Flavescence Dorée Phytoplasma Has Multiple ftsH Genes that Are Differentially Expressed in Plants and Insects
Abstract
1. Introduction
2. Results
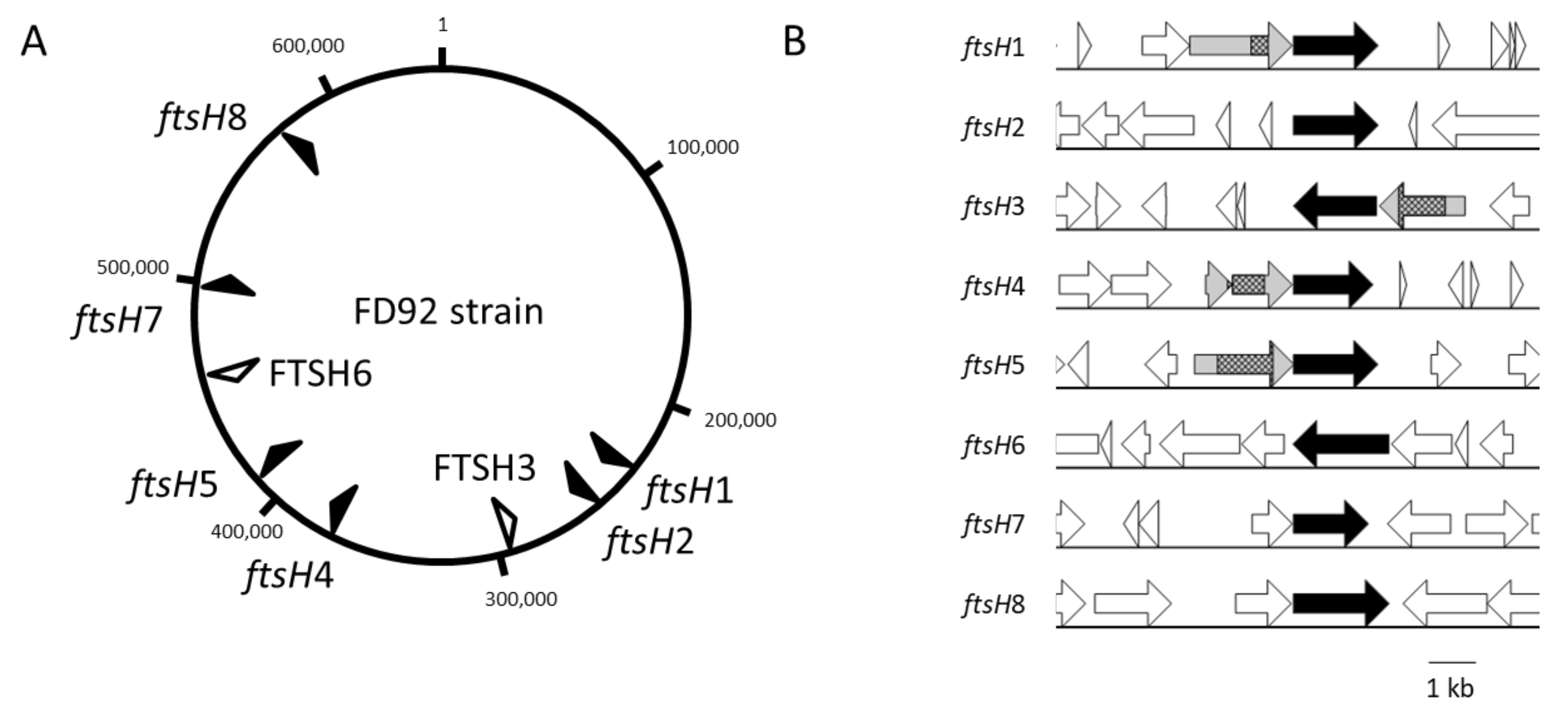
2.1. Eight ftsH Gene Sequences Were Identified in the FDP Genome
2.2. Phylogenetic Analysis of Bacterial and Phytoplasma FtsH Proteins Reveals Ancient Duplication of ftsH Genes
2.3. FtsH Proteins Are Predicted to Be Differentially Oriented in the Membrane
2.4. Substrate-Binding Proteins Are Encoded by Genes Upstream of Several ftsH Genes
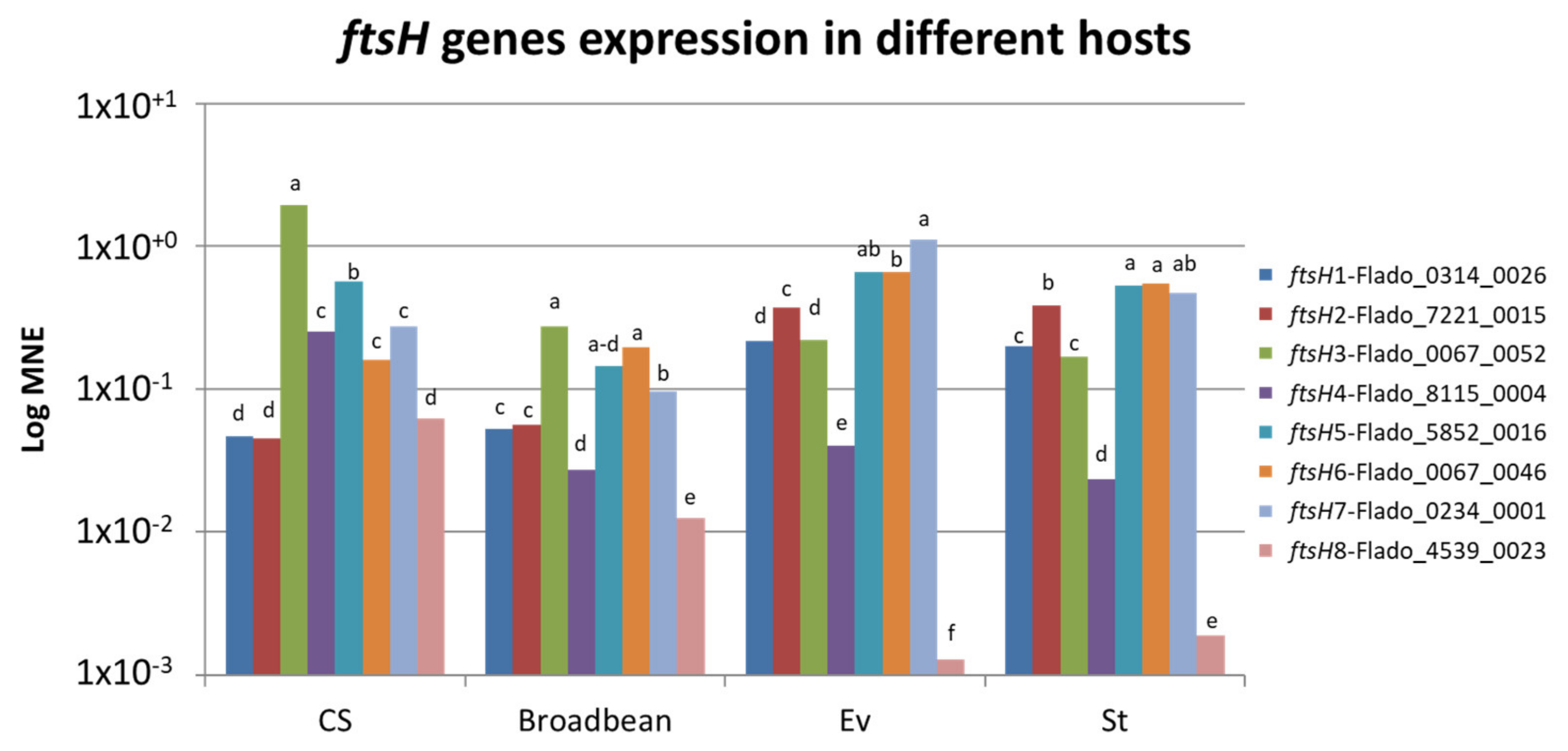
2.5. Phytoplasma ftsH Genes Are Differentially Expressed Based on the Host
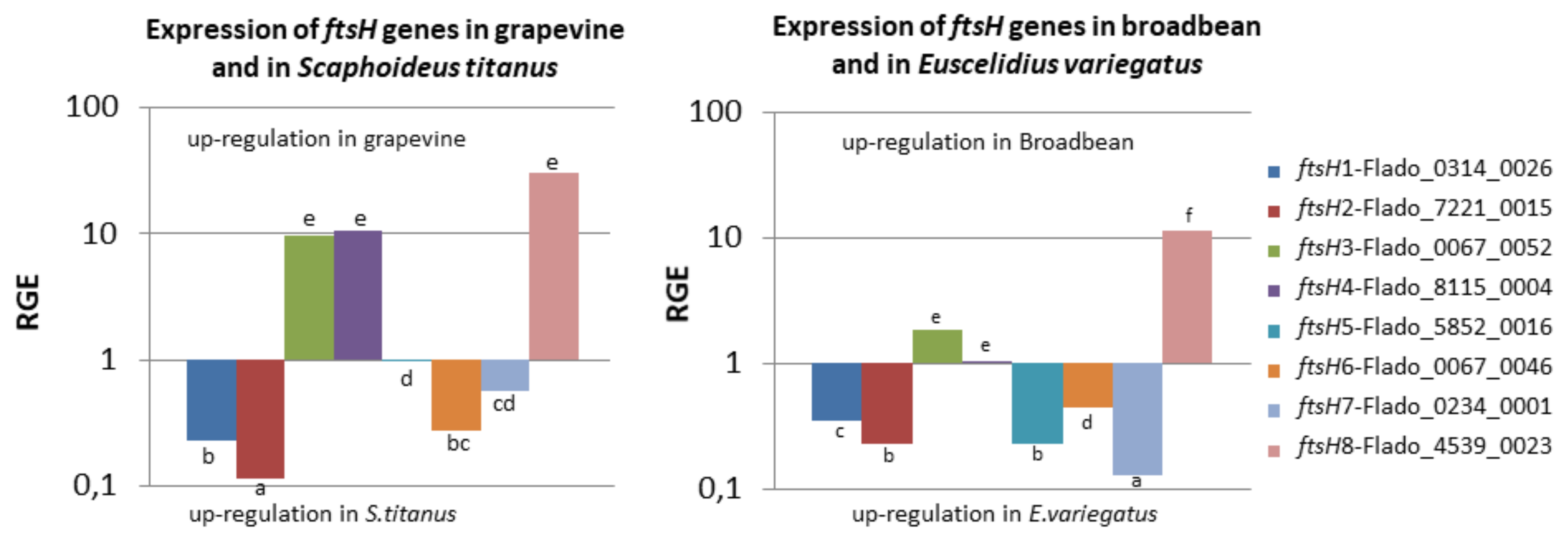
2.6. Three ftsH Genes Are More Expressed in Grapevine than in S. titanus
2.7. ftsH8 is More Expressed in Broad Bean than in E. variegatus
3. Discussion
4. Materials and Methods
4.1. Phytoplasma Isolates, Plants, and Insects
4.2. Nucleic Acid Extraction
4.3. Phytoplasma Quantification
4.4. In Silico Sequence Analysis of FtsH
4.5. cDNA Synthesis and Quantitative Real-Time Reverse Transcription PCR (RT-PCR)
4.6. Data Analysis
4.7. Molecular Phylogenetic Analysis by the Maximum Likelihood Method
4.8. Sequence Data
