded from growth cones until synaptogenesis is concluded [8]. Upon release, eCBs have been shown to act in both autocrine and paracrine fashion and can cross cell membranes. In contrast to the primarily autocrine mechanism of action in the immature CNS, in the adult synapses it has been shown that eCBs act as retrograde signals (released post-synaptically, and acts on pre-synaptic termini to modulate neurotransmitter release) through a paracrine mechanism.
Following binding of eCBs, phyto-CBs and synthetic CBs to CB1R, a variety of transduction pathways are activated in a time- and region-dependent way, depending on the cell type and receptor repertoires. In particular, eCBs have been proposed to activate Rho-class of guanosine tri-phosphate phosphatases (Rho-GTPases) and to control the neuronal actomyosin cytoskeleton [9]. It has been demonstrated that [10] direct interaction of CB1 with the WAVE1/RAC1 complex, which acts on actin-binding proteins, results in actin remodeling and growth cone dynamics. Activated CB1R can also modulate microtubule dynamics at the growth cone, via the c-Jun NH(2)-terminal kinases (JNKs) pathway [11,12,13]. By modulating the actomyosin network and microtubule dynamics at the growth cone [9], in addition to axon growth and guidance, CB1 receptors have been shown to participate in the control of synaptogenesis and synaptic plasticity in various experimental models [14,15,16].
Data from the medical literature support the role of CB1R in brain wiring during early development. Fetuses exposed to cannabis during embryonic life have increased risk of cognitive deficit [17,18], attention deficit [19] and anxiety and depression [20], as well as defects in neuronal migration and axonal pathfinding in several brain districts [2,21]. Most experimental studies have focused mainly on the cerebral cortex, the hippocampus and the visual system in mice, where CB1R regulates axon guidance and map formation [22,23,24,25,26,27,28]. Importantly, exposure of pregnant mice to ∆9-THC results in life-long circuit modification and altered wiring in the offspring [29]. In zebrafish embryos, interfering with CB1R signaling has been shown to result in abnormal axonal growth and fasciculation in the anterior and posterior commissures of the forebrain. Moreover, exuberant axons of reticulospinal neurons in the hindbrain were found to cross the midline or to deviate from their trajectory and turn backward [30]. Similarly, the selective pharmacological inhibition of CB1R and CB2R in early embryos resulted in altered development of the locomotor system at later stages [31], and treatment with ∆9-THC or CBD resulted in altered motor neurons morphology, synaptic activity at the neuromuscular junction and locomotor responses to sound [32]. Reducing the level of 2-AG via knockdown of Daglα resulted in abnormal behaviors, characterized by stereotyped movement and altered motion perception [33].
Few reports have linked the eCB/CB1R signaling functions to the maturation and physiology of the hypothalamic neuroendocrine systems controlling reproduction and appetite [34,35,36]. The gonadotropin releasing hormone (GnRH) neurons, which are central for the control of sexual maturation, fertility and reproduction, have been extensively studied. In this context, eCBs have been shown to modulate the input and the firing activity of GnRH neurons [36], and CB1R activation inhibits presynaptic GABA release [37,38]. Accordingly, the literature reports a link between perturbations of CB1R (either due to consumption of phyto-cannabinoids or exposure to synthetic cannabinoids) and altered functionality of the neuroendocrine gonadotropic system, puberty and fertility deficits [35,36].
Another important prominent effect of the eCB system is the regulation of appetite and energy balance. The hypothalamic network of peptidergic neurons controlling food intake/energy metabolism, enhancing feeding behavior, includes mainly neurons expressing pro-opiomelanocortin (POMC), neuropeptide Y (NPY) and agouti-related peptide (AgRP) [39]. This system is highly conserved among vertebrates. Pharmacological blockade of CB1R suppresses hunger and induces hypophagia, and the CB1R-antagonist Rimonabant in the past has been proposed for the treatment of obesity [40,41,42]. Importantly, eCB/CB1R signaling has been shown to have an impact on AgRP/NPY neurons [43] and indeed its perturbation has been associated with altered appetite control, energy storage/consumption and metabolic conditions [44].
While most previous works have focused on postnatal and adult hypothalamic functions, little is known on the role of CB1R and its misactivation for the correct development of hypothalamic neuroendocrine neurons and their connectivity needed to attain control of sexual maturation and/or reproduction and appetite/energy storage. This question is highly relevant, to fully assess the impact of exposure to these (and other molecules) during embryonic and early infancy on human health, and the possible link with adult pathologies, such as reduced fertility and altered food intake. Furthermore, these studies aimed to elucidate the role of eCB ligands and the mechanisms underlying axonal miswiring upon altered CB1R activation [7].
We have investigated the possible role of CB1R altered activity on the extension, fasciculation and pathfinding of GnRH3 and AgRP axons during forebrain development in zebrafish. Using transgenic lines in which GnRH3 and AgRP neurons are fluorescently labeled, we show that pharmacological and genetic manipulation of CB1R resulted in abnormal guidance, fasciculation and routing of GnRH3 and AgRP1 axons during embryonic development, indicating that CB1R is a possible regulator of the development of reproduction- and food intake-related neurons.
2. Results
2.1. Survival and Hatching Rates of Zebrafish Embryos Treated with CB1R Ligands
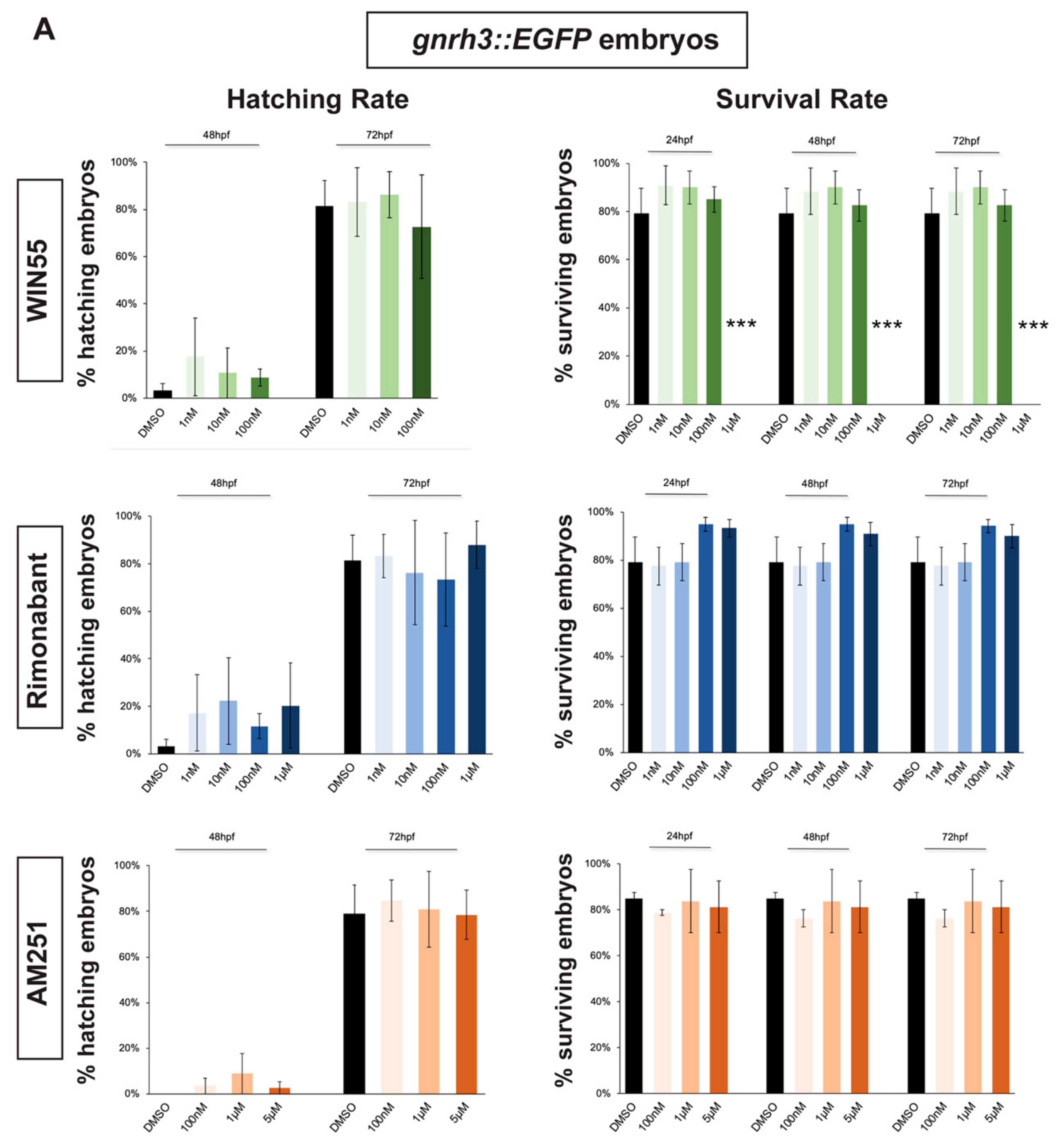
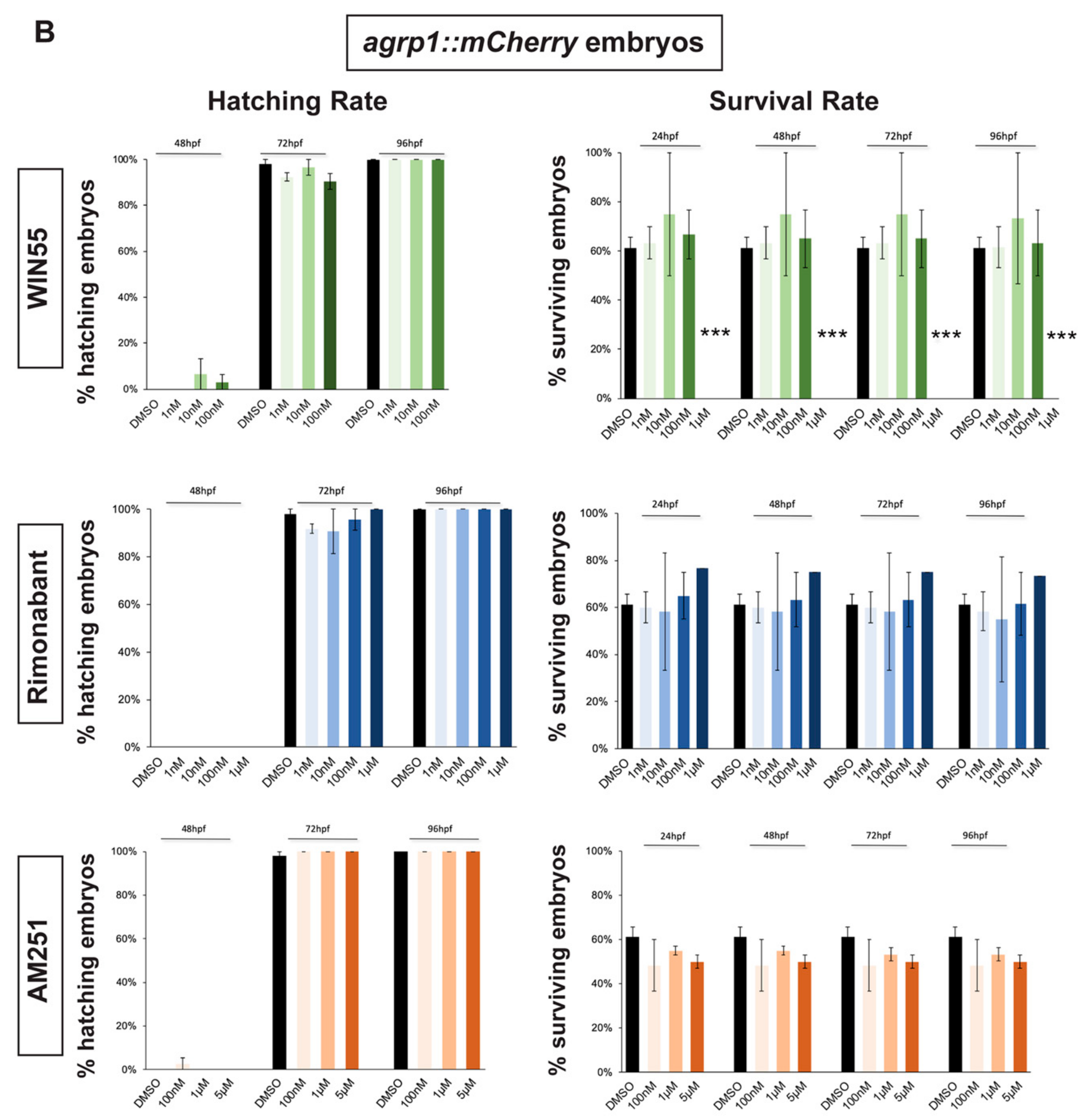
In order to select a range of non-toxic concentrations and non-teratogenic doses of CB1R agonists and antagonists, we initially determined their effects on the survival and hatching rates of zebrafish embryos. gnrh3::EGFP [45,46] and agrp1::mCherry [47] zebrafish embryos were exposed to different compounds starting at 0 hours post fertilization (hpf). Hatching rate was examined at 48 hpf and 72 hpf for gnrh3::EGFP embryos and also at 96 hpf for agrp1::mCherry embryos. The survival rate was examined at 24 hpf, 48 hpf and 72 hpf for gnrh3::EGFP embryos and at 24 hpf, 48 hpf, 72 hpf and 96 hpf for agrp1::mCherry embryos. Survival rate of agrp1::mCherry embryos at 96 hpf was analyzed as well because at 72 hpf the fluorescence signal of the AgRP1 fibers was still too weak to be reliably evaluated by fluorescence microscopy. The survival rate was calculated as the percentage of live embryos over the total number of embryos (30 embryos/well for each concentration); the hatching rate was calculated as the percentage of hatched embryos over the remaining living embryos.