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CS | Grapevine (Cabernet Sauvignon) |
| Ev | Euscelidius variegatus |
| FDP | Flavescence dorée phytoplasma |
| PMU | Potential Mobile Unit |
| RGE | Relative Gene Expression |
| TM | Transmembrane |
References
- Boudon-Padieu, E. Flavescence dorée of the grapevine, knowledge and new developments in epidemiology, etiology and diagnosis. ATTI Giornate Fitopatol. 2002, 1, 15–34. [Google Scholar]
- EFSA Panel on Plant Health (PLH). Scientific Opinion on pest categorisation of Grapevine Flavescence dorée: Grapevine Flavescence dorée pest categorisation. EFSA J. 2014, 12, 3851. [Google Scholar] [CrossRef]
- Schvester, D.; Carle, P.; Moutous, G. Sur la transmission de la flavescence dorée des vignes par une cicadelle. C. R. Acad. Agric. Fr. 1961, 47, 1021–1024. [Google Scholar]
- Caudwell, A.; Giannotti, J.; Kuszala, C.; Larrue, J. Etude du rôle de particules de type “mycoplasme” dans l’étiologie de la flavescence dorée de la vigne. Examen cytologique des plantes. Ann. Phytopathol. 1971, 3, 107–123. [Google Scholar]
- Caudwell, A. Deux années d’étude sur la flavescence dorée, nouvelle maladie grave de la vigne. Ann. Amélior. Plantes 1957, 4, 359–363. [Google Scholar]
- Pavan, F.; Mori, N.; Bigot, G.; Zandigiacomo, P. Border effect in spatial distribution of Flavescence dorée affected grapevines and outside source of Scaphoideus titanus vectors. Bull. Insectol. 2012, 65, 281–290. [Google Scholar]
- Chuche, J.; Thiéry, D. Biology and ecology of the Flavescence dorée vector Scaphoideus titanus: A review. Agron. Sustain. Dev. 2014, 34, 381–403. [Google Scholar] [CrossRef]
- Jeger, M.; Bragard, C.; Caffier, D.; Candresse, T.; Chatzivassiliou, E.; Dehnen-Schmutz, K.; Gilioli, G.; Miret, J.A.J.; MacLeod, A.; Navarro, M.N.; et al. Risk to plant health of Flavescence dorée for the EU territory. EFSA J. 2016, 14, 04603. [Google Scholar]
- Lee, I.M.; Davis, R.E.; Gundersen-Rindal, D.E. Phytoplasma: Phytopathogenic mollicutes. Annu. Rev. Microbiol. 2000, 54, 221–255. [Google Scholar] [CrossRef]
- Weintraub, P.G.; Beanland, L. Insect Vectors of Phytoplasmas. Annu. Rev. Entomol. 2006, 51, 91–111. [Google Scholar] [CrossRef]
- Christensen, N.M.; Nicolaisen, M.; Hansen, M.; Schulz, A. Distribution of Phytoplasmas in Infected Plants as Revealed by Real-Time PCR and Bioimaging. Mol. Plant Microbe Interact. 2004, 17, 1175–1184. [Google Scholar] [CrossRef] [PubMed]