As agonist of the CB1 receptor, we chose WIN55,212-2 (renamed WIN55 for simplicity), which has been reported to mimic endocannabinoids (eCBs) signaling in several studies [22,48,49,50]. Four concentration of WIN55 were tested: 1 nM, 10 nM, 100 nM and 1 µM [48,50]. As antagonist/inhibitors of CB1R we chose Rimonabant and AM251, known to act on CB1R as inverse agonist or antagonist, respectively [9,22,23,48]. Four concentrations for Rimonabant (1 nM, 10 nM, 100 nM and 1 µM) and three concentrations for AM251 (100 nM, 1 µM and 5 µM) were tested.
Hatching, normally occurring between 48 and 72 hpf, was not affected by the exposure to any concentration of CB1R ligand in both gnrh3::EGFP and agrp1::mCherry embryos (Figure 1A,B). Concerning the survival rate, we observed that the highest WIN55 concentration (1 µM) caused complete embryonic lethality for both gnrh3::EGFP and agrp1::mCherry embryos, already at 24 h, while the survival of both transgenic lines following different concentrations of AM251 and Rimonabant was unchanged as compared to the DMSO-treated control embryos (Figure 1A,B).
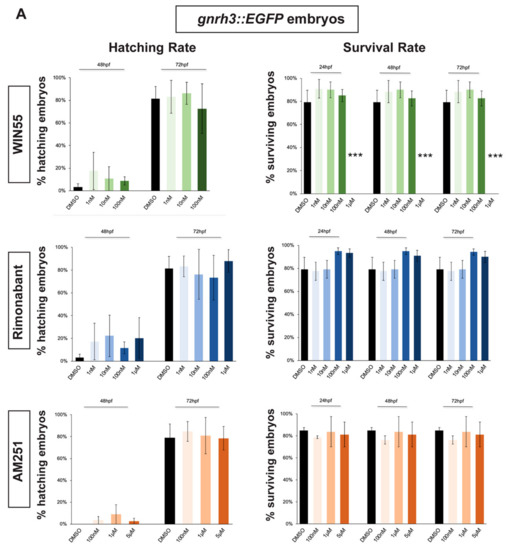
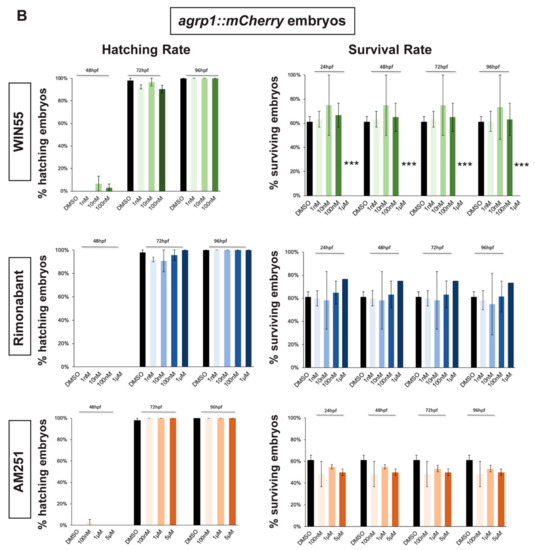
Figure 1.
Survival and hatching rates in zebrafish embryos treated with CB1R ligands. (A) Hatching rate (panels on the left) and survival rate (panels on the right) of gnrh3::EGFP zebrafish embryos following treatments with WIN55, Rimonabant or AM251, at the indicated doses, examined at the indicated embryonic age. Results are expressed as % over the total number of embryos examined. (B) Same as in (A), but using agrp1::mCherry embryos. *** = p < 0.001.
2.2. Effects of CB1R Ligands on Zebrafish GnRH3 Neurons
In fish embryos, GnRH3 neurons are first seen at 24 hpf in the olfactory placode region, associated with the terminal nerve. By 48 hpf, axons extending from GnRH3 neurons present in the nasal region project dorsally and ventrally into the forebrain and enter the anterior commissure (AC) and the postoptic commissure. In addition, two tracts extend along the optic nerve and innervate the retina, a tract extends dorsally towards the region of the pineal gland and a tract extends posteriorly towards the hypothalamus and preoptic area. Between 3 and 15 days post-fertilization (dpf), soma migration takes place in an axonophilic manner along the terminal nerve and the ventral telencephalon towards the ventral hypothalamus [45]. Some GnRH3 neurons are thought to migrate to the hypothalamus, while others remain along the path [46].
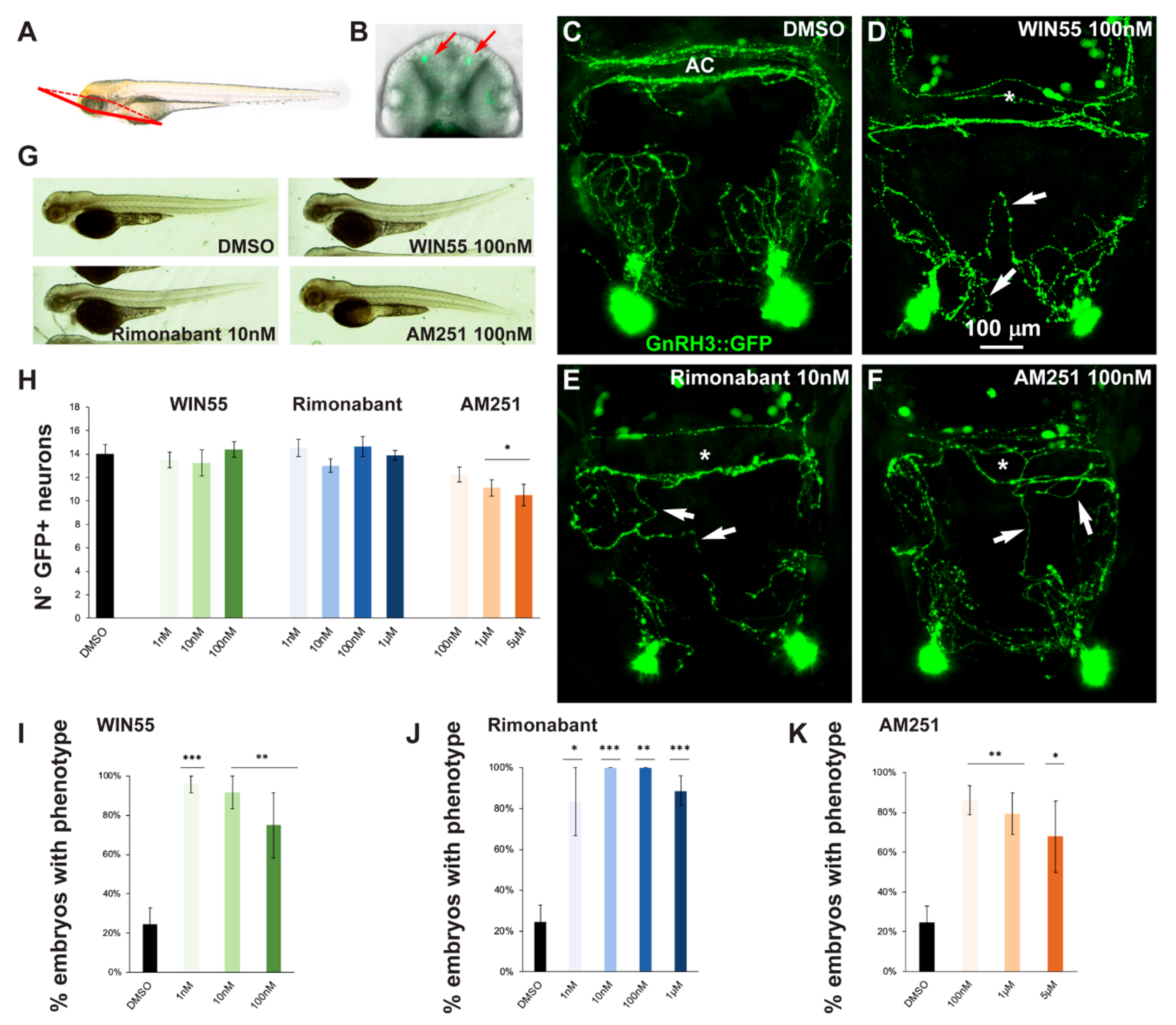
In order to assess possible effects of altered CB1R function on the development of the GnRH3 system in the olfactory region and anterior commissure, gnrh3::EGFP embryos at the 1-cell stage were treated with various concentrations of CB1R-ligands for 72 h after which the EGFP+ embryos were selected and examined by confocal microscopy. A proper view plane of the transgenic fish was adopted to allow the correct visualization of both GnRH3 neurons present in the olfactory region and GnRH3 fibers extending towards the anterior commissure (shown in Figure 2A,B). As pointed out in the previous section, with all the ligands and all concentrations used in this study we found no evident morphological changes or developmental delay, as shown by low magnification bright field whole embryos examination (Figure 2G).
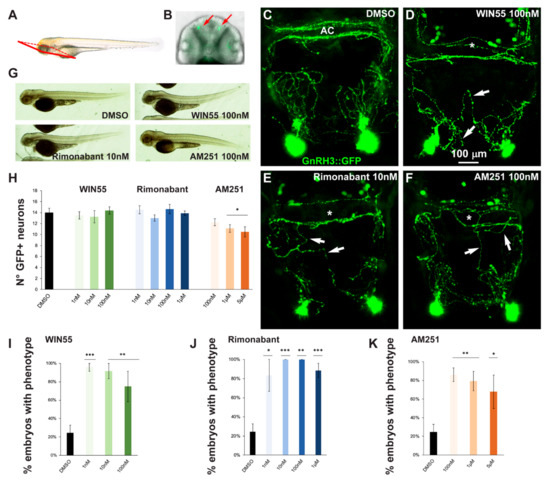
Figure 2.
Pharmacological manipulation of CB1R on GnRH3 neurons in zebrafish embryos. (A) Scheme showing the observation plane used for the micrographs in (C–F). (B) Low magnification of the head piece of gnrh3::EGFP embryos, viewed in combined fluorescence and bright field illumination. Red arrows indicate the EGFP+ cells associated to the terminal nerve. (C–F) Representative images of EGFP+ neurons and their projections present in the nasal and basal forebrain regions of embryos at 72 hpf, treated either with DMSO only (negative control; C), with WIN55 (D), Rimonabant (E) or AM251 (F), at the indicated doses. Scale bar is reported in D. White arrows indicate misguided EGFP+ projections, and white asterisks indicate altered organization of EGFP+ fibers at the anterior commissure. (G) Bright-field low magnification images of whole embryos treated with DMSO only, WIN55, Rimonabant or AM251, showing a normal general morphology and growth. (H) Quantification of the number of EGFP+ neurons in the nasal region of fish embryos treated with DMSO only (black bar), or with WIN55, Rimonabant or AM251 at the indicated doses. The color code is the same as in Figure 1. No significant difference in the number of EGFP+ neurons was observed following these treatments. (I–K) Quantification of the misguidance and altered commissural phenotypes, expressed as % of the number of embryos presenting the phenotype, upon treatment with WIN55 (I), Rimonabant (J) or AM251 (K). Color code as in Figure 1. Data are expressed as means ± SEM from three independent experiments. * = p < 0.05; ** = p < 0.01; *** = p < 0.001. AC, anterior commissure.
The number, neurite extension and commissural fasciculation of GnRH3 neurons and fibers were examined and compared to control embryos treated with DMSO only (Figure 2C). We found that ligand exposure led to abnormal axonal pathfinding, with fibers that did not extend with proper directionality (Figure 2D–F, white arrows) and a failure in fasciculation of axonal bundles at the level of the anterior commissure (Figure 2D–F, asterisks). For quantification purposes, we defined as “phenotype” the condition in which we observe a) altered orientation and position of EGFP+ fibers; b) altered fasciculation of EGFP+ fibers at the commissures; or c) both of these together. With this definition, the percentage of embryos showing this phenotype was calculated for each ligand concentration tested (WIN55: 1 nM, n = 10 embryos; 10 nM, n = 10; 100 nM, n = 12; Rimonabant: 1 nM, n = 10; 10 nM, n = 13; 100 nM, n = 10; 1 μM, n = 20; AM251: 100 nM, n = 12; 1 μM, n = 14; 5 μM, n = 11) and for the control embryos (DMSO only, n = 30). The results were then expressed as the number of embryos with the phenotype in the various conditions, as percentage over the total number of embryos examined, and reported in Figure 2I–K. We observed an increase in the proportion of embryos showing the phenotype after drug treatment (Figure 2I–K). Thus, treatment with either a CB1R-agonist (WIN55) or CB1R-antagonist/inverse agonist (AM251 and Rimonabant) resulted in miswiring of GnRH3 fibers, indicating that either the activation or de-activation of CB1R has similar negative consequences on axonal fasciculation and navigation.
At higher magnification, we also counted the number of EGFP+ cells present in the olfactory region (Figure 2H). We observed no difference between the control embryos and embryos treated with WIN55 and Rimonabant (Figure 2I–J). Conversely, a decrease in cell number was observed with high concentrations of AM251 (Figure 2K), suggesting that the inhibition of CB1R signaling may interfere with GnRH3 cell proliferation or accelerate the migration of GnRH3 neurons to the hypothalamus.