- Oshima, K.; Kakizawa, S.; Nishigawa, H.; Jung, H.-Y.; Wei, W.; Suzuki, S.; Arashida, R.; Nakata, D.; Miyata, S.; Ugaki, M.; et al. Reductive evolution suggested from the complete genome sequence of a plant-pathogenic phytoplasma. Nat. Genet. 2004, 36, 27–29. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Zhang, J.; Ewing, A.; Miller, S.A.; Jancso Radek, A.; Shevchenko, D.V.; Tsukerman, K.; Walunas, T.; Lapidus, A.; Campbell, J.W.; et al. Living with Genome Instability: The Adaptation of Phytoplasmas to Diverse Environments of Their Insect and Plant Hosts. J. Bacteriol. 2006, 188, 3682–3696. [Google Scholar] [CrossRef]
- Hogenhout, S.A.; Oshima, K.; Ammar, E.-D.; Kakizawa, S.; Kingdom, H.N.; Namba, S. Phytoplasmas: Bacteria that manipulate plants and insects. Mol. Plant Pathol. 2008, 9, 403–423. [Google Scholar] [CrossRef] [PubMed]
- Sugio, A.; Hogenhout, S.A. The genome biology of phytoplasma: Modulators of plants and insects. Curr. Opin. Microbiol. 2012, 15, 247–254. [Google Scholar] [CrossRef]
- Kube, M.; Mitrovic, J.; Duduk, B.; Rabus, R.; Seemüller, E. Current View on Phytoplasma Genomes and Encoded Metabolism. Available online: https://www.hindawi.com/journals/tswj/2012/185942/ (accessed on 2 April 2019).