2.3. Effects of CB1R Knockdown on Zebrafish GnRH3 Neurons
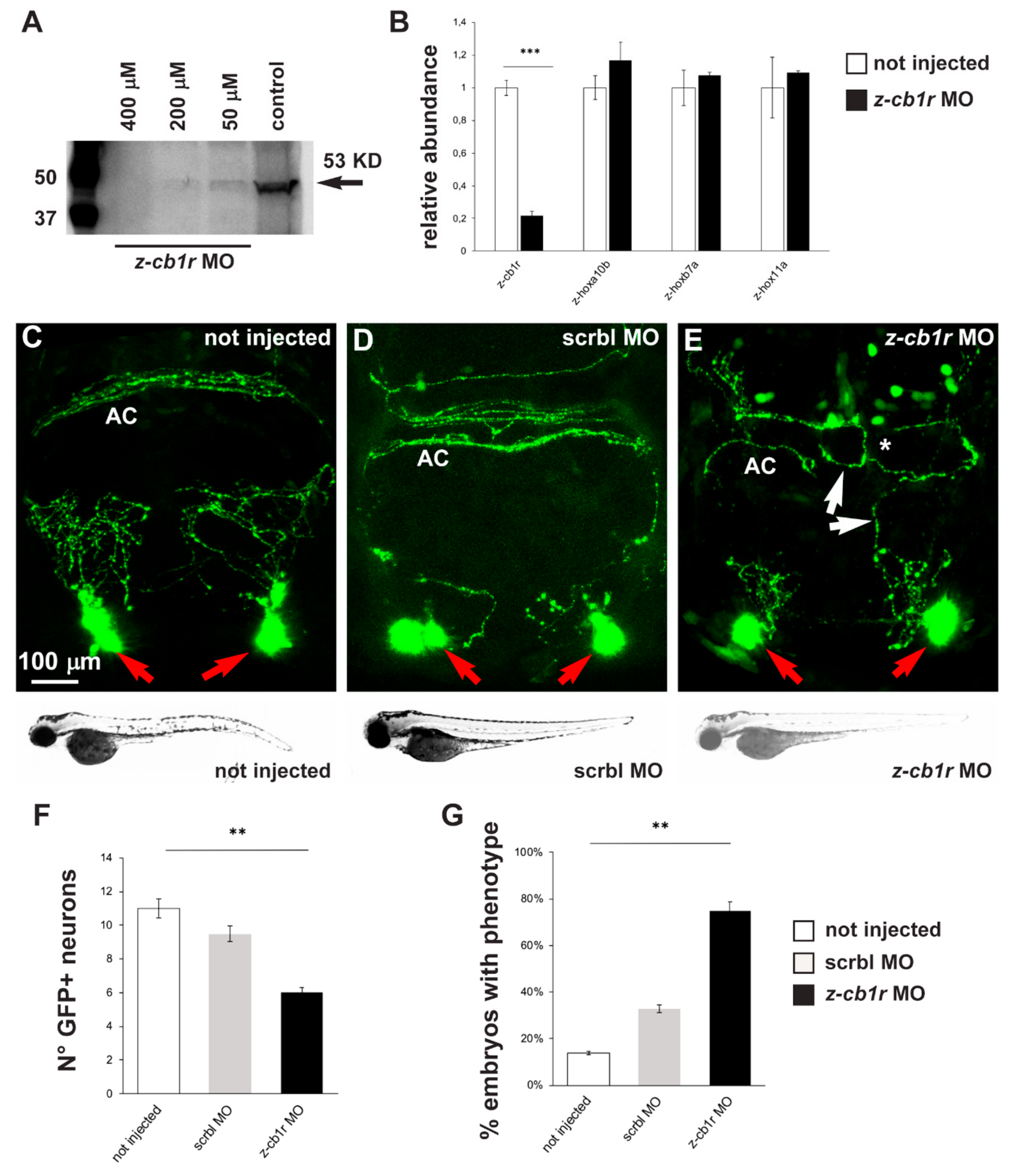
In order to verify if down-modulation of CB1R could lead to phenotypes similar to the ones observed after treatment with CB1R ligands, we utilized a conventional antisense morpholino oligonucleotide (MO)-mediated strategy. gnrh3::EGFP embryos were injected at the 1-cell stage with a previously published z-cb1r morpholino, whose sequence was designed to bind to a region 5′ of the ATG start codon and was shown to block z-cb1r mRNA translation [30]. Uninjected embryos and embryos injected with a scrambled-sequence MO (Mismatch_PPFIA) were used as controls.
First, the efficiency of the z-cb1r MO to deplete the endogenous CB1R protein was tested by Western blot analysis on total proteins, extracted from z-cb1r MO-injected embryos and from controls (uninjected embryos) at 72 hpf, using an anti-CB1R antibody. A significant dose-dependent reduction of the endogenous level of z-CB1R protein was observed in z-cb1r MO-injected embryos, with the most efficient concentration being 400 µM z-cb1r MO (Figure 3A). Real-time qPCR analyses on RNA samples extracted from embryos injected with 400 µM z-cb1r MO showed that the amount of z-cb1r mRNA was reduced in knockdown embryos compared to control uninjected embryos (Figure 3B). The expression levels of three developmental genes, z-hoxb7a, z-hoxa10b and z-hox11a [51], whose expression is age dependent, were quantified by real-time qPCR and employed to exclude any developmental defects of the embryos caused by the injection of z-cb1r MO. No significant differences were observed between the z-cb1r MO-injected versus the uninjected control embryos (Figure 3B), indicating that the depletion of CB1R did not globally affect the rate of embryonic development.
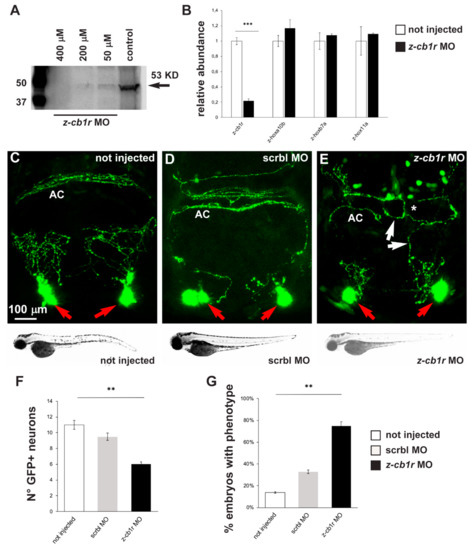
Figure 3.
Effects of MO-mediated CB1R knockdown on GnRH3 neurons in zebrafish embryos. (A) Western blot analyses of protein extracts from uninjected or z-cb1r MO-injected zebrafish embryos. A significant depletion of the CB1R protein was detected in the treated embryos. (B) Real-time qPCR analyses of z-cb1r and three z-hox genes on RNA samples extracted from uninjected (open bars) or z-cb1r MO-injected (solid black bars) embryos. Expression is shown relative to the expression of the housekeeping gene mRNA (z-β-actin). The abundance of the control RNA is set as 1. A depletion of the z-cb1r mRNA in embryos injected with z-cb1r MO is observed, as compared to control ones. No change in the expression of z-hox genes was observed, indicating unaltered developmental progression. (C–E) Representative images of EGFP GnRH3 neurons and projections in the nasal and the basal forebrain regions of zebrafish embryos, at the age 72 hpf. The observation plane is the same as in Figure 2. Scale bar is reported in C. Red arrows indicate EGFP+ neurons, white arrows indicate altered EGFP+ projections, and white asterisks indicate altered organization and fasciculation at the anterior commissure. Below each micrograph, the bright-field images show the general morphology and growth of the corresponding whole animal, with no difference between the three conditions. (F) Quantification of the number of EGFP+ neurons in the nasal region of uninjected (open bars), scrmbl MO-injected (solid grey bars) or z-cb1r MO-injected (solid black bars) embryos. A significant decrease was observed in embryos injected with the z-cb1r MO, compared to control ones. (G) Quantification of the misguidance and altered commissural phenotype, expressed as % of the number of embryos observed. A significant increase was observed in embryos injected with the z-cb1r MO, compared to control ones. Data are expressed as means ± SEM from three independent experiments. ** = p < 0.01; *** = p < 0.001. AC, anterior commissure.
Having proven the efficiency of z-cb1r MO, 1-cell stage gnrh3::EGFP embryos were injected with the z-cb1r MO at the highest concentration (400 μM) or with scrambled (scrbl)-MO as control, and examined at 72 hpf to determine the number, neurite arborization and commissural organization by confocal microscopy (Figure 3C–E). Upon depletion of CB1R, the number of terminal nerve-associated GnRH3 neurons was significantly reduced in comparison to the controls (average 6 EGFP+ cells in z-cb1r MO-treated embryos vs. 10.8 in controls; Figure 3F). We then examined the extension, orientation and fasciculation of EGFP+ fibers in the nasal region and in the basal forebrain. EGFP+ axons displayed an abnormal position, altered direction or aberrant fasciculation within the anterior commissure. We defined as “phenotype” the condition in which we observe a) altered orientation/position of EGFP+ fibers; b) altered fasciculation of EGFP+ fibers; or c) both of these together, as we previously observed with CB1-ligands. The results were then expressed as the number of embryos presenting the phenotype in the various conditions (z-cb1r MO n = 17; scrbl MO n = 17; uninjected n = 19) over the total number of embryos examined. As reported in Figure 3G, upon depletion of CB1R, we observed a significant increase in the fraction of embryos with the indicated phenotype compared to the controls (78% in z-cb1r MO-injected embryos vs. 14% in uninjected controls).
2.4. Expression of CB1R in Developing Zebrafish Brain
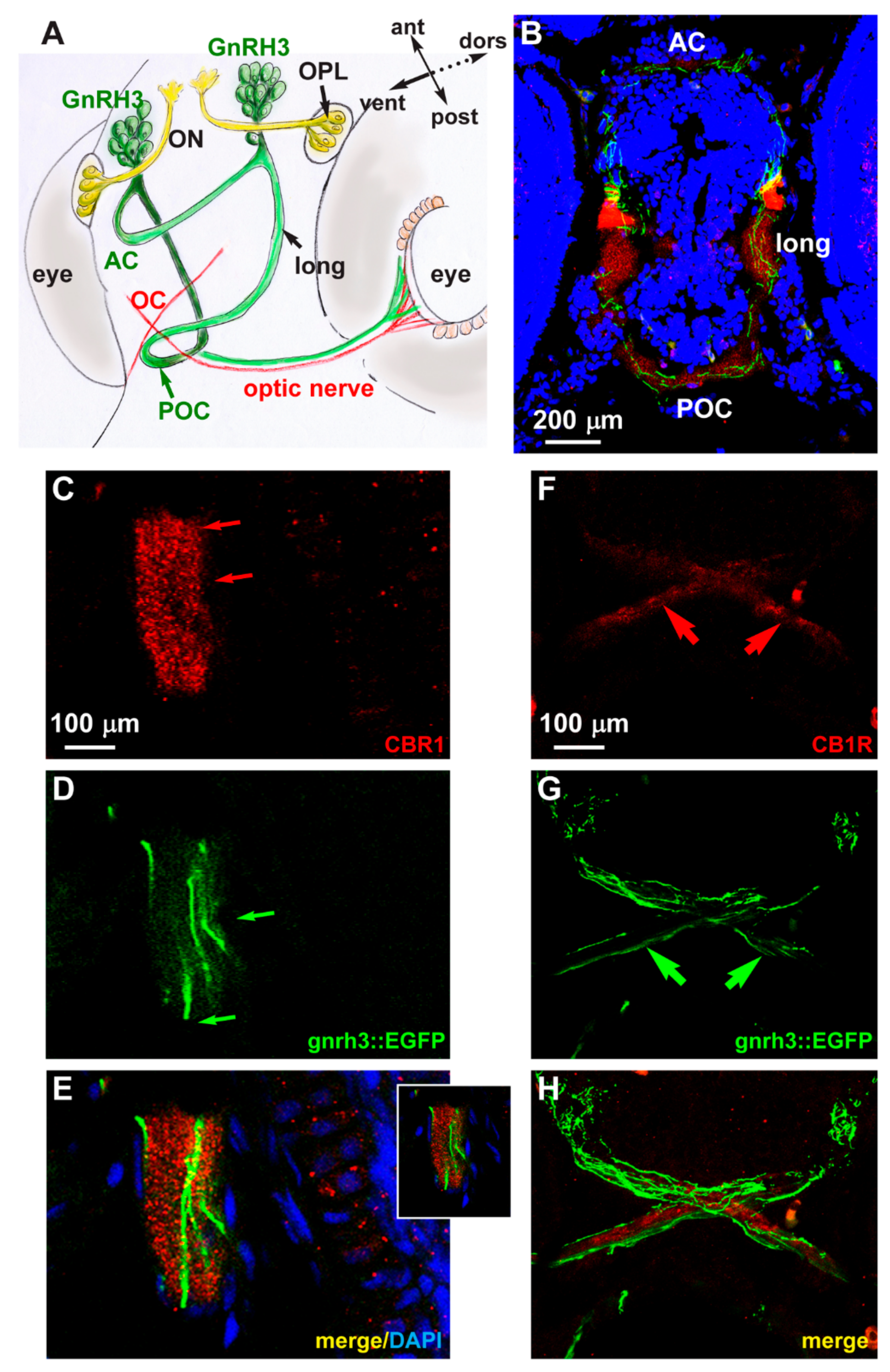
We examined the endogenous expression of the CB1 receptor in zebrafish embryos at the age 72 hpf. We carried out fluorescent immunohistochemistry with anti-CB1R antibody on cryostatic sections obtained from gnrh3::EGFP embryos. CB1R immunoreactivity (red fluorescence) was detected in various regions of the basal forebrain, in particular in the anterior and postoptic commissures, in the optic chiasm and in the longitudinal tracts. In the same regions, EGFP+ axonal extensions are clearly seen in close proximity to the CB1R signal (Figure 4B). The CB1R immunoreactivity mainly appears as punctate staining (Figure 4C,F). No physical overlap between CB1R immunoreactivity and the GnRH3 axons along the optic tracts and nerves (Figure 4F–H) and no CB1R immunoreactivity in the GnRH3+ cell bodies in the nasal-terminal nerve region were detected (not shown). Colocalization of CB1R immunoreactivity and EGFP fluorescence was instead found in longitudinal fibers and the anterior commissure (Figure 4B–E). These results suggest that CB1R is expressed by GnRH3 fibers in commissures and the longitudinal tracts, while CB1R+ fibers and GnRH3 fibers are mostly distinct, although running in close association along the optic fibers. It should be noted that GnRH3 fibers entering the optic nerve run in an opposite direction to retinofugal CB1R+ axons. Nonetheless, cross talk among GnRH3- and CB1R-containing fibers could occur by means of paracrine and/or cell adhesion signaling.