- Carle, P.; Malembic-Maher, S.; Arricau-Bouvery, N.; Desque, D.; Eveillard, S.; Carrere, S.; Foissac, X. “Flavescence doree” phytoplasma genome: A metabolism oriented towards glycolysis and protein degradation. Bull. Insectol. 2011, 64, S13–S14. [Google Scholar]
- Ogura, T.; Wilkinson, A.J. AAA+ superfamily ATPases: Common structure–diverse function. Genes Cells 2001, 6, 575–597. [Google Scholar] [CrossRef]
- Ito, K.; Akiyama, Y. Cellular Functions, Mechanism of Action, and Regulation of Ftsh Protease. Annu. Rev. Microbiol. 2005, 59, 211–231. [Google Scholar] [CrossRef]
- Staats, C.C.; Boldo, J.; Broetto, L.; Vainstein, M.; Schrank, A. Comparative genome analysis of proteases, oligopeptide uptake and secretion systems in Mycoplasma spp. Genet. Mol. Biol. 2007, 30, 225–229. [Google Scholar] [CrossRef][Green Version]
- Kube, M.; Siewert, C.; Migdoll, A.M.; Duduk, B.; Holz, S.; Rabus, R.; Seemüller, E.; Mitrovic, J.; Müller, I.; Büttner, C.; et al. Analysis of the Complete Genomes of Acholeplasma brassicae, A. palmae and A. laidlawii and Their Comparison to the Obligate Parasites from ‘Candidatus Phytoplasma’. J. Mol. Microbial. Biotechnol. 2014, 24, 19–36. [Google Scholar] [CrossRef]
- Kube, M.; Schneider, B.; Kuhl, H.; Dandekar, T.; Heitmann, K.; Migdoll, A.M.; Reinhardt, R.; Seemüller, E. The linear chromosome of the plant-pathogenic mycoplasma “Candidatus Phytoplasma mali”. BMC Genom. 2008, 9, 306. [Google Scholar] [CrossRef] [PubMed]
- Seemüller, E.; Sule, S.; Kube, M.; Jelkmann, W.; Schneider, B. The AAA+ ATPases and HflB/FtsH proteases of “Candidatus Phytoplasma mali”: Phylogenetic diversity, membrane topology and relationship to strain virulence. Mol. Plant Microbe Interact. 2013, 26, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Seemüller, E.; Kampmann, M.; Kiss, E.; Schneider, B. HflB gene-based phytopathogenic classification of “Candidatus phytoplasma mali” strains and evidence that strain composition determines virulence in multiply infected apple trees. Mol. Plant Microbe Interact. 2011, 24, 1258–1266. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Schneider, B.; Sule, S.; Jelkmann, W.; Seemüller, E. Suppression of Aggressive Strains of ‘Candidatus Phytoplasma mali’ by Mild Strains in Catharanthus roseus and Nicotiana occidentalis and Indication of Similar Action in Apple Trees. Phytopathology 2014, 104, 453–461. [Google Scholar] [CrossRef]
- Seemüller, E.; Schneider, B. Differences in Virulence and Genomic Features of Strains of ‘Candidatus Phytoplasma mali’, the Apple Proliferation Agent. Phytopathology 2007, 97, 964–970. [Google Scholar] [CrossRef]
- Seemüller, E.; Kiss, E.; Sule, S.; Schneider, B. Multiple Infection of Apple Trees by Distinct Strains of ‘Candidatus Phytoplasma mali’ and Its Pathological Relevance. Phytopathology 2010, 100, 863–870. [Google Scholar] [CrossRef]
- Wang, J.; Song, L.; Jiao, Q.; Yang, S.; Gao, R.; Lu, X.; Zhou, G. Comparative genome analysis of jujube witches’-broom Phytoplasma, an obligate pathogen that causes jujube witches’-broom disease. BMC Genom. 2018, 19, 689. [Google Scholar] [CrossRef]
- Chung, W.-C.; Chen, L.-L.; Lo, W.-S.; Lin, C.-P.; Kuo, C.-H. Comparative Analysis of the Peanut Witches’-Broom Phytoplasma Genome Reveals Horizontal Transfer of Potential Mobile Units and Effectors. PLoS ONE 2013, 8, 62770. [Google Scholar] [CrossRef]
- Fischer, A.; Santana-Cruz, I.; Wambua, L.; Olds, C.; Midega, C.; Dickinson, M.; Kawicha, P.; Khan, Z.; Masiga, D.; Jores, J.; et al. Draft Genome Sequence of “Candidatus Phytoplasma oryzae” Strain Mbita1, the Causative Agent of Napier Grass Stunt Disease in Kenya. Genome Announc 2016, 4, e00297-16. [Google Scholar] [CrossRef]
- Firrao, G.; Martini, M.; Ermacora, P.; Loi, N.; Torelli, E.; Foissac, X.; Carle, P.; Kirkpatrick, B.C.; Liefting, L.; Schneider, B.; et al. Genome wide sequence analysis grants unbiased definition of species boundaries in “Candidatus Phytoplasma”. Syst. Appl. Microbiol. 2013, 36, 539–548. [Google Scholar] [CrossRef]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Käll, L.; Krogh, A.; Sonnhammer, E.L.L. A Combined Transmembrane Topology and Signal Peptide Prediction Method. J. Mol. Biol. 2004, 338, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- Kakizawa, S.; Oshima, K.; Namba, S. Diversity and functional importance of phytoplasma membrane proteins. Trends Microbiol. 2006, 14, 254–256. [Google Scholar] [CrossRef] [PubMed]
- Eveillard, S.; Jollard, C.; Labroussaa, F.; Khalil, D.; Perrin, M.; Desqué, D.; Salar, P.; Razan, F.; Hévin, C.; Bordenave, L.; et al. Contrasting Susceptibilities to Flavescence Dorée in Vitis vinifera, Rootstocks and Wild Vitis Species. Front. Plant Sci. 2016, 7, 1762. [Google Scholar] [CrossRef]
- Langklotz, S.; Baumann, U.; Narberhaus, F. Structure and function of the bacterial AAA protease FtsH. Biochim. Biophys. Acta 2012, 1823, 40–48. [Google Scholar] [CrossRef]
- Shotland, Y.; Shifrin, A.; Ziv, T.; Teff, D.; Koby, S.; Kobiler, O.; Oppenheim, A.B. Proteolysis of Bacteriophage λ CII by Escherichia coli FtsH (HflB). J. Bacteriol. 2000, 182, 6. [Google Scholar] [CrossRef]
- Rapp, M.; Granseth, E.; Seppälä, S.; von Heijne, G. Identification and evolution of dual-topology membrane proteins. Nat. Struct. Mol. Biol. 2006, 13, 112–116. [Google Scholar] [CrossRef]
- von Heijne, G. A day in the life of Dr K. or how I learned to stop worrying and love lysozyme: A tragedy in six acts. J. Mol. Biol. 1999, 293, 367–379. [Google Scholar] [CrossRef][Green Version]
- Sääf, A.; Johansson, M.; Wallin, E.; von Heijne, G. Divergent evolution of membrane protein topology: The Escherichia coli RnfA and RnfE homologues. Proc. Natl. Acad. Sci. USA 1999, 96, 8540–8544. [Google Scholar] [CrossRef]
- Leonhard, K.; Herrmann, J.M.; Stuart, R.A.; Mannhaupt, G.; Neupert, W.; Langer, T. AAA proteases with catalytic sites on opposite membrane surfaces comprise a proteolytic system for the ATP-dependent degradation of inner membrane proteins in mitochondria. EMBO J. 1996, 15, 4218–4229. [Google Scholar] [CrossRef] [PubMed]
- Davidson, A.L.; Chen, J. ATP-binding cassette transporters in bacteria. Annu. Rev. Biochem. 2004, 73, 241–268. [Google Scholar] [CrossRef] [PubMed]
- Abbà, S.; Galetto, L.; Carle, P.; Carrère, S.; Delledonne, M.; Foissac, X.; Palmano, S.; Veratti, F.; Marzachì, C. RNA-Seq profile of flavescence dorée phytoplasma in grapevine. BMC Genom. 2014, 15, 1088. [Google Scholar] [CrossRef] [PubMed]
- Oshima, K.; Ishii, Y.; Kakizawa, S.; Sugawara, K.; Neriya, Y.; Himeno, M.; Minato, N.; Miura, C.; Shiraishi, T.; Yamaji, Y.; et al. Dramatic Transcriptional Changes in an Intracellular Parasite Enable Host Switching between Plant and Insect. PLoS ONE 2011, 6, 23242. [Google Scholar] [CrossRef] [PubMed]
- Kiran, M.; Chauhan, A.; Dziedzic, R.; Maloney, E.; Mukherji, S.K.; Madiraju, M.; Rajagopalan, M. Mycobacterium tuberculosis ftsH expression in response to stress and viability. Tuberculosis 2009, 89, S70–S73. [Google Scholar] [CrossRef]