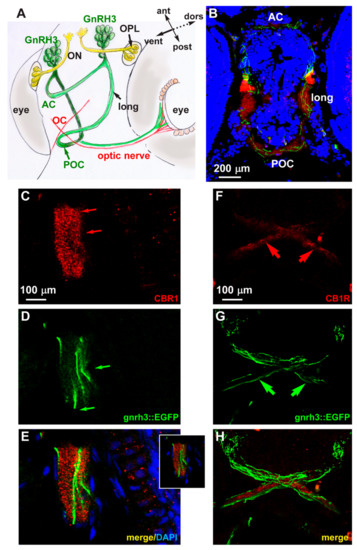
Figure 4.
Expression of CB1R in developing zebrafish brain. (A) Scheme illustrating the position of the GnRH3 neurons relative to the position and orientation of the main forebrain commissure in the zebrafish brain (redrawn from [52]). (B) Low magnification double-fluorescent images of GnRH3::EGFP neurons (green) and IFL with an anti-CB1R antibody (red) on cryostatic sections of the zebrafish head at the age 72 hpf. (C–E) Higher magnification double-fluorescence images of the anterior commissure, showing overlapping localization of CB1R punctate staining (red) with EGFP+ fibers (green). Sections were counterstained with DAPI (blue). The merged signal is shown in E. Inset in E shows a single stack of the same image. Arrows indicate the fluorescence signal. (F–H) Higher magnification double-fluorescence images of the optic chiasm, immunostained for CB1R (red), as in panels C–E. Adjacent but non-overlapping expression was observed in these fibers. Arrows indicate the fluorescence signal. Scale bars are reported in panels B, in C (for C,D) and in F (for F–H). AC, anterior commissure; ant, anterior; dors, dorsal; long, longitudinal tract; OC, optic chiasm; ON, olfactory nerve; OPL, olfactory placode; POC, postoptic commissure; post, posterior; RGC, retinal ganglion cells; ven, ventral.
2.5. Effect of Exposure to CB1R Ligands on AgRP1 Neurons
In order to assess possible effects of altered CB1R function on the development of a well-defined group of hypothalamic neurons involved in the control of food-intake, we used a zebrafish strain in which the AgRP1 neurons are genetically marked with mCherry, a red fluorescence reporter [47]. We selected this neuronal population for the following reasons: (1) the neuronal axons and dendrite extend at approximately the same developmental age and for a similar length as the GnRH3 fibers; (2) the AgRP1 axons cross the midline in the anterior and the postoptic commissures, as some of the GnRH3 axons do; and (3) in combination with POMC neurons, the AgRP1 neurons participate in the neuroendocrine control of appetite and energy storage [53]. AgRP1 neurons have cell bodies located in the ventral periventricular hypothalamus with fibers that project towards the rostral, intermediate and dorsal hypothalamus, as well as the preoptic area (POA), the anterior commissure (AC), the post-optic commissure (POC) and the ventral tegmental commissure (VTC) [47].
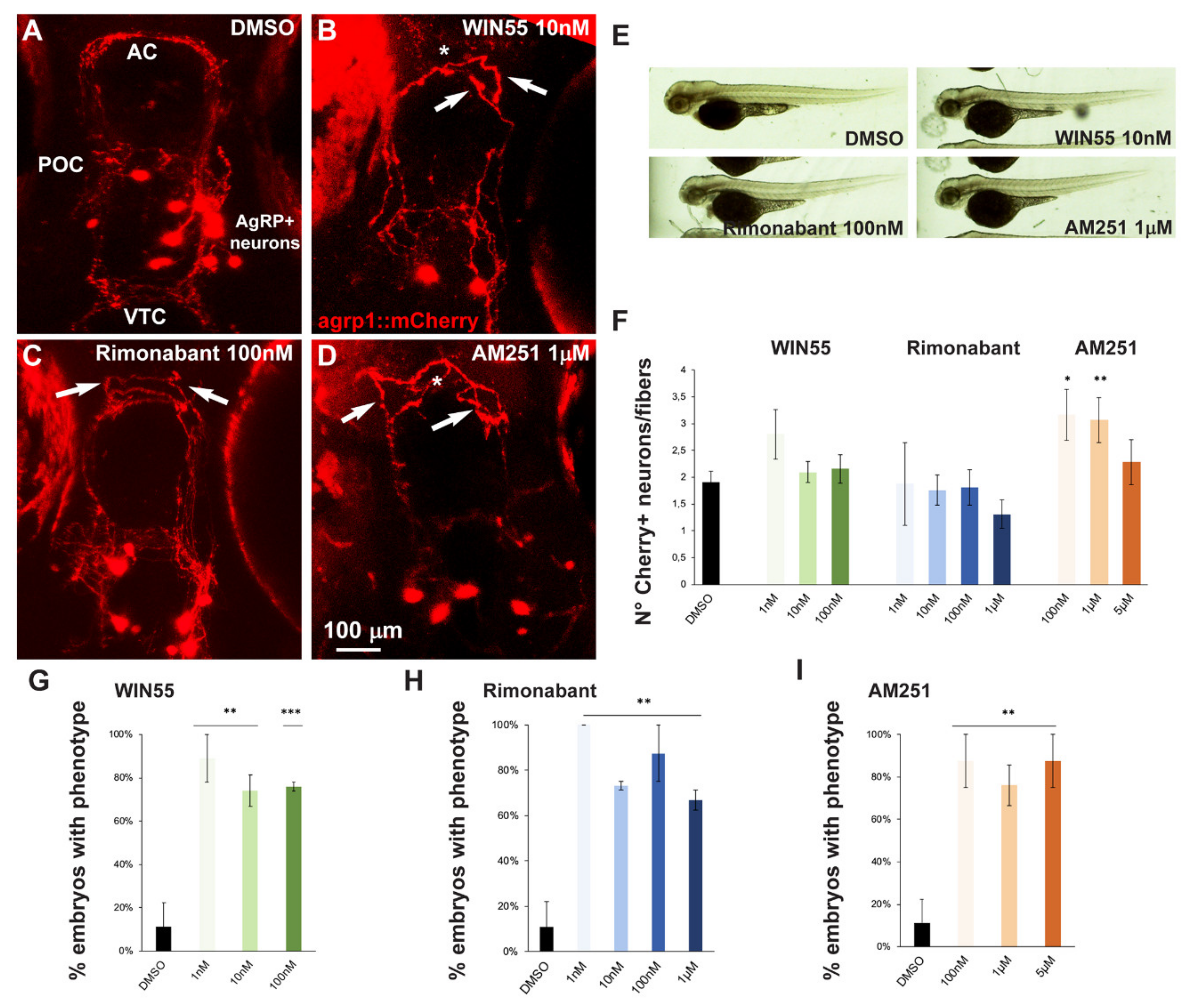
For our purposes, we treated embryos derived from breeding between the agrp1:Gal4 and the UAS:nfsB-mCherry. The resultant offspring, agrp1::mCherry, were treated with the CB1R ligands WIN55, Rimonabant or AM251 for 96 h as described above. No evident morphological embryos anomalies (Figure 5E) or developmental delays were observed. However, treatment with these compounds resulted in shorter or misoriented mCherry+ fibers, as well as their altered organization at the AC (Figure 5B–D) compared to the control shown in Figure 5A. For quantification purposes we defined as “phenotype” the condition with altered orientation of mCherry+ fibers, or altered fasciculation of mCherry+ fibers, or both of these together. The results were then expressed as the fraction of embryos showing the phenotype over the total number of embryos examined, in percent and reported in Figure 5G–I. The treatment with all three ligands with all doses employed resulted in a significant increase in the fraction of embryos with the indicated phenotype, compared to the controls (Figure 5G–I).
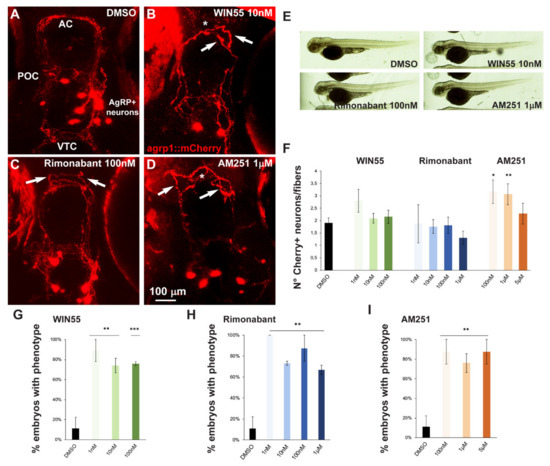
Figure 5.
Effect of pharmacological manipulation of CB1R on AgRP1 neurons in zebrafish embryos. (A–D) Representative images of mCherry+ neurons and projections in the basal forebrain of embryos, at the age 96 hpf, treated with DMSO only as control (A), WIN55 (B), Rimonabant (C) or AM251 (D) at the indicated doses. The observation plane is the same as in Figure 2. Scale bar is reported in (D). White asterisks indicate altered fasciculation at the anterior commissure, and white arrows indicate misguided mCherry+ projections. (E) Bright-field, low-magnification images of whole fish embryos treated with DMSO only or with the indicated ligands. A normal general morphology and growth was observed. (F) Quantification of the ratio of mCherry+ neurons/mCherry+ fibers in embryos treated with DMSO only (solid black bar) or treated with WIN55, Rimonabant or AM251 at the indicated doses. Color code is the same as in Figure 1. Treatment with AM251 resulted in a higher neurons/fibers ratio. (G–I) Quantification of the misguidance and altered commissural phenotype, expressed as % of the number of embryos observed upon treatment with WIN55 (G), Rimonabant (H) or AM251 (I). Color code as in Figure 1. Data are expressed as means ± SEM from three independent experiments. * = p < 0.05; ** = p < 0.01; *** = p < 0.001. AC, anterior commissure; POC, postoptic commissure; VTC, ventral tegmental commissure.
We also determined the number of mCherry+ cells in the treated versus the control embryos, and calculated the ratio between the number of cells and the number of visible fibers; this was done in order to normalize the value between embryos, considering that the absolute number of mCherry+ cells is variable as a consequence of the genetics of the two parent lines [47]. Treatment with AM251, but not with WIN55 or Rimonabant, resulted in a significant increase in the cell number/cell fiber ratio (Figure 5F), indicating a possible reduction in neurite formation.
2.6. Search for Genes Functionally Related to CB1R Involved in Axon Fasciculation and Guidance
In order to identify genes related to the CB1R signaling pathway that might participate in axonal growth and fasciculation during development, we resorted to bioinformatic tools and publicly available datasets. First, we selected the following genes known to be involved in axon fasciculation and guidance by a literature search: ROBO1, PAX6, BRCA1, AKT1, PI3KCA, SLIT2, L1CAM and GAP43 [50,54,55,56]. Using these eight genes plus the CB1R gene as entry, we searched the Human Protein Reference Database for protein::protein interactions and selected those interactors appearing in the lists of at least two of the entry genes. This search yielded 27 genes (Table S1). Next, we entered 23 of these genes (four were not present in the database) + CB1R into the coexpression dataset FuncPred [57], reasoning that genes involved in the same cellular function are significantly coexpressed, and their coexpression is significantly maintained across species [58]. This search yielded a global coexpression network of 625 genes, which was then intersected with a dataset of genes differentially expressed during development of the mouse hypothalamus (Affymetrix MOE430 microarrays, dataset GSE21278 of the GEO repository) [59]. From this last dataset we specifically compared samples of the hypothalamus and POA of C57Bl/6 mouse embryos at E10.5 with the same samples from CD-1 embryos at E18.5, sex-independently. Only genes with p < 0.001 and logFC > 1 for upregulated genes, and logFC < −1 for downregulated ones, were considered. This intersection yielded a total of 269 “best candidates”, 207 of which were upregulated and 62 of which were downregulated during hypothalamus development (Table S2). Candidates resulting from this intersection were then functionally annotated and the following relevant over-represented categories were detected: “cytoskeleton”, 13%; “synapse”, 12%; “neuron projection”, 10%.
Among the genes appearing in relevant functional classes we noted the presence of semaphorins, cell adhesion molecules, kinesins, GABA and glutamate receptors. Finally, from the list of relevant genes we further selected them by a literature search and came up with a set of 8 genes putatively interacting with CB1R, or sharing a common expression pattern, and potentially involved in axon elongation–fasciculation-guidance: Glycoprotein M6A (GPM6A), N-deacetylase and N-sulfotransferase-3 (NDST3, also known as HSST3), Neural growth regulator-1(NEGR1), Sodium voltage-gated channel alpha subunit-2 (SCN2A, also known as Na(v)1.2), Seizure related gene-6 (SEZ6), HECT, C2 and WW domain containing E3 ubiquitin protein ligase-1 (HECW1, also known as NEDL1), Septin-6 (SEPT6) and Stathmin-2 (STMN2, also known as SCG10).
For further analyses, we focused on z-stmn2a, z-stmn2b, negr1, z-sez6a and z-sez6b, since they have been reported as regulator of axons growth and cell migration [60,61,62,63]. We decided to define whether the expression of these selected candidates changes following treatment with the CB1R ligands WIN55, Rimonabant and AM251, or following depletion of endogenous CB1R by MO injection.
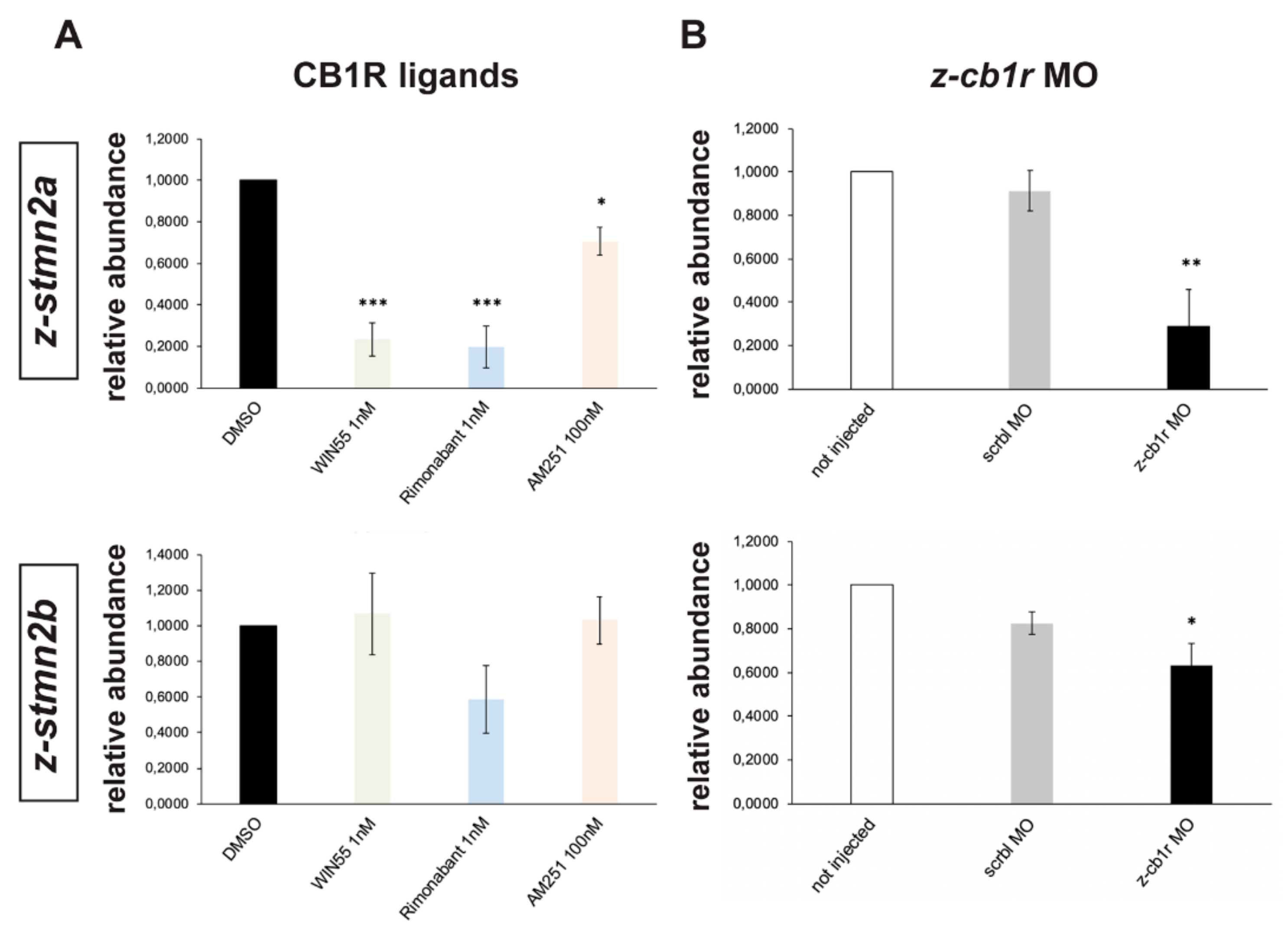
Real-time qPCR was used to compare the abundance of z-stmn2a, z-stmn2b, negr1, z-sez6a and z-sez6b mRNAs in samples extracted from embryos treated with WIN55 (1 nM), Rimonabant (1 nM) or AM251 (100 nM), or treated with DMSO only as control. All treatments resulted in a significant reduction in the abundance of z-stmn2a relative to the control (Figure 6A), while the expression of z-stmn2b (Figure 6A) and of the other targets (data not shown) did not significantly change.
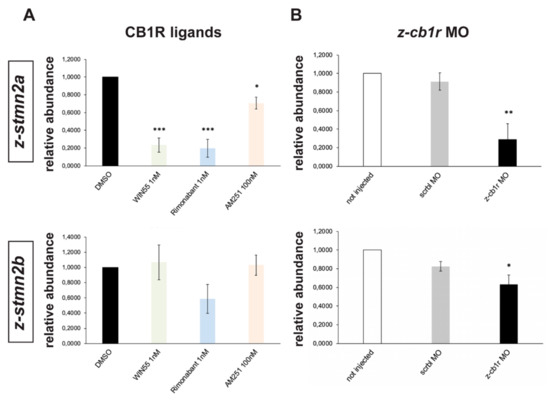
Figure 6.
Expression of z-stmn2a and z-stmn2b upon treatment with CB1R ligands or CB1R knockdown in early zebrafish embryos. (A) Real-time qPCR analyses of z-stmn2a (top histogram) and z-stmn2b (bottom histogram) on RNA samples obtained from embryos treated with the indicated CB1R ligands, at the indicated doses. Expression is shown relative to the expression of the housekeeping mRNA (z-β-actin). The abundance of the control RNA (DMSO) was set to 1. A reduced abundance of z-stmn2a mRNA was observed in embryos treated with the ligands, as compared to embryos treated with DMSO only. (B) Same analysis as above, on RNA samples obtained from uninjected (open bars), scrambled MO-injected (solid grey bars) or z-cb1r MO-injected (solid black bars) embryos. A reduced expression of z-stmn2a and z-stmn2b mRNAs was observed in embryos injected with z-cb1r MO as compared to the controls. * = p < 0.05; ** = p < 0.01; *** = p<0.001.
Similarly, expression levels of z-stmn2a and z-stmn2b was analyzed in z-cb1r MO, scrbl MO or uninjected zebrafish embryos. In CB1R-depleted embryos we observed a significant reduction in the abundance of z-stmn2a and z-stmn2b relative to the controls (Figure 6B), while the expression of the other candidates did not change (data not shown).
In conclusion, the expression of the z-stmn2a gene in zebrafish embryos is consistently modulated by treatments reducing or interfering with the CB1 receptor.
3. Discussion
In the present paper we show that pharmacological manipulation and gene knockdown of CB1R in early zebrafish embryos cause profound alterations in the ability of GnRH3 and AgRP1 axons to organize in discrete and well-fasciculated commissures, accompanied by misrouting and an altered neuron number. We also provide protein coexpression analyses that confirm, in early embryos, the localization of CB1 receptors in commissural structures and longitudinal tracts associated with GnRH3 fibers. Thus, we propose that CB1R is involved in the fasciculation and guidance of neuroendocrine GnRH3 and AgRP1 axons during forebrain development, implying high sensitivity of these neuroendocrine systems to neurodevelopmental exposure to exogenous cannabinoids.
During brain development, axonal projections elongate, fasciculate and defasciculate in distinctive pathways, often crossing the midline and reaching the appropriate targets guided by the orchestrated interactions between axon tracts and the environment at distinct domains, as well as homo- and hetero-philic interactions among axonal fibers. Several signaling and adhesion molecules involved in axon guidance and fasciculation have been identified [64,65]. Recent reports suggest that CB1R signaling is likely to modulate one or more of these signaling cascades during neural circuit formation, however most studies have focused on a few brain regions and neuronal types, in particular corticofugal and retinofugal neurons [22,23,24,25,66,67,68,69]. Here we show a novel role of eCBs signaling via CB1R on the development of two well-characterized neuroendocrine systems in zebrafish, the GnRH3 hypophysiotropic system and the AgRP1 system, involved in the central control of reproduction and food intake, respectively.
In order to gain relevant information on the developmental effects of the synthetic CB1R ligands employed in this study (WIN55, Rimonabant and AM251) we tested the possible toxic effects of their chronic exposure on zebrafish embryos from 0 to 72 or 96 hpf using a wide range of concentrations. Embryo survival was affected only by the highest agonist concentration (1 μM WIN55), and therefore this concentration was excluded from subsequent experiments. The selected ligands concentrations did not interfere with the overall embryo development and the hatching rate. These results partly agree with a previous study [70], in which AM251 treatment (10 and 20 nM) on zebrafish embryos for 72–96 hpf did not lead to morphological changes but decreased the hatching rate.
We analyzed the impact of chronic CB1R ligand exposure on the developing GnRH3 and AgRP1 systems, taking advantage of the fluorescently labelled neurons present in the gnrh3::EGFP and the agrp1::mCherry strains [47,71]. Pharmacological interference with CB1R in gnrh3::EGFP embryos clearly showed abnormal fasciculation of GnRH3 axons in the anterior commissure, with some ectopic axons detaching from the commissure itself. Defective phenotypes resulted from exposures to all three tested drugs (CB1R agonist, inverse agonist and antagonist) even at the lowest concentrations, indicating that interfering with the endogenous eCB system, either through over-activation or inhibition, equally disrupts the modulatory control exerted by CB1R on axonal navigation and fasciculation. We obtained similar results by knocking down the CB1R with a morpholino strategy, confirming previous observations made on the anterior commissure [30]. Furthermore, by immunocytochemistry we show co-expression of CB1R and GnRH3::EGFP in several fiber systems, although evidence of co-localization of the two labeling in exactly the same axons was found only in the anterior commissure and in longitudinal fiber tracts. Together these data suggest that at least part of the axons crossing the anterior commissure are affected by CB1R ligand exposure, including CB1R-expressing GnRH3 fibers.
In addition to the defects observed in fiber tracts, we found a significant decrease in the number of GnRH3 cell bodies (terminal nerve-associated GnRH3 neurons) following both antagonist (AM251) exposure and CB1R knockdown. These results are consistent with the previously reported role of CB1R as a regulator of progenitor niches [7,23]. Alternatively, CB1R could be involved in the timing of GnRH3 neuronal migration. A role of CB1R in neuronal migration has been previously reported during radial migration of immature pyramidal neurons in the cortex [23,72], and eCB signaling has been shown to promote migration of newborn neurons along the rostral migratory stream in the postnatal mouse brain [14]. Further experiments are needed to define a possible role of CB1R in GnRH3 neuronal migration.
Pharmacological interference of CB1R in agrp1::mCherry zebrafish embryos resulted in anterior commissure defects similar to those observed in gnrh3::EGFP embryos, suggesting that a common mechanism affected both GnRH3- and AgRP1-containing fibers. We found an increase in the number of AgRP1+ cell bodies in the hypothalamus in AM251-treated embryos, however this result could be affected by the methodology we chose for counting cells. It is possible that the apparent augmented number of AgRP1 cells depends on a decrease in fiber number, which would be in line with an axonal defect.
In our experiments, we observed similar phenotypes when applying CB1R agonists or antagonist/inverse agonists. This is consistent with the fact that different studies reported opposite effects of CB1R activation on axonal growth. Indeed, in some cases CB1R-activation has been reported to induce collapse of the growth cone and axon retraction, while its inhibition resulted in axon elongation [9,10,22,25,49]; other studies reported the opposite effects [23,73]. It should be kept in mind that while eCBs can reach very high local concentrations due to spatially and temporally coordinated activity of their synthetic and degrading enzymes, exogenous CB1R agonists and antagonists engage their targets indiscriminately, possibly impacting the spatial- and chrono-dynamics of eCB signaling in developing axons in a similar fashion.
Multiple mechanisms downstream of CB1R activation in developing neurons have been suggested. It has been reported that CB1R acts via the WAVE complex and Rac1-GTPases, and via Rho-GTPase and the Rho-dependent ROCK activation [9,10]; these pathways appear to mainly control the dynamics of the actin cytoskeleton. Notably, these intracellular signaling pathways are essential for neuronal “motility” during embryo development, including migration, neuritogenesis, spinogenesis and synaptogenesis [74].