- Deuerling, E.; Mogk, A.; Richter, C.; Purucker, M.; Schumann, W. The ftsH gene of Bacillus subtilis is involved in major cellular processes such as sporulation, stress adaptation and secretion. Mol. Microbiol. 1997, 23, 921–933. [Google Scholar] [CrossRef]
- Sanders, J.W.; Venema, G.; Kok, J. Environmental stress responses in Lactococcus lactis. FEMS Microbiol. Rev. 1999, 23, 483–501. [Google Scholar] [CrossRef]
- Deuerling, E.; Paeslack, B.; Schumann, W. The ftsH gene of Bacillus subtilis is transiently induced after osmotic and temperature upshift. J. Bacteriol. 1995, 177, 4105–4112. [Google Scholar] [CrossRef]
- Bove, P.; Capozzi, V.; Garofalo, C.; Rieu, A.; Spano, G.; Fiocco, D. Inactivation of the ftsH gene of Lactobacillus plantarum WCFS1: Effects on growth, stress tolerance, cell surface properties and biofilm formation. Microbiol. Res. 2012, 167, 187–193. [Google Scholar] [CrossRef]
- Oshima, K.; Maejima, K.; Namba, S. Genomic and evolutionary aspects of phytoplasmas. Front. Microbiol. 2013, 4, 230. [Google Scholar] [CrossRef]
- Lepka, P.; Stitt, M.; Moll, E.; Seemüller, E. Effect of phytoplasmal infection on concentration and translocation of carbohydrates and amino acids in periwinkle and tobacco. Physiol. Mol. Plant Pathol. 1999, 55, 59–68. [Google Scholar] [CrossRef]
- Musetti, R.; Marabottini, R.; Badiani, M.; Martini, M.E.; di Toppi, L.S.; Borselli, S.; Borgo, M.; Osler, R. On the role of H2O2 in the recovery of grapevine (Vitis vinifera cv. Prosecco) from Flavescence dorée disease. Funct. Plant Biol. 2007, 34, 750–758. [Google Scholar] [CrossRef]
- Musetti, R.; Buxa, S.V.; De Marco, F.; Loschi, A.; Polizzotto, R.; Kogel, K.-H.; van Bel, A.J.E. Phytoplasma-triggered Ca(2+) influx is involved in sieve-tube blockage. Mol. Plant Microbe Interact. 2013, 26, 379–386. [Google Scholar] [CrossRef] [PubMed]
- Pagliari, L.; Buoso, S.; Santi, S.; Bel, A.J.E.V.; Musetti, R. What Slows Down Phytoplasma Proliferation? Speculations on the Involvement of AtSEOR2 Protein in Plant Defence Signalling. Plant Signal. Behav. 2018, 13, 1473666. [Google Scholar] [CrossRef]
- van Bel, A.J.E.; Musetti, R. Sieve-element biology provides leads for research on phytoplasma lifestyle in plant hosts. J. Exp. Bot. 2019. [Google Scholar] [CrossRef]
- Caudwell, A.; Kuszala, C.; Bachelier, J.C.; Larrue, J. Transmission de la Flavescence doree de la vigne aux plantes herbacees par l’allongement du temps d’utilisation de la cicadelle Scaphoideus littoralis Ball et l’etude de sa survie sur en grand nombre d’especes vegetales. Ann. Phytopathol. 1970, 2, 415–428. [Google Scholar]
- Caudwell, A.; Kuszala, C.; Larrue, J.; Bachelier, J. Transmission de la Flavescence doree de la feve a la feve par des cicadelles des genres Euscelis et Eusceliditis. Intervention possible de ces insectes dans l’epidemiologie du bois noir en Bourgogne. Ann. Phytopathol. 1972, 181–189. [Google Scholar]
- Maixner, M.; Ahrens, U.; Seemüller, E. Detection of the German grapevine yellows (Vergilbungskrankheit) MLO in grapevine, alternative hosts and a vector by a specific PCR procedure. Eur. J. Plant Pathol. 1995, 101, 241–250. [Google Scholar] [CrossRef]
- Cookson, S.J.; Moreno, M.J.C.; Hevin, C.; Mendome, L.Z.N.; Delrot, S.; Trossat-Magnin, C.; Ollat, N. Graft union formation in grapevine induces transcriptional changes related to cell wall modification, wounding, hormone signalling, and secondary metabolism. J. Exp. Bot. 2013, 64, 2997–3008. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Kaell, L.; Krogh, A.; Sonnhammer, E.L.L. Advantages of combined transmembrane topology and signal peptide prediction—the Phobius web server. Nucleic Acids Res. 2007, 35 (Suppl. S2), W429–W432. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Simon, P. Q-Gene: Processing quantitative real-time RT-PCR data. Bioinformatics 2003, 19, 1439–1440. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucl. Acids Res. 2001, 29, 45. [Google Scholar] [CrossRef]