Another reported mechanism involves the stathmin family of proteins and the fine modulation of microtubule dynamics at the growth cone. Stathmin-2, also known as SCG10 (superior cervical ganglia neural-specific 10) is a neuron-specific protein highly expressed in the developing forebrain, able to stabilize the plus ends both at steady state and early during polymerization by increasing the rate and extent of growth and facilitating microtubule extension into filopodia [13]. Overexpression of stathmin-2 strongly enhances neurite outgrowth, while its phosphorylation by the JNKs negatively regulates its microtubule destabilizing activity, suggesting that the JNK-stathmin-2 pathway may link extracellular signals to the rearrangement of the neuronal cytoskeleton [11,12]. By applying a bioinformatic analysis based on conserved coexpression and transcriptomic datasets from the developing hypothalamus, we report the identification of stathmin-2 as most likely to be involved in the effect of CB1R on axon elongation/fasciculation/guidance. Recently, stathmin-2 has been shown to be a specific molecular target for a CB1R-mediated effect of ∆9-THC in mouse and possibly in human fetal nervous systems [29]. By restoring stathmin-2 expression in the context of ∆9-THC exposure and consequent CB1R activation, the authors were able to acutely correct the altered cytoskeleton dynamics [29]. Highly relevant for neuroendocrine neurons, stathmin expression has been shown to modulate the migratory properties of mammalian GnRH neurons in vitro [60]. Thus, several evidence including ours indicate that stathmin-2/SCG10 participates in a regulatory cascade that is influenced by the activity and/or misactivity of the CB1R and controls cytoskeleton dynamics during neurite elongation and possibly neuronal migration.
4. Materials and Methods
4.1. Zebrafish Strains and Treatments
All procedures using zebrafish (Danio rerio) were authorized by the Ethical Committee of the University of Torino and the Italian Ministry of Health (approval code: N. 425/2016-PR; 27 April 2016). The wild-type fish strain TL-AB was used. The fish strain Tg(gnrh3::EGFP) [45,46,71] was obtained from Prof. Y. Zohar (Univ. Maryland Biotechnology Institute, Baltimore, MD, USA). The fish strain named, for simplicity, agrp1::mCherry was obtained by breeding the TgBAC(agrp:GAL4-VP16) with the UAS::nfsB-mCherry strains, followed by selection of the reporter fishes by quick fluorescent examination at low magnification [47]. Adult fish were routinely maintained under a 14 h light and 10 h dark photoperiod at approximately 28 °C, bred and genotyped according to standard procedures. Allelic transmission followed the expected mendelian ratios. Eggs were generated by natural mating, and following fertilization were collected, treated and maintained under a 12 h light and 12 h dark photoperiod, incubated at 28 °C.
The Tg(gnrh3::EGFP) embryos were grown in the presence of 0.003% 1-phenyl-2-thiourea (PTU) to prevent the formation of melanin pigment, which could interfere with the visualization of fluorescence in neurons and fibers; the agrp1::mCherry embryos were grown in fish water without PTU, because the pigmentation in this case did not affect the analysis under microscope.
4.2. Evaluation of Survival Rate and Hatching Rate
The endpoints used to assess developmental toxicity comprised embryos survival (percentage of live embryos/total) and hatching rate (percentage of hatching embryos/living embryos), observed every 24 h during the whole exposure period (72 or 96 hpf). Dead embryos were removed without solution renewal.
4.3. Morpholino Injections
To down-modulate CB1R we utilized a conventional antisense morpholino oligonucleotide (MO)-mediated strategy (GeneTools, LLC, Philomat, OR, USA). The z-cb1r MO was designed as previously published [30]. The MOs used for injection were
| z-cb1r MO: | 5′ CTAGAGGAAACCTGTCGGAGGAAAT 3′ |
| Mismatch_PPFIA scrambled MO (control): | 5′ TCGTGGCCATCAACTCGAACA 3′. |
Zygotes were collected at the 1-cell stage and injected under stereological examination with 50 µM, 200 µM or 400 µM of z-cb1r MO (or the scrambled MO as control), in the presence of Phenol Red for subsequent selection of the injected embryos. Uninjected embryos were also analyzed.
4.4. Western Blot
Zebrafish embryonic heads at 72 hpf were lysed manually in modified RIPA buffer composed as follows: 150 mM NaCl, 50 mM Tris pH 7.5 with 1% Triton X-100, 0.5% Na deoxycholate, 0.1% SDS, 2 mM EDTA, 1 mM sodium orthovanadate, 1 mM sodium fluoride and protease inhibitors. Homogenates were centrifuged at 3000 rpm for 5 min at 4 °C, extracts were quantified with BCA method (Bradford). A total of 30 µg of proteins/lane were separated in 10% acrylamide gel and transferred to a PVDF membrane (Millipore, Burlington, MA, USA). Membranes were saturated with 5% BSA in 0.3% TBS-Tween-20, washed in TBS-Tween, incubated overnight at 4 °C with an anti-CB1R primary antibody (diluted 1:800), then washed in TBS-Tween, incubated with an anti-rabbit IgG HRP-conjugated secondary antibody (diluted 1:3000; BioRad, Hercules, CA, USA), washed and developed with chemo-luminescence reagent Luminata TM Forte Western HRP Substrate (Millipore). Images were acquired with the ChemiDoc system (Bio-Rad), exported and analyzed.
4.5. Immunostaining
gnrh3::EGFP fish embryos were collected at 72 hpf and processed as described above. Embryos were sacrificed with a tricaine overdose, fixed in 4% PFA overnight, then washed in PBS. Solution was replaced with 0.1M PB and 7% sucrose at 4 °C for 12 h, then with 0.1M PB and 30% sucrose at 4 °C overnight. Samples were then embedded in OCT (Bio Optica, Mila, Italy) and sectioned at 14 μm thickness. Sections were washed in 0.01M PBS pH 7.4 for 10 min and incubated in normal goat serum (diluted 1:100 in PBS-Triton-X 100), then incubated with the anti-CB1R antibody (diluted 1:600 in PBS-Triton-X 100-normal goat serum) O/N. Slices were then washed in 0.01M PBS pH 7.4 for 10 min and incubated with anti-rabbit IgG CY3-conjugated secondary antibody (diluted 1:800 in PBS-Triton-X 100; Jackson ImmunoResearch Laboratories, Philadelphia, PA, USA) for 45 min. For cellular nuclei visualization, slices were incubated with DAPI (4′,6-diamidine-2′-phenylindole dihydrochloride), then washed in PBS and mounted in DABCO. The fluorescent signal was visualized using a Leica TCS SP5 (Leica Microsystem, Wetzlar, Germany).
4.6. Bioinformatic Searches and Statistical Analyses
Known and putative protein::protein interactions were searched for using the Human Protein Reference Database (http://www.hprd.org). Only those which appear in the list of at least two entry genes were further considered. Conserved coexpression in transcriptomic datasets was searched for using FuncPred (https://www.mbc.unito.it/en/notizie/funcpred) [57,58]. Conversion of the “gene ID” from human to mouse was done using the BioMart tool (http://www.biomart.org).
As a reference dataset of differentially expressed genes in the hypothalamus, we used the dataset GSE21278 present in Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) [59]. This data set provides extensive gene expression data on developing mouse hypothalamic tissues, done using the Affymetrix MOE430 microarray.
Functional annotation and definition of the over-represented categories was done using the tools available at http://geneontology.org/page/go-enrichment-analysis and at http://amp.pharm.mssm.edu/Enrichr/.
All data were statistically evaluated using commercially available software (SPSS version 18.0 for Windows, SPSS Inc., Chicago, IL, USA; and Microsoft Excel 2011, Microsoft Corporation, Albuquerque, NM, USA). Significance was calculated using a t-test and one-way ANOVA followed by Bonferroni’s post hoc test for multiple comparisons.
4.7. Real-Time qPCR Analysis
Total RNA was extracted from 72 hpf embryo heads with the TRIzol reagent (Life Technologies, Carlsbad, CA, USA), following the manufacturer’s instructions. Genomic DNA was destroyed by passing each sample into a 22G needle connected to a 1 mL syringe. Chloroform was added and, after 15–20 min of centrifugation at 12,000× g at 4 °C, RNA was precipitated in isopropanol. Samples were centrifuged at 12,000× g at 4 °C for 15 min and the RNA was then washed with 75% ethanol. The RNA pellet was briefly air-dried, resuspended in 20 µL sterile water and stored at −20 °C. Samples were quantified using a NanoDrop1000 spectrophotometer (Nanodrop Technologies, Inc., Wilmington, DE, USA). Real-time qPCR was performed using Superscript III Platinum One-step qRT-PCR system (Invitrogen, Carlsbad, CA, USA) and the thermal cycler Rotor Gene Q (Qiagen, Hilden, Germany). Each RNA sample was analyzed in three technical replicates containing 50 ng of total RNA. Primers sequences were
| z-β-actin | F | CCCACATAGGAGTCTTTCTG |
| R | TCCCCTTGTTCACAATAACC | |
| z-cb1r | F | CAGAACAGATCATACACCATGA |
| R | TGGTCTTATTCATCATCTACGC | |
| z-hoxb7a | F | GCTGGCGATCTCTGTAAAGC |
| R | TTTTGATGGTAGCCCCTCTG | |
| z-hoxa10b | F | GAGCTAAGGGGGTCCACTG |
| R | CACTTTTGGAATCTCCTGCTTT | |
| z-hox11a | F | CAGCAAACAGTCGACACCAC |
| R | CGGTCGCTCCTTTCCTTC | |
| z-stmn2a | F | TCCATGCTCTCGCTTATTTG |
| R | GGAGGATGTAGAGATGTGCT | |
| z-stmn2b | F | ATGGAGCAGATCAAGGAGAA |
| R | GGAGGATGTAGAGATGTGCT | |
| z-sez6a | F | AGTTTAGCAGTGAACGAACC |
| R | GAAGACCAGTGACTGAAACC | |
| z-sez6b | F | GTCCATTTTCATCCCTGTGG |
| R | GGCTGCTTTGAATCATAGGT | |
| z-negr1 | F | TTTTCGAGTGGTACAAGGGA |
| R | GGGTTGGATGGATTCAGAGG |
Quantification of mRNA abundance in each sample was done using a standard curve, built with several dilution of the samples. The abundance of the z-β-actin housekeeping mRNA was used for normalization. The mRNA expression was calculated relative to the one of control embryos (=1).
4.8. CB1 Ligands Exposure and Confocal Microscope Analysis
Upon fertilization, at the 1-cell stage, embryos were treated with the following ligands (Tocris Bioscience, Bristol, UK): CB1R-agonist WIN 55,212-2 (renamed WIN55 for simplicity, 1 nM, 10 nM and 100 nM); CB1R-inverse agonist Rimonabant (SR141716A) (1 nM, 10 nM, 100 nM and 1 μM); CB1R-antagonist AM251 (100 nM, 1 μM and 5 μM). All these compounds were dissolved in 0.1% dimethyl-sulfoxide (DMSO) and added to the fish water for 72 h (gnrh3::EGFP) or 96 h (agrp1::mCherry) without solution replacement for the entire exposure period. At 24 hpf 0.003% PTU was added to the water for gnrh3::EGFP fish embryos. Embryos were grown in 6-well Petri dishes, 30 embryos/well. Negative control embryos were treated with 0.1% DMSO alone.
For whole mount visualization, the gnrh3::EGFP and agrp1::mCherry embryos were collected at 72 hpf or 96 hpf, sacrificed with a tricaine overdose, fixed with 4% paraformaldehyde (PFA) at 4 °C O/N, washed in PBS and embedded in 4% low melting agarose. The apical portion of the head was manually dissected from the rest of the embryo. Confocal microscopy examination was carried out at 20× and 40× magnification with a Leica TCS SP5 or Leica TCS SP8 (Leica Microsystems). Images were acquired as Z-stacks of 5 μm-thick optical sections. For counting, stacks of Z-slices at 40× were used, images were processed and analyzed with the assistance of ImageJ software (NIH, Bethesda, MD, USA) after adjustment of contrast and brightness.
4.9. Statistical Analyses
Data are expressed as mean ± standard error of the mean (SEM) from three independent experiments. The results of pharmacological treatments/morpholino injections were compared to control (DMSO-treated/uninjected embryos). Statistical analyses were performed using commercially available software (SPSS version 18.0 for Windows and Microsoft Excel 2011). Significance was calculated using Fisher’s exact test for the analysis of altered phenotypes and one-way analysis of variance (ANOVA) for all other experiments, followed by Bonferroni’s and Tuckey’s post hoc tests for multiple comparisons. p ≤ 0.05 was taken as minimum level of significance.
5. Conclusions
The impact of exogenous cannabinoids on the gonadotropic neurons and the orexigenic/anorexigenic neurons has long been investigated [36,75,76]. Most previous work has focused on the adolescent or adult brain, while the effect of interfering with CB1R in early development of neuroendocrine neurons has remained poorly investigated. Based on our results, exposure to low doses of CB1R ligands during critical phases of development could alter axon pathfinding of GnRH3 and AgRP1 neurons, and possibly other neurons, potentially leading to a range of long-term chronic deficits of sexual maturation/reproduction and/or in the control of food intake/energy accumulation. Importantly, the hypothalamic neurons attaining the control of sexual maturation/reproduction and the control of food intake, are synaptically and functionally linked, as indicated by the fact that alterations of food intake control often lead to sexual disturbances, in mice as well as in humans [44]. These issues deserve further studies in other animal models, in order to attest the realistic possibility that embryo exposure to exogenous cannabinoid ligands could impact on the development of GnRH and AgRP neuroendocrine systems.
Supplementary Materials
Supplementary Materials can be found at https://www.mdpi.com/1422-0067/21/1/168/s1.