| Protein | Number of Amino Acids | Number of Transmembrane Domains Predicted | C-tail Orientation (Phobius), out (%) |
|---|---|---|---|
| FtsH1-Flado_0314_0026 | 595 | 2 | 9 |
| FtsH2-Flado_7221_0015 | 596 | 2 | 8 |
| FtsH3-Flado_0067_0052 | 584 | 2 | 10 |
| FtsH4-Flado_8115_0004 | 599 | 2 | 1 |
| FtsH5-Flado_5852_0016 | 611 | 2 | 3 |
| FtsH6-Flado_0067_0046 | 674 | 2 | 74 |
| FtsH7-Flado_0234_0001 | 528 | 2 | 86 |
| FtsH8-Flado_4539_0023 | 673 | 2 | 25 |
| Gene Name | Final Concentration (µM) | Length (bp) | Tm (°C)/Number of Cycle | Efficiency | ||
|---|---|---|---|---|---|---|
| ftsH1 | Forward primer 5′–3′ | TTCTGAATTTGTTGAAATGTACG | 1 | 156 | 60/36 | 100% |
| Reverse primer 5′–3′ | TTTTCTTGAGATCCTCCTGAGA | |||||
| ftsH 2 | Forward primer 5′–3′ | TAGCTGGGGAATCAGGAGTT | 0.1 | 553 | 60/40 | 90% |
| Reverse primer 5′–3′ | ACAGCTTCTTCTAATTCTATCAT | |||||
| ftsH 3 | Forward primer 5′–3′ | TGGTTCTGAATTTATGGATAGA | 1 | 157 | 62/36 | 99% |
| Reverse primer 5′–3′ | GTACTTCCATCATCATTTGC | |||||
| ftsH 4 | Forward primer 5′–3′ | AATCGGCATTAAACTCCCTCG | 1 | 207 | 60/36 | 100% |
| Reverse primer 5′–3′ | TCATAAGCACCACTAGAAACT | |||||
| ftsH 5 | Forward primer 5′–3′ | GTGTTGGTGCTTCTCGTGTC | 0.1 | 291 | 60/36 | 90% |
| Reverse primer 5′–3′ | CACGTGCTTTAACATCTGGC | |||||
| ftsH 6 | Forward primer 5′–3′ | GTTTTGGACCTCTTTTATCTGC | 0.1 | 274 | 60/36 | 93% |
| Reverse primer 5′–3′ | GGAATTCTAGCACCCATCAAAT | |||||
| ftsH 7 | Forward primer 5′–3′ | CTGTATCTGGTTCTGAATTTGA | 0.5 | 161 | 60/36 | 90% |
| Reverse primer 5′–3′ | TTGAGATTCTCCTGAAAAACC | |||||
| ftsH 8 | Forward primer 5′–3′ | TTGTTGAAAGGTATGTCGGCGTC | 0.1 | 213 | 56/40 | 90% |
| Reverse primer 5′–3′ | TAATAATTCCTTTAGACGAAGAA | |||||
| FD-gyrA | Forward primer 5′–3′ | CTAGAATTGTCGGTGATGTTATGG | 0.1 | 338 | 64/36 | 99% |
| Reverse primer 5′–3′ | AGCCATTCCTACAGCTATACCCG | |||||
| FD-dnaB | Forward primer 5′–3′ | CTTTATCAACCTTTAATAGGTTTAGG | 1 | 355 | 62/36 | 99% |
| Reverse primer 5′–3′ | TTTCTAATATTTTTTGTTCTTCGTCG |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jollard, C.; Foissac, X.; Desqué, D.; Razan, F.; Garcion, C.; Beven, L.; Eveillard, S. Flavescence Dorée Phytoplasma Has Multiple ftsH Genes that Are Differentially Expressed in Plants and Insects. Int. J. Mol. Sci. 2020, 21, 150. https://doi.org/10.3390/ijms21010150
Jollard C, Foissac X, Desqué D, Razan F, Garcion C, Beven L, Eveillard S. Flavescence Dorée Phytoplasma Has Multiple ftsH Genes that Are Differentially Expressed in Plants and Insects. International Journal of Molecular Sciences. 2020; 21(1):150. https://doi.org/10.3390/ijms21010150
Chicago/Turabian StyleJollard, Camille, Xavier Foissac, Delphine Desqué, Frédérique Razan, Christophe Garcion, Laure Beven, and Sandrine Eveillard. 2020. "Flavescence Dorée Phytoplasma Has Multiple ftsH Genes that Are Differentially Expressed in Plants and Insects" International Journal of Molecular Sciences 21, no. 1: 150. https://doi.org/10.3390/ijms21010150
APA StyleJollard, C., Foissac, X., Desqué, D., Razan, F., Garcion, C., Beven, L., & Eveillard, S. (2020). Flavescence Dorée Phytoplasma Has Multiple ftsH Genes that Are Differentially Expressed in Plants and Insects. International Journal of Molecular Sciences, 21(1), 150. https://doi.org/10.3390/ijms21010150