Author Contributions
Conceptualization, P.B. and G.R.M.; Data curation, G.Z., I.D. and P.P.; Formal analysis, G.Z., I.D., E.C. and P.P.; Funding acquisition, P.B. and G.R.M.; Investigation, G.Z., I.D., E.C., I.S. and Y.G.; Project administration, P.B. and G.R.M.; Resources, K.M., I.S. and Y.G.; Software, P.P.; Supervision, Y.G., P.P., P.B. and G.R.M.; Visualization, G.R.M.; Writing—Original draft, P.B. and G.R.M.; Writing—Review and editing, G.Z., I.D., E.C., K.M., Y.G. and P.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Fondazione CRT Torino, Italy (grant number RF 2014.0814 to G.R.M.), MIUR Ministry of University and Research (PRIN, grant number 2010W87LBJ_005 to P.B.) and “Fondazione Camis de Fonseca” Torino, Italy to G.Z.
Acknowledgments
We are indebted with E. Abraham and Y. Zohar (Univ. Maryland Biotechnology Institute, Baltimore, USA) for providing the gnrh3::EGFP strain. We thank Massimo Santoro for helpful comments on the manuscript and Zohar Ben-Moshe Livne, Flavio Barberis, Valentina Panero, Valentina Pomatto, Ilaria Corrado and Mattia Camera for their technical help.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| ∆9-THC | Δ9-tetrahydrocannabinol |
| 2 AG | 2-arachidonoylglycerol |
| AC | anterior commissure |
| AgRP | agouti-related peptide |
| AgRP1 | agouti-related peptide 1 |
| AM251 | (N-(Piperidin-1-yl)-5-(4-iodophenyl)-1-(2,4-dichlorophenyl)-4-methyl-1H-pyrazole-3-carboxamide) |
| CB1R | cannabinoid receptor type 1 |
| CB2R | cannabinoid receptor type 2 |
| CBD | cannabidiol |
| DAGL | diacylglycerol lipase |
| DMSO | dimethyl sulfoxide |
| dpf | days post fertilization |
| eCB | endocannabinoid |
| GnRH | gonadotropin-releasing hormone |
| GTPase | guanosine triphosphatase |
| Hox | homeobox gene |
| hpf | hours post fertilization |
| JNK1 | c-Jun N-terminal kinase 1 |
| MAGL | monoacylglycerol lipase |
| MO | morpholino oligonucleotide |
| Negr1 | neuronal growth regulator 1 |
| POA | pre-optic area |
| POC | post-optic commissure |
| Rimonabant | SR141716, (N-(Piperidin-1-yl)-5-(4-chlorophenyl)-1-(2,4-dichlorophenyl)4-methyl-1H-pyrazole-3-carboxamide hydrochloride) |
| Sez6 | Seizure protein 6 |
| Stmn2/SCG10 | Stathmin2/Superior Cervical Ganglion-10 Protein |
| VTC | ventral tegmental commissure |
| WIN55 | WIN 55,212-2; (R)-( + )-[2,3-Dihydro-5-methyl-3-(4-morpholinylmethyl)pyrrolo [1,2,3-de]-1,4-benzoxazin-6-yl]-1-naphthalenylmethanone mesylate |
References
- Grotenhermen, F. Pharmacokinetics and Pharmacodynamics of Cannabinoids. Clin. Pharmacokinet. 2003, 42, 327–360. [Google Scholar] [CrossRef] [PubMed]
- Scheyer, A.F.; Melis, M.; Trezza, V.; Manzoni, O.J.J. Consequences of Perinatal Cannabis Exposure. Trends Neurosci. 2019, 42, 871–884. [Google Scholar] [CrossRef] [PubMed]
- Howlett, A.C. The cannabinoid receptors. Prostaglandins Other Lipid Mediat. 2002, 68, 619–631. [Google Scholar] [CrossRef]
- Kano, M.; Ohno-Shosaku, T.; Hashimotodani, Y.; Uchigashima, M.; Watanabe, M. Endocannabinoid-Mediated Control of Synaptic Transmission. Physiol. Rev. 2009, 89, 309–380. [Google Scholar] [CrossRef]
- Pertwee, R.G.; Howlett, A.C.; Abood, M.E.; Alexander, S.P.H.; Di Marzo, V.; Elphick, M.R.; Greasley, P.J.; Hansen, H.S.; Kunos, G.; Mackie, K.; et al. International Union of Basic and Clinical Pharmacology. LXXIX. Cannabinoid Receptors and Their Ligands: Beyond CB1 and CB2. Physiol. Rev. 2010, 62, 588–631. [Google Scholar] [CrossRef]
- Di Marzo, V. Endocannabinoid signaling in the brain: Biosynthetic mechanisms in the limelight. Nat. Neurosci. 2011, 14, 9–15. [Google Scholar] [CrossRef]
- Maccarone, M.; Guzman, M.; Mackie, K.; Doherty, P.; Harkany, T. Programming and reprogramming neural cells by (endo-)cannabinoids: From physiological rules to emerging therapies. Nat. Rev. Neurosci. 2014, 91, 165–171. [Google Scholar]
- Keimpema, E.; Barabas, K.; Morozov, Y.M.; Tortoriello, G.; Torii, M.; Cameron, G.; Yanagawa, Y.; Watanabe, M.; Mackie, K.; Harkany, T. Differential Subcellular Recruitment of Monoacylglycerol Lipase Generates Spatial Specificity of 2-Arachidonoyl Glycerol Signaling during Axonal Pathfinding. J. Neurosci. 2010, 30, 13992–14007. [Google Scholar] [CrossRef]
- Roland, A.B.; Ricobaraza, A.; Carrel, D.; Jordan, B.M.; Rico, F.; Simon, A.; Humbert-Claude, M.; Ferrier, J.; McFadden, M.H.; Scheuring, S.; et al. Cannabinoid-induced actomyosin contractility shapes neuronal morphology and growth. eLife 2014, 3, e03159. [Google Scholar] [CrossRef]
- Njoo, C.; Agarwal, N.; Lutz, B.; Kuner, R. The Cannabinoid Receptor CB1 Interacts with the WAVE1 Complex and Plays a Role in Actin Dynamics and Structural Plasticity in Neurons. PLoS Biol. 2015, 13, e1002286. [Google Scholar] [CrossRef]
- Grenningloh, G.; Soehrman, S.; Bondallaz, P.; Ruchti, E.; Cadas, H. Role of the microtubule destabilizing proteins SCG10 and stathmin in neuronal growth. J. Neurobiol. 2004, 58, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Tararuk, T.; Östman, N.; Li, W.; Björkblom, B.; Padzik, A.; Zdrojewska, J.; Hongisto, V.; Herdegen, T.; Konopka, W.; Courtney, M.J.; et al. JNK1 phosphorylation of SCG10 determines microtubule dynamics and axodendritic length. J. Cell Biol. 2006, 173, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Manna, T.; Grenningloh, G.; Miller, H.P.; Wilson, L. Stathmin Family Protein SCG10 Differentially Regulates the Plus and Minus End Dynamics of Microtubules at Steady State In Vitro: Implications for Its Role in Neurite Outgrowth. Biochemistry 2007, 46, 3543–3552. [Google Scholar] [CrossRef] [PubMed]
- Oudin, M.J.; Hobbs, C.; Doherty, P. DAGL-dependent endocannabinoid signalling: Roles in axonal pathfinding, synaptic plasticity and adult neurogenesis: DAGL-dependent endocannabinoid signalling. Eur. J. Neurosci. 2011, 34, 1634–1646. [Google Scholar] [CrossRef] [PubMed]
- Keimpema, E.; Mackie, K.; Harkany, T. Molecular model of cannabis sensitivity in developing neuronal circuits. Trends Pharmacol. Sci. 2011, 32, 551–561. [Google Scholar] [CrossRef] [PubMed]
- Harkany, T.; Keimpema, E.; Barabás, K.; Mulder, J. Endocannabinoid functions controlling neuronal specification during brain development. Mol. Cell. Endocrinol. 2008, 286, S84–S90. [Google Scholar] [CrossRef]
- Huizink, A.C.; Mulder, E.J.H. Maternal smoking, drinking or cannabis use during pregnancy and neurobehavioral and cognitive functioning in human offspring. Neurosci. Biobehav. Rev. 2006, 30, 24–41. [Google Scholar] [CrossRef]
- Calvigioni, D.; Hurd, Y.L.; Harkany, T.; Keimpema, E. Neuronal substrates and functional consequences of prenatal cannabis exposure. Eur. Child Adolesc. Psychiatry 2014, 23, 931–941. [Google Scholar] [CrossRef]
- Leech, S.L.; Richardson, G.A.; Goldschmidt, L.; Day, N.L. Prenatal Substance Exposure: Effects on Attention and Impulsivity of 6-year-olds. Neurotoxicol. Teratol. 1999, 21, 109–118. [Google Scholar] [CrossRef]
- Leech, S.L.; Larkby, C.A.; Day, R.; Day, N.L. Predictors and Correlates of High Levels of Depression and Anxiety Symptoms Among Children at Age 10. J. Am. Acad. Child Adolesc. Psychiatry 2006, 45, 223–230. [Google Scholar] [CrossRef]
- Hwang, H.M.; Ku, R.Y.; Hashimoto-Torii, K. Prenatal Environment That Affects Neuronal Migration. Front. Cell Dev. Biol. 2019, 7, 138. [Google Scholar] [CrossRef] [PubMed]
- Berghuis, P.; Rajnicek, A.M.; Morozov, Y.M.; Ross, R.A.; Mulder, J.; Urban, G.M.; Monory, K.; Marsicano, G.; Matteoli, M.; Canty, A.; et al. Hardwiring the Brain: Endocannabinoids Shape Neuronal Connectivity. Science 2007, 316, 1212–1216. [Google Scholar] [CrossRef] [PubMed]
- Mulder, J.; Aguado, T.; Keimpema, E.; Barabas, K.; Ballester Rosado, C.J.; Nguyen, L.; Monory, K.; Marsicano, G.; Di Marzo, V.; Hurd, Y.L.; et al. Endocannabinoid signaling controls pyramidal cell specification and long-range axon patterning. Proc. Natl. Acad. Sci. USA 2008, 105, 8760–8765. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-S.; Zhu, J.; Wager-Miller, J.; Wang, S.; O’Leary, D.; Monory, K.; Lutz, B.; Mackie, K.; Lu, H.-C. Requirement of cannabinoid CB1 receptors in cortical pyramidal neurons for appropriate development of corticothalamic and thalamocortical projections: CB1R in the development of sensory networks. Eur. J. Neurosci. 2010, 32, 693–706. [Google Scholar] [CrossRef] [PubMed]
- Argaw, A.; Duff, G.; Zabouri, N.; Cecyre, B.; Chaine, N.; Cherif, H.; Tea, N.; Lutz, B.; Ptito, M.; Bouchard, J.-F. Concerted Action of CB1 Cannabinoid Receptor and Deleted in Colorectal Cancer in Axon Guidance. J. Neurosci. 2011, 31, 1489–1499. [Google Scholar] [CrossRef]
- Duff, G.; Argaw, A.; Cecyre, B.; Cherif, H.; Tea, N.; Zabouri, N.; Casanova, C.; Ptito, M.; Bouchard, J.-F. Cannabinoid Receptor CB2 Modulates Axon Guidance. PLoS ONE 2013, 8, e70849. [Google Scholar] [CrossRef]
- Abbas Farishta, R.; Robert, C.; Turcot, O.; Thomas, S.; Vanni, M.P.; Bouchard, J.-F.; Casanova, C. Impact of CB1 Receptor Deletion on Visual Responses and Organization of Primary Visual Cortex in Adult Mice. Investig. Opthalmol. Vis. Sci. 2015, 56, 7697. [Google Scholar] [CrossRef][Green Version]
- Diaz-Alonso, J.; Aguado, T.; Wu, C.-S.; Palazuelos, J.; Hofmann, C.; Garcez, P.; Guillemot, F.; Lu, H.-C.; Lutz, B.; Guzman, M.; et al. The CB1 Cannabinoid Receptor Drives Corticospinal Motor Neuron Differentiation through the Ctip2/Satb2 Transcriptional Regulation Axis. J. Neurosci. 2012, 32, 16651–16665. [Google Scholar] [CrossRef]
- Tortoriello, G.; Morris, C.V.; Alpar, A.; Fuzik, J.; Shirran, S.L.; Calvigioni, D.; Keimpema, E.; Botting, C.H.; Reinecke, K.; Herdegen, T.; et al. Miswiring the brain: 9-tetrahydrocannabinol disrupts cortical development by inducing an SCG10/stathmin-2 degradation pathway. EMBO J. 2014, 33, 668–685. [Google Scholar] [CrossRef]
- Watson, S.; Chambers, D.; Hobbs, C.; Doherty, P.; Graham, A. The endocannabinoid receptor, CB1, is required for normal axonal growth and fasciculation. Mol. Cell. Neurosci. 2008, 38, 89–97. [Google Scholar] [CrossRef]
- Sufian, M.S.; Amin, M.R.; Kanyo, R.; Allison, W.T.; Ali, D.W. CB1 and CB2 receptors play differential roles in early zebrafish locomotor development. J. Exp. Biol. 2019, 222, jeb206680. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, K.T.; Amin, M.R.; Shah, P.; Ali, D.W. Motor neuron development in zebrafish is altered by brief (5-hr) exposures to THC (∆9-tetrahydrocannabinol) or CBD (cannabidiol) during gastrulation. Sci. Rep. 2018, 8, 10581. [Google Scholar] [CrossRef] [PubMed]
- Martella, A.; Sepe, R.M.; Silvestri, C.; Zang, J.; Fasano, G.; Carnevali, O.; De Girolamo, P.; Neuhauss, S.C.F.; Sordino, P.; Di Marzo, V. Important role of endocannabinoid signaling in the development of functional vision and locomotion in zebrafish. FASEB J. 2016, 30, 4275–4288. [Google Scholar] [CrossRef] [PubMed]
- Tasker, J.G.; Chen, C.; Fisher, M.O.; Fu, X.; Rainville, J.R.; Weiss, G.L. Endocannabinoid Regulation of Neuroendocrine Systems. In International Review of Neurobiology; Elsevier: Amsterdam, The Netherlands, 2015; Volume 125, pp. 163–201. ISBN 978-0-12-801278-9. [Google Scholar]
- Hillard, C.J. Endocannabinoids and the Endocrine System in Health and Disease. In Endocannabinoids; Pertwee, R.G., Ed.; Springer International Publishing: Cham, Switzerland, 2015; Volume 231, pp. 317–339. ISBN 978-3-319-20824-4. [Google Scholar]
- Bovolin, P.; Cottone, E.; Pomatto, V.; Fasano, S.; Pierantoni, R.; Cobellis, G.; Meccariello, R. Endocannabinoids are Involved in Male Vertebrate Reproduction: Regulatory Mechanisms at Central and Gonadal Level. Front. Endocrinol. 2014, 5, 54. [Google Scholar] [CrossRef] [PubMed]
- Glanowska, K.M.; Moenter, S.M. Endocannabinoids and prostaglandins both contribute to GnRH neuron-GABAergic afferent local feedback circuits. J. Neurophysiol. 2011, 106, 3073–3081. [Google Scholar] [CrossRef] [PubMed]
- Farkas, I.; Kalló, I.; Deli, L.; Vida, B.; Hrabovszky, E.; Fekete, C.; Moenter, S.M.; Watanabe, M.; Liposits, Z. Retrograde Endocannabinoid Signaling Reduces GABAergic Synaptic Transmission to Gonadotropin-Releasing Hormone Neurons. Endocrinology 2010, 151, 5818–5829. [Google Scholar] [CrossRef]
- Wu, Q.; Clark, M.S.; Palmiter, R.D. Deciphering a neuronal circuit that mediates appetite. Nature 2012, 483, 594–597. [Google Scholar] [CrossRef]
- Horn, H.; Böhme, B.; Dietrich, L.; Koch, M. Endocannabinoids in Body Weight Control. Pharmaceuticals 2018, 11, 55. [Google Scholar] [CrossRef]
- Stefanucci, A.; Macedonio, G.; Dvorácskó, S.; Tömböly, C.; Mollica, A. Novel Fubinaca/Rimonabant hybrids as endocannabinoid system modulators. Amino Acids 2018, 50, 1595–1605. [Google Scholar] [CrossRef]
- Dimmit, M.P.; Stefanucci, A.; Pieretti, S.; Minosi, P.; Dvorácskó, S.; Tömböly, C.; Zengin, G.; Mollica, A. Mollica Discovery of Orexant and Anorexant Agents with Indazole Scaffold Endowed with Peripheral Antiedema Activity. Biomolecules 2019, 9, 492. [Google Scholar] [CrossRef]
- Morozov, Y.M.; Koch, M.; Rakic, P.; Horvath, T.L. Cannabinoid type 1 receptor-containing axons innervate NPY/AgRP neurons in the mouse arcuate nucleus. Mol. Metab. 2017, 6, 374–381. [Google Scholar] [CrossRef] [PubMed]
- Manfredi-Lozano, M.; Roa, J.; Tena-Sempere, M. Connecting metabolism and gonadal function: Novel central neuropeptide pathways involved in the metabolic control of puberty and fertility. Front. Neuroendocrinol. 2018, 48, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Abraham, E.; Palevitch, O.; Gothilf, Y.; Zohar, Y. The zebrafish as a model system for forebrain GnRH neuronal development. Gen. Comp. Endocrinol. 2009, 164, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Abraham, E.; Palevitch, O.; Gothilf, Y.; Zohar, Y. Targeted Gonadotropin-Releasing Hormone-3 Neuron Ablation in Zebrafish: Effects on Neurogenesis, Neuronal Migration, and Reproduction. Endocrinology 2010, 151, 332–340. [Google Scholar] [CrossRef]
- Shainer, I.; Buchshtab, A.; Hawkins, T.A.; Wilson, S.W.; Cone, R.D.; Gothilf, Y. Novel hypophysiotropic AgRP2 neurons and pineal cells revealed by BAC transgenesis in zebrafish. Sci. Rep. 2017, 7, 44777. [Google Scholar] [CrossRef]
- Antonelli, T.; Tanganelli, S.; Tomasini, M.C.; Finetti, S.; Trabace, L.; Steardo, L.; Sabino, V.; Carratu, M.R.; Cuomo, V.; Ferraro, L. Long-term effects on cortical glutamate release induced by prenatal exposure to the cannabinoid receptor agonist (r)-(+)-[2,3-dihydro-5-methyl-3-(4-morpholinyl-methyl) pyrrolo [1,2,3-de]-1,4-benzoxazin-6-yl]-1-naphthalenylmethanone: An in vivo microdialysis study in the awake rat. Neuroscience 2004, 124, 367–375. [Google Scholar]
- Berghuis, P.; Dobszay, M.B.; Wang, X.; Spano, S.; Ledda, F.; Sousa, K.M.; Schulte, G.; Ernfors, P.; Mackie, K.; Paratcha, G.; et al. Endocannabinoids regulate interneuron migration and morphogenesis by transactivating the TrkB receptor. Proc. Natl. Acad. Sci. USA 2005, 102, 19115–19120. [Google Scholar] [CrossRef]
- Alpár, A.; Tortoriello, G.; Calvigioni, D.; Niphakis, M.J.; Milenkovic, I.; Bakker, J.; Cameron, G.A.; Hanics, J.; Morris, C.V.; Fuzik, J.; et al. Endocannabinoids modulate cortical development by configuring Slit2/Robo1 signalling. Nat. Commun. 2014, 5, 4421. [Google Scholar] [CrossRef]
- Garaffo, G.; Conte, D.; Provero, P.; Tomaiuolo, D.; Luo, Z.; Pinciroli, P.; Peano, C.; D’Atri, I.; Gitton, Y.; Etzion, T.; et al. The Dlx5 and Foxg1 transcription factors, linked via miRNA-9 and-200, are required for the development of the olfactory and GnRH system. Mol. Cell. Neurosci. 2015, 68, 103–119. [Google Scholar] [CrossRef]
- Barresi, M.J.F. Hedgehog regulated Slit expression determines commissure and glial cell position in the zebrafish forebrain. Development 2005, 132, 3643–3656. [Google Scholar] [CrossRef]
- Shainer, I.; Michel, M.; Marquart, G.D.; Bhandiwad, A.A.; Zmora, N.; Ben-Moshe Livne, Z.; Zohar, Y.; Hazak, A.; Mazon, Y.; Förster, D.; et al. Agouti-Related Protein 2 Is a New Player in the Teleost Stress Response System. Curr. Biol. 2019, 29, e7. [Google Scholar] [CrossRef] [PubMed]
- Benowitz, L.I.; Routtenberg, A. GAP-43: An intrinsic determinant of neuronal development and plasticity. Trends Neurosci. 1997, 20, 84–91. [Google Scholar] [CrossRef]
- Bromberg, K.D.; Iyengar, R.; He, J.C. Regulation of neurite outgrowth by G(i/o) signaling pathways. Front. Biosci. 2008, 13, 4544–4557. [Google Scholar] [CrossRef] [PubMed]
- Wiencken-Barger, A.E.; Mavity-Hudson, J.; Bartsch, U.; Schachner, M.; Casagrande, V.A. The role of L1 in axon pathfinding and fasciculation. Cereb. Cortex 2004, 14, 121–131. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Perron, U.; Provero, P.; Molineris, I. In silico prediction of lncRNA function using tissue specific and evolutionary conserved expression. BMC Bioinform. 2017, 18, 144. [Google Scholar] [CrossRef] [PubMed]
- Ala, U.; Piro, R.M.; Grassi, E.; Damasco, C.; Silengo, L.; Oti, M.; Provero, P.; Di Cunto, F. Prediction of Human Disease Genes by Human-Mouse Conserved Coexpression Analysis. PLoS Comput. Biol. 2008, 4, e1000043. [Google Scholar] [CrossRef] [PubMed]
- Shimogori, T.; Lee, D.A.; Miranda-Angulo, A.; Yang, Y.; Wang, H.; Jiang, L.; Yoshida, A.C.; Kataoka, A.; Mashiko, H.; Avetisyan, M.; et al. A genomic atlas of mouse hypothalamic development. Nat. Neurosci. 2010, 13, 767–775. [Google Scholar] [CrossRef]
- Giampietro, C.; Luzzati, F.; Gambarotta, G.; Giacobini, P.; Boda, E.; Fasolo, A.; Perroteau, I. Stathmin Expression Modulates Migratory Properties of GN-11 Neurons in Vitro. Endocrinology 2005, 146, 1825–1834. [Google Scholar] [CrossRef][Green Version]
- Szczurkowska, J.; Pischedda, F.; Manago, F.; Haas, C.; Papaleo, F.; Schäfer, M.; Piccoli, G.; Cancedda, L. Negr1 is required for transition of migrating pyramidal neurons from layer V to layer II/III of the mouse cerebral cortex. Int. J. Dev. Neurosci. 2015, 47, 16. [Google Scholar] [CrossRef]
- Kim, M.H.; Gunnersen, J.M.; Tan, S.-S. Localized expression of the seizure-related gene SEZ-6 in developing and adult forebrains. Mech. Dev. 2002, 118, 171–174. [Google Scholar] [CrossRef]
- Osaki, G.; Mitsui, S.; Yuri, K. The distribution of the seizure-related gene 6 (Sez-6) protein during postnatal development of the mouse forebrain suggests multiple functions for this protein: An analysis using a new antibody. Brain Res. 2011, 1386, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Leyva-Díaz, E.; López-Bendito, G. In and out from the cortex: Development of major forebrain connections. Neuroscience 2013, 254, 26–44. [Google Scholar] [CrossRef] [PubMed]
- Stoeckli, E.T. Understanding axon guidance: Are we nearly there yet? Development 2018, 145, dev151415. [Google Scholar] [CrossRef] [PubMed]
- Vitalis, T.; Lainé, J.; Simon, A.; Roland, A.; Leterrier, C.; Lenkei, Z. The type 1 cannabinoid receptor is highly expressed in embryonic cortical projection neurons and negatively regulates neurite growth in Vitro. Eur. J. Neurosci. 2008, 28, 1705–1718. [Google Scholar] [CrossRef] [PubMed]
- Keimpema, E.; Tortoriello, G.; Alpar, A.; Capsoni, S.; Arisi, I.; Calvigioni, D.; Hu, S.S.-J.; Cattaneo, A.; Doherty, P.; Mackie, K.; et al. Nerve growth factor scales endocannabinoid signaling by regulating monoacylglycerol lipase turnover in developing cholinergic neurons. Proc. Natl. Acad. Sci. USA 2013, 110, 1935–1940. [Google Scholar] [CrossRef] [PubMed]
- De Salas-Quiroga, A.; Díaz-Alonso, J.; García-Rincón, D.; Remmers, F.; Vega, D.; Gómez-Cañas, M.; Lutz, B.; Guzmán, M.; Galve-Roperh, I. Prenatal exposure to cannabinoids evokes long-lasting functional alterations by targeting CB1 receptors on developing cortical neurons. Proc. Natl. Acad. Sci. USA 2015, 112, 13693–13698. [Google Scholar] [CrossRef]
- Hedrich, J.; Angamo, E.A.; Conrad, A.; Lutz, B.; Luhmann, H.J. Cell type specific impact of cannabinoid receptor signaling in somatosensory barrel map formation in mice. J. Comp. Neurol. 2020, 528, 7–17. [Google Scholar] [CrossRef]
- Migliarini, B.; Carnevali, O. A novel role for the endocannabinoid system during zebrafish development. Mol. Cell. Endocrinol. 2009, 299, 172–177. [Google Scholar] [CrossRef]
- Abraham, E.; Palevitch, O.; Ijiri, S.; Du, S.J.; Gothilf, Y.; Zohar, Y. Early Development of Forebrain Gonadotrophin-Releasing Hormone (GnRH) Neurones and the Role of GnRH as an Autocrine Migration Factor. J. Neuroendocrinol. 2008, 20, 394–405. [Google Scholar] [CrossRef]
- Díaz-Alonso, J.; de Salas-Quiroga, A.; Paraíso-Luna, J.; García-Rincón, D.; Garcez, P.P.; Parsons, M.; Andradas, C.; Sánchez, C.; Guillemot, F.; Guzmán, M.; et al. Loss of Cannabinoid CB1 Receptors Induces Cortical Migration Malformations and Increases Seizure Susceptibility. Cereb. Cortex 2017, 27, 5303–5317. [Google Scholar]
- Williams, E.-J.; Walsh, F.S.; Doherty, P. The FGF receptor uses the endocannabinoid signaling system to couple to an axonal growth response. J. Cell Biol. 2003, 160, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Gomez, T.M.; Letourneau, P.C. Actin dynamics in growth cone motility and navigation. J. Neurochem. 2014, 129, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Brents, L.K. Marijuana, the Endocannabinoid System and the Female Reproductive System. Yale J. Biol. Med. 2016, 89, 175–191. [Google Scholar] [PubMed]
- Koch, M. Cannabinoid Receptor Signaling in Central Regulation of Feeding Behavior: A Mini-Review. Front. Neurosci. 2017, 11, 293. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).